Abstract
Cereal cyst nematode (CCN, Heterodera avenae) presents severe challenges to wheat (Triticum aestivum L.) production worldwide. An investigation of the interaction between wheat and CCN can greatly improve our understanding of how nematodes alter wheat root metabolic pathways for their development and could contribute to new control strategies against CCN. In this study, we conducted transcriptome analyses of wheat cv. Wen 19 (Wen19) by using RNA-Seq during the compatible interaction with CCN at 1, 3 and 8 days past inoculation (dpi). In total, 71,569 transcripts were identified, and 10,929 of them were examined as differentially expressed genes (DEGs) in response to CCN infection. Based on the functional annotation and orthologous findings, the protein phosphorylation, oxidation-reduction process, regulation of transcription, metabolic process, transport, and response process as well as many other pathways previously reported were enriched at the transcriptional level. Plant cell wall hydrolysis and modifying proteins, auxin biosynthesis, signalling and transporter genes were up-regulated by CCN infection to facilitate penetration, migration and syncytium establishment. Genes responding to wounding and jasmonic acid stimuli were enriched at 1 dpi. We found 16 NBS-LRR genes, 12 of which were down-regulated, indicating the repression of resistance. The expression of genes encoding antioxidant enzymes, glutathione S-transferases and UDP-glucosyltransferase was significantly up-regulated during CCN infection, indicating that they may play key roles in the compatible interaction of wheat with CCN. Taken together, the results obtained from the transcriptome analyses indicate that the genes involved in oxidation-reduction processes, induction and suppression of resistance, metabolism, transport and syncytium establishment may be involved in the compatible interaction of Wen 19 with CCN. This study provides new insights into the responses of wheat to CCN infection. These insights could facilitate the elucidation of the potential mechanisms of wheat responses to CCN.
Introduction
Wheat (Triticum aestivum L.) is one of the most widely grown staple crops, providing a major source of energy and dietary fiber for humans1. Cereal cyst nematode (CCN, Heterodera avenae) is one of the most devastating sedentary endoparasitic pests and causes large yield declines in wheat production worldwide2. In China, the CCN occurs in 80% of the wheat growing areas and causes annual yield losses of 20–30%3. During the CCN life cycle, second-stage juveniles (J2) hatch from eggs at an optimum temperature of 16 °C after a low temperature (approximately 4 °C) treatment for at least 40 days. The J2 larvae invade from an elongation zone and migrate intracellularly in the wheat root with the help of their secreted cell wall softening enzymes4. The J2 larvae then select a cell adjacent to the host vascular tissues and establish a nematode feeding site (NFS) approximately 3 to 4 days past infection5,6. Subsequently, the NFS develops and lesds to the formation of a syncytium, which serves as the sole nutrient source for the sedentary life stages7. The J2s undergo three moults to develop into male or female adults, prior to fertilization and completion of their reproductive cycle2,8.
These nematodes have evolved a special capacity to secrete effectors and dramatically manipulate the host functions for their own development9. Compatible plant-nematode interactions have revealed a large number of genes and pathways associated with nematode infection, which greatly helps us understand the parasitic mechanism of the nematode. Several high-throughput methods have been employed to analyse the transcriptional regulation of the host genes by nematode parasitism including microarrays10–12, proteomic analysis13 and RNA-Seq6,14. Microarray analysis has been used to study the compatible interactions of soybean cyst nematode (SCN, H. glycines) with soybean (Glycine max L.). The results revealed that the genes encoding trehalose phosphate synthase and phospholipase D and the genes involved in metabolism, transport and resistance were induced in susceptible soybean plants12. The compatible interaction of Musa acuminata and root-knot nematode (RKN, Meloidogyne incognita) led to early host defence responses including reactive oxygen species and jasmonate/ethylene plant hormone signalling15. Auxin metabolism and cell wall modification genes were also induced15. Host transcriptional profiling of Phaseolus vulgaris in response to RKN during compatible interactions showed that 797 host genes were differentially expressed; the resistance was repressed, and reactive oxygen species were reduced16. The transcriptome response of rice to sedentary endoparasitic RKN and migratory root rot nematode (RRN, Hirschmanniella oryzae) showed that RKN suppress the salicylic acid and ethylene pathways, stimulate host metabolism, and strongly induce the expression of plant development-associated hormones, while the migratory nematode (RRN) induces program cell death and oxidative stress17. In addition, the previous studies conducted on fungal diseases, e.g., compatible interactions between wheat and Septoria tritici18 and between tomato and Phytophthora infestans19, broadened our knowledge of the infection process. Analyses of resistant and susceptible host genotypes in response to CCN infection has shown that the resistance does not disturb the penetration stages of CCN but does affect it through the developmental stages; the phospholipases may play a role in defence responses, and there is a strong burst of reactive oxygen species in resistant wheat after infection by CCN6. Comparative transcriptome analyses of soybean-SCN interactions has shown that ethylene, protein degradation, and phenylpropanoid pathways are important for resistance20. An investigation on the interaction of wild soybean with SCN interactions has shown that plant hormone signalling, MAPK signalling and defence signalling were affected in both resistant and susceptible genotypes21.
