Abstract
The ALK tyrosine kinase receptor is oncogenically activated in neuroblastoma. Whereas numerous ALK fusion genes have been reported in different malignancies, in neuroblastoma ALK is mainly activated through point mutations. Three hotspot residues (F1174, F1245, and R1275) account for 85% of mutant ALK seen in neuroblastoma. In a cohort of 105 Swedish neuroblastoma cases of all stages, these hotspot regions were re-sequenced (>5000X). ALK mutations were detected in 16 of 105 patients (range of variant allele fraction: 2.7–60%). Mutations at the F1174 and F1245 hotspot were observed in eleven and three cases respectively. ALK mutations were also detected at the I1171 and L1240 codons in one tumor each. No mutations were detected at R1275. Sanger sequencing could confirm ALK status for all mutated samples with variant allele fraction above 15%. Four of the samples with subclonal ALK mutation fraction below this would have gone undetected relying on Sanger sequencing only. No distinct mutation spectrum in relation to neuroblastoma tumours genomic subtypes could be detected although there was a paucity of ALK mutations among 11q-deleted tumors. As ALK mutations status opens up an excellent opportunity for application of small molecule inhibitors targeting ALK, early and sensitive detection of ALK alterations is clinically important considering its potential role in tumour progression.
Introduction
Neuroblastoma (NB), the most common extracranial solid cancer in childhood, displays unique heterogeneity in terms of both genomic and clinical behavior1, ranging from children with complete spontaneous tumour regression to children with wide spread metastatic disease resistant to treatment. Although there is treatment available for high-risk NB cases, long-term survival for this patient group is less than 50% despite aggressive treatment. The adverse outcome of high-risk NB constitutes a major clinical problem, mainly attributed to insufficient means to treat refractory or relapsed disease2. Consequently, one of the most important practical utilities of studying NB tumour heterogeneity lies in its implications for improving therapeutic strategy and ultimately, increased survival.
In-depth molecular characterization of cancer specimens can provide prognostic or diagnostic information and identify molecular therapeutic targets3. Importantly, not all mutations detected in a cancer cell are helpful for increasing knowledge regarding the mechanism of transformation. Whereas some genetic defects provide selective advantage for cancer development and/or progression, others are mere passengers without functional relevance and thus, not valuable as targetable candidates4. In NB, only a limited number of recurrent somatic mutations have been reported, these include mutations in ALK (Anaplastic Lymphoma Kinase), and a set of genes involved in chromatin-remodelling and neuritogenesis. Apart from the hotspot mutations in ALK most variants identified are private5,6. Activating germline mutations in ALK are the major cause of hereditary NB although somatically acquired ALK alterations are observed in 6–12% of sporadic cases7–9. Activating point mutations are mainly seen at three “hot spot” residues; F1174 (mutated to L, S, I, C or V), F1245 (mutated to L, I, V, or C), and R1275 (mutated to Q or L), all localized within the kinase domain of ALK and together accounting for 85% of all mutant ALK in NB10. Less frequent observed are substitutions at I1170 (to N or S) and I1171 (to N)11. Besides activating missense mutations, ALK can also be activated by amplification or rare translocation events, further supporting the necessity of this tyrosine kinase receptor for NB tumourigenesis9,12,13. The recent discovery of the ALK ligand, ALKAL2 (FAM150B/AUGα),14,15 with a genetic locus in the genomic proximity of ALK, makes it tempting to speculate a role for ligand-dependent activation of the wildtype ALK receptor in NB pathogenesis. This makes ALK an attractive therapeutic target in NB. A number of small molecule ALK inhibitors are currently under clinical or preclinical investigation for treatment of ALK-positive malignancies. Single drug treatment with the small molecule ALK/MET inhibitor crizotinib has shown very promising results in adult non-small cell lung cancer and in large cell anaplastic lymphoma that harbour ALK translocations16. However, inhibition of ALK mutation in the context of the full-length receptor is complex, and crizotinib has proven to be less clinically effective in treatment of NB10,17. Biochemical studies of ALK mutations have shown that the different variants display both variable kinase activity and variable inhibitor susceptibility10 but this could possibly be surmountable through more high affinity inhibitors recently developed11,18. Second and third generation ALK specific inhibitors such as ceritinib (LDK378)19, brigatinib (AP26113)20,21, alectinib (CH-5424802)22, and lorlatinib (PF-06463922)23,24 might significantly improve NB treatment options. We recently reported on a child with underlying Fanconi anemia (FA) and ALK mutant high-risk NB responding strongly to precision therapy with the ALK TKI ceritinib25.
More sensitive detection of ALK mutations, either already present at time of diagnosis or acquired during disease progression, is of major clinical importance for NB patients as this may lead to new therapeutic possibilities26,27. However, only a few comprehensive studies have explored the genomic landscape of relapsing NB28–30. We recently showed that ALK alterations are enriched at relapse and that these mutations can be detected as minor subclones in the primary tumour that subsequently emerge as the major relapse clone31. It is highly likely that these initial subclones present at diagnosis confer a selective advantage after first line treatment with chemotherapy, ultimately leading to clonal expansion and tumour relapse. These data strongly suggest that precise molecular characterization of ALK mutations should be included in clinical diagnostics of NB tumours not only at diagnosis but also continuously throughout the clinical management of the patient.
To investigate this rigorously we have now undertaken a study of a series of 105 NB tumours to explore ALK copy number gain as well as the frequency and type of ALK mutations that may remain undetected using Sanger-based sequencing methodology. To do this we have employed deep parallel DNA sequencing techniques that allow detection of very low frequency of mutations and we compare this method with previously used Sanger sequencing methodology. We further intend to define the subclonal heterogeneity of the tumour and their potential role in clonal evolution and progression of NB.
