Abstract
Shoot branching is a key process for plant growth and fitness. Newly produced axes result from axillary bud outgrowth, which is at least partly mediated through the regulation of BRANCHED1 gene expression (BRC1/TB1/FC1). BRC1 encodes a pivotal bud-outgrowth-inhibiting transcription factor belonging to the TCP family. As the regulation of BRC1 expression is a hub for many shoot-branching-related mechanisms, it is influenced by endogenous (phytohormones and nutrients) and exogenous (light) inputs, which involve so-far only partly identified molecular networks. This review highlights the central role of BRC1 in shoot branching and its responsiveness to different stimuli, and emphasizes the different knowledge gaps that should be addressed in the near future.
Keywords: TCP transcription factors, hormones, nutrients, light, regulation, shoot branching
Introduction
Plants are sessile organisms that need to adjust their shape to suit the diversity of the changing environmental conditions in which they are growing. The regulation of shoot branching is a relevant strategy for plant survival and space occupancy, and involves an intricate regulatory network. Shoot branching depends on the status of bud dormancy, which is a temporary and reversible state (Shimizu and Mori, 1998). Shoot branching patterns, considered here as the distribution of branches along a parent stem, are generated during plant postembryonic development (Domagalska and Leyser, 2011). They depend on the ability of axillary vegetative buds located at the axil of each leaf to remain inactive or to produce a new branch in response to variable stimuli (Shinohara et al., 2013; Rameau et al., 2015; Wang and Jiao, 2018).
Shoot branching is an important feature of plant architecture that determines the interface between the plant and the surrounding environment. Shoot branching contributes to essential processes such as the establishment of leaf area and distribution that determine light interception and photosynthesis, which in turn influence the number of flowers and fruits, fruit filling and yield (Jiang and Egli, 1993; Richards, 2000). Branching also influences the plant competitiveness against weeds or the propagation of pests (Lemerle et al., 1996; Zhao et al., 2006; Simon et al., 2011). In ornamental plants, branching also determines plant visual quality, which drives consumers’ preferences (Ta et al., 1987; Boumaza et al., 2009, 2010; Garbez et al., 2015).
Extensive studies have been undertaken for several decades to find out the mechanisms involved in branching. The currently accepted idea supports that endogenous, developmental, and environmental inputs converge into bud-located integrators, which are at the head of a network of mechanisms governing the ability of buds to grow out. Among these inputs, hormones, sugar, nitrogen, light, and water play a determining role in shoot branching regulation (McSteen, 2009; González-Grandío et al., 2013; Niwa et al., 2013; Li-Marchetti et al., 2015; Rameau et al., 2015; Teichmann and Muhr, 2015; Corot et al., 2017; Le Moigne et al., 2018). Those factors may influence shoot branching via various physiological and molecular mechanisms, targeting different branching-related genes and acting synergistically or antagonistically. BRC1 (BRANCHED 1) is well known to act locally in buds and is considered to be an important hub of different signals controlling the ability of a bud to grow out in many species (Aguilar-Martínez et al., 2007; Dun et al., 2009; Leyser, 2009; Beveridge and Kyozuka, 2010; Rameau et al., 2015). Arabidopsis thaliana harbors two BRANCHED genes, namely BRANCHED 1 (BRC1) and BRANCHED 2 (BRC2); they encode TCP transcription factors closely related to TEOSINTE BRANCHED1 (TB1) in maize and FINE CULM 1 (FC1) in rice. In addition, they are conserved in many species of the plant kingdom (Table 1). The corresponding mutants show an altered branching phenotype as compared to the wild type (Aguilar-Martínez et al., 2007; González-Grandío et al., 2013). This review addresses the molecular identity of BRC1, its involvement in shoot branching, and its regulation in response to endogenous inputs (hormones and nutrients) and exogenous cues (light). We also discuss how BRC1 can mechanistically govern bud outgrowth, and raise a few questions about future investigations.
Table 1.
The publication of BRC1 homolog genes in different species.
| Species | Name of the gene | Reference | |
|---|---|---|---|
| Monocots | Zea mays | TB1 | Doebley et al., 1997 |
| Oryza sativa | Ostb1/FC1 | Takeda et al., 2003 | |
| Sorghum bicolor | SbTB1 | Kebrom et al., 2006 | |
| Hordeum vulgare | INTERMEDIUM-C | Ramsay et al., 2011 | |
| Triticum aestivum | TB-D1 | Dixon et al., 2018 | |
| Eudicots | Solanum tuberosum | StBRC1 | Nicolas et al., 2015 |
| Pisum sativum | PsBRC1 | Braun et al., 2012 | |
| Dendranthema grandiflora | DgBRC1 | Chen et al., 2013 | |
| Arabidopsis thaliana | AtBRC1 | Aguilar-Martínez et al., 2007 | |
| Solanum lycopersicum | SlBRC1 | Martín-Trillo et al., 2011 | |
| Rosa hybrida | RhBRC1 | Barbier et al., 2015 | |
| Nicotiana tabacum | NtBRC1a; NtBRC1b; NtBRC1c; NtBRC1d | Chen et al., 2016 | |
| Populus canescens | PcBRC1 | Muhr et al., 2018 | |
BRC1 Belongs to the TCP Transcription Factor Family
AtBRC1 (also called AtTCP18) contains an open reading frame (ORF) made of ca.1,290-bp that encodes a protein with a TCP domain and an R domain. It belongs to the TCP gene family, an evolutionarily conserved family that first appeared in freshwater algae of the Charophyta family (Navaud et al., 2007). The TCP gene family was first described by Cubas et al. (1999) and is represented by four ‘founding members’: TEOSINTE BRANCHED1 (TB1), CYCLOIDEA (CYC), PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1 (PCF1), and PCF2, all identified on the basis of their functions in plant development or their DNA-binding capacities (for a review see Li, 2015; Danisman, 2016). In Arabidopsis, the TCP family comprises 24 genes encoding predicted proteins with a TCP domain (Cubas et al., 1999; Kosugi and Ohashi, 2002; Palatnik et al., 2003; Cubas, 2004) and categorized into two classes: class I (also known as PCF or TCP-P) is made up of 13 predicted proteins related to the PCF rice factors (Kosugi and Ohashi, 1997), and class II (also known as TCP-C) is made up of 11 predicted proteins related to the Antirrhinum CYC and CIN genes and to the Zea mays TB1 gene (Luo et al., 1996; Doebley et al., 1997; Nath et al., 2003; Palatnik et al., 2003). All these transcription factors have the so-called TCP domain, a 59-amino-acid basic helix–loop–helix (bHLH), in common (Martín-Trillo and Cubas, 2010). Such a motif allows for DNA binding and protein–protein interactions in cells. The TCP domain is also necessary for nuclear localization (Kosugi and Ohashi, 1997; Cubas et al., 1999), and some TCP proteins can be targeted to the nucleus in heterologous systems (Suzuki et al., 2001; Qin et al., 2004).
Besides the TCP domain, a few class-II TCPs, including BRC1, display a functionally unknown arginine-rich motif, the R-domain, which is predicted to mediate protein interactions (Lupas et al., 1991; Cubas et al., 1999). The R domain may involve the phosphorylation process of BRC1 by a cAMP-dependent protein kinase (Dulhanty and Riordan, 1994; Martín-Trillo and Cubas, 2010). Additionally, most members of the CYC/TB1 subclass, to which BRC1 belongs, contain a conserved ECE (glutamic acid-cysteine-glutamic acid) motif that remains functionally uncharacterized and is located between their TCP and R domains (Howarth and Donoghue, 2006).
The TCP proteins of various species regulate many biological processes, including seed germination, plant branching, lateral organ development, floral asymmetry, gametophyte development, leaf senescence, circadian rhythms, and defense responses (for a review see Li, 2015; Danisman, 2016). These TCP-dependent regulations could occur directly through their binding to the promoter of target genes or indirectly via their interactions with plant hormones (Schommer et al., 2008; Guo et al., 2010; Danisman et al., 2012; Li and Zachgo, 2013; Nicolas and Cubas, 2016). In Arabidopsis, the CYC/TB1 clade consists of AtBRC1, AtBRC2 (also called AtTCP12) and AtTCP1, and is mainly involved in the development of axillary meristems, giving rise to either flowers or lateral shoots (Martín-Trillo and Cubas, 2010).
BRC1 Is a Central Actor of Shoot Branching
The shoot axillary meristem produces a branch when the appropriate endogenous and exogenous inputs occur, so as to adapt plant architecture to environmental conditions. In monocots, TB1 from Z. mays (Doebley et al., 1997) and homologs of TB1 in Oryza sativa (OsTB1/FC1, Takeda et al., 2003) and Sorghum bicolor (SbTB1, Kebrom et al., 2006) promote bud arrest locally, without affecting the number of buds, and thus lead to reduced tillering. Consistently, TB1 and OsTB1 are mainly expressed in axillary bud meristems (Hubbard et al., 2002; Takeda et al., 2003), and their mutants tb1 and fc1 exhibit over-tillering phenotypes (Doebley et al., 1997; Wang et al., 1999; Takeda et al., 2003). The barley TB1 ortholog, INT-C, has been shown to act mainly in the control of spike architecture, with a minor role in tillering (Ramsay et al., 2011). Moreover, modern maize displays less branching than the wild teosinte ancestor due to increased TB1 expression (Studer et al., 2011; Zhou et al., 2011). However, the int-c loss-of-function mutant showed less tillers in barley, whose phenotype is opposite to the recessive tb1 mutant in maize (Liller et al., 2015; Dong et al., 2019).