Little is known about compatible wheat responses to CCN parasitism during invasion, the establishment and maintenance of feeding sites. Information about CCN-responsive genes and pathways is lacking. In this study, we applied RNA-Seq technology to investigate the compatible interaction of wheat and CCN at three early time points. This approach was used not only to characterize the differentiallly expressed genes (DEGs) during CCN penetration but also to examine the gene response to feeding site establishment and maintenance. Our study provides novel data on comprehensive gene expression profiling of the compatible wheat and CCN interaction, which will be important to reveal the underlying infection mechanism and to effectively manage CCN in the future.
Results
Identification and functional annotation of differentially expressed genes
In our previous studies, to identify CCN resistance candidate genes, the transcriptome of Wen 19 in response to CCN was sequenced by RNA-Seq6. To generate comprehensive information during the compatible interaction between T. aestivum L. and H. avenae, this dataset was used to explore compatible transcriptional analysis of Wen 19 in response to the CCN at three time points: 1 dpi, 3 dpi and 8 dpi. Similar numbers of clean reads were obtained at each time point: 214,092,190, 215,995,184 and 215,455,900, respectively (Table S1). The DEGs between the control (before nematode infection) and nematode-infected roots were defined as two-fold up or down-regulated genes with FDR correction and a P value ≤ 0.001. In this manner, we found the largest numbers of DEGs (6,029) at the time point of feeding site establishment (3 dpi), including 4,467 up-regulated and 1,563 down-regulated genes, followed by the penetration stage (1 dpi), at which 3,649 DEGs were detected including 2,513 up-regulated and 1,136 down-regulated genes. At the development stage of the feeding site (8 dpi), only 1,251 significant DEGs were found, including 945 up-regulated and 306 down-regulated genes (Table S1), suggesting that the Wen 19 may have a strong reaction during the early infection stages of CCN (1 dpi and 3 dpi) and that the CCN was able to inhibit the wheat responses in the compatible interaction at 8 dpi (Table S1). To search the primary functional groups of the DEGs, we performed BLASTx alignments to protein databases Nr, Swiss-Prot, KEGG and COG with a cut-off E-value of 1e-5. This indicates that a total of 10,929 (81.72%), 9,247 (79.55%), 7,270 (51.06%) and 5,437 (49.75%) DEGs had significant matches in the Nr, Swiss-Prot, KEGG and COG databases, respectively (Table 1).
Table 1.
Functional annotation of the differentially expressed genes (DEGs) of Wen 19 in response to the CCN.
| CCN response | No. of DEGs | No. of DEGs in NR | No. of DEGs in Swiss-Prot | No. of DEGs in KEGG | No. of DEGs in COG |
|---|---|---|---|---|---|
| 1 dpi | 3649 | 2982 | 2377 | 1136 | 1507 |
| 3 dpi | 6029 | 5228 | 4103 | 3747 | 2553 |
| 8 dpi | 1251 | 1037 | 790 | 697 | 477 |
| Total | 10929 | 9247 (81.72%) | 7270 (79.55%) | 5580 (51.06%) | 5437 (49.75%) |
Among the 7,925 up-regulated genes, 240 genes were up-regulated at all three time points, 1,564 genes at two time points (1,241 genes at 1 and 3 dpi, 265 at 1 and 8 dpi, and 538 at 3 and 8 dpi) and 4,557 at one time point (1247, 2928 and 382 at 1 dpi, 3 dpi and 8 dpi, respectively). The numbers of exclusive genes belonging to were (Fig. 1a). Three genes were significantly down-regulated at all three time points (Fig. 1b), while 242 genes were down-regulated at two time points (202 genes at 1 and 3 dpi, 6 genes at 1 and 8 dpi and 40 genes at 3 and 8 dpi) and 2570 genes were down-regulated at one time point (931, 1,323 and 263 at 1 dpi, 3 dpi and 8 dpi, respectively) (Fig. 1b).