Materials and Methods
Patient material
NB tumour samples were obtained through surgery/biopsying after informed consent from parents/guardians. The tumours were graded according the international NB staging system (INSS) and international NB risk group (INRG). All samples belong to a Swedish cohort of patients with NB tumours of all stages, Table 1. Tumours were included in the study only if containing >50% tumour cells as judged by Single Nucleotide Polymorphism (SNP) array (Affymetrix Inc., Santa Clara, CA) or through histopathological examination.
Table 1.
Patients used in the study, ALK mutations and clinical data.
| Case | ALK mut. status - Sanger | ALK mut. status - Deep seq (% of mutated allele) | Effect on ALK | Outcome | SNP array genomic profile | aad (age at diagnosis) | INRG |
|---|---|---|---|---|---|---|---|
| 1 | Not done | Neg | NED > 5 y | Other segm. | 18 | L | |
| 2 | Neg | Neg | DOD | NMA | 27 | M | |
| 3 | Neg | Neg | DOD | 11q-del | 76 | L | |
| 4 | Not done | Neg | NED > 5 y | 17q-gain | 14 | L | |
| 5 | Neg | Neg | NED > 5 y | 17q-gain | 102 | L | |
| 6 | Not done | Neg | NED > 5 y | Num only | 32 | L | |
| 7 | Neg | Neg | NED > 5 y | Other segm. | 12 | L | |
| 8 | Neg D1160D | Neg | NED > 5 y | Num only | 21 | L | |
| 9 | Neg | Neg | DOD | NMA | 19 | M | |
| 10 | Neg | Neg | DOD | Num only | 48 | M | |
| 11 | Not done | Neg | NED > 5 y | 11q-del | 30 | L | |
| 12 | F1174L | F1174L (24,7%) | GOF | DOD | NMA | 15 | M |
| 13 | Neg | Neg | NED > 5 y | Num only | 30 | L | |
| 14 | Neg | Neg | NED > 5 y | 17q-gain | 19 | L | |
| 15 | Neg | Neg | DOD | 11q-del | 80 | L | |
| 16 | Neg | Neg | DOD | NMA | 18 | M | |
| 17 | F1174C | F1174C (19%) | GOF | NED > 5 y | Num only | 25 | L |
| 18 | Neg | I1171T (2,7%) | GOF | NED > 5 y | NMA | 29 | M |
| 19 | Neg | Neg | DOD | NMA | 37 | L | |
| 20 | Neg | Neg | NED > 5 y | Other segm. | 21 | L | |
| 21 | Neg | Neg | NED > 5 y | 11q-del | 9 | L | |
| 22 | Neg | Neg | DOD | 11q-del | 80 | M | |
| 23 | Neg | Neg | DOD | NMA | 8 | L | |
| 24 | Neg | Neg | NED > 5 y | NMA | 35 | L | |
| 25 | Neg | Neg | NED > 5 y | Num only | 43 | L | |
| 26 | F1245I | F1245I (53%) | GOF | AWD | Other segm. | 5 | L |
| 27 | Neg | Neg | NED > 5 y | Num only | 0 | L | |
| 28 | Neg | Neg | DOD | NMA | 21 | M | |
| 29 | Neg | Neg | NED > 5 y | 11q-del | 74 | M | |
| 30 | Neg | Neg | DOD | 11q-del | 6 | M | |
| 31 | Neg | Neg | NED > 5 y | 11q-del | 43 | M | |
| 32 | Neg | Neg | DOD | 17q-gain | 10 | L | |
| 33 | Not done | Neg | NED > 5 y | Other segm. | 12 | L | |
| 34 | Neg | Neg | DOD | 11q-del | 170 | L | |
| 35 | Neg | Neg | DOD | NMA | 37 | M | |
| 36 | Neg | Neg | NED > 5 y | Other segm. | 4 | L | |
| 37 | Neg | Neg | NED > 5 y | Other segm. | 63 | L | |
| 38 | F1245C | F1245C (52,4%) | GOF | NED > 5 y | 17q-gain | 92 | M |
| 39 | Neg | Neg | NED > 5 y | Num only | 11 | L | |
| 40 | 1st Neg/2nd F1147S | 2nd F1174S (58,7) and F1174I (7,7%) | GOF & GOF | DOD | 17q-gain | 12 | M |
| 41 | Not done | Neg | NED > 5 y | 17q-gain | 6 | L | |
| 42 | F1174I | F1174I (22%) | GOF | NED > 5 y | Other segm. | 27 | L |
| 43 | F1174L | F1174L (22%) | GOF | NED > 5 y | Other segm. | 90 | M |
| 44 | Neg | Neg | DOD | NMA | 12 | M | |
| 45 | Not done | Neg | Amp. | DOD | NMA ( + ALK-amp) | 11 | M |
| 46 | Neg | Neg | DOD | Other segm. | 33 | M | |
| 47 | Neg | F1245I (14%) | GOF | DOD | NMA | 21 | M |
| 48 | Neg | Neg | DOD | 17q-gain | 55 | M | |
| 49 | Not done | Neg | NED > 5 y | NMA | 15 | M | |
| 50 | 1st Neg /2nd F1174L | F1174L (25,9%) | GOF | DOD | NMA | 29 | M |
