Abstract
Renal Cysts and Diabetes Syndrome (RCAD) is an autosomal dominant disorder caused by mutations in the HNF1B gene encoding for the transcriptional factor hepatocyte nuclear factor-1B. RCAD is characterized as a multi-organ disease, with a broad spectrum of symptoms including kidney abnormalities (renal cysts, renal hypodysplasia, single kidney, horseshoe kidneys, hydronephrosis), early-onset diabetes mellitus, abnormal liver function, pancreatic hypoplasia and genital tract malformations. In the present study, using capillary electrophoresis coupled to mass spectrometry (CE-MS), we investigated the urinary proteome of a pediatric cohort of RCAD patients and different controls to identify peptide biomarkers and obtain further insights into the pathophysiology of this disorder. As a result, 146 peptides were found to be associated with RCAD in 22 pediatric patients when compared to 22 healthy age-matched controls. A classifier based on these peptides was generated and further tested on an independent cohort, clearly discriminating RCAD patients from different groups of controls. This study demonstrates that the urinary proteome of pediatric RCAD patients differs from autosomal dominant polycystic kidney disease (PKD1, PKD2), congenital nephrotic syndrome (NPHS1, NPHS2, NPHS4, NPHS9) as well as from chronic kidney disease conditions, suggesting differences between the pathophysiology behind these disorders.
Introduction
Renal Cysts and Diabetes (RCAD) syndrome is caused by heterozygous mutations in the HNF1B gene, encoding the transcriptional factor hepatocyte nuclear factor-1B. RCAD syndrome (RCAD, OMIM #137920)1 can also be referred as MODY5 (Maturity Onset Diabetes of the Young type 5)2. The wide spectrum of clinical features in RCAD patients is due to the multisystem role of HNF1B, which is involved in normal morphogenesis of several organs, including kidneys, pancreas, liver, and genitourinary tract3. Consistent with its broad developmental expression pattern4,5, studies on fetuses carrying HNF1B mutations revealed a fundamental function during kidney, urogenital tract and pancreas development4,6–8.
The RCAD disease is inherited in an autosomal dominant pattern9. To date, more than 150 mutations have been described in the HNF1B gene. Half of the RCAD patients characterized up to date present missenses, nonsenses, frameshifts, splice site mutations, and insertions/deletions while the other half of patients present whole gene deletions10.
The most prominent clinical feature in HNF1B-associated syndrome is the renal disease, usually characterized by renal cysts, renal dysplasia, solitary or horseshoe kidney, hydronephrosis, and hyperuricaemic nephropathy11. The renal abnormalities of HNF1B mutant carriers have also been related to the congenital anomalies of the kidney and the urinary tract (CAKUT)11. Moreover, recently, HNF1B-mutations have been associated in some patients to autosomal dominant tubulointerstitial kidney disease (ADTKD-HNF1B)12. In addition, several cases of unknown chronic kidney disease (CKD) have been reported both in children13 and adults14,15. Tubular dysfunction manifesting by hypomagnesemia, hypocalciuria16–18, and hyperuricemia14,19 has also been described. Extrarenal features comprise maturity-onset diabetes of the young, pancreatic hypoplasia, abnormal liver function, and genital tract malformations. The phenotype of HNF1B mutant carriers is indeed highly variable within and between families20. These observations led to the hypothesis that non-allelic factors, as well as stochastic variation in temporal HNF1B gene expression and environmental factors, could cause the strong intrafamilial variability of RCAD patients3,21.
Urinary proteomics is increasingly being employed in kidney disease research. Several studies have demonstrated that capillary electrophoresis coupled to mass spectrometry (CE-MS) enables the identification and validation of several biomarkers or peptide signatures classifying the diagnosis and prognosis of various kidney diseases22–25. In addition to their diagnostic and prognostic usefulness, proteomics derived biomarkers may advance the understanding of the molecular pathways involved in the pathogenesis of a specific disorder or condition.
In this study, we aimed to obtain more insights into the renal pathophysiology of the RCAD syndrome by applying a proteomic approach to investigate changes at urinary peptides level that can be used to characterize RCAD patients.
Results
Study setup and patient data
In total, 244 urine samples were included in this study: 44 samples were used for discovery and 200 samples were used as a validation set (Fig. 1A). In the discovery set, we included 22 RCAD patients and 22 healthy controls. Subsequently, we used a wider population comprising healthy patients (n = 20), RCAD patients (n = 24), autosomal dominant polycystic kidney disease (ADPKD) patients (n = 55), CKD patients (n = 55), and patients with nephrotic syndrome (n = 46) as a validation set. The urinary proteome data for all the samples were previously measured and derived from the Human Urinary Proteome Database26–28 with the exception of all RCAD urine samples, which were analyzed by CE-MS for this specific study. The 46 RCAD patients were divided into discovery and validation set. The RCAD samples used in the discovery were matched with healthy controls based on age and gender. Furthermore, we divided both sets considering similar phenotypes. The clinical features of the used RCAD patients are described in Table 1 and Supplementary Table 1.
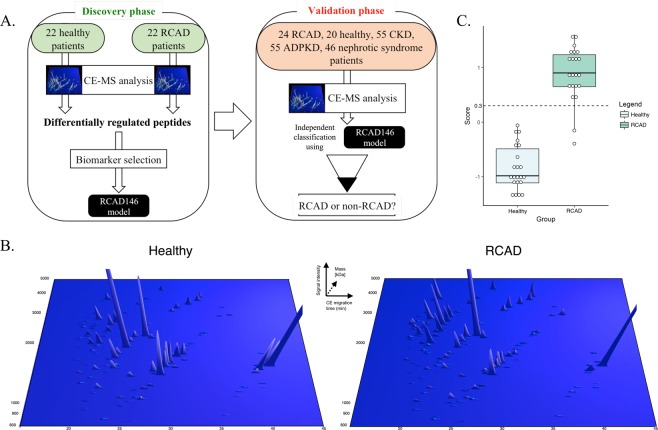
Figure 1.