In dicots, genes closely related to TB1 have been studied in a variety of species. In Arabidopsis, AtBRC1 and AtBRC2 both negatively regulate the branching process (Aguilar-Martínez et al., 2007; Poza-Carrión et al., 2007). However, AtBRC1 seems to play a more pivotal role in axillary bud development than AtBRC2. The AtBRC1 gene is predominantly expressed during the development of axillary buds (axillary meristems, bud leaf primordia and subtending vascular tissue). AtBRC1 expression is inversely correlated with bud outgrowth and brc1 mutant phenotypes are non-pleiotropic, while constitutive overexpression of AtBRC1 reduces the growth of the whole plant (Aguilar-Martínez et al., 2007). Moreover, many AtBRC1-homologous genes have also been found to be involved in shoot branching suppression (Table 1). In addition, repressed buds in pea have been found to be as metabolically active as growing buds, so BRC1 growth repression may not involve metabolism (Stafstrom and Sussex, 1988). Recent data demonstrate that AtBRC1 is not always necessary for the complete inhibition of all buds in Arabidopsis (Seale et al., 2017).
Genomic sequences of Solanum species, including potato and tomato, also contain the BRC1-like gene, where it occurs under two forms (Brewer, 2015). More interestingly, in Solanum tuberosum, the BRANCHED1a (StBRC1a) gene encompasses an alternative splice site leading to the generation of two BRC1a protein isoforms, BRC1aLong and BRC1aShort, with distinct C-terminal regions (Martín-Trillo et al., 2011; Nicolas et al., 2015). The BRC1aLong C-terminal region has a strong activation domain and moves to the nucleus, whereas the BRC1aShort C-terminal region lacks an activation domain, which prevents the nuclear targeting of the protein (Nicolas et al., 2015). These different splice variants of AtBRC1 have also been found in Arabidopsis (data not shown), but whether the mechanism mentioned above exists in Arabidopsis is still unknown. A central role of BRC1 in shoot branching has also been revealed in pea (PsBRC1, Braun et al., 2012), Chrysanthemum (DgBRC1, Chen et al., 2013), and poplar (PcBRC1, Muhr et al., 2016 and 2018). In Rosa sp., Li-Marchetti et al. (2017) carried out a Quantitative Trait Loci (QTL) analysis of the plant architecture, using a segregating, recurrent blooming population called ‘The Fairy’ × ‘Old Blush’. They showed that the branching angle of order 2 long axes, the number of short axes (the type of axis that comprises one to four internodes), and stem elongation were correlated, with QTL located in the genomic region of RhBRC1, and assumed a pleiotropic role of RhBRC1 in the establishment of the bushy shape of Rosa sp. Further work will be required to more accurately define the role of BRC1 in the establishment of the plant complex architecture.
BRC1 Is an Integrator of Diverse Hormonal Signaling Networks
Auxin, cytokinins (CK), and strigolactones (SL) are implicated in the hormonal regulation of BRC1 expression. In this regulation network, auxin and SL act as inducers while CK act as repressors (Rameau et al., 2015; Teichmann and Muhr, 2015). According to Ferguson and Beveridge (2009), this kind of regulation could be involved in various metabolism pathways such as feedback regulation, long-distance hormone transport, and the interplay of plant hormone metabolism and signaling.
In apical dominance, the polar auxin transport (PAT) stream in the main stem, which is mediated by the PIN (PIN-FORMED) auxin-efflux facilitators located in xylem-associated cells (Petrášek and Friml, 2009), inhibits axillary bud outgrowth (Morris, 1977; Li and Bangerth, 1999; Zhang et al., 2007; Balla et al., 2011). Auxin cannot directly regulate BRC1 expression because it is not transported from the stem to the buds in great enough amounts (Hall and Hillman, 1975). It is hypothesized that PAT prevents the establishment of auxin canalization from axillary buds to the stem, and that this might be necessary for the buds to grow out (Li and Bangerth, 1999; Domagalska and Leyser, 2011; Chabikwa et al., 2019). The characterization of the auxin-resistant Arabidopsis mutant axr1 indicated that such an auxin effect occurred after axillary meristem initiation through the inhibition of bud outgrowth (Stirnberg et al., 1999).
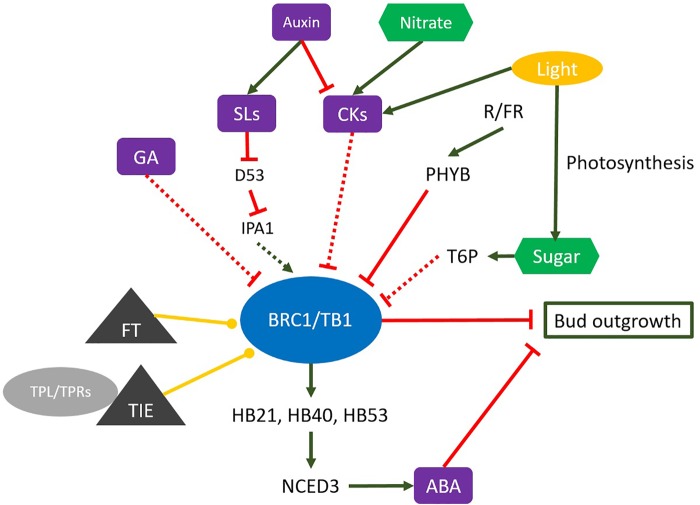
Auxin can indirectly promote BRC1 expression in the bud (Aguilar-Martínez et al., 2007). Furthermore, auxin-mediated BRC1 regulation through the control of two antagonistic factors, CK and SL, fine-tunes BRC1 expression inside buds (Rameau et al., 2015). The role of CK, a collection of adenine-related compounds, in bud outgrowth was evidenced decades ago, when CK application to dormant buds was shown to promote bud outgrowth (Wickson and Thimann, 1958; Sachs and Thimann, 1967; Bangerth, 1994; Tanaka et al., 2006). In parallel, auxin indirectly inhibits bud outgrowth by decreasing systemic and local CK levels, which determines the CK supply to the buds (Miyawaki et al., 2004; Nordström et al., 2004; Tanaka et al., 2006; Müller and Leyser, 2011). CK can act to promote branching partly by promoting PIN3,4,7 cross-stem auxin transport between the bud and the adjoining stem, thereby potentially acting partly independently of AtBRC1 repression directly in the bud (Waldie and Leyser, 2018). High CK levels in axillary buds lead to the activation of axillary buds through downregulation of BRC1 expression (Braun et al., 2012), although Psbrc1 (a pea BRC1 mutant) remained sensitive to CK application. These findings might indicate that the branch-promoting hormone CK partly controls shoot branching by negatively regulating BRC1 at the transcriptional level. In rice, transcript levels of OsTB1/FC1 also decreased in a CK-dose-dependent manner (Minakuchi et al., 2010), and similar down-regulation of DgBRC1 was reported in Chrysanthemum (Dierck et al., 2016). This CK-dependent BRC1 regulation can be part of the light intensity-dependent bud outgrowth regulation in Rosa sp. (Roman et al., 2016; Corot et al., 2017). The Arabidopsis altered meristem program1 (amp1) mutants are characterized by higher levels of CK, more bud outgrowth, more axillary meristems, and reduced BRC1 expression (Helliwell et al., 2001). Although CK are a powerful repressor of BRC1/TB1/FC1 expression, the molecular mechanisms driven by this CK-dependent regulation still remain an open question (Figure 1).
FIGURE 1.
Many factors influence the expression of BRC1, including developmental, positional, genetic, hormonal, sugar signal and environmental factors. Auxin, cytokinin (CK), and strigolactone (SL) are implicated in the hormonal regulation of BRC1 expression; auxin and SLs as promoters of BRC1 and CKs as an inhibitor of BRC1. The red line, inhibition effect; the green arrow, stimulation effect; the yellow bullet-end lines, protein interaction; the violet element, plant hormones; the green element, plant nutrition; the green element, the yellow element, exogenous influence factor; the gray triangle, the proteins that interact with BRC1/TB1; D53, DWARF 53; HB21, HOMEOBOX PROTEIN 21; HB40, HOMEOBOX PROTEIN 40; HB53, HOMEOBOX PROTEIN 53; IPA1, IDEAL PLANT ARCHITECTURE1; NCED3, NINE-CIS-EPOXICAROTENOID DIOXIGENASE 3; PHYB, PHYTOCHROME B; T6P, trehalose-6 phosphate.