| 51 | Not done | Neg | NED > 5 y | 11q-del | 33 | M | |
| 52 | Neg | Neg | DOD | 11q-del | 32 | M | |
| 53 | F1174I, const. | F1174I (61,2%) | GOF | DOD | Other segm. | 1 | M |
| 54 | Neg | Neg | DOD | Num only | 0 | MS | |
| 55 | Neg | Neg | DOD | 11q-del | 21 | M | |
| 56 | Neg | Neg | DOD | NMA | 14 | M | |
| 57 | Neg | Neg | Amp. | DOD | NMA ( + ALK-amp) | 31 | M |
| 58 | Not done | Neg | NED > 5 y | 11q-del | 10 | L | |
| 59 | Not done | Neg | NED > 5 y | 17q gain | 98 | L | |
| 60 | Neg | Neg | Amp. | DOD | NMA ( + ALK-amp) | 25 | M |
| 61 | F1174L | F1174L (18%) | GOF | DOD | NMA | 41 | L |
| 62 | Neg | Neg | NED > 5 y | 11q-del | 92 | M | |
| 63 | Not done | Neg | NED < 5 y | Other segm. | 6 | L | |
| 64 | Neg | Neg | NED < 5 y | Other segm. | 5 | L | |
| 65 | Not done | Neg | NED < 5 y | NMA | 28 | M | |
| 66 | Not done | Neg | NED < 5 y | Other segm. | 32 | L | |
| 67 | Not done | Neg | NED < 5 y | Num only | 2 | L | |
| 68 | Not done | Neg | NED < 5 y | 17q gain | 37 | L | |
| 69 | Not done | Neg | DOD | NMA | 39 | M | |
| 70 | Neg | Neg | DOD | NMA + 11q-del | 28 | M | |
| 71 | Not done | Neg | NED < 5 y | NMA | 18 | L | |
| 72 | Neg | Neg | NED < 5 y | 11q-del | 18 | L | |
| 73 | Not done | Neg | NED < 5 y | 11q-del | 26 | L | |
| 74 | Not done | Neg | NED < 5 y | Other segm. | 11 | M | |
| 75 | Neg | Neg | NED < 5 y | 11q-del | 70 | L | |
| 76 | Not done | Neg | NED < 5 y | Num only | 1 | L | |
| 77 | Not done | Neg | NED < 5 y | Num only | 18 | L | |
| 78 | Neg | Neg | NED < 5 y | Other segm. | 0 | L | |
| 79 | Neg | Neg | DOD | NMA + 11q-del | 30 | M | |
| 80 | Neg | Neg | NED < 5 y | Other segm. | 37 | L | |
| 81 | Neg | Neg | NED < 5 y | NMA | 13 | L | |
| 82 | Neg | Neg | NED < 5 y | NMA | 10 | L | |
| 83 | Neg | Neg | NED < 5 y | 11q-del | 18 | L | |
| 84 | Neg | F1174L (15,1%) | GOF | NED < 5 y | NMA | 23 | M |
| 85 | Neg | Neg | NED < 5 y | Num only | 6 | L | |
| 86 | Neg | Neg | NED < 5 y | 11q-del | 158 | M | |
| 87 | Neg | Neg | NED < 5 y | Other segm. | 12 | L | |
| 88 | Neg | Neg | NED < 5 y | NMA | 33 | M | |
| 89 | Neg | Neg | Amp. | NED < 5 y | NMA ( + ALK-amp) | 39 | M |
| 90 | Neg | Neg | NED < 5 y | Num only | 2 | L | |
| 91 | Neg | Neg | DOD | NMA + 11q-del | 11 | M | |
| 92 | Neg | F1174L (14%) | GOF | NED < 5 y | 17q gain | 37 | L |
| 93 | Neg | Neg | NED < 5 y | 11q-del | 124 | L | |
| 94 | Neg | Neg | NED < 5 y | 11q-del | 51 | L | |
| 95 | Neg | Neg | NED < 5 y | 11q-del | 13 | L | |
| 96 | Neg | Neg | NED < 5 y | 11q-del | 29 | L | |
| 97 | F1174L | F1174L (22,1%) | GOF | NED < 5 y | NUM only | 8 | L |
| 98 | Neg | Neg | NED < 5 y | 11q-del | 50 | M | |
| 99 | L1240V | L1240V (57,4%) | GOF | DOD | 17q gain + chrom.tr | 44 | M |
| 100 | Not done | Neg | NED < 5 y | Other segm. | 2 | L | |
| 101 | Not done | Neg | NED < 5 y | NMA | 27 | L | |
| 102 | Not done | Neg | NED < 5 y | 17q-gain | 66 | M | |
| 103 | Not done | Neg | NED < 5 y | Num only | 8 | L | |
| 104 | Not done | Neg | NED < 5 y | NMA | 15 | L | |
| 105 | Not done | Neg | AWD | NMA | 37 | M |
MYCN status and tumour copy number aberrations has been determined earlier, using SNP arrays and/or FISH32. Clinical data and genomic profiles of included patients are summarized in Table 1. Patients were treated according to relevant treatment protocols. Ethics approval of treatment protocols was obtained according to national guidelines and the study was authorized by the local ethical committees (Karolinska Institutet and Karolinska University Hospital). The methods were carried out in accordance with the relevant guidelines and regulations.