Study design and urinary CE-MS analysis of patients with RCAD. (A) The analysis was performed in two phases: a discovery phase, where the urinary proteome of 44 pediatric patients (22 healthy, 22 RCAD) was analyzed leading to the identification of 146 sequenced urinary peptides that were modelled in a SVM classifier called RCAD146. In the next step, the validation phase, we studied the discriminatory ability of the panel RCAD146 panel in new RCAD patients (n = 24) and individuals with CKD or patients carrying monogenic mutations associated with different renal diseases. (B) Representation of the 146 urinary peptides significantly modified between RCAD and healthy controls. Normalized molecular mass (kDa) was plotted against normalized capillary electrophoresis (CE)-migration time (min). Mean signal intensity was given in 3-dimensional depiction. (C) Cross-validation score of the RCAD146 model from the analysis of the discovery cohort along with the definition of the cut-off 0.3 (dashed line).
Table 1.
Clinical characteristics of children with RCAD syndrome.
| All | |
|---|---|
| n | 46 |
| Male, n (%) | 37 (80.4) |
| Female, n (%) | 9 (19.6) |
| Age, years | 8.4 ± 5.7 |
| eGFR (ml/min/1.73 m2) | 86.74 ± 41.15 |
| U-protein (g/L) | 0.12 ± 0.35 |
| Renal phenotypes | |
| kidney cysts, n (%) | 34 (73.9) |
| bilateral hyperechoic kidneys, n (%) | 24 (52.1) |
| hypo/dysplastic kidneys, n (%) | 21 (45.6) |
| single kidneys, n (%) | 3 (6.5) |
| vesicoureteral reflux, n (%) | 2 (4.3) |
| horseshoe kidneys, n (%) | 1 (2.1) |
| chronic renal failure, n (%) | 1 (2.1) |
| Extrarenal phenotypes | |
| diabetes, n (%) | 3 (6.5) |
| pancreatic hypoplasia, n (%) | 3 (6.5) |
| uterine malformations, n (%) | 2 (4.3) |
| unilateral ectopic testis, n (%) | 2 (4.3) |
| hyperechoic liver, n (%) | 1 (2.1) |
| cholestasis, n (%) | 1 (2.1) |
| megabladder, n (%) | 1 (2.1) |
| hyperuricemia, n (%) | 5 (10.8) |
Identification of RCAD-related urinary peptides and development of a urinary peptide-based classifier
For the identification of significant urinary peptides related to the RCAD syndrome, we compared the urinary proteome profiles of 22 patients carrying HNF1B heterozygous mutations with 22 age- and gender-matched healthy controls (Table 2A). The statistical analysis was adjusted for multiple testing following the concept described by Benjamini and Hochberg29 and defined in the clinical proteomics guidelines30. This led to the identification of 294 differentially excreted peptides (corrected p < 0.05) between these two groups. For 146 out of the 294, high-confidence sequence information could be assigned. Fragments of uromodulin (UMOD), protein unc-119 homolog A (UNC119), and mucin (MUC1), as well as a large number of collagen fragments, were identified. Moreover, peptides associated with calcium binding were also detected. Amongst them, peptides such as sarcalumenin (SRL), and annexin A1 (ANXA1) were downregulated. In contrast, peptide fragments such as gelsolin (GSN), short transient receptor potential channel 4-associated protein (TRPC4AP) and the direct target of HNF1B - osteopontin (SPP1)31 were upregulated. All relevant details, encompassing the sequence information as well as the fold-change, are described in Supplementary Table 2. The difference in abundance of these 146 peptides between RCAD patients and healthy controls is shown in Fig. 1B and Table 3. These proteome plots are showing the mean abundance of the significant peptide-biomarker in urine of RCAD patients and healthy individuals. The 146 sequenced peptides were combined into a classifier termed “RCAD146” using a support vector machine (SVM), which was optimized for the classification of patients in the discovery cohort. Based on a cut-off score of 0.30 (Fig. 1C), the RCAD classifier discriminated RCAD from healthy controls with 90.9% sensitivity and 100% specificity and an AUC of 0.99 in the discovery cohort.
Table 2.
Baseline characteristics of the subjects used in the A. discovery set and B. validation set.
| Group of patients | Mutation | Sample size (n) | Gender | Age (years) | eGFR (ml/min/1.73 m2) | U-albumin (mg/l) | U-protein (g/l) | |
|---|---|---|---|---|---|---|---|---|
| Male | Female | |||||||
| A. Discovery Set | ||||||||
| RCAD | HNF1B | 22 | 17 | 5 | 9.4 ± 6.4 | 81.0 ± 43.67 | 0.18 ± 0.50 | |
| Healthy | — | 22 | 17 | 5 | 9.3 ± 4.1 | 126.33 ± 30.03 | — | — |
| Patients’ condition | Mutation | Sample size (n) | Gender | Age (years) | Mean eGFR (ml/min/1.73 m2) | U-albumin (mg/l) | U-protein (g/l) | |
| Male | Female | |||||||
| B. Validation Set | ||||||||
| RCAD | HNF1B | 24 | 20 | 4 | 7.5 ± 5.1 | 92.03 ± 38.88 | 0.07 ± 0.08 | |
| Healthy | — | 20 | 15 | 5 | 9.3 ± 2.4 | 118.5 ± 38.32 | — | — |
| CKD | — | 55 | 28 | 27 | 12.0 ± 6.1 | 69.79 ± 23.94 | 867.65 ± 1673.49 | — |
| ADPKD | PKD1 | 46 | 20 | 26 | 34.3 ± 6.0 | 77.33 ± 18.08 | — | |
| PKD2 | 9 | 3 | 6 | 40.2 ± 3.3 | — | |||
| Nephrotic Syndrome | NPHS1 | 2 | 2 | — | 6.5 ± 8.8 | 108.50 ± 80.95 | 244.14 ± 398.23 | — |
| NPHS2 | 35 | 14 | 21 | 10.5 ± 6.4 | ||||
| NPHS4 (WT1) | 6 | 1 | 5 | 13.3 ± 3.7 | ||||
| NPHS9 (ADCK4) | 3 | 1 | 2 | 13.6 ± 3.1 | ||||
Table 3.