Strigolactones (SL), a group of carotenoids derived from terpenoid lactones (Lin et al., 2009; Alder et al., 2012), act as endogenous shoot branching inhibitors (Gomez-Roldan et al., 2008; Leyser, 2008; Wang et al., 2013). Direct application of GR24 – an SL analog – on buds inhibited outgrowth on intact and decapitated plants (Brewer et al., 2009), and auxin application elevated the transcription levels of SL biosynthesis genes (Sorefan et al., 2003; Foo et al., 2005; Johnson et al., 2006; Zou et al., 2006; Arite et al., 2007, 2009; Hayward et al., 2009). These findings support that auxin-mediated bud outgrowth inhibition involves the promotion of systemic and local SL synthesis in the stem and thereby of SL levels inside buds. Consistently, different SL mutants exhibited a highly branched phenotype in pea [ramosus (rms)], petunia [decreased apical dominance (dad)], and Arabidopsis [more axillary growth (max)] (Crawford et al., 2010). A role for BRC1 downstream of SL was first reported in Arabidopsis and pea, where BRC1 expression was upregulated by SL, and shoot branching in the brc1 mutant was insensitive to SL (Aguilar-Martínez et al., 2007; Dun et al., 2012; Revel et al., 2015). However, SL application did not change the transcriptional activation of OsTB1/FC1 expression in rice (Minakuchi et al., 2010). Recent investigations showed that DWARF 53 (D53)/SUPPRESSOR OF MAX2 1-LIKE genes (SMXL6, 7, 8) acted downstream of SL as repressors of SL-dependent BRC1 upregulation and thereby promoted shoot branching (Jiang et al., 2013; Zhou et al., 2013; Kong et al., 2014; Wang et al., 2015). Mutants deficient in D53-like genes indeed displayed constitutive BRC1 upregulation (Soundappan et al., 2015; Wang et al., 2015; Seale et al., 2017). Moreover, SL perception by D14 (α/β hydrolase) and the recruitment of the SCF complex resulted in the polyubiquitination and 26S-proteasome–mediated degradation of D53 (Kerr and Beveridge, 2017; Waters et al., 2017). D53 physically interacts with IPA1 (IDEAL PLANT ARCHITECTURE1), a repressor of shoot branching, and prevents it from upregulating TB1 expression (Figure 1) (Song et al., 2017). IPA1, also named OsSPL14, is a member of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family of plant-specific transcription factors (Miura et al., 2010) that directly binds to the TB1 promoter in rice and activates TB1transcriptional activity (Figure 1; Jiao et al., 2010; Lu et al., 2013). Further support for the relevance of the “IPA-1-related genes and TB1” module in shoot branching comes from a study in wheat, where TaD53 physically interacted with TaSPL3 and prevented TaSPL3 upregulation of TaTB1 gene expression (Liu et al., 2017). Although the Arabidopsis homologs of IPA1 have been identified as being SPL9/15, further work will be required to confirm whether this mechanism is involved in the SL-dependent regulation of AtBRC1.
Besides auxin, CK, and SL, gibberellin (GA) might also be involved in the regulation of BRC1 expression, even if the mechanism is still unknown (Lantzouni et al., 2017). GAs (diterpenoid tetracyclin molecules) are plant hormones that regulate various developmental processes, including stem elongation, germination, dormancy, flowering, flower development, and leaf and fruit senescence (Hedden and Sponsel, 2015). In Rosa sp., GA biosynthesis strongly increases during bud outgrowth (Choubane et al., 2012). In the perennial woody plant Jatropha curcas, GA and CK synergistically promote lateral bud outgrowth, and both hormones negatively influence BRC1 and BRC2 expression (Ni et al., 2015). Simultaneously altered GA and SL levels positively influenced the expression of the GA2 OXIDASE2 gene which encodes a GA-catabolic enzyme, and the expression of BRC1 (Figure 1) (Lantzouni et al., 2017). Furthermore, GA is required for CK-mediated axillary bud outgrowth in A. thaliana (Jasinski et al., 2005; Lo et al., 2008).
BRC1 Expression Is Regulated by Light
Shoot branching is negatively affected by low light intensity and low ratios of red/far red (R:FR) light in many species (Kebrom et al., 2006; Finlayson et al., 2010; Su et al., 2011; Revel et al., 2015). In this process, light acts both as a driver of photosynthesis for the supply of sugars to axillary buds and as a photomorphogenic signal (Su et al., 2011). The signaling role of light in plant branching was first unraveled by Kebrom et al. (2006). In 2006 and 2010, these authors showed that active PHYB suppressed the expression of the SbTB1 gene in sorghum, leading to high plant branching, whereas environmental conditions that inactivate phyB (low R/FR ratio) increased SbTB1 expression and in turn repressed bud outgrowth. Additional experiments carried out in Arabidopsis confirmed these findings: a low R/FR ratio favored AtBRC1 upregulation through the PHYB pathway, which is required for shoot branching reduction (Figure 1; González-Grandío et al., 2013). This effect seems to be reversible, as evidenced by the rapid and local downregulation of AtBRC1 after increasing the R/FR ratio (Holalu and Finlayson, 2017). Such a response may contribute to the rapid adaptation of plants to fluctuations in the R/FR light ratio.
Besides light quality, a slight decrease of the photosynthetic leaf area is associated with a stimulation of TB1 expression in sorghum seedlings and consequently a lower propensity of tiller buds to grow out (Kebrom and Mullet, 2015). In addition, darkness-exposed Rosa sp. exhibited no bud outgrowth and higher levels of RhBRC1 transcripts than plants placed under light (Roman et al., 2016). All these findings indicate that BRC1 expression is very sensitive to both light intensity and quality. However, this regulation may involve distinct mechanisms (Kebrom et al., 2010).
BRC1 Is Regulated by Nutrients
Sugars are well known to promote bud outgrowth in many species (Leduc et al., 2014; Rameau et al., 2015; Kebrom, 2017; Tarancón et al., 2017; Ferreira et al., 2018), and the relationship between sugars and bud outgrowth has been investigated for years (Maurel et al., 2004; Chao et al., 2007; Girault et al., 2010; Kebrom et al., 2010, 2012; Henry et al., 2011; Rabot et al., 2012; Mason et al., 2014; Barbier et al., 2015; Fichtner et al., 2017). Sugar effects are seemingly dependent on environmental conditions (Corot et al., 2017). Sugars not only serve as a carbon source for plant metabolism, but also as an important signaling entity that affects many developmental processes including BRC1 gene expression (Price et al., 2004; Hellmann and Smeekens, 2014; Barbier et al., 2015; Sakr et al., 2018). In an interesting study, Mason et al. (2014) demonstrated that the initial signal responsible for the release of bud outgrowth after decapitation in pea was an increase in sugar availability rather than a decrease in apically supplied auxin, as traditionally thought. This is in line with the earlier proposal by Morris and collaborators (Morris et al., 2005), who assumed the existence of an auxin-independent “fast-decapitation signal” leading to bud outgrowth initiation after decapitation. Furthermore, Mason et al. (2014) also reported that the timing of the increase of the sugar flux inside buds and bud outgrowth tightly coincided with the downregulation of BRC1 expression. In this process, sugar acts more likely as a signaling entity, because many non-metabolizable sugar analogs can trigger bud outgrowth (Rabot et al., 2012) and repress BRC1 expression (Barbier et al., 2015). In addition, this effect of sugar on BRC1 transcription could be mediated indirectly via sugar regulation of CK biosynthesis and SL signaling (Barbier et al., 2015) and/or directly (irrespective of hormonal action). Decapitation led to a rapid and sustained rise in trehalose-6 phosphate (T6P) levels in axillary buds and a decreased expression level of BRC1, which supports that T6P could partly mediate the sugar-dependent down-regulation of BRC1 (Figure 1) (Fichtner et al., 2017). Further works are required to further unravel this molecular regulatory network. In the present state of knowledge, we cannot rule out that the transcriptional regulation of BRC1 in response to sugars could involve many sugar-signaling pathways and also that BRC1 expression is sensitive to the plant carbon status and/or energy levels (Martín-Fontecha et al., 2018).
Mineral nutrition influences tiller bud outgrowth in barley (Fletcher and Dale, 1974). In wheat, phosphorus deficiency directly altered the normal pattern of tiller emergence by reducing the rate of tiller emergence for each tiller (Rodríguez et al., 1999). Although several links exist between phosphate and the branching-related hormones (auxin, SL and CK), no direct effect of the phosphate status on BRC1/TB1/FC1 gene expression is documented. Low-phosphate growth conditions enhance SL production in many species (Yoneyama et al., 2007; López-Ráez et al., 2008; Umehara et al., 2008; Domagalska and Leyser, 2011; Kohlen et al., 2011; Yamada et al., 2014). This situation leads to the repression of shoot branching (Umehara et al., 2008; Kohlen et al., 2011), but also to the stimulation of lateral root formation for soil foraging (Yoneyama et al., 2007; Ruyter-Spira et al., 2011). In contrast to SL, low levels of inorganic phosphate reduce CK production, which correlates with a reduced number of branches (Horgan and Wareing, 1980).