Figure 1.
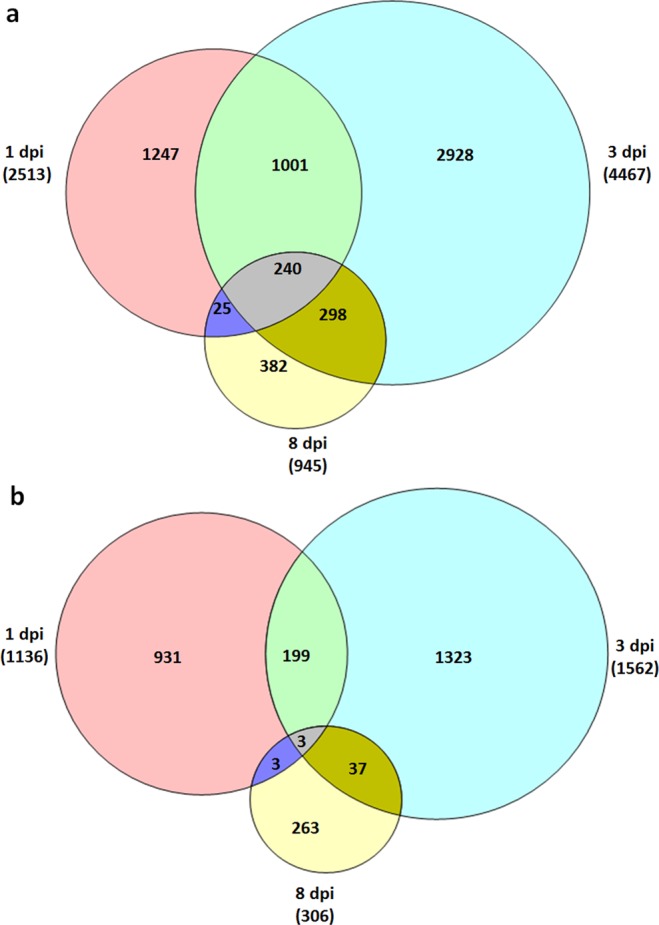
Venn diagram of the numbers of up-regulated (a) and down-regulated (b) genes at 1 dpi, 3 dpi and 8 dpi.
Transcriptome changes in the root tissue at 1 dpi
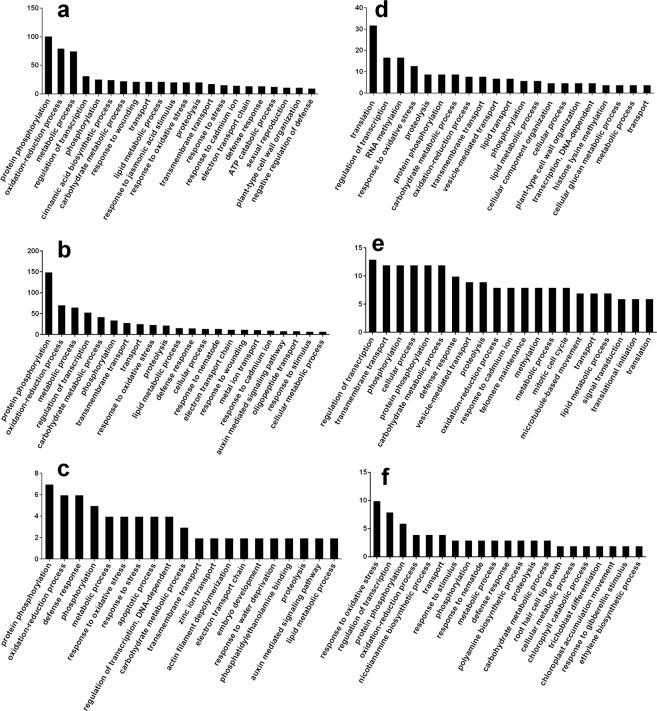
At 1 dpi when the CCN had penetrated the root, 2,178 DEGs (FDR <0.001) were exclusively found. Of these, 1,247 genes were up-regulated and 931 genes were down-regulated (Fig. 1; Table S1). Gene enrichment analysis showed 101 genes involved in protein phosphorylation, 75 genes involved in metabolic processes, 40 genes involved in transporter and transmembrane transport, and 32 genes involved in the regulation of transcription were up-regulated at 1 dpi (Fig. 2a). Plant cell wall modification may facilitate penetration and movement. We found that 12 genes in plant type cell wall organization were induced by the CCN (Fig. 2a). Genes involved in the oxidation-reduction process, in response to wounding and jasmonic acid stimuli were greatly up-regulated, which may reflect the induced host resistance at the early stage of the CCN invasion (Fig. 2a). The strongest up-regulated genes were the genes encoding aromatic-L-amino-acid decarboxylase, beta-sesquiphellandrene synthase, UDP-glycosyltransferase 91B1, nodulation-signalling pathway 2 protein, and probably linoleate 9S-lipoxygenase 4 (Table S2). The genes for the GO terms of translation, regulation of transcription, RNA methylation, response to oxidative stress, proteolysis, vesicle-mediated transport, and lipid metabolic processes were down-regulated in the infected roots (Fig. 2d). The genes that were strongly down-regulated were two genes encoding a dehydration-responsive element-binding protein and a senescence-induced receptor-like serine/threonine-protein kinase (Table S2).