Proteins origin of the 146 differentially excreted urinary peptides obtained by the comparison between RCAD and healthy patients.
| Protein name | Gene symbol | N° of protein fragments | P-value (adjusted) | Mean fold change |
|---|---|---|---|---|
| Mucin-3a | MUC3A | 1 | 1.10E-05 | −56.9 |
| Collagen alpha-1 (XXVI) chain | COL26A1 | 1 | 1.72E-03 | −11.2 |
| Protein unc-119 homolog A | UNC119 | 1 | 4.95E-03 | −7.05 |
| Collagen alpha-3 (V) chain | COL5A3 | 1 | 7.70E-04 | −6.25 |
| Uromodulin | UMOD | 4 | 4.82E-02 | −5.7 (±2.33) |
| Collagen alpha-1 (XXVII) chain | COL27A1 | 1 | 2.04E-02 | −5.66 |
| Cystatin-A | CSTA | 1 | 5.88E-03 | −3.67 |
| Hemoglobin subunit delta | HBD | 1 | 3.65E-02 | −3.38 |
| Annexin A1 | ANXA1 | 1 | 6.44E-03 | −2.68 |
| Sarcalumenin | SRL | 1 | 1.05E-02 | −2.21 |
| Collagen alpha-1 (XVII) chain | COL17A1 | 1 | 3.14E-03 | −2.03 |
| Ig lambda-2 chain C regions | IGLC2 | 1 | 1.45E-02 | −1.92 |
| Beta-2-microglobulin | B2M | 1 | 1.04E-02 | −1.17 |
| Serum amyloid A protein | SAA1 | 2 | 4.87E-02 | −0.81 (±2.95) |
| Collagen alpha-1 (XI) chain | COL11A1 | 1 | 4.21E-03 | 1.19 |
| Collagen alpha-1 (VIII) chain | COL8A1 | 1 | 3.61E-02 | 1.29 |
| Kininogen-1 | KNG1 | 1 | 1.69E-02 | 1.53 |
| Collagen alpha-1 (I) chain | COL1A1 | 52 | 4.97E-02 | 1.7 (±3.42) |
| Short transient receptor potential channel 4-associated protein | TRPC4AP | 1 | 1.74E-02 | 1.98 |
| Collagen alpha-2 (V) chain | COL5A2 | 3 | 1.67E-02 | 2.23 (±2.9) |
| Collagen alpha-1 (XVI) chain | COL16A1 | 1 | 3.53E-02 | 2.24 |
| Mucin-1 subunit alpha | MUC1 | 1 | 9.90E-03 | 2.4 |
| Ig kappa chain C region | IGKC | 1 | 1.68E-02 | 2.64 |
| Collagen alpha-5 (IV) chain | COL4A5 | 1 | 2.88E-02 | 2.86 |
| Collagen alpha-6 (IV) chain | COL4A6 | 1 | 5.61E-04 | 3 |
| Retinol binding protein 4 | RBP4 | 1 | 7.78E-03 | 3.11 |
| Protein scribble homolog | SCRIB | 1 | 3.51E-02 | 3.15 |
| Collagen alpha-2 (I) chain | COL1A2 | 20 | 4.84E-02 | 3.28 (±6.86) |
| Actin, cytoplasmic 1 | ACTB | 1 | 2.88E-02 | 3.3 |
| Neurosecretory protein VGF | VGF | 1 | 3.80E-02 | 3.45 |
| Collagen alpha-1 (III) chain | COL3A1 | 21 | 4.67E-02 | 3.47 (±4.54) |
| Osteopontin | OPN | 2 | 1.56E-02 | 3.85 (±2.21) |
| Collagen alpha-4 (IV) chain | COL4A4 | 1 | 1.46E-03 | 4 |
| Collagen alpha-1 (II) chain | COL2A1 | 7 | 3.05E-02 | 4.45 (±5.37) |
| Fibrinogen alpha chain | FGA | 2 | 5.87E-03 | 4.79 (±0.48) |
| Ephrin-A1 | EFNA1 | 1 | 5.46E-03 | 5.55 |
| Interleukin-1 receptor-associated | IL-1R | 1 | 1.27E-04 | 43.2 |
| Immunoglobulin kappa variable 4-1 | IGKV4-1 | 1 | 1.88E-04 | 56.7 |
| Gelsolin | GSN | 1 | 1.66E-05 | 105.4 |
| Microfibrillar-associated protein 5 | MFAP5 | 1 | 4.32E-05 | 308.3 |
| Collagen alpha-1 (V) chain | COL5A1 | 2 | 4.36E-02 | 180.32 (±253) |
Proteins origin of the 146 differentially excreted urinary peptides obtained by the comparison between RCAD and healthy patients. An extended detailed version of the table can be found in Supplementary Table 2. Data described the number of significant peptides related to each protein, the lowest observed P-value and the mean fold change (±standard deviation).
Validation of the RCAD146 classifier in an independent group
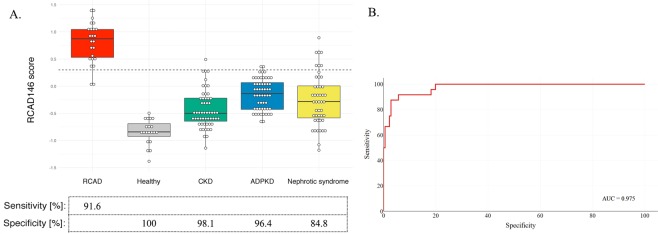
The RCAD146 classifier was validated in an independent group of samples (Table 2B), consisting of 24 RCAD and 20 healthy controls. The analysis revealed an AUC of 1.00 [0.92 to 1.00 (95% CI); p < 0.0001]. At the pre-defined cut-off level of 0.30 based on the discovery cohort, the classifier displayed a sensitivity of 91.67% and specificity of 100%. To obtain confirmation about how RCAD146 performed in differentiating the pediatric RCAD urinary proteome from other kidney diseases, we further selected a group of patients with chronic kidney disease (CKD) (n = 55). This control group of children was particularly interesting as (i) 40% of adults with HNF1B mutations develop CKD14, (ii) it represents a condition with severe chronic kidney damage and, (iii) confirms that the performance of the RCAD146 classifier is independent of proteinuria in RCAD patients. This analysis showed a specificity of 98.18% and an AUC of 0.987 [0.931 to 1.00 (95% CI); p < 0.0001] for the classification of children with CKD as non-RCAD. To further evaluate the specificity and validity of the pediatric RCAD urinary proteomic pattern, the classifier was subsequently tested in patients with different monogenic kidney diseases, including ADPKD and nephrotic syndrome. In the case of ADPKD, 53 out of 55 patients were scored as non-RCAD corresponding to a specificity of 96.4% [AUC: 0.974] (Fig. 2A). In the group of patients with nephrotic syndrome, 39 of 46 patients were scored by RCAD146 as non-RCAD displaying a specificity of 84.8% [AUC: 0.952] (Fig. 2A).