In herbaceous and woody plants, high levels of nitrogen fertilization (nitrate and/or ammonium) result in (i) a large number of outgrowing buds (Lortie and Aarssen, 1997; Médiène et al., 2002; Cline et al., 2006; Emarat-Pardaz et al., 2013; Pal et al., 2013; Furet et al., 2014), and (ii) improved secondary axis elongation (Thitithanakul, 2012; Thitithanakul et al., 2012). Luo et al. (2017) confirmed that nitrogen deficiency did not affect the initiation of tiller buds, but suppressed tiller bud outgrowth in O. sativa. In Arabidopsis, low nitrate delayed axillary bud activation, and this process involved an effect of the plant nitrogen status rather than a direct nitrate-signaling pathway (De Jong et al., 2014). Recent results demonstrated a relationship between nitrogen fertilization and BRC1 expression in rice (Li et al., 2016). They showed that high ammonium nitrate intake in the root environment induced a reduction of apical dominance through overexpression of miRNA393 in the buds; miRNA393 inhibits the expression of the genes involved in auxin synthesis and signaling (OsTIR1, OsAFB2, and OsIAA6) as well as OsTB1. In Arabidopsis, the brc1-2/brc2-1 double mutant exhibited a higher number of branches than the wild type, but low availability of nitrate reduced this effect (Seale et al., 2017). As root nitrate is widely known to induce CK biosynthesis and signaling events in the whole plant (Crawford, 1995; Sakakibara et al., 1998; Takei et al., 2001, 2002; Forde, 2002a,b), and CK repress BRC1 expression, we cannot exclude that nitrate may affect BRC1 expression through CK modulation. In rice, the supply of a CK analog (BAP) or ammonium nitrate regulated SL amounts in the stem and the bud within 3 h after treatment, but nothing has been reported regarding BRC1 expression (Xu et al., 2015).
In Rosaceae as in many woody plants, nitrate is reduced and assimilated into amino acids directly in the roots; consequently, asparagine, arginine, aspartate, and glutamine are the main forms of nitrogen translocated to the buds via the xylem sap (Millard et al., 1998; Malaguti et al., 2001; Grassi et al., 2002; Guak et al., 2003; Le Moigne et al., 2018). In rose, asparagine is a major nitrogen form involved in bud outgrowth (Le Moigne et al., 2018); this is in accordance with previous data showing that application of asparagine on the soil of olive trees or on the leaves of poplar trees contributed to enhance bud outgrowth and secondary axis elongation (Proietti and Tombesi, 1996; Cline et al., 2006). In rice, a lack of cytosolic glutamine synthetase1;2 in the vascular tissues of axillary buds severely reduced their outgrowth (Funayama et al., 2013; Ohashi et al., 2015) independently of the SL level (Ohashi et al., 2015). In rose bush, sucrose, glucose, and fructose had to be associated to asparagine to allow for the buds to grow out in vitro (Le Moigne et al., 2018). This effect involved the upregulation of IPT3 gene expression in the stem and in the vicinity of the bud (Le Moigne et al., 2018) and the downregulation of BRC1 (Barbier et al., 2015). In addition to a nutritional role, asparagine might also be a signal representing the nitrogen status of the plant, so as to counteract BRC1 expression through CK stimulation.
A BRC1-Related Regulatory Mechanism
Many studies ascribe an inhibitory function of mitotic cell activity to BRC1 (Poza-Carrión et al., 2007; Kieffer et al., 2011). This is because early results of EMSA (Electrophoresis Mobility Shift Assay) revealed the capacity of the TCP domain to associate specifically with the promoter element of the rice proliferating cell nuclear antigen (PCNA) gene (Kosugi and Ohashi, 1997, 2002). These cis-regulatory modules are indispensable for the transcriptional activation of the PCNA gene in rice meristem tissues (Kosugi and Ohashi, 1997), which seems to be an ancient and prevalent role of TCP transcription factors (Ortiz-Ramírez et al., 2016).
BRC1-mediated branching is repressed by the regulation of abscisic acid (ABA) metabolism (Figure 1). ABA is a plant hormone that plays important roles in many phases of the plant life cycle (Seo and Koshiba, 2002; Hayes, 2018; Wang et al., 2018). Evidence for a role of ABA in regulating bud growth comes from the positive correlation between a reduction of ABA levels in buds and their release from dormancy (Cline, 1991). Moreover, the Arabidopsis era1 (ENHANCED RESPONSE TO ABA 1) mutant exhibited high sensitivity to ABA and reduced branching (Pei et al., 1998). In brc1 Arabidopsis mutants, the ABA-signaling pathway showed a significantly reduced response as compared to the wild type. Additional data revealed that the expression levels of two ABA markers, ABA-RESPONSE PROTEIN (ABR) and UDP-GLUCOSYL TRANSFERASE 74D1 (UGT74D1), were significantly upregulated in the wild type but not in brc1 mutants treated with low R:FR light (González-Grandío et al., 2013). González-Grandío and Cubas (2014) support a model in which ABA acts rather downstream of BRC1, because ABRE-BINDING FACTOR 3 (ABF3) and ABA INSENSITIVE 5 (ABI5), two key regulators of the ABA response that contain TCP-binding sites in their promoters (Finkelstein and Lynch, 2000; Yoshida et al., 2010; González-Grandío et al., 2013; Nicolas and Cubas, 2016), are upregulated in axillary buds upon BRC1 induction (González-Grandío and Cubas, 2014). They also indicated that BRC1 bound to and positively regulated three transcription factors: HOMEOBOX PROTEIN 21 (HB21), HOMEOBOX PROTEIN 40 (HB40), and HOMEOBOX PROTEIN 53(HB53). These three proteins, together with BRC1, enhanced NINE-CIS-EPOXICAROTENOID DIOXIGENASE 3 (NCED3) expression, the main ABA-biosynthesis enzyme, leading in turn to ABA accumulation in buds (González-Grandío et al., 2017). This finding demonstrates a direct relationship between BRC1 and ABA signaling, and places ABA downstream of BRC1. Consistently, BRC1 expression was found to be insensitive to exogenous ABA application (Yao and Scott, 2015).
The TCP genes generally act by positively or negatively regulating the cell cycle (Sarvepalli and Nath, 2018). As a transcription factor, BRC1 could bind to the promoter region of various genes to regulate the branching process and participate to many regulatory mechanisms (González-Grandío et al., 2013). In maize, TB1 can directly activate the tassels replace upper ears1 (tru1) gene that encodes an ankyrin-repeat-domain protein by binding to the promoter region of tru1 (Dong et al., 2017). In Arabidopsis, bioinformatic analysis indicates that the promoter sequences of 1,950 genes expressed in the shoot bear the TCP-cis regulatory motif (5′-RRVMMMV-3′) and could be putatively regulated by AtBRC1. Based on Gene Ontology (GO) enrichment analysis, these putative target genes are thought to be mainly involved in metabolic processes, including amino acid metabolism [e.g., ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1 (AOAT1); HYDROXYPYRUVATE REDUCTASE (ATHPR1)] and sulfur (e.g., sulfate transmembrane transporter (MOT2), sulfate transporter 1;2 (SULTR1;2)] (data not shown). We can therefore speculate that BRC1/TB1 might control bud outgrowth via various pathways, such as stimulating the ABA-signaling pathway and inhibiting cell division and cell metabolism.
Conclusion and Perspectives
BRC1/TB1/FC1 is an integrator gene involved in shoot branching, which fits well with the ability of BRC1/TB1/FC1 expression to integrate many endogenous and exogenous inputs (Figure 1). However, the detailed mechanism whereby these stimuli regulate BRC1 expression is still puzzling, and many mechanistic scenarios are plausible. Many questions are thus still open and include how CK and SL, the main two branching-related hormones, antagonistically regulate BRC1 expression, and which molecular actors could be involved. Similar questions concern the sugar-mediated downregulation of BRC1, and the molecular mechanism behind the combined effect of nutrients and hormones on BRC1 expression (Sakr et al., 2018). In addition, the regulation of gene expression includes many aspects, such as epigenetic regulation, transcriptional regulation, post-transcriptional regulation, translational and post-translational regulation. The relevance of these mechanisms in the regulation of BRC1 expression deserves to be investigated in different biological contexts. Recent data showed that the protein interaction process also influences BRC1 expression. For example, the florigen proteins FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) influence axillary meristem development via their interaction with AtBRC1 (Niwa et al., 2013); TIE1 (TCP interactor containing EAR motif protein 1), a transcriptional repressor identified as involved in the control of leaf development, controls shoot branching by interacting with BRC1 (Yang et al., 2018). Additional protein partners may also interact with BRC1, including those related to the energy and nutrient statuses [Sucrose non-fermenting-related kinase (SnRK1)/Target of rapamycin (TOR kinase)] (Martín-Fontecha et al., 2018). Meanwhile, our knowledge about the molecular network governing the BRC1-dependent reduction of plant branching is still limited, and the only available data report that BRC1 action could be related to different biological functions such as cell proliferation, cell metabolism, hormone biosynthesis, ribosome biosynthesis, etc. All these findings indicate that further work is required to fully investigate the regulatory network behind the regulation and function of BRC1 in shoot branching.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.rosaceae.org.