Figure 2.
Gene ontology (GO) biological process of Wen 19 during the compatible interaction with the CCN at 1 dpi (a,d), 3 dpi (c,e) and 8 dpi (c,f). Numbers of up-regulated (a–c) and down-regulated (d–f) genes are illustrated in the y-axis.
Transcriptome changes in the root tissue at 3 dpi
A total of 4,251 DEGs (FDR <0.001) were found exclusively at 3 dpi when the feeding sites were established. Of these, 2,928 genes were up-regulated, and 1,323 genes were down-regulated (Fig. 1, Table S1). GO terms enrichment analysis showed that genes involved in protein phosphorylation, the oxidation-reduction process, regulation of transcription, metabolic processes, and transmembrane transport were significantly enriched in up- and down-regulated genes at 3 dpi (Fig. 2b,d). Auxin is a key element for syncytium formation22. In our dataset, the genes in the auxin-mediated signalling pathway were up-regulated (Fig. 2b). The genes encoding glutathione S-transferase GSTF1, bidirectional sugar transporter SWEET12, ethylene-responsive transcription factor RAP2-2, indole-2-monooxygenase, and peptide transporter PTR2 were strongly up-regulated (Table S2), while those involved in vesicle-mediated transport, signal transduction and translation were down-regulated in the infected root (Fig. 2d). The genes encoding ethylene-responsive transcription factor TINY, transcription factor HY5, and phytochrome-associated serine/threonine-protein phosphatase were strongly down-regulated (Table S2).
Transcriptome changes in root tissue at 8 dpi
In total, 645 DEGs (FDR <0.001) were exclusively found at 8 dpi. Of these, 382 genes were up-regulated and 263 genes were down-regulated (Fig. 1, Table S1). GO enrichment revealed that the genes involved in protein phosphorylation, response to stress, the oxidation-reduction process, defence response and metabolic process, transport, and response to stimulus were significantly affected upon CCN infection (Fig. 2c,f), and the genes involved in the regulation of transcription and the nicotiana mine biosynthetic process were down-regulated at 8 dpi (Fig. 2f). Alternatively, genes involved in zinc ion transport and the auxin mediated signalling pathway were up-regulated at 8 dpi (Fig. 2c). The strongest up-regulated genes were those encoding proline-rich receptor-like protein kinase PERK10, ethylene responsive transcription factor 6, and cytosolic sulfotransferase 12. The strongest down-regulated genes were those encoding beta-galactosidase 7, elongation factor 1-alpha C and uncharacterized proteins (Table S2).