Figure 2.
Blinded validation of urinary peptide classifier RCAD146 in a separate RCAD patient population together with healthy controls and patients suffering from other kidney diseases. (A) Box-and-Whisker plot for the classification of all patient cohorts (RCAD subjects, non-RCAD groups) of the validation set according to the RCAD146 scores. (B) ROC curve for the RCAD146 model based on all samples used in the validation cohort.
When evaluating all data sets combined, the overall sensitivity and specificity were 91.67% and 94.32%, respectively. Furthermore, this ROC analysis including all datasets revealed an AUC of 0.975 [0.943 to 0.992 (95% CI); p < 0.0001] (Fig. 2B).
Discussion
This is the first study showing a unique proteome profile that distinguishes children with RCAD from healthy controls and patients suffering from different renal diseases.
The most prominent finding of the study was the identification of 294 differentially regulated peptides potentially related to RCAD syndrome, where sequence information was obtained for 146 peptides. Similar to a previous study on ADPKD and urinary peptides32, the majority of peptides enriched in the urine of RCAD patients were collagen type I or type III fragments. This may reflect active extracellular matrix (ECM) remodelling, which could be related to ECM modifications due to cyst expansion33. The abundance of collagen and osteopontin fragments in RCAD children displayed an opposite tendency to previous findings described in several studies for different kidney diseases22,23,34. Collagens are characterized as the most abundant urinary peptides as well as the main elements of the interstitial ECM, being involved in different biological functions as cell adhesion, tissue development and tensile strength35,36. Along these lines, osteopontin-derived peptides were also identified increased in RCAD patients, due to the involvement of osteopontin in the remodelling of the ECM34. Therefore, the increased abundance of collagen and osteopontin fragments in the RCAD urinary samples may reflect the cystic phenotype and the still non-fibrotic status of patients’ kidneys, whereas tubulointerstitial fibrosis determines the peptide excretion pattern in CKD37. Additionally, an early decline in kidney function may be predicted by the decreased excretion of uromodulin (UMOD) fragments38, which has also been found reduced in cases with tubular atrophy and fibrosis39. Another peptide fragment deregulated in the present study was mucin 1 (MUC1), an extracellular protein expressed in the renal tubular epithelium. Recently, MUC1 was described to be a predictor of renal impairment40, along with its increase in mice and human was correlated with the development of fibrosis41. It is important to notice that mutations in UMOD, MUC1, and HNF1B are responsible for ADTKD, showing a strict correlation between these proteins and RCAD along with ADTKD phenotypes42. Considering the acknowledged role of HNF1B in regulating kidney transports and also calcium-sensing receptor CaSR16,43, it was interesting to notice that several peptides associated with calcium binding or calcium regulating properties might be changed in RCAD patients. The disruption of multiple calcium regulators may be one of the bases of the renal cysts formation as observed previously44,45. Furthermore, the protein unc-119 homolog A (UNC119) that plays a crucial role in the proper ciliary targeting of the cystic gene nephrocystin-346, was decreased.
The RCAD146 classifier correctly identified most of CKD patients as non-RCAD. Since CKD is a rare condition in RCAD children13, it would be of interest testing a cohort of adult RCAD patients suffering from CKD, in order to investigate the performance of the RCAD146 classifier.
The other disorders used as disease controls in this study were biologically related (e.g. ADPKD) or non-related (e.g. nephrotic syndrome) to the RCAD syndrome. A group of patients affected by ADPKD appears relevant as they display phenotypic correlations with the RCAD syndrome. HNF1B was shown to regulate Pkd2 in the mouse47 and mutations in HNF1B can mimic polycystic kidney disease especially in the prenatal setting and early childhood48,49. Notably, the RCAD146 classifier precisely discriminated RCAD from ADPKD.
When compared with monogenic mutations sharing a common nephrotic syndrome phenotype, the RCAD146 classifier also identified subjects carrying mutations in NPHS1, NPHS2, WT1 (NPHS4), and ADCK4 (NPHS9) as non-RCAD. This group of patients may confirm, as the CKD group, that RCAD urinary proteome is not reflecting just proteinuria.
Interestingly, in a parallel test, the urinary proteome of a two year old PAX2 mutant carrier was misrecognized by RCAD146 (data not shown). This observation suggests that the RCAD pediatric proteome could potentially be closer to patients with mutations in the gene encoding the transcription factor PAX2, known to cooperate with HNF1B in kidney morphogenesis and ureter differentiation50, than patients with either polycystic or nephrotic syndrome. Additional samples are required to further validate this common feature.
A limitation of the current study is that the recruited ADPKD patients were not children, but young adults. This is due to the difficulty in recruiting children with ADPKD because the average age at the diagnosis is 30 years old51. Another shortcoming is that there was no information available related to the respective albuminuria/proteinuria values of the ADPKD patients. Moreover, this study included a post hoc analysis, due to the selection of the diseased control population from previous studies37,52–57. However, all the samples were analysed according to the same rules and identical conditions (sample preparation and proteomic platform). No discrepancy between the data of the measured RCAD samples and the stored data is to be expected, because the normalization procedure protects the data from aberration of the intensity of the peptide signals. Furthermore, we controlled all measurements with a urine standard sample to identify unforeseeable technical aspects over time58.
Overall, the study, performed in agreement with the guidelines of clinical proteomics, demonstrates a significant value of the urinary proteome analysis in the detection of RCAD highlighting some proteins that potentially participate in the development of cysts and that may be useful for early diagnosis.