Author Contributions
All the authors listed have contributed significantly to this manuscript. MW, J-MD, and SS managed this review and contributed to different sections. M-DP-G and LO contributed to the TCP transcription factor section. JB, SD-M, LH, and LC contributed to both hormonal and environmental factors sections, and M-ALM contributed to the nutrient section.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the program of China Scholarships Council (No. 201506320203) and by the ANR (Agence Nationale de la Recherche) project Labcom, called ESTIM (Evaluation de stimulateurs de vitalité des plantes).
References
- Aguilar-Martínez J. A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 458–472. 10.1105/tpc.106.048934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., et al. (2012). The path from ββ-carotene to carlactone, a strigolactone-like plant hormone. Science 335 1348–1351. 10.1126/science.1218094 [DOI] [PubMed] [Google Scholar]
- Arite T., Iwata H., Ohshima K., Maekawa M., Nakajima M., Kojima M., et al. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51 1019–1029. 10.1111/j.1365-313X.2007.03210.x [DOI] [PubMed] [Google Scholar]
- Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., et al. (2009). d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50 1416–1424. 10.1093/pcp/pcp091 [DOI] [PubMed] [Google Scholar]
- Balla J., Kalousek P., Reinöhl V., Friml J., Procházka S. (2011). Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 65 571–577. 10.1111/j.1365-313X.2010.04443.x [DOI] [PubMed] [Google Scholar]
- Bangerth F. (1994). Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 194 439–442. 10.1007/BF00197546 [DOI] [Google Scholar]
- Barbier F., Peron T., Lecerf M., Perez-Garcia M.-D., Barriere Q., Rolčik J., et al. (2015). Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 66 2569–2582. 10.1093/jxb/erv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C. A., Kyozuka J. (2010). New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 13 34–39. 10.1016/j.pbi.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Boumaza R., Demotes-Mainard S., Huché-Thellier L., Guérin V. (2009). Visual characterization of the esthetic quality of the rosebush. J. Sens. Stud. 24 774–796. 10.1111/j.1745-459X.2009.00238.x [DOI] [Google Scholar]
- Boumaza R., Huché-Thélier L., Demotes-Mainard S., Le Coz E., Leduc N., Pelleschi-Travier S., et al. (2010). Sensory profiles and preference analysis in ornamental horticulture: the case of the rosebush. Food Qual. Prefer. 21 987–997. 10.1016/j.foodqual.2010.05.003 [DOI] [Google Scholar]
- Braun N., de Saint Germain A., Pillot J. P., Boutet-Mercey S., Dalmais M., Antoniadi I., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158 225–238. 10.1104/pp.111.182725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer P. B. (2015). Plant architecture: the long and the short of branching in potato. Curr. Biol. 25 R724–R725. 10.1016/j.cub.2015.06.066 [DOI] [PubMed] [Google Scholar]
- Brewer P. B., Dun E. A., Ferguson B. J., Rameau C., Beveridge C. A. (2009). Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 150 482–493. 10.1104/pp.108.134783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabikwa T. G., Brewer P. B., Beveridge C. A. (2019). Initial bud outgrowth occurs independent of auxin flow out of buds. Plant Physiol. 179 55–65. 10.1104/pp.18.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W. S., Foley M. E., Horvath D. P., Anderson J. V. (2007). Signals regulating dormancy in vegetative buds. Int. J. Plant Dev. Biol. 1 49–56. [Google Scholar]
- Chen X., Zhou X., Xi L., Li J., Zhao R., Ma N., et al. (2013). Roles of DgBRC1 in regulation of lateral branching in chrysanthemum (Dendranthema × grandiflora cv. Jinba). PLoS One 8:e61717. 10.1371/journal.pone.0061717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. J., Xia X. J., Guo X., Zhou Y. H., Shi K., Zhou J., et al. (2016). Apoplastic H2O2 plays a critical role in axillary bud outgrowth by altering auxin and cytokinin homeostasis in tomato plants. New Phytol. 211 1266–1278. 10.1111/nph.14015 [DOI] [PubMed] [Google Scholar]
- Choubane D., Rabot A., Mortreau E., Legourrierec J., Péron T., Foucher F., et al. (2012). Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J. Plant Physiol. 169 1271–1280. 10.1016/j.jplph.2012.04.014 [DOI] [PubMed] [Google Scholar]
- Cline M. G. (1991). Apical dominance. Bot. Rev. 57 318–358. 10.1007/BF02858771 [DOI] [Google Scholar]
- Cline M. G., Thangavelu M., Dong-Il K. (2006). A possible role of cytokinin in mediating long-distance nitrogen signaling in the promotion of sylleptic branching in hybrid poplar. J. Plant Physiol. 163 684–688. 10.1016/j.jplph.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Corot A., Roman H., Douillet O., Autret H., Perez-Garcia M. D., Citerne S., et al. (2017). Cytokinins and abscisic acid act antagonistically in the regulation of the bud outgrowth pattern by light intensity. Front. Plant Sci. 8:1724. 10.3389/fpls.2017.01724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N. M. (1995). Nitrate: nutrient and signal for plant growth. Plant Cell 7 859–868. 10.1105/tpc.7.7.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S., Shinohara N., Sieberer T., Williamson L., George G., Hepworth J., et al. (2010). Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137 2905–2913. 10.1242/dev.051987 [DOI] [PubMed] [Google Scholar]
- Cubas P. (2004). “Role of TCP genes in the evolution of morphological characters in angiosperms,” in Developmental Genetics and Plant Evolution, eds Cronk Q. C. B., Bateman R. M., Hawkins J. A. (Boca Raton, FL: CRC Press; ), 262–281. [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18 215–222. 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Danisman S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7:1930. 10.3389/fpls.2016.01930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S., Van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159 1511–1523. 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong F., Thodey K., Lejay L. V., Bevan M. W. (2014). Glucose elevates NITRATE TRANSPORTER2. 1 protein levels and nitrate transport activity independently of its HEXOKINASE1-mediated stimulation of NITRATE TRANSPORTER2. 1 expression. Plant Physiol. 164 308–320. 10.1104/pp.113.230599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierck R., De Keyser E., De Riek J., Dhooghe E., Van Huylenbroeck J., Prinsen E., et al. (2016). Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in Chrysanthemum. PLoS One 11:e0161732. 10.1371/journal.pone.0161732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L. E., Greenwood J. R., Bencivenga S., Zhang P., Cockram J., Mellers G., et al. (2018). TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum L.). Plant Cell 30 563–581. 10.1105/tpc.17.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Domagalska M. A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12 211–221. 10.1038/nrm3088 [DOI] [PubMed] [Google Scholar]
- Dong Z., Alexander M., Chuck G. (2019). Understanding grass domestication through maize mutants. Trends Genet. 35 118–128. 10.1016/j.tig.2018.10.007 [DOI] [PubMed] [Google Scholar]
- Dong Z., Li W., Unger-Wallace E., Yang J., Vollbrecht E., Chuck G. (2017). Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. U.S.A. 114 E8656–E8664. 10.1073/pnas.1714960114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulhanty A. M., Riordan J. R. (1994). Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry 33 4072–4079. 10.1021/bi00179a036 [DOI] [PubMed] [Google Scholar]
- Dun E. A., Brewer P. B., Beveridge C. A. (2009). Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci. 14 364–372. 10.1016/j.tplants.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Dun E. A., de Saint Germain A., Rameau C., Beveridge C. A. (2012). Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158 487–498. 10.1104/pp.111.186783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emarat-Pardaz J., Shakiba M. R., Toorchi M., Mohammadinasab A. D. (2013). The influence of light intensities and nitrogen on growth of Hypericum perforatum L. Int. J. Agric. 3 775–781. [Google Scholar]
- Ferguson B. J., Beveridge C. A. (2009). Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 149 1929–1944. 10.1104/pp.109.135475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D. A., Soldi M. C. M., Cheavegatti Gianotto A., Carneiro M. S., Amadeu R. R., Aricetti J. A., et al. (2018). Metabolite profiles of sugarcane culm reveal the relationship among metabolism and axillary bud outgrowth in genetically related sugarcane commercial cultivars. Front. Plant Sci. 9:857. 10.3389/fpls.2018.00857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner F., Barbier F. F., Feil R., Watanabe M., Annunziata M. G., Chabikwa T. G., et al. (2017). Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J. 92 611–623. 10.1111/tpj.13705 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Lynch T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson S. A., Krishnareddy S. R., Kebrom T. H., Casal J. J. (2010). Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 152 1914–1927. 10.1104/pp.109.148833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. M., Dale J. E. (1974). Growth of tiller buds in barley: effects of shade treatment and mineral nutrition. Ann. Bot. 38 63–76. 10.1093/oxfordjournals.aob.a084802 [DOI] [Google Scholar]
- Foo E., Bullier E., Goussot M., Foucher F., Rameau C., Beveridge C. A. (2005). The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17 464–474. 10.1105/tpc.104.026716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B. G. (2002a). Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53 203–224. 10.1146/annurev.arplant.53.100301.135256 [DOI] [PubMed] [Google Scholar]