Following DNA extraction using standard procedures, mutations of the receptor tyrosine kinase (RTK) domain of the ALK gene were analyzed by two different deep parallel sequencing methods (i) the HaloPlex™ target enrichment system and (ii) an amplicon-based assay for sequencing of three hotspot regions in ALK (exon 23, 24 and 25) using Illumina MiSeq sequencing platform (Illumina, San Diego, CA). In order to ensure the potential of our method in detection and discrimination between ALK three hotspot residues F1174, F1245 and R1275, three serial dilutions (undiluted, 1:10, and 1:40) from each of the three hotspot amplicons (ALK exon 23, 24, 25) were generated using 20 ng DNA of three patients each carrying the relevant mutation and dilute it with 20 ng of unmatched germline DNA. Subsequently, eight germline genomic DNAs from healthy donors and three tumours with known ALK mutation served as controls to quantify background abundances.
HaloPlex™ target enrichment system
HaloPlex (Agilent Technologies, Santa Clara, CA) is a custom in-solution capture hybridization method that was used for deep sequencing of ALK gene. Custom probe design for the ALK region of interest (exon 21–25) was created with SureDesign (Agilent). In brief, 225 ng of genomic DNA from each sample were fragmented using eight seperate restriction enzyme digestion reactions and denatured prior hybridization to the HaloPlex probe library. The HaloPlex Target Enrichment System Protocol (Version D.5) was followed without modification to perform the library preparation and target capture using the HaloPlex Exome capture baits. We assessed library quality using Agilent’s 2200 TapeStation system. Enriched and indexed library were prepared for 150-bp paired-end sequencing (2 × 150 bp) on an Illumina MiSeq sequencing system (Illumina, San Diego, CA).
Amplicon-based assay for targeted sequencing of relevant ALK exonic regions
For sequencing library construction, 12 ng of genomic DNA were used to amplify the ALK regions of interest containing hotspots F1174 (exon 23), F1245 (exon 24), and R1275 (exon 25). During the PCR, using KAPA HiFi HotStart DNA Polymerase (Kapa Biosystems, Wilmington, MA), the region-specific primers including Illumina sequencing adapters overhang were used. After amplicon purification with Ampure magnetic beads (Beckman Coulter Inc., Brea, CA), dual indices are attached using Nextera XT Index Kit (Illumina). Following another clean-up and quality control, the final libraries were pooled equimolarly, denatured prior pair-end sequencing (2 × 150 bp) using MiSeq v.3 reagent kit (Illumina). The number of libraries sequenced per flow cell was adjusted to achieve a depth of coverage of at least 5000x for the amplicons in each sample.
Detection of variants
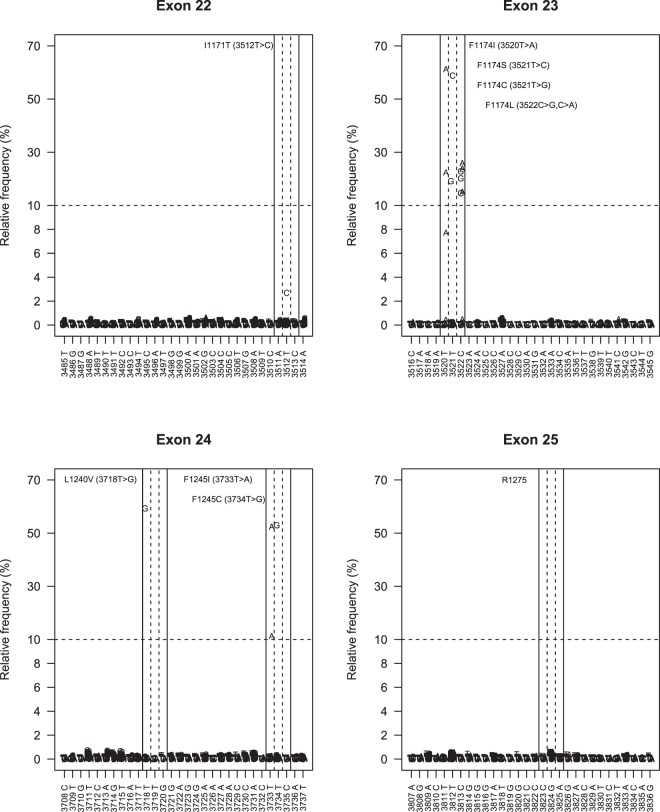
Using MultiQC33, quality of reads was assessed through Fastqc showing that majority of reads never dropped below a phred score of 30 at any position (Supplementary Figure 1). Demultiplexing of pooled libraries was performed using the unique indexes introduced during sample preparation. All reads were quality trimmed and adopters were cut with Cutadapt34. Paired reads (2 × 150 bp) were mapped against Human genome build hg19 (Human Genome Browser, http://genome.ucsc.edu/; hg19) with BWA-MEM default settings35. We did variant calling with the tool UnifiedGenotyper with the option to EMIT_ALL_CONFIDENT_SITES offered in the Genome Analysis Toolkit (GATK)36. This option was used to be able to predict low frequency alleles, since it gives the allele count for each allele at each position (Supplementary Table 1). We focused on hg19 coordinates exon 21 chr2:29445383-29445473, exon 22 chr2:29445210-29445274, exon 23 chr2:29443572-29443701, exon 24 chr2:29436850-29436947, and exon 25 chr2:29432652-29432744 (Human Genome Browser; hg19), to analyze mutation status of ALK tyrosine kinase domain. The most frequently observed sporadic ALK substitutions at amino acids positions F1174 (exon 23), F1245 (exon 24) and R1275 (exon 25) were studied in detail (Fig. 1).
Figure 1.