The urinary peptide signature of pediatric RCAD patients is mainly characterized by the increase of collagen peptides (especially type I or type III fragments), and osteopontin, along with the decrease of uromodulin. Including the 146 peptides differentially excreted between RCAD and healthy patients in a diagnostic biomarker classifier, we demonstrated that RCAD pediatric urinary proteome is different from patients with Pkd1-2 and Nphs1-2-4-9 mutations, as well as from CKD patients. Future studies will be conducted to evaluate the performance of the RCAD146 panel in additional pediatric cohorts of disorders more related to RCAD such as autosomal recessive polycystic kidney disease (ARPKD), ADTKD or diabetic patients. Moreover, follow-up clinical data of the patients described in this study will be addressed to estimate the performance of this classifier to predict the progression of RCAD. These analyses together are expected to provide further insights into the pathophysiology and disease evolution of RCAD patients.
Methods
Patient recruitment
RCAD urine samples were collected from three different clinical centres: Children’s Hospital, CHU-Toulouse (France, n = 33), University Children Hospital, Heidelberg (Germany; n = 11), Clinical Research Center for Rare Diseases Aldo e Cele Daccò, Ranica (Italy, n = 2). RCAD patients’ average age was 8.4 years. Furthermore, 56.5% of the RCAD patients had a normal renal function (estimated glomerular filtration rate (eGFR) > 90 ml/min/1.73 m2). For patients under 20 years, baseline eGFR (mL/min/1.73 m2) was estimated using the creatinine-based “Bedside Schwartz” equation59. On the other hand, for patients over 20 years (e.g. ADPKD cohort), the CKD-EPI formula was used to calculate the eGFR values60. After collection, urine samples were stored at −20 °C and shipped frozen for subsequent proteome analysis. In addition, all non-RCAD samples were retrieved from the Human Urinary Proteome database26–28. This group of samples included healthy patients (n = 42), and patients suffering from kidney diseases and carrying different genetic mutations, such as: PKD1 (n = 46); PKD2 (n = 9), NPHS1 (n = 2), NPHS2 (n = 35), WT1 (n = 6), ADCK4 (n = 3). Additionally, a group of samples from a pediatric cohort with chronic kidney disease was tested with different etiologies, like focal segmental glomerulosclerosis, IgA nephropathy, membranous glomerulonephritis, mesangioproliferative glomerulonephritis, diabetic nephropathy, vasculitis, and Henoch-Schönlein purpura nephritis (n = 55). This wider group of negative controls (non-RCAD) presented an average age of 16.5 years. RCAD and healthy patients were selected by similar age and gender; CKD and nephrotic syndrome cohorts were age-matched and ADPKD patients were phenotypic-matched for the presence of cysts. Characteristics of all individuals included in this study are extended in Supplementary Table 1.
This study was designed and performed in compliance with all the regulations regarding the protection of subjects participating in medical research. Collection, storage and analysis of urine samples have been approved by the local ethics committees of the three participating centres (Comité de Protection des Personnes Sud-Ouest et Outre Mer III, Ethikkommission der Medizinischen Fakultät Heidelberg, and Comitato Etico di Bergamo respectively). All participating subjects or legal guardians of patients provided written informed consent to the use of urine samples. This study was performed in accordance with the Helsinki Declaration.
Urine sample preparation and CE-MS analysis
Urine sample collection and CE-MS analysis were performed as reported previously61,62. Briefly, immediately before preparation, urine samples aliquots stored at −20 °C were thawed and 700 μl were diluted with the same volume of 2 M urea, 10 mM NH4OH comprising 0.02% SDS. Then, samples were filtered via a Centristat 20-kDa cut-off centrifugal filter device (Sartorius, Goettingen, Germany) at 2,600 g for one hour at 4 °C in order to remove high molecular weight compounds. The obtained filtrate was desalted using a PD-10 column (GE Healthcare, Sweden) equilibrated in 0.01% aqueous NH4OH to eliminate urea, electrolytes and salts. Finally, samples were lyophilized and stored at 4 °C prior to CE-MS analysis. The samples were re-suspended in 10 µL of HPLC-grade H2O shortly before CE-MS analysis, as described62. CE-MS analyses were accomplished using the P/ACE MDQ capillary electrophoresis system (Beckman Coulter, Fullerton, USA) online coupled to a MicroTOF MS (BrukerDaltonic, Bremen, Germany)62. The electro-ionization sprayer (Agilent Technologies) was grounded, and the ion spray interface potential was defined between −4 and −4.5 kV. Spectra were accumulated every 3 s along with over a range of m/z to 350–3000. Detailed information on accuracy, precision, selectivity, sensitivity, reproducibility and stability of the CE-MS method have been described previously62.
CE-MS data processing
A proprietary software (MosaiquesVisu) was used to deconvolute mass spectral ion peaks demonstrating identical molecules at different charge states into single masses63. The achieved peak list allows the characterization of each polypeptide according to its CE-migration time (in minutes), molecular mass (in Daltons), and ion signal intensity. Subsequently, normalization of the amplitude of the urinary peptides was conducted on twenty-nine ‘housekeeping’ peptides (peptides varied slightly between samples, generally present in at least 90% of all urine), similarly to previous studies64. These 29 ‘housekeeping’ peptides are commonly used for normalization in all studies. Furthermore, these peptides are consistently reported in urine and to date, they do not appear to be significantly associated with any diseases investigated64. All detected peptides were deposited, clustered, matched and annotated in a Microsoft SQL database26–28, allowing further statistical analysis. All normalized amplitudes of the analysed samples are included in Supplementary Table 3.
Peptide sequencing
Candidate peptides for the RCAD-classifier were identified and sequenced by the use of tandem mass spectrometry (MS/MS) analysis and searched against human entries in the UniProt database, as previously described65,66. Briefly, to acquire the sequence information, urine samples were separated on a Dionex Ultimate 3000 RSLC nano flow system (Dionex, Camberly, UK) or a Beckman CE systems (PACE MDQ) coupled to an Orbitrap Velos MS instrument (Thermo Fisher Scientific)65,66. Thereafter, data files were examined against the UniProt human non-redundant database using Proteome Discoverer 1.2 (Thermo) and the SEQUEST search engine. No fixed modifications were selected, hydroxylation of proline and lysine and oxidation of methionine were enabled as an optional modification, no enzyme specificity was specified in the settings65. The matching of the peptide sequence obtained by MS/MS analysis to the CE-MS peaks was based on molecular mass [Da] and theoretical migration time, calculated using the number of basic amino acids67. Peptides were accepted only if they had a mass deviation below ±5 ppm and <50 mDa for the fragment ions.