- Forde B. G. (2002b). The role of long-distance signalling in plant responses to nitrate and other nutrients. J. Exp. Bot. 53 39–43. 10.1093/jexbot/53.366.39 [DOI] [PubMed] [Google Scholar]
- Funayama K., Kojima S., Tabuchi-Kobayashi M., Sawa Y., Nakayama Y., Hayakawa T., et al. (2013). Cytosolic glutamine synthetase1; 2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 54 934–943. 10.1093/pcp/pct046 [DOI] [PubMed] [Google Scholar]
- Furet P. M., Lothier J., Demotes-Mainard S., Travier S., Henry C., Guérin V., et al. (2014). Light and nitrogen nutrition regulate apical control in Rosa hybrida L. J. Plant Physiol. 171 7–13. 10.1016/j.jplph.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Garbez M., Galopin G., Sigogne M., Favre P., Demotes-Mainard S., Symoneaux R. (2015). Assessing the visual aspect of rotating virtual rose bushes by a labeled sorting task. Food Qual. Prefer. 40(Part B), 287–295. 10.1016/j.foodqual.2014.06.008 [DOI] [Google Scholar]
- Girault T., Abidi F., Sigogne M., Pelleschi-Travier S. A. N. D. R. I. N. E., Boumaza R., Sakr S., et al. (2010). Sugars are under light control during bud burst in Rosa sp. Plant Cell Environ. 33 1339–1350. 10.1111/j.1365-3040.2010.02152.x [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pagès V., Dun E. A., Pillot J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- González-Grandío E., Cubas P. (2014). Identification of gene functions associated to active and dormant buds in Arabidopsis. Plant Signal. Behav. 9:e27994. 10.4161/psb.27994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E., Pajoro A., Franco-Zorrilla J. M., Tarancón C., Immink R. G., Cubas P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. U.S.A. 114 E245–E254. 10.1073/pnas.1613199114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E., Poza-Carrión C., Sorzano C. O. S., Cubas P. (2013). BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25 834–850. 10.1105/tpc.112.108480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi G., Millard P., Wendler R., Minotta G., Tagliavini M. (2002). Measurement of xylem sap amino acid concentrations in conjunction with whole tree transpiration estimates spring N remobilization by cherry (Prunus avium L.) trees. Plant Cell Environ. 25 1689–1699. 10.1046/j.1365-3040.2002.00949.x [DOI] [Google Scholar]
- Guak S., Neilsen D., Millard P., Wendler R., Neilsen G. H. (2003). Determining the role of N remobilization for growth of apple (Malus domestica Borkh.) trees by measuring xylem-sap N flux. J. Exp. Bot. 54 2121–2131. 10.1093/jxb/erg228 [DOI] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E. B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22 1161–1173. 10.1105/tpc.109.069203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. M., Hillman J. R. (1975). Correlative inhibition of lateral bud growth in Phaseolus vulgaris L. Timing of bud growth following decapitation. Planta 123 137–143. 10.1007/BF00383862 [DOI] [PubMed] [Google Scholar]
- Hayes S. (2018). Revealing the invisible: a synthetic reporter for ABA. Plant Physiol. 177 1346–1347. 10.1104/pp.18.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A., Stirnberg P., Beveridge C., Leyser O. (2009). Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 151 400–412. 10.1104/pp.109.137646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Sponsel V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34 740–760. 10.1007/s00344-015-9546-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C. A., Chin-Atkins A. N., Wilson I. W., Chapple R., Dennis E. S., Chaudhury A. (2001). The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13 2115–2125. 10.1105/TPC.010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H. A., Smeekens S. (2014). Sugar sensing and signaling in plants. Front. Plant Sci. 5:113. 10.3389/fpls.2014.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C., Rabot A., Laloi M., Mortreau E., Sigogne M., Leduc N., et al. (2011). Regulation of RhSUC2, a sucrose transporter, is correlated with the light control of bud burst in Rosa sp. Plant Cell Environ. 34 1776–1789. 10.1111/j.1365-3040.2011.02374.x [DOI] [PubMed] [Google Scholar]
- Holalu S. V., Finlayson S. A. (2017). The ratio of red light to far red light alters Arabidopsis axillary bud growth and abscisic acid signalling before stem auxin changes. J. Exp. Bot. 68 943–952. 10.1093/jxb/erw479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan J. M., Wareing P. F. (1980). Cytokinins and the growth responses of seedlings of Betula pendula Roth. and Acer pseudoplatanus L. to nitrogen and phosphorus deficiency. J. Exp. Bot. 31 525–532. 10.1093/jxb/31.2.525 [DOI] [Google Scholar]
- Howarth D. G., Donoghue M. J. (2006). Phylogenetic analysis of the “ECE”(CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U.S.A. 103 9101–9106. 10.1073/pnas.0602827103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L., McSteen P., Doebley J., Hake S. (2002). Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Jiang H., Egli D. B. (1993). Shade induced changes in flower and pod number and flower and fruit abscission in soybean. Agron. J. 85 221–225. 10.2134/agronj1993.00021962008500020011x [DOI] [Google Scholar]
- Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504 401–405. 10.1038/nature12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42 541–544. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Johnson X., Brcich T., Dun E. A., Goussot M., Haurogné K., Beveridge C. A., et al. (2006). Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiol. 142 1014–1026. 10.1104/pp.106.087676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T., Chandler P., Swain S., King R., Richards R., Spielmeyer W. (2012). Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 160 308–318. 10.1104/pp.112.197954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T. H. (2017). A growing stem inhibits bud outgrowth-the overlooked theory of apical dominance. Front. Plant Sci. 8:1874. 10.3389/fpls.2017.01874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T. H., Brutnell T. P., Finlayson S. A. (2010). Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ. 33 48–58. 10.1111/j.1365-3040.2009.02050.x [DOI] [PubMed] [Google Scholar]
- Kebrom T. H., Burson B. L., Finlayson S. A. (2006). Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140 1109–1117. 10.1104/pp.105.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T. H., Mullet J. E. (2015). Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant Cell Environ. 38 1471–1478. 10.1111/pce.12500 [DOI] [PubMed] [Google Scholar]
- Kerr S. C., Beveridge C. A. (2017). IPA1: a direct target of SL signaling. Cell Res. 27 1191–1192. 10.1038/cr.2017.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68 147–158. 10.1111/j.1365-313X.2011.04674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M. A., Beguerie S., et al. (2011). Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155 974–987. 10.1104/pp.110.164640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Zhang M., Ding Z. (2014). D53: the missing link in strigolactone signaling. Mol. Plant 7 761–763. 10.1093/mp/ssu016 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 1607–1619. 10.1105/tpc.9.9.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 337–348. 10.1046/j.1365-313X.2002.01294.x [DOI] [PubMed] [Google Scholar]
- Lantzouni O., Klermund C., Schwechheimer C. (2017). Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings. Plant J. 92 924–938. 10.1111/tpj.13729 [DOI] [PubMed] [Google Scholar]
- Le Moigne M. A., Guérin V., Furet P. M., Billard V., Lebrec A., Spíchal L., et al. (2018). Asparagine and sugars are both required to sustain secondary axis elongation after bud outgrowth in Rosa hybrid. J. Plant Physiol. 222 17–27. 10.1016/j.jplph.2017.12.013 [DOI] [PubMed] [Google Scholar]
- Leduc N., Roman H., Barbier F., Péron T., Huché-Thélier L., Lothier J., et al. (2014). Light signaling in bud outgrowth and branching in plants. Plants 3 223–250. 10.3390/plants3020223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemerle D., Verbeek B., Cousens R. D., Coombes N. E. (1996). The potential for selecting wheat varieties strongly competitive against weeds. Weed Res. 36 505–513. 10.1111/j.1365-3180.1996.tb01679.x [DOI] [Google Scholar]
- Leyser O. (2008). Strigolactones and shoot branching: a new trick for a young dog. Dev. Cell 15 337–338. 10.1016/j.devcel.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2009). The control of shoot branching: an example of plant information processing. Plant Cell Environ. 32 694–703. 10.1111/j.1365-3040.2009.01930.x [DOI] [PubMed] [Google Scholar]
- Li C. J., Bangerth F. (1999). Autoinhibition of indoleacetic acid transport in the shoots of two-branched pea (Pisum sativum) plants and its relationship to correlative dominance. Physiol. Plant. 106 415–420. 10.1034/j.1399-3054.1999.106409.x [DOI] [Google Scholar]
- Li S. (2015). The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal. Behav. 10:e1044192. 10.1080/15592324.2015.1044192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zachgo S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76 901–913. 10.1111/tpj.12348 [DOI] [PubMed] [Google Scholar]