ALK mutations in neuroblastoma patients. Frequency distribution of mutated ALK allele at the ALK F1245, L1240, F1174 and T1171 hotspots detected in 16 samples. The x-axis represents the genomic coordinates of the targeted region, the y-axis represents the percentage of high-quality reads supporting the mutated base. The base corresponding to the reference genome sequence is reported below the graph, for the forward strand. The mutated base as well as the amino acid change from the reference is indicated above the graph. The circeled-x represent the same tumuor that showed two different mutations in ALK.
Data analysis
In order to evaluate the level of signal vs. noise in the mutation detection system, mutations were determined by visually inspecting nucleotide frequencies at relevant hotspots (Fig. 2). Nucleotide frequencies larger than five standard deviations from background noise were classified as mutations.
Figure 2.
Analysis of detection level and background noise. Considerations concerning the problem of detection of low frequency alleles. Frequency distribution of mutated ALK allele at the ALK exon 21 to 25 detected in 16 samples. The x-axis represents the coding sequence (CDS) position of the targeted region (the base corresponding to the reference genome sequence is reported below the graph, for the reverse strand), the y-axis represents the fraction of high quality reads supporting each base. Sequencing analysis reveals the presence of the ALK mutation with a statistically significant difference from the background variability. The lowest level of mutation detected were in exon 22 for case #18, where the level of mutated alleles were 2,7%. As seen in this figure this low frequency differs distinctly from the background level.
The datasets analysed during the current study are available from the corresponding author on request.
Results
In this study we performed ultra-deep sequencing in order to determine the frequency of ALK hotspot mutations that might be missed with conventional Sanger sequencing due to different degrees of sensitivity. This was done through using two different sequencing strategies, either the hybridization-based HaloPlex target enrichment method or amplicon-sequencing, in a series of totally 105 NB samples. Total aligned reads across overlapping amplicons translated into a minimum coverage depth per position of 5000x (range 5000–65000x). The percentages of bases at coordinates that correspond to positions of ALK hotspots mutations were studied in detail. The background variation frequency at the relevant coordinates was determined in the controls and compared to cases followed by analysis of the mutational status of ALK exon 21–25 in the hybridization-based assay and exon 23–25 in the amplicon-based assay. ALK mutations with a variant allele frequency less than 20% were detected in six tumours while ten other cases showed ALK mutations with higher variant allele frequency (between 20–60%). At residue I1171 in the ALK locus (chr2:29445213), an ALK variant was detected in one case with the frequency of mutated allele as low as 2.7%. This was the lowest variant frequency level detected by deep-targeted sequencing in our cohort. At the F1174 hotspot (chr2:29443695-29443697), alterations were observed in eleven cases: seven cases harbored a mutation leading to the amino acid change F1174L, two cases with F1174I, one each of the F1174C, and F1174S substitutions, with the mutated allele fractions ranging from 14% to 60%. Interestingly the patient harbouring F1174S mutation with 58% frequency also exhibited a subclone with an F1174I mutation detected at 8% frequency of the mutated allele (Fig. 3). At the F1245 hotspot (chr2:29436858-29436860), alterations were detected in 3 tumours: two samples showed a F1245I mutation while the third case carried the F1245C mutation, with frequencies of 14%, 51% and 52%, respectively. At the L1240 locus (chr2:29436875) a L1240V substitution were detected in one patient with a mutated allele fraction at 57%. In total, sixteen out of the 105 tumour samples (15.2%) were ALK mutant positive (Fig. 1, Table 1). We have not observed any mutations at hotspot mutation site R1275. No additional variants outside previously reported mutational sites were observed in the targeted regions in our cohort.
Figure 3.
NB case with two mutations. Aligned sequencing reads visualized in the Integrative Genomics Viewer DNA reference standards with ALK wildtype and two positive A > G (F1174S, MAF 58%) and A > T (F1174I, MAF 8%) variant calls at cis and trans alleles observed in one NB patient. MAF, mutant allele frequency.
ALK mutations were observed in tumors from all clinical stages. No distinct preference of F1174 or F1245 mutations in relation to different genomic subtypes (numerical only, other segmental, 17q-gained or MYCN-amplified) could be identified (Fig. 4). However, no ALK mutations were detected in any of the tumours, in this set, carrying segmental 11q-deletion while in patients without 11q deletion, 20% presented ALK point mutations (n = 27; P value 0.01; Fig. 4). No statistical difference in overall survival was observed when comparing NB patients whose tumor harbour an ALK mutation with cases without an ALK mutation. The comparison of survival of patients with ALK wild-type or ALK mutation, with or without MYCN amplification showed a poorer survival in patients whose tumours harbour MYCN amplification, with or without ALK mutation (Fig. 5). The presence of the F1174 ALK mutation was not found to have an influence on survival when comparing patients with ALK F1174 mutated tumours versus all other patients (data not shown).
Figure 4.
The NB tumours presenting mutations sorted by genome profile and outcome. Note that most ALK mutations are present in the MYCN amplified cases while no 11q cases showed ALK mutations in this set. Also the four cases with ALK amplification are included.
Figure 5.

Kaplan-Meier survival comparisons of cases with ALK mutation vs non-ALK mutation cases. Kaplan-Meier (overall survival) analysis according to the presence or absence of somatic ALK aberrations in 105 neuroblastoma tumours (a) ALK mutated versus non-ALK mutated cases (b) OS according to MYCN and ALK status (MYCN amplification vs. no amplification; ALK alteration versus no alteration). ALK mutational status does not add prognostic information to the independent clinical parameter MYCN status. Pos, positive for ALK alteration (ALK mutations and/or amplification). Wt, wildtype. OS, overall survival.