Plant cell wall, plant hormone auxin and jasmonic acid play important roles in CCN infection
Both root penetration and syncytium establishment depend on the degradation and modification of the host cell wall23,24. In our dataset, the genes involved in plant cell wall hydrolysis and modifying proteins such as xyloglucan endotransglycosylase/hydrolase protein 32, expansins, endoglucanase 2 and endoglucanase 4 were up-regulated by CCN infection at 1 dpi and 3 dpi (Table 2). The genes encoding expansin-B7, expansin-A3 and probable pectate lyase 8 were up-regulated by CCN infection at 1 dpi, 3 dpi and 8 dpi (Table 2). The genes encoding xyloglucan endotransglycosylase/hydrolase protein 8 and pectate lyase were up-regulated at 3 dpi, while the genes encoding probable xyloglucan endotransglucosylase/hydrolase and expansin-A9 were down-regulated at 1 dpi and 3 dpi (Table 2). In jasmonic acid (JA) biosynthesis and signalling, the probable linoleate 9S-lipoxygenase 4 and linoleate 9S-lipoxygenase 1 genes were intensely induced at 1 dpi but not in the later infection stages. The genes encoding allene oxide cyclase 3 and pathogenesis-related protein PRB1–2 were only induced at 3 dpi (Table 3). The genes encoding a lipoxygenase 6 and a linoleate 9S-lipoxygenase 2 were up-regulated at 1 and 3 dpi, while another linoleate 9S-lipoxygenase 2 gene was down-regulated at 3 and 8 dpi. Lipoxygenase 8 and probable linoleate 9S-lipoxygenase 5 genes were up-regulated at 1 dpi, 3 dpi and 8 dpi (Table 3). The gene set of the auxin-mediated signalling pathway was up-regulated at 3 dpi and 8 dpi (Fig. 2b,c). Genes involved in auxin biosynthesis, such as those encoding flavin-containing monooxygenase YUCCA9, indole-3-acetic acid-amido synthetase, aromatic-L-amino-acid decarboxylase and auxin response and induced proteins were up-regulated by CCN at 1 dpi and 3 dpi (Table 3). A gene encoding an auxin influx carrier was significantly up-regulated, but one encoding the auxin efflux carrier component 1c was down-regulated (Table 3).
Table 2.
Expression of genes involved in cell wall hydrolysis and encoding modifying proteins in the infected Wen19 compared to the non-infected Wen 19. FC: fold change (CCN infection vs non-infected samples);–: not differentially expressed.
| GenBand-ID | Annotation | Log2FC 1 dpi | Log2FC 3 dpi | Log2FC 8 dpi |
|---|---|---|---|---|
| MH883021 | Expansin-B2 | 1.4162 | 2.4476 | 1.0598 |
| MH908106 | Expansin-B4 | 2.738 | 3.8566 | — |
| MH908107 | Expansin-B3 | 2.326 | 1.8641 | — |
| MH908108 | Expansin-B2 | 2.6025 | 3.5502 | — |
| MH908109 | Expansin-B15 | 1.2109 | 1.415 | — |
| MH908110 | Expansin-A8 | 2.8025 | 2.2812 | — |
| MH908111 | Expansin-A7 | 1.7415 | 1.9023 | — |
| MH908112 | Expansin-A3 | 3.0243 | 3.7292 | 2.1655 |
| MH908113 | Expansin-A2 | 1.717 | 2.246 | — |
| MH908114 | Expansin-A1 | 1.7882 | 2.2398 | — |
| MH908115 | Xyloglucan endotransglycosylase/hydrolase protein 8 | — | 1.2156 | — |
| MH908116 | Probable xyloglucan endotransglucosylase/hydrolase protein 32 | 2.3702 | 1.4671 | — |
| MH908117 | Pectate lyase | — | 1.0949 | — |
| MH908118 | Endonuclease 4 | 1.3031 | 1.3742 | — |
| MH908119 | Endonuclease 2 | 1.7503 | 1.2929 | — |
| MH908120 | Probable xyloglucan endotransglucosylase/hydrolase | −2.6012 | −1.7502 | — |
| MH908121 | Expansin-A9 | −1.3792 | −1.3956 | — |
Table 3.
Expression of the genes involved in jasmonic acid and auxin signalling pathways in the infected Wen19 compared to the non-infected Wen 19. FC: fold change (CCN infection vs non-infected samples);–: not differentially expressed.