- Li X., Xia K., Liang Z., Chen K., Gao C., Zhang M. (2016). MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 6:32158. 10.1038/srep32158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liller C. B., Neuhaus R., Von Korff M., Koornneef M., Van Esse W. (2015). Mutations in barley row type genes have pleiotropic effects on shoot branching. PLoS One 10:e0140246. 10.1371/journal.pone.0140246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Marchetti C., Le Bras C., Chastellier A., Relion D., Morel P., Sakr S., et al. (2017). 3D phenotyping and QTL analysis of a complex character: rose bush architecture. Tree Genet. Genomes 13:112 10.1007/s11295-017-1194-0 [DOI] [Google Scholar]
- Li-Marchetti C., Le Bras C., Relion D., Citerne S., Huché-Thélier L., Sakr S., et al. (2015). Genotypic differences in architectural and physiological responses to water restriction in rose bush. Front. Plant Sci. 6:355. 10.3389/fpls.2015.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., et al. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21 1512–1525. 10.1105/tpc.109.065987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cheng X., Liu P., Sun J. (2017). miR156-targeted SBP-Box transcription factors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in bread wheat. Plant Physiol. 174 1931–1948. 10.1104/pp.17.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S. F., Yang S. Y., Chen K. T., Hsing Y. I., Zeevaart J. A., Chen L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20 2603–2618. 10.1105/tpc.108.060913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez J. A., Charnikhova T., Gómez-Roldán V., Matusova R., Kohlen W., De Vos R., et al. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178 863–874. 10.1111/j.1469-8137.2008.02406.x [DOI] [PubMed] [Google Scholar]
- Lortie C. J., Aarssen L. W. (1997). Apical dominance as an adaptation in Verbascum thapsus: effects of water and nutrients on branching. Int. J. Plant Sci. 158 461–464. 10.1086/297456 [DOI] [Google Scholar]
- Lu Z., Yu H., Xiong G., Wang J., Jiao Y., Liu G., et al. (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25 3743–3759. 10.1105/tpc.113.113639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 794–799. 10.1038/383794a0 [DOI] [PubMed] [Google Scholar]
- Luo L., Pan S., Liu X., Wang H., Xu G. (2017). Nitrogen deficiency inhibits cell division-determined elongation, but not initiation, of rice tiller buds. Isr. J. Plant Sci. 64 32–40. 10.1080/07929978.2016.1275367 18208511 [DOI] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252 1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Malaguti D., Millard P., Wendler R., Hepburn A., Tagliavini M. (2001). Translocation of amino acids in the xylem of apple (Malus domestica Borkh.) trees in spring as a consequence of both N remobilization and root uptake. J. Exp. Bot. 52 1665–1671. 10.1093/jexbot/52.361.1665 [DOI] [PubMed] [Google Scholar]
- Martín-Fontecha E. S., Tarancón C., Cubas P. (2018). To grow or not to grow, a power-saving program induced in dormant buds. Curr. Opin. Plant Biol. 41 102–109. 10.1016/j.pbi.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Grandío E. G., Serra F., Marcel F., Rodríguez-Buey M. L., Schmitz G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67 701–714. 10.1111/j.1365-313X.2011.04629.x [DOI] [PubMed] [Google Scholar]
- Mason M. G., Ross J. J., Babst B. A., Wienclaw B. N., Beveridge C. A. (2014). Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. U.S.A. 111 6092–6097. 10.1073/pnas.1322045111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel K., Sakr S., Gerbe F., Guilliot A., Bonhomme M., Rageau R., et al. (2004). Sorbitol uptake is regulated by glucose through the hexokinase pathway in vegetative peach-tree buds. J. Exp. Bot. 55 879–888. 10.1093/jxb/erh087 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2009). Hormonal regulation of branching in grasses. Plant Physiol. 149 46–55. 10.1104/pp.108.129056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médiène S., Pagès L., Jordan M. O., Le Bot J., Adamowicz S. (2002). Influence of nitrogen availability on shoot development in young peach trees [Prunus persica (L.) Batsch]. Trees 16 547–554. 10.1007/s00468-002-0204-4 12490426 [DOI] [Google Scholar]
- Millard P., Wendler R., Hepburn A., Smith A. (1998). Variations in the amino acid composition of xylem sap of Betula pendula Roth. trees due to remobilization of stored N in the spring. Plant Cell Environ. 21 715–722. 10.1046/j.1365-3040.1998.00313.x [DOI] [Google Scholar]
- Minakuchi K., Kameoka H., Yasuno N., Umehara M., Luo L., Kobayashi K., et al. (2010). FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51 1127–1135. 10.1093/pcp/pcq083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X. J., Ito M., Asano K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42 545–549. 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37 128–138. 10.1046/j.1365-313X.2003.01945.x [DOI] [PubMed] [Google Scholar]
- Morris D. A. (1977). Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.). Planta 136 91–96. 10.1007/BF00387930 [DOI] [PubMed] [Google Scholar]
- Morris S. E., Cox M. C., Ross J. J., Krisantini S., Beveridge C. A. (2005). Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 138 1665–1672. 10.1104/pp.104.058743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhr M., Paulat M., Awwanah M., Brinkkötter M., Teichmann T. (2018). CRISPR/Cas9-mediated knockout of Populus BRANCHED1 and BRANCHED2 orthologs reveals a major function in bud outgrowth control. Tree Physiol. 38 1588–1597. 10.1093/treephys/tpy088 [DOI] [PubMed] [Google Scholar]
- Muhr M., Prüfer N., Paulat M., Teichmann T. (2016). Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytol. 212 613–626. 10.1111/nph.14076 [DOI] [PubMed] [Google Scholar]
- Müller D., Leyser O. (2011). Auxin, cytokinin and the control of shoot branching. Ann. Bot. 107 1203–1212. 10.1093/aob/mcr069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B. C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299 1404–1407. 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65 23–33. 10.1007/s00239-006-0174-z [DOI] [PubMed] [Google Scholar]
- Ni J., Gao C., Chen M. S., Pan B. Z., Ye K., Xu Z. F. (2015). Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 56 1655–1666. 10.1093/pcp/pcv089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Cubas P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33 33–41. 10.1016/j.pbi.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Nicolas M., Rodríguez-Buey M. L., Franco-Zorrilla J. M., Cubas P. (2015). A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr. Biol. 25 1799–1809. 10.1016/j.cub.2015.05.053 [DOI] [PubMed] [Google Scholar]
- Niwa M., Daimon Y., Kurotani K. I., Higo A., Pruneda-Paz J. L., Breton G., et al. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25 1228–1242. 10.1105/tpc.112.109090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A., Tarkowski P., Tarkowska D., Norbaek R., Åstot C., Dolezal K., et al. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. U.S.A. 101 8039–8044. 10.1073/pnas.0402504101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M., Ishiyama K., Kusano M., Fukushima A., Kojima S., Hanada A., et al. (2015). Lack of cytosolic glutamine synthetase1; 2 in vascular tissues of axillary buds causes severe reduction in their outgrowth and disorder of metabolic balance in rice seedlings. Plant J. 81 347–356. 10.1111/tpj.12731 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramírez C., Hernandez-Coronado M., Thamm A., Catarino B., Wang M., Dolan L., et al. (2016). A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 9 205–220. 10.1016/j.molp.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Pal P. K., Prasad R., Pathania V. (2013). Effect of decapitation and nutrient applications on shoot branching, yield, and accumulation of secondary metabolites in leaves of Stevia rebaudiana Bertoni. J. Plant Physiol. 170 1526–1535. 10.1016/j.jplph.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Pei Z. M., Ghassemian M., Kwak C. M., McCourt P., Schroeder J. I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290. 10.1126/science.282.5387.287 [DOI] [PubMed] [Google Scholar]
- Petrášek J., Friml J. (2009). Auxin transport routes in plant development. Development 136 2675–2688. 10.1242/dev.030353 [DOI] [PubMed] [Google Scholar]
- Poza-Carrión C., Aguilar-Martínez J. A., Cubas P. (2007). Role of TCP gene BRANCHED1 in the control of shoot branching in Arabidopsis. Plant Signal. Behav. 2 551–552. 10.4161/psb.2.6.4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J., Laxmi A., Martin S. K. S., Jang J. C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150. 10.1105/tpc.104.022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti P., Tombesi A. (1996). Effects of gibberellic acid, asparagine and glutamine on flower bud induction in olive. J. Hortic. Sci. 71 383–388. 10.1080/14620316.1996.11515418 [DOI] [Google Scholar]
- Qin L. J., Guo X. Z., Feng X. Z., Weng L., Yan J., Hu X. H., et al. (2004). Cloning of LjCYC1 gene and nuclear localization of LjCYC1 protein in Lotus japonicus. J. Plant Physiol. Mol. Biol. 30 523–532. [PubMed] [Google Scholar]
- Rabot A., Henry C., Baaziz K. B., Mortreau E., Azri W., Lothier J., et al. (2012). Insight into the role of sugars in bud burst under light in the rose. Plant Cell Physiol. 53 1068–1082. 10.1093/pcp/pcs051 [DOI] [PubMed] [Google Scholar]
- Rameau C., Bertheloot J., Leduc N., Andrieu B., Foucher F., Sakr S., et al. (2015). Multiple pathways regulate shoot branching. Front. Plant Sci. 5:741. 10.3389/fpls.2014.00741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L., Comadran J., Druka A., Marshall D. F., Thomas W. T., Macaulay M., et al. (2011). INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43 169–172. 10.1038/ng.745 [DOI] [PubMed] [Google Scholar]