In this study, tumour samples with a mix of up to 50% normal cells were included which may underestimate the mutation frequencies of heterogeneous tumour samples. Thus it would be expected that subclonal mutation events would represent a greater fraction of the tumour cell population in the actual tumour mass as compared to analyzed DNA fragments. In this regard, our detection of ten ALK variants with mutated allele fraction above 20% might be interpreted as clonal, whereas the six ALK mutations observed with a fraction below 20% might be interpreted as subclonal events that might be unrecognized through Sanger sequencing. All ALK mutated samples were also tested by Sanger sequencing which confirmed all ALK mutations occurring with a mutated allele fraction above 15% leaving four samples with a mutated allele fraction below 15% undetected by Sanger sequencing in our hands.
In addition, four of the 105 tumours included also had ALK gene amplification detected from previously performed SNP microarrays. All four cases also had co-amplification of MYCN and no ALK mutation was detected in any of these samples. Focal low copy number gain for ALK was also detected in one MYCN-amplified patient who also had a chromothripsis pattern for chromosome 2 (case #57). Partial chromosome 2p gain that includes the ALK locus was observed in 26 patients while gain of whole chromosome 2 was seen in 11 cases. Interestingly, all patients (n = 4) with partial 2p gain that also carry an ALK mutation are dead of disease (Table 1). In the tumours with partial 2p gain, the extra chromosomal segments varied in size from 25 to 130 Mb and included both ALK and MYCN except in 6 patients (data available on request). In these six cases, the MYCN amplification segment was present with a breakpoint proximal to the start genomic position of gained region. The partial 2p gains encompassing the ALK locus were present in only 4 of 16 mutated tumours indicating that 2p gain is not a common mechanism for increasing mutated ALK copy number.
Discussion
With the emergence of targeted therapeutic strategies, full characterization of genetic alterations becomes crucial to improve treatment and patient outcome. Intratumour genetic heterogeneity has been reported in number of human malignancies and provides insights into clonal evolution and pathogenesis37,38. This type of heterogeneity may thereby hinder therapeutic strategies that depend on results from single tumour-biopsy samples and provides a risk to render clinically important variants undetected if using less sensitive detection methods.
In order to determine the frequency of ALK mutations in exon 23, 24 and 25 including those also present at subclonal level, 105 diagnostic NB samples were analyzed through ultra-deep sequencing that provides increased sensitivity to that of conventional Sanger sequencing (Table 1). Ultra-deep sequencing (>5000x) enables discovery of rare variants present at frequencies as low as 1%. In this study, we employed two different strategies, the HaloPlex enrichment kit (Agilent) and an amplicon PCR-based approach, both of which were sequenced on a MiSeq Illumina platform. Coverage, uniformity and variant calling ability did not differ between the methods that were evaluated in parallel on a subset of tumours in our cohort. The results are excellent with low overall background noise in both techniques although the HaloPlex method is somewhat more costly and slightly more labor intensive. The amplicon approach requires less DNA as compared to HaloPlex (12 ng vs. 225 ng) and also easier processing when analyzing few samples per run that might be more common in a clinical setting. Other approaches can be applied; Positive mutant–specific amplification can be done by Droplet Digital PCR (ddPCR) which is able to detect a mutant allele frequency of less than 1%.
ALK point mutations were detected in 16 samples while gene amplification was identified in an additional four samples, giving a total of ALK mutational events in 20 of 105 NB samples (19%). Variant allele fraction of observed ALK point mutations in this study ranged from 2.7% to 60% of all NB cases, with a subclonal (mutated allele fraction <20%) observed in 6 of 16 ALK-mutated samples. This is in concordance with the work of Bellini et al. who performed deep sequence of exons 23 and 25 of ALK in a series of 276 NB samples and showed that more than half of the identified point mutations were subclonal events that might have gone undetected by Sanger sequencing. This highlights the importance of deep sequencing techniques for the identification of mutations in NB, especially when targeted therapy is a clinical consideration31,39. All of the NB -associated ALK mutations, observed in our dataset are known to confer strong gain of function effect to ALK in vivo assays, and are therefore known as drivers in the progression of NB tumours10,11,40. The potent ligand-independent F1174L mutation was present in 7 cases of which 4 had MYCN-amplification whereas the other three displayed other genomic profiles. Three of four patients with MYCN amplification and ALK F1174L co-occurrence showed adverse outcome (Table 1, Fig. 4) suggesting a cooperative effect between the two aberrations. It has previously been suggested that the F1174L mutation might contribute to a particular growth advantage for MYCN-amplified NBs41,42. We observed that the majority of MYCN-amplified tumours (4 of 6) contained subclonal (<20% variant allele fraction) ALK aberrations while in MYCN-nonamplified tumours the majority of ALK mutations (8 of 10) were clonal. This implies that MYCN-nonamplified tumours are dependent on the presence of this constitutive active kinase in a higher percentage of the cell population. In addition to mutations, ALK activation can also result from high-level gene amplification. In keeping with previous studies, amplification of ALK almost exclusively occurs in MYCN-amplified tumours43. Interestingly, we do not detect any mutations among the 11q-deleted genomic subgroup of NB tumours (Fig. 4) in this set of tumours. Finally, we show that chromosome 2p is not frequently gained in tumours with ALK mutations, indicating that mutated ALK alleles are not selected for high expression by copy number gain. However, 2p gains are present in as much as 19.3% of all NBs with segmental imbalances44. This raises the possibility that NB tumours with 2p gain may instead benefit from extra copies of MYCN and ALK wildtype receptor in collaboration with its recently described ligand, ALKAL2 located at 2p25.