| GenBank-ID | Annotation | Log2FC 1 dpi | Log2FC 3 dpi | Log2FC 8 dpi |
|---|---|---|---|---|
| Jasmonic acid signalling pathway | ||||
| MH891149 | Probable lipoxygenase 8, chloroplastic | 2.6551 | 3.7291 | 1.7409 |
| MH891150 | Probable linoleate 9S-lipoxygenase 5 | 4.3855 | 5.9062 | 1.3881 |
| MH891151 | Probable lipoxygenase 6 | 1.4287 | 1.3063 | — |
| MH891153 | Allene oxide cyclase 3, chloroplastic | 1.3774 | 1.6346 | — |
| MH891152 | Pathogenesis-related protein PR1–2 | — | 1.9079 | — |
| MH891154 | Linoleate 9S-lipoxygenase 1 | 2.1548 | — | — |
| Auxin signalling pathway | ||||
| MH891155 | Aromatic-L-amino-acid decarboxylase | 6.0844 | 4.9628 | 2.3992 |
| MH891156 | Flavin-containing monooxygenase FMO | — | 1.7399 | — |
| MH891157 | Flavin-containing monooxygenase YUCCA9 | 1.2702 | 2.4058 | — |
| MH891158 | Indole-3-acetic acid-induced protein ARG7 | — | 1.4409 | — |
| MH891159 | Auxin-induced in root cultures protein 12 | 2.4763 | 3.6903 | — |
| MH891160 | Auxin response factor 5 | — | 1.3689 | — |
| MH891161 | Auxin-repressed 12.5 kDa protein | — | 1.4916 | — |
| MH891162 | Auxin-responsive protein IAA21 | — | 1.1678 | — |
| MH891163 | Auxin-induced protein X15 | — | 1.474 | — |
| MH891164 | Auxin-induced protein 10A5 | 1.2925 | 1.4842 | — |
| MH891165 | WAT1-related protein | 2.2927 | 2.7679 | 1.688 |
| MH891166 | Auxin influx carrier (AUX1 LAX family) | — | 2.015 | 1.6046 |
| MH891167 | Probable auxin efflux carrier component 1c | — | −1.6004 | — |
CCN-responsive genes in antioxidant and detoxification processes
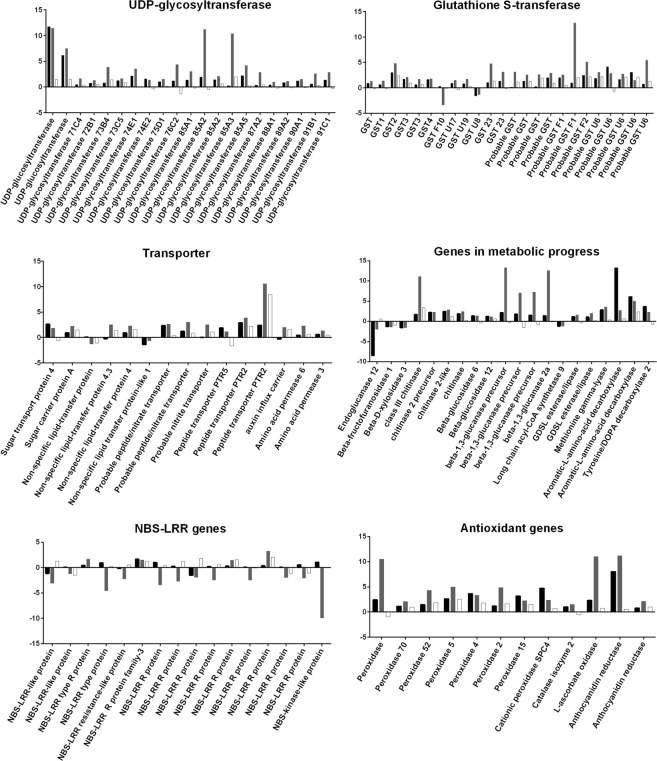
The GO biological process enrichment showed that genes involved in the oxidation-reduction process and in response to oxidative stress were enriched in both up- and down-regulated genes at all three time points tested. After pathogen recognition, reactive oxygen species (ROS) are rapidly induced and accumulated6. In this study we found that eight respiratory burst oxidase genes were induced by CCN infection and were especially highly expressed at 3 dpi (Table S3). Genes encoding peroxidase and the antioxidant enzymes ascorbate and anthocyanidin reductase were induced (Fig. 3; Table S3). Detoxification genes, such as those encoding UDP-glycosyltransferase and glutathione S-transferases (GST) were highly expressed at the early stages. We found 24 DEGs in the RNA-Seq data which were annotated to encode glutathione S-transferases in the Nr or Swissprot database. Of these, 12, 22 and 11 were significantly up-regulated at 1 dpi, 3 dpi and 8dpi respectively (Fig. 3; Table S4). Among the differentially expressed UDP-glycosyltransferase genes, 11 and 14 were significantly up-regulated at 1 dpi and 3 dpi, respectively (Fig. 3; Table S4). In addition, 17 NBS-LRR genes were differentially expressed during the CCN infection, and 12 of them were down-regulated (Fig. 3; Table S5).