- Revel S. M., Bart J. J., Luo Z., Oplaat C., Susan E. L., Mark W. W., et al. (2015). Environmental control of branching in petunia. Plant Physiol. 168 735–751. 10.1104/pp.15.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards R. A. (2000). Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 51 447–458. 10.1093/jexbot/51.suppl_1.447 [DOI] [PubMed] [Google Scholar]
- Rodríguez D., Andrade F. H., Goudriaan J. (1999). Effects of phosphorus nutrition on tiller emergence in wheat. Plant Soil 209 283–295. 10.1023/A:1004690404870 [DOI] [Google Scholar]
- Roman H., Girault T., Barbier F., Péron T., Brouard N., Pencik A., et al. (2016). Cytokinins are initial targets of light in the control of bud outgrowth. Plant Physiol. 172 489–509. 10.1104/pp.16.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another below-ground role for strigolactones? Plant Physiol. 155 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T., Thimann K. V. (1967). The role of auxins and cytokinins in the release of buds from dominance. Am. J. Bot. 54 136–144. 10.1002/j.1537-2197.1967.tb06901.x [DOI] [Google Scholar]
- Sakakibara H., Suzuki M., Takei K., Deji A., Taniguchi M., Sugiyama T. (1998). A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 14 337–344. 10.1046/j.1365-313X.1998.00134.x [DOI] [PubMed] [Google Scholar]
- Sakr S., Wang M., Dédaldéchamp F., Perez-Garcia M. D., Ogé L., Hamama L., et al. (2018). The sugar-signaling hub: overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 19:E2506. 10.3390/ijms19092506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K., Nath U. (2018). CIN-TCP transcription factors: transiting cell proliferation in plants. IUBMB Life 70 718–731. 10.1002/iub.1874 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale M., Bennett T., Leyser O. (2017). BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144 1661–1673. 10.1242/dev.145649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7 41–48. 10.1016/S1360-1385(01)02187-2 [DOI] [PubMed] [Google Scholar]
- Shimizu S., Mori H. (1998). Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol. 39 255–262. 10.1093/oxfordjournals.pcp.a029365 [DOI] [PubMed] [Google Scholar]
- Shinohara N., Taylor C., Leyser O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11:e1001474. 10.1371/journal.pbio.1001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Morel K., Durand E., Brevalle G., Girard T., Lauri P. -É. (2011). Aphids at crossroads: when branch architecture alters aphid infestation patterns in the apple tree. Trees 26 273–282. 10.1007/s00468-011-0629-8 [DOI] [Google Scholar]
- Song X., Lu Z., Yu H., Shao G., Xiong J., Meng X., et al. (2017). IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 27 1128–1141. 10.1038/cr.2017.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., et al. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 1469–1474. 10.1101/gad.256603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., et al. (2015). SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27 3143–3159. 10.1105/tpc.15.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom J. P., Sussex I. M. (1988). Patterns of protein synthesis in dormant and growing vegetative buds of pea. Planta 176 497–505. 10.1007/BF00397656 [DOI] [PubMed] [Google Scholar]
- Stirnberg P., Chatfield S. P., Leyser H. O. (1999). AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121 839–847. 10.1104/pp.121.3.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A., Zhao Q., Ross-Ibarra J., Doebley J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43 1160–1163. 10.1038/ng.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Abernathy S. D., White R. H., Finlayson S. A. (2011). Photosynthetic photon flux density and phytochrome B interact to regulate branching in Arabidopsis. Plant Cell Environ. 34 1986–1998. 10.1111/j.1365-3040.2011.02393.x [DOI] [PubMed] [Google Scholar]
- Suzuki T., Sakurai K., Ueguchi C., Mizuno T. (2001). Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in his-to-asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol. 42 37–45. 10.1093/pcp/pce011 [DOI] [PubMed] [Google Scholar]
- Ta T. C., MacDowall F. D. H., Faris M. A. (1987). Utilization of carbon from shoot photosynthesis and nodule CO2 fixation in the fixation and assimilation of nitrogen by alfalfa root nodules. Can. J. Bot. 65 2537–2541. 10.1139/b87-344 [DOI] [Google Scholar]
- Takeda T., Suwa Y., Suzuki M., Kitano H., Ueguchi-Tanaka M., Ashikari M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33 513–520. 10.1046/j.1365-313X.2003.01648.x [DOI] [PubMed] [Google Scholar]
- Takei K., Sakakibara H., Taniguchi M., Sugiyama T. (2001). Nitrogen-dependent accumulation of cytokinins in root and thetranslocation to leaf: implication of cytokinin species that induces geneexpression of maize responseregulator. Plant Cell Physiol. 42 85–93. 10.1093/pcp/pce009 [DOI] [PubMed] [Google Scholar]
- Takei K., Takahashi T., Sugiyama T., Yamaya T., Sakakibara H. (2002). Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J. Exp. Bot. 53 971–977. 10.1093/jexbot/53.370.971 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Takei K., Kojima M., Sakakibara H., Mori H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45 1028–1036. 10.1111/j.1365-313X.2006.02656.x [DOI] [PubMed] [Google Scholar]
- Tarancón C., González-Grandío E., Oliveros J. C., Nicolas M., Cubas P. (2017). A conserved carbon starvation response underlies bud dormancy in woody and herbaceous species. Front. Plant Sci. 8:788. 10.3389/fpls.2017.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann T., Muhr M. (2015). Shaping plant architecture. Front. Plant Sci. 6:233. 10.3389/fpls.2015.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitithanakul S. (2012). Effect of Nitrogen Supply Before Bud Break on Early Development of the Young Hybrid Poplar. Doctoral dissertation, Université Blaise Pascal, Clermont-Ferrand. [Google Scholar]
- Thitithanakul S., Pétel G., Chalot M., Beaujard F. (2012). Supplying nitrate before bud break induces pronounced changes in nitrogen nutrition and growth of young poplars. Funct. Biol. 39 795–803. 10.1071/FP12129 [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Waldie T., Leyser O. (2018). Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 177 803–818. 10.1104/pp.17.01691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., et al. (2015). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27 3128–3142. 10.1105/tpc.15.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhao Y., Li Z., Hsu C. C., Liu X., Fu L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69 100–112. 10.1016/j.molcel.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. L., Stec A., Hey J., Lukens L., Doebley J. (1999). The limits of selection during maize domestication. Nature 398 236–239. 10.1038/18435 [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiao Y. (2018). Axillary meristem initiation-a way to branch out. Curr. Opin. Plant Biol. 41 61–66. 10.1016/j.pbi.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. (2013). Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 27 681–688. 10.1016/j.devcel.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Waters M. T., Gutjahr C., Bennett T., Nelson D. C. (2017). Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68 291–322. 10.1146/annurev-arplant-042916-040925 [DOI] [PubMed] [Google Scholar]
- Wickson M., Thimann K. V. (1958). The antagonism of auxin and kinetin in apical dominance. Physiol. Plant. 11 62–74. 10.1111/j.1399-3054.1958.tb08426.x [DOI] [Google Scholar]
- Xu J., Zha M., Li Y., Ding Y., Chen L., Ding C., et al. (2015). The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep. 34 1647–1662. 10.1007/s00299-015-1815-8 [DOI] [PubMed] [Google Scholar]
- Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S., Umehara M. (2014). Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240 399–408. 10.1007/s00425-014-2096-0 [DOI] [PubMed] [Google Scholar]
- Yang Y., Nicolas M., Zhang J., Yu H., Guo D., Yuan R., et al. (2018). The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 14:e1007296. 10.1371/journal.pgen.1007296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C., Scott A. F. (2015). Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169 611–626. 10.1104/pp.15.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Yoneyama K., Takeuchi Y., Sekimoto H. (2007). Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225 1031–1038. 10.1007/s00425-006-0410-1 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Zhang Z. B., Yang G., Arana F., Chen Z., Li Y., Xia H. J. (2007). Arabidopsis inositol polyphosphate 6-/3-kinase (AtIpk2β) is involved in axillary shoot branching via auxin signaling. Plant Physiol. 144 942–951. 10.1104/pp.106.092163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D. L., Atlin G. N., Bastiaans L., Spiertz J. H. J. (2006). Developing selection protocols for weed competitiveness in aerobic rice. Field Crops Res. 97 272–285. 10.1016/j.fcr.2005.10.008 [DOI] [Google Scholar]
- Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., et al. (2013). D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504 406–410. 10.1038/nature12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhang J., Yan J., Song R. (2011). Two transposable element insertions are causative mutations for the major domestication gene teosinte branched 1 in modern maize. Cell Res. 21 1267–1270. 10.1038/cr.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Zhang S., Zhang W., Li G., Chen Z., Zhai W., et al. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48 687–698. 10.1111/j.1365-313X.2006.02916.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.rosaceae.org.