This study of 105 NBs shows that ALK mutations can be observed in tumours of various clinical stages and of various genomic profiles. We have previously shown through ultra-deep sequencing that subclonal ALK mutations can be detected in NB tumours at diagnosis and that they are enriched at relapse, suggesting that ALK alterations may contribute to progression and a more aggressive disease31. This is further supported by another study showing that NB patients with tumours that harbor ALK alterations have a decreased 5-year overall survival as compared to those with wildtype ALK39. We fail to show any statistically significant ALK-dependent difference in overall survival in our material (Fig. 5) but this could be due to smaller sample size in our study. Interestingly, we observed a case containing at the same time both F1174S and F1174I mutations with 58% and 8% allele frequencies respectively. Our group has previously performed Sanger sequence analysis for this patient revealing a homozygous missense mutation for F1174S45. At this time, the deep sequencing revealed a heterozygous F1174S mutation cooperating with a secondary subclone harbouring another amino acid substitution, I instead of S, for the same locus (Fig. 3). This case provides evidence that extra information can be achieved with NGS methods that are impossible to gain by Sanger approach alone. In the near future, additional information emerging from these deep sequencing studies will be useful for guiding treatment decisions.
Treating NB is still a therapeutic challenge despite recent advances in pediatric oncology. However, due to the recognition of oncogenic forms of ALK as driver in NB malignancy, ALK have emerged as a tractable therapeutic target. Small molecule ALK inhibitors may become the gold standard therapy in NB treatment, making diagnostic high sensitive detection of ALK mutations a necessary step in identifying optimal treatment modalities. We recently reported a NB patient with novel ALK-I1171T variant who revealed complete clinical remission upon treatment with ceritinib25. However, a large-scale analysis of the spectrum of ALK mutations, their clinical significance, and their biochemical properties in NB is essential to direct preclinical and clinical studies of ALK inhibitors and to identify patients likely to benefit from ALK inhibition in NB. In our previous study31 we showed that some NB tumours have subclones harbouring ALK mutations at diagnosis that may contribute to tumour evolution and to relapse. Thus, deep-targeted sequencing would enable us to monitor changes in clonal and subclonal composition of cancer cells during disease progression to orient treatment decisions and to benefit of the patient.
Despite the encouraging development of next generation ALK inhibitors, both intrinsic and acquired resistance may occur when using single drug treatment for receptor tyrosine kinase inhibitors. One mechanism of resistance is acquirement of secondary genetic changes that reduce the accessibility of the hydrophobic inhibitor-binding pocket for small molecule inhibitor by increasing the affinity for ATP instead. A number of these resistant promoting gatekeeper mutations have been demonstrated in non-small cell lung cancer patients treated with crizotinib or alectinib46,47. The recent discovery of the ALK ligand, ALKAL2 (FAM150B/AUGα)14,15 with a genetic locus in the genomic proximity of ALK and MYCN, may aid in further development of therapeutic tools for intervention in NB development in the near future.
Conclusion
ALK is the most frequently mutated gene in NB and has provided new hope for NB treatment using ALK-specific inhibitors. As ALK mutations may contribute to disease progression and relapse, early detection and eradication of these subclones is of uttermost importance as they may improve patient prognosis. We show here that through ultra-deep sequencing, ALK point mutations were detected in 16 samples whereof 6 were subclonal that may remain undetected through conventional sequencing methods. This proves that ultra-deep sequencing could become crucial in clinical analysis as it allows sensitive detection of activating ALK mutations as well as of resistance promoting mutations present at subclonal level.
Supplementary information
Acknowledgements
This work was supported by grants from the Swedish Cancer Society (CAN2015/794 TM), the Swedish Research Council, (521-2014-3031, TM), the Swedish Children’s Cancer Foundation (PR2016-0147, T.M.; 15-0061 S.F., 14-0064, S.F.), the Swedish foundation for Strategic research (RB13-0204), Lions Cancerfond West, Fondkistan, The Assar Gabrielsson foundation, and the Swedish state through the LUA/ALF agreement. This funding provided the necessary resources for the experimental phase of the study, and for data analysis.