Figure 3.
Expression profiles of UDP-glucosyltransferase, GST (glutathione S-transferase), transporter, metabolic progress, and NBS-LRR genes from the RNA-Seq data. Units are normalized to the numbers of reads. Log2FC between nematode-infected and non-infected (control) samples at each time point is indicated in the y-axis. FC: fold change (CCN infection vs control samples).  : 1 dpi
: 1 dpi  : 3 dpi
: 3 dpi  : 8 dpi.
: 8 dpi.
Genes associated with metabolic and transporter progress were strongly affected by CCN infection
In this study, we found that the expression profiles of many genes that function in metabolic pathways were significantly affected by the CCN. For example, we observed that the genes encoding glucan endo-1,3-beta-glucosidase, beta-glucosidase, chitinase and acidic endochitinase in the carbohydrate metabolic process were up-regulated at 1 dpi and 3 dpi, while beta-fructofuranosidase, endoglucanase 12, beta-fructofuranosidase and sucrose: sucrose 1-fructosyltransferase genes were down-regulated. In the fatty acid biosynthetic process, the 3-ketoacyl-CoA synthase 11 gene was up-regulated while the long chain acyl-CoA synthetase 9 gene was down-regulated. Genes involved in the lipid metabolic process were up-regulated at 3 dpi. The tyrosine/DOPA decarboxylase 2, methionine gamma-lyase, and aromatic-L-amino-acid decarboxylase genes which are involved in the amino acid metabolic process were greatly up-regulated at 1 and 3 dpi (Fig. 3; Table S6). In addition, transporter genes were also greatly enriched in both up- and down-regulated genes, but the vesicle-mediated transport genes were only enriched in the down-regulated group at 1 dpi and 3 dpi. For example, the gene encoding sugar transport protein 4 was up-regulated at 1 and 3 dpi, and the gene encoding sugar carrier protein A was up-regulated at 3 dpi and 8 dpi. Genes encoding amino-acid permease BAT1, probable peptide/nitrate transporter, and peptide transporter PTR5 were significantly up-regulated at 1 dpi and 3 dpi. Genes encoding peptide transporter PTR2, aquaporin NIP1-1, probable purine permease 5, probable potassium transporter, auxin transporter-like protein 2 and probable auxin efflux carrier component 4 were significantly up-regulated at 1 dpi, 3 dpi and 8 dpi (Fig. 3; Table S7).
GO enrichment of the up-regulated genes at all three time points
There are some genes that were up-regulated at all three time points: 1 dpi, 3 dpi and 8 dpi. In total, 240 up-regulated transcripts were obtained. GO enrichment showed that a large number of the oxidation-reduction process, translation, protein phosphorylation, response to biotic stimulus, metabolic process and cell wall modification genes were found in the up-regulated group (Fig. 4a). Cellular component enrichment showed that the largest numbers of genes products were found in the cytoplasmic membrane-bounded vesicle (Fig. 4b). The first set of candidates related to wheat susceptibility to CCN were selected from 3 GO terms: protein phosphorylation, oxidation-reduction process, metabolic process (Table 4). The second set of candidates was selected among the strongest up-regulated genes including those for: vegetative cell wall protein, elongation factor 1-alpha, ent-kaurenoic acid oxidase 1, and the cytoskeleton related genes: actin, tubulin alpha chain, and tubulin beta chain (Table 4).
Figure 4.
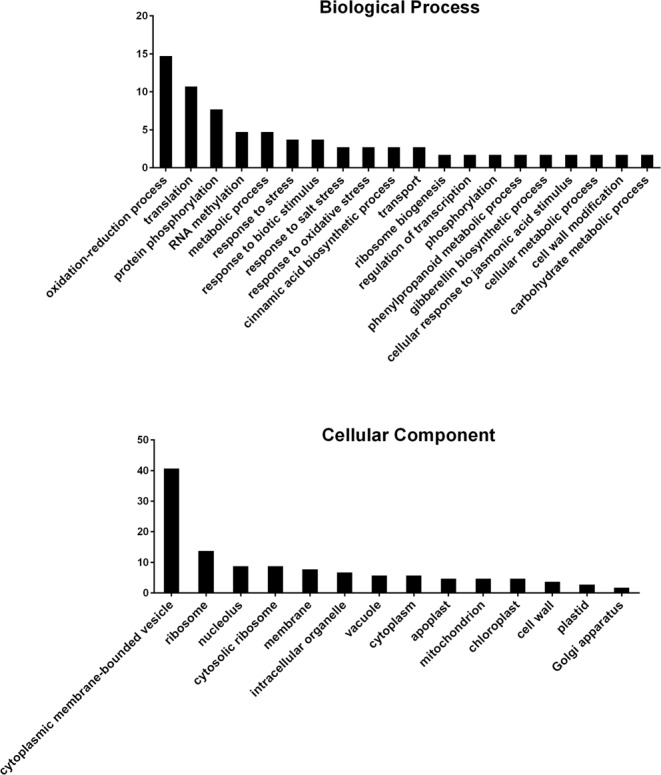
GO biological process and cellular component definition of up-regulated genes at all three time points (1 dpi, 3 dpi and 8 dpi). Y-axis indicates the numbers of genes.