Peptide identification and statistical analysis
For the identification of potential HNF1B-related urinary peptide biomarkers, a comparison between RCAD cases and healthy controls was performed. Only peptides that were detected in at least 70% (frequency threshold) of the samples in at least one of the two groups were further considered for statistical analysis. Using the Wilcoxon rank-sum test followed by adjustment for multiple testing with the false-discovery rate method presented by Benjamini and Hochberg29, adjusted P-values were calculated based on the comparison between RCAD cases and healthy controls. Only peptides with a P-value less than 0.05 were considered as statistically significant.
The RCAD146 classifier is developed as SVM classification model68,69, based on the amplitudes of the significant urinary peptides related to RCAD, which allows the calculation of specific classification scores. These classification scores were further used for statistical analysis, e.g. ROC curves. In more detail, the sensitivity and specificity assessed for the RCAD146 classifier were calculated based on the number of correctly classified subjects. The receiver operating characteristic (ROC) plots and the confidence intervals (95% CI) were based on exact binomial calculations. The area under the curve (AUC), and sensitivity and specificity values of the ROC of the classifier were determined using R-based statistical software (version 3.3.3) and confirmed using MedCalc version 12.7.5.0 (MedCalc Software bvba, Ostend, Belgium). Graphs related to ROC curves and Box-and-Whisker plot were generated with R-based statistic software (packages ggplot2, plotly).
Supplementary information
Detailed clinical data from the discovery and validation cohort
Detailed information on the 146 RCAD-related peptides
Acknowledgements
The research presented in this manuscript was supported by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No. 642937 (RENALTRACT; MSCA-ITN-2014-642937). Furthermore, J.D. and L.P. were supported by the NEOCYST consortium, which is funded by the German Federal Ministry of Research and Education (BMBF, grant 01GM1515).
Author Contributions
P.R. and P.M. wrote the main manuscript text, performed CE-MS analysis, data analysis and interpretation, and prepared figures and tables. M.K. contributed to data interpretation, statistical analysis and preparation of figures and tables. M.P., I.B. and L.G. contributed to data analysis and results interpretation. E.D., M.R.C., G.R., L.P. and J.D. contributed to clinical data and urine sample collection, and supported results interpretation. S.D., F.S. and J.P.S. contributed to clinical data and urine sample collection as well as for the writing of the manuscript along with supervising the study. H.M., M.U., S.C. and P.Z. supervised experiments and results interpretation, provided critical reading and editing of the manuscript, and directed the study. All authors reviewed and approved the manuscript.
Data Availability
The raw data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
H.M. is the founder and co-owner of Mosaiques Diagnostic GmbH, who developed the CE-MS technology for clinical application. P.M., M.K., M.P., I.B., and P.Z. are employees of Mosaiques Diagnostic GmbH.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pierbruno Ricci and Pedro Magalhães contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-38713-5.
References
- 1.Bingham CBM, et al. Mutations in the hepatocyte nuclear factor-1beta gene are associated with familial hypoplastic glomerulocystic kidney disease. Am J Hum Genet Jan. 2001;68(1):219–24. doi: 10.1086/316945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horikawa Y, et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nature Genetics. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 3.Clissold RL, Hamilton AJ, Hattersley AT, Ellard S, Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nature Reviews Nephrology. 2015;11:102–112. doi: 10.1038/nrneph.2014.232. [DOI] [PubMed] [Google Scholar]
- 4.Haumaitre C, et al. Severe pancreas hypoplasia and multicystic renal dysplasia in two human fetuses carrying novel HNF1 beta/MODY5 mutations. Human Molecular Genetics. 2006;15:2363–2375. doi: 10.1093/hmg/ddl161. [DOI] [PubMed] [Google Scholar]
- 5.Kato N, Motoyama T. Expression of Hepatocyte Nucler Factor-1 beta in Human Urogenital Tract During the Embryonic Stage. Analytical and Quantitative Cytology and Histology. 2009;31:34–40. [PubMed] [Google Scholar]
- 6.Body-Bechou D, et al. TCF2/HNF-1beta mutations: 3 cases of fetal severe pancreatic agenesis or hypoplasia and multicystic renal dysplasia. Prenatal Diagnosis. 2014;34:90–93. doi: 10.1002/pd.4264. [DOI] [PubMed] [Google Scholar]
- 7.Duval H, et al. Fetal anomalies associated with HNF1B mutations: report of 20 autopsy cases. Prenatal Diagnosis. 2016;36:744–751. doi: 10.1002/pd.4858. [DOI] [PubMed] [Google Scholar]
- 8.Haldorsen IS, et al. Lack of pancreatic body and tail in HNF1B mutation carriers. Diabetic Medicine. 2008;25:782–787. doi: 10.1111/j.1464-5491.2008.02460.x. [DOI] [PubMed] [Google Scholar]
- 9.Bingham C, Renal HA. cysts and diabetes syndrome resulting from mutations in hepatocyte nuclear factor-1beta. Nephrol Dial Transplant. Nov. 2004;19(11):2703–8. doi: 10.1093/ndt/gfh348. [DOI] [PubMed] [Google Scholar]
- 10.Stenson PD, et al. Human gene mutation database (HGMD (R)): 2003 update. Human Mutation. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama M, et al. HNF1B alterations associated with congenital anomalies of the kidney and urinary tract. Pediatric Nephrology. 2010;25:1073–1079. doi: 10.1007/s00467-010-1454-9. [DOI] [PubMed] [Google Scholar]
- 12.Eckardt KU, et al. Autosomal dominant tubulointerstitial kidney disease: diagnosis, classification, and management–A KDIGO consensus report. Kidney Int. 2015;88:676–683. doi: 10.1038/ki.2015.28. [DOI] [PubMed] [Google Scholar]