Author Contributions
N.J., S.F., P.K. and T.M. conceived the experiments. N.J., R.M.S. and A.D. conducted the experiments. N.J. and T.M. wrote the manuscript. S.N. and K.T. performed statistical analyses of the data. T.M. and P.K. collected tumour material and corresponding clinical information. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37240-z.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Chand D, et al. Cell culture and Drosophila model systems define three classes of anaplastic lymphoma kinase mutations in neuroblastoma. Dis Model Mech. 2013;6:373–382. doi: 10.1242/dmm.010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 6.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caren H, Abel F, Kogner P, Martinsson T. High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem J. 2008;416:153–159. doi: 10.1042/BJ20081834. [DOI] [PubMed] [Google Scholar]
- 8.Janoueix-Lerosey I, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 9.De Brouwer S, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res. 2010;16:4353–4362. doi: 10.1158/1078-0432.CCR-09-2660. [DOI] [PubMed] [Google Scholar]
- 10.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 11.Bresler SC, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26:682–694. doi: 10.1016/j.ccell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazes A, et al. Characterization of rearrangements involving the ALK gene reveals a novel truncated form associated with tumor aggressiveness in neuroblastoma. Cancer Res. 2013;73:195–204. doi: 10.1158/0008-5472.CAN-12-1242. [DOI] [PubMed] [Google Scholar]
- 13.Fransson S, et al. Intragenic anaplastic lymphoma kinase (ALK) rearrangements: translocations as a novel mechanism of ALK activation in neuroblastoma tumors. Genes Chromosomes Cancer. 2015;54:99–109. doi: 10.1002/gcc.22223. [DOI] [PubMed] [Google Scholar]
- 14.Guan J, et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. Elife. 2015;4:e09811. doi: 10.7554/eLife.09811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reshetnyak AV, et al. Augmentor alpha and beta (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc Natl Acad Sci USA. 2015;112:15862–15867. doi: 10.1073/pnas.1520099112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosse KR, Maris JM. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016;122:20–33. doi: 10.1002/cncr.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bresler SC, et al. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med. 2011;3:108ra114. doi: 10.1126/scitranslmed.3002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsilje TH, et al. Synthesis, structure-activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulf onyl)phenyl)pyrimidine-2,4-diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J Med Chem. 2013;56:5675–5690. doi: 10.1021/jm400402q. [DOI] [PubMed] [Google Scholar]
- 20.Siaw JT, et al. Brigatinib, an anaplastic lymphoma kinase inhibitor, abrogates activity and growth in ALK-positive neuroblastoma cells, Drosophila and mice. Oncotarget. 2016;7:29011–29022. doi: 10.18632/oncotarget.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mologni L. Current and future treatment of anaplastic lymphoma kinase-rearranged cancer. World J Clin Oncol. 2015;6:104–108. doi: 10.5306/wjco.v6.i5.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, et al. The second-generation ALK inhibitor alectinib effectively induces apoptosis in human neuroblastoma cells and inhibits tumor growth in a TH-MYCN transgenic neuroblastoma mouse model. Cancer Lett. 2017;400:61–68. doi: 10.1016/j.canlet.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan J, et al. The ALK inhibitor PF-06463922 is effective as a single agent in neuroblastoma driven by expression of ALK and MYCN. Dis Model Mech. 2016;9:941–952. doi: 10.1242/dmm.024448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Infarinato NR, et al. The ALK/ROS1 Inhibitor PF-06463922 Overcomes Primary Resistance to Crizotinib in ALK-Driven Neuroblastoma. Cancer Discov. 2016;6:96–107. doi: 10.1158/2159-8290.CD-15-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan, J. et al. Clinical response of the novel activating ALK-I1171T mutation in neuroblastoma to the ALK inhibitor ceritinib. Cold Spring Harb Mol Case Stud, 10.1101/mcs.a002550 (2018). [DOI] [PMC free article] [PubMed]
- 26.Carpenter EL, Mosse YP. Targeting ALK in neuroblastoma–preclinical and clinical advancements. Nat Rev Clin Oncol. 2012;9:391–399. doi: 10.1038/nrclinonc.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosse YP, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children’s Oncology Group phase 1 consortium study. Lancet Oncol. 2013;14:472–480. doi: 10.1016/S1470-2045(13)70095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleiermacher G, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–3130. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 29.Schramm A, et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet. 2015;47:872–877. doi: 10.1038/ng.3349. [DOI] [PubMed] [Google Scholar]
- 30.Padovan-Merhar OM, et al. Enrichment of Targetable Mutations in the Relapsed Neuroblastoma Genome. PLoS Genet. 2016;12:e1006501. doi: 10.1371/journal.pgen.1006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schleiermacher G, et al. Emergence of new ALK mutations at relapse of neuroblastoma. J Clin Oncol. 2014;32:2727–2734. doi: 10.1200/JCO.2013.54.0674. [DOI] [PubMed] [Google Scholar]
- 32.Caren H, et al. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci USA. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads EMBnet journal17, pp. 10–12, 10.14806/ej.17.1.200 (2012).
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 38.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 39.Bellini A, et al. Deep Sequencing Reveals Occurrence of Subclonal ALK Mutations in Neuroblastoma at Diagnosis. Clin Cancer Res. 2015;21:4913–4921. doi: 10.1158/1078-0432.CCR-15-0423. [DOI] [PubMed] [Google Scholar]
- 40.Ni Z, Wang X, Zhang T, Jin RZ. Molecular dynamics simulations reveal the allosteric effect of F1174C resistance mutation to ceritinib in ALK-associated lung cancer. Comput Biol Chem. 2016;65:54–60. doi: 10.1016/j.compbiolchem.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Berry T, et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell. 2012;22:117–130. doi: 10.1016/j.ccr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu S, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362–373. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Thiele CJ. ALK and MYCN: when two oncogenes are better than one. Cancer Cell. 2012;21:325–326. doi: 10.1016/j.ccr.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caren H, et al. High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: four cases of homozygous deletions of the CDKN2A gene. BMC Genomics. 2008;9:353. doi: 10.1186/1471-2164-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinsson T, et al. Appearance of the novel activating F1174S ALK mutation in neuroblastoma correlates with aggressive tumor progression and unresponsiveness to therapy. Cancer Res. 2011;71:98–105. doi: 10.1158/0008-5472.CAN-10-2366. [DOI] [PubMed] [Google Scholar]
- 46.Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol. 2016;27(Suppl 3):iii4–iii15. doi: 10.1093/annonc/mdw301. [DOI] [PubMed] [Google Scholar]
- 47.Ou SH, et al. I1171 missense mutation (particularly I1171N) is a common resistance mutation in ALK-positive NSCLC patients who have progressive disease while on alectinib and is sensitive to ceritinib. Lung Cancer. 2015;88:231–234. doi: 10.1016/j.lungcan.2015.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.