- 13.Verbitsky M, et al. Genomic imbalances in pediatric patients with chronic kidney disease. Journal of Clinical Investigation. 2015;125:2171–2178. doi: 10.1172/jci80877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubois-Laforgue DCE, Saint-Martin C, Coste J, Bellanné-Chantelot C, Timsit J. Diabetes, Associated Clinical Spectrum, Long-term Prognosis, and Genotype/Phenotype Correlations in 201 Adult Patients With Hepatocyte Nuclear Factor 1B (HNF1B) Molecular Defects. Diabetes Care. Nov. 2017;40(11):1436–1443. doi: 10.2337/dc16-2462. [DOI] [PubMed] [Google Scholar]
- 15.Musetti, C. Q. M. et al. Chronic renal failure of unknown origin is caused by HNF1B mutations in 9% of adult patients: a single centre cohort analysis. Nephrology (Carlton). 19(4), 202–9 (2014). [DOI] [PubMed]
- 16.Adalat S, et al. HNF1B Mutations Associate with Hypomagnesemia and Renal Magnesium Wasting. Journal of the American Society of Nephrology. 2009;20:1123–1131. doi: 10.1681/asn.2008060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidet L, et al. Spectrum of HNF1B Mutations in a Large Cohort of Patients Who Harbor Renal Diseases. Clinical Journal of the American Society of Nephrology. 2010;5:1079–1090. doi: 10.2215/cjn.06810909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhave JCBA, Wetzels JF. Nijenhuis T2. Hepatocyte Nuclear Factor 1β-Associated Kidney Disease: More than Renal Cysts and Diabetes. J Am Soc Nephrol. Feb. 2016;27(2):345–53. doi: 10.1681/ASN.2015050544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bingham, C. et al Atypical familial juvenile hyperuricemic nephropathy associated with a hepatocyte nuclear factor-1beta gene mutation. Kidney Int. 63(5), 1645–51 (2003). [DOI] [PubMed]
- 20.Decramer S, et al. Anomalies of the TCF2 gene are the main cause of fetal bilateral hyperechogenic kidneys. Journal of the American Society of Nephrology. 2007;18:923–933. doi: 10.1681/asn.2006091057. [DOI] [PubMed] [Google Scholar]
- 21.Edghill EL, Bingham C, Ellard S, Hattersley AT. Mutations in hepatocyte nuclear factor-1 beta and their related phenotypes. Journal of Medical Genetics. 2006;43:84–90. doi: 10.1136/jmg.2005.032854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drube J, et al. Urinary Proteome Analysis to Exclude Severe Vesicoureteral Reflux. Pediatrics. 2012;129:E356–E363. doi: 10.1542/peds.2010-3467. [DOI] [PubMed] [Google Scholar]
- 23.Good DM, et al. Naturally Occurring Human Urinary Peptides for Use in Diagnosis of Chronic Kidney Disease. Molecular & Cellular Proteomics. 2010;9:2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kistler, A. D. et al. Urinary Proteomic Biomarkers for Diagnosis and Risk Stratification of Autosomal Dominant Polycystic Kidney Disease: A Multicentric Study. Plos One8, 10.1371/journal.pone.0053016 (2013). [DOI] [PMC free article] [PubMed]
- 25.Klein, J. et al. Fetal Urinary Peptides to Predict Postnatal Outcome of Renal Disease in Fetuses with Posterior Urethral Valves (PUV). Science Translational Medicine5, 10.1126/scitranslmed.3005807 (2013). [DOI] [PubMed]
- 26.Coon JJ, et al. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clinical Applications. 2008;2:964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siwy J, Mullen W, Golovko I, Franke J, Zuerbig P. Human urinary peptide database for multiple disease biomarker discovery. Proteomics Clinical Applications. 2011;5:367–374. doi: 10.1002/prca.201000155. [DOI] [PubMed] [Google Scholar]
- 28.Stalmach A, Albalat A, Mullen W, Mischak H. Recent advances in capillary electrophoresis coupled to mass spectrometry for clinical proteomic applications. Electrophoresis. 2013;34:1452–1464. doi: 10.1002/elps.201200708. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 30.Mischak H, et al. Clinical proteomics: A need to define the field and to begin to set adequate standards. Proteomics Clinical Applications. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 31.Senkel S, Lucas B, Klein-Hitpass L, Ryffel GU. Identification of target genes of the transcription factor HNF1 beta and HNF1 alpha in a human embryonic kidney cell line. Biochimica Et Biophysica Acta-Gene Structure and Expression. 2005;1731:179–190. doi: 10.1016/j.bbaexp.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Kistler AD, et al. Identification of a unique urinary biomarker profile in patients with autosomal dominant polycystic kidney disease. Kidney International. 2009;76:89–96. doi: 10.1038/ki.2009.93. [DOI] [PubMed] [Google Scholar]
- 33.Norman J. Fibrosis and progression of Autosomal Dominant Polycystic Kidney Disease (ADPKD) Biochimica Et Biophysica Acta-Molecular Basis of Disease. 2011;1812:1327–1336. doi: 10.1016/j.bbadis.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drube J, et al. Urinary proteome pattern in children with renal Fanconi syndrome. Nephrology Dialysis Transplantation. 2009;24:2161–2169. doi: 10.1093/ndt/gfp063. [DOI] [PubMed] [Google Scholar]
- 35.Genovese F, Manresa AA, Leeming DJ, Karsdal MA, Boor P. The extracellular matrix in the kidney: a source of novel non-invasive biomarkers of kidney fibrosis? Fibrogenesis Tissue Repair. 2014;7:4. doi: 10.1186/1755-1536-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossing K, et al. The urinary proteome in diabetes and diabetes-associated complications: New ways to assess disease progression and evaluate therapy. Proteomics Clin Appl. 2008;2:997–1007. doi: 10.1002/prca.200780166. [DOI] [PubMed] [Google Scholar]
- 37.Magalhães, P. et al. Association of kidney fibrosis with urinary peptides: a path towards non-invasive liquid biopsies? Sci Rep. 20 5, 7(1), 16915 (2017). [DOI] [PMC free article] [PubMed]
- 38.Ovrehus, M. A., Zurbig, P., Vikse, B. E. & Hallan, S. I. Urinary proteomics in chronic kidney disease: diagnosis and risk of progression beyond albuminuria. Clinical Proteomics12, 10.1186/s12014-015-9092-7 (2015). [DOI] [PMC free article] [PubMed]
- 39.El-Achkar TM, Wu XR. Uromodulin in Kidney Injury: An Instigator, Bystander, or Protector? American Journal of Kidney Diseases. 2012;59:452–461. doi: 10.1053/j.ajkd.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang ZY, et al. A Urinary Fragment of Mucin-1 Subunit alpha Is a Novel Biomarker Associated With Renal Dysfunction in the General Population. Kidney Int Rep. 2017;2:811–820. doi: 10.1016/j.ekir.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie YS, et al. Expression, roles, receptors, and regulation of osteopontin in the kidney. Kidney International. 2001;60:1645–1657. doi: 10.1046/j.1523-1755.2001.00032.x. [DOI] [PubMed] [Google Scholar]
- 42.Chan SC, et al. Mechanism of Fibrosis in HNF1B-Related Autosomal Dominant Tubulointerstitial Kidney Disease. J Am Soc Nephrol. Oct. 2018;29(10):2493–2509. doi: 10.1681/ASN.2018040437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kompatscher A, et al. Transcription factor HNF1beta regulates expression of the calcium-sensing receptor in the thick ascending limb of the kidney. Am J Physiol Renal Physiol. 2018;315:F27–F35. doi: 10.1152/ajprenal.00601.2017. [DOI] [PubMed] [Google Scholar]
- 44.Kuo IY, et al. Cyst formation following disruption of intracellular calcium signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14283–14288. doi: 10.1073/pnas.1412323111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdul-Majeed S, Nauli SM. Calcium-mediated mechanisms of cystic expansion. Biochimica Et Biophysica Acta-Molecular Basis of Disease. 2011;1812:1281–1290. doi: 10.1016/j.bbadis.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright KJ, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gresh L, et al. A transcriptional network in polycystic kidney disease. Embo Journal. 2004;23:1657–1668. doi: 10.1038/sj.emboj.7600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergmann C. ARPKD and early manifestations of ADPKD: the original polycystic kidney disease and phenocopies. Pediatric Nephrology. 2015;30:15–30. doi: 10.1007/s00467-013-2706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faguer S, et al. The HNF1B score is a simple tool to select patients for HNF1B gene analysis. Kidney International. 2014;86:1007–1015. doi: 10.1038/ki.2014.202. [DOI] [PubMed] [Google Scholar]
- 50.Paces-Fessy M, Fabre M, Lesaulnier C, Cereghini S. Hnf1b and Pax2 cooperate to control different pathways in kidney and ureter morphogenesis. Human Molecular Genetics. 2012;21:3143–3155. doi: 10.1093/hmg/dds141. [DOI] [PubMed] [Google Scholar]
- 51.Dell KM. The Spectrum of Polycystic Kidney Disease in Children. Advances in Chronic Kidney Disease. 2011;18:339–347. doi: 10.1053/j.ackd.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pejchinovski M, et al. Urine peptidome analysis predicts risk of end-stage renal disease and reveals proteolytic pathways involved in autosomal dominant polycystic kidney disease progression. Nephrology Dialysis Transplantation. 2017;32:487–497. doi: 10.1093/ndt/gfw243. [DOI] [PubMed] [Google Scholar]
- 53.Siwy J, et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrology Dialysis Transplantation. 2017;32:2079–2089. doi: 10.1093/ndt/gfw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haubitz M, et al. Identification and Validation of Urinary Biomarkers for Differential Diagnosis and Evaluation of Therapeutic Intervention in Anti-neutrophil Cytoplasmic Antibody-associated Vasculitis. Molecular & Cellular Proteomics. 2009;8:2296–2307. doi: 10.1074/mcp.M800529-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Decramer S, et al. Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nature Medicine. 2006;12:398–400. doi: 10.1038/nm1384. [DOI] [PubMed] [Google Scholar]
- 56.Julian BA, et al. Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis. 2007;28:4469–4483. doi: 10.1002/elps.200700237. [DOI] [PubMed] [Google Scholar]
- 57.Trautmann A, et al. Spectrum of Steroid-Resistant and Congenital Nephrotic Syndrome in Children: The PodoNet Registry Cohort. Clinical Journal of the American Society of Nephrology. 2015;10:592–600. doi: 10.2215/cjn.06260614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mischak H, et al. Comprehensive human urine standards for comparability and standardization in clinical proteome analysis. Proteomics Clinical Applications. 2010;4:464–478. doi: 10.1002/prca.200900189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartz GJ, et al. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20:629–637. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levey AS, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metzger J, et al. Urine proteomic analysis differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Gut. 2013;62:122–130. doi: 10.1136/gutjnl-2012-302047. [DOI] [PubMed] [Google Scholar]
- 62.Mischak H, Vlahou A, Ioannidis JPA. Technical aspects and inter-laboratory variability in native peptide profiling: The CE-MS experience. Clinical Biochemistry. 2013;46:432–443. doi: 10.1016/j.clinbiochem.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 63.Kaiser THA, Kielstein JT. Conference: 16th International Symposium on Microscale Separation and Analysis Location: SAN DIEGO, CALIFORNIA. Journal of Chromatography. 2003;1013:157–171. doi: 10.1016/S0021-9673(03)00712-X. [DOI] [PubMed] [Google Scholar]
- 64.Jantos-Siwy J, et al. Quantitative Urinary Proteome Analysis for Biomarker Evaluation in Chronic Kidney Disease. Journal of Proteome Research. 2009;8:268–281. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- 65.Klein J, Papadopoulos T, Mischak H, Mullen W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis. 2014;35:1060–1064. doi: 10.1002/elps.201300327. [DOI] [PubMed] [Google Scholar]
- 66.Magalhães, P. et al. Comparison of Urine and Plasma Peptidome Indicates Selectivity in Renal Peptide Handling. Proteomics. Clinical applications, e1700163-e1700163, 10.1002/prca.201700163 (2018). [DOI] [PubMed]
- 67.Zurbig P, et al. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis. 2006;27:2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
- 68.Girolami MMH, Krebs R. Analysis of complex, multidimensional datasets. Drug Discov Today Technol. Spring. 2006;3(1):13–9. doi: 10.1016/j.ddtec.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 69.Yang ZR, Chou KC. Bio-support vector machines for computational proteomics. Bioinformatics. 2004;20:735–U549. doi: 10.1093/bioinformatics/btg477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed clinical data from the discovery and validation cohort
Detailed information on the 146 RCAD-related peptides
Data Availability Statement
The raw data generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.