Table 4.
Selected genes that were continuously up-regulated in the infected Wen 19 compared to the non-infected Wen 19 at all the time points (1 dpi, 3d pi and 8 dpi). FC: fold change (CCN infection vs non-infected samples).
| GenBank-ID | Annotation | Log2FC 1 dpi | Log2FC 3 dpi | Log2FC 8 dpi |
|---|---|---|---|---|
| MH908122 | Wall-associated receptor kinase 5 | 1.9181 | 4.234 | 1.756 |
| MH908123 | Wall-associated receptor kinase 3 | 1.5391 | 3.7122 | 2.0208 |
| MH908124 | Peroxidase 4 | 3.4673 | 4.3622 | 1.6753 |
| MH908125 | Peroxidase 23 | 3.853 | 2.3877 | 1.5927 |
| MH908126 | Probable flavin-containing monooxygenase 1 | 2.5031 | 10.894 | 8.3219 |
| MH908127 | 2-oxoglutarate/Fe(II)-dependent dioxygenase | 12.893 | 3.5903 | 1.9144 |
| MH908128 | Leucoanthocyanidin dioxygenase | 11.7895 | 10.5855 | 2.3999 |
| MH908129 | Bifunctional monodehydroascorbate reductase | 12.7432 | 11.7372 | 5.9681 |
| MH908130 | 1-aminocyclopropane-1-carboxylate oxidase homologue 2 | 2.9182 | 10.5613 | 7.6675 |
| MH908131 | UDP-glucosyltransferase | 11.7396 | 11.4498 | 1.4929 |
| MH908132 | Probable glutathione S-transferase GSTU6 | 1.836 | 3.0525 | 1.4407 |
| MH908133 | glutathione S-transferase 23 | 2.977 | 4.7771 | 2.3581 |
| MH908134 | Probable glutathione S-transferase GSTF2 | 2.4487 | 5.059 | 2.1934 |
| MH908135 | Elongation factor 1-alpha | 11.1322 | 12.1807 | 11.0826 |
| MH908136 | Ent-kaurenoic acid oxidase 1 | 13.2378 | 11.1729 | 5.0316 |
| MH908137 | Actin | 7.3949 | 11.5589 | 13.9831 |
| MH908138 | Tubulin alpha chain | 10.3013 | 12.4728 | 11.6972 |
| MH908139 | Tubulin beta chain | 9.1306 | 11.3262 | 10.623 |
Discussion
In this study, we analysed the gene transcription levels in susceptible wheat Wen 19 during interaction with the CCN at 1 dpi, 3 dpi and 8 dpi which correspond to the stages of nematode penetration, feeding site establishment and maintenance, respectively5,6. We found 3649, 6029 and 1251 significant DEGs from evenly clean reads of 1 dpi, 3 dpi and 8 dpi samples, respectively (Table S1), suggesting that the Wen19 may have a strong reaction during the early infection stages (1 dpi and 3 dpi), but the nematode is able to inhibit the plant responses in the compatible interaction at 8dpi.
At 1 dpi, the J2s have penetrated the root6. Both the penetration and migration of J2 are dependent on the degradation and loosening of the host cell wall16. At 3 dpi, the syncytium is established5,6, possibly as a result of the modulation of the plant cell wall and hormones. For instance, cell wall modification and hydrolysis-related genes were induced, and auxin concentration, transportation and distribution were altered9,22. In this study, the gene set of plant-type cell wall was enriched at 1 dpi (Fig. 2a). Genes encoding xyloglucan endotransglycosylase/hydrolase protein, expansins, endoglucanase 2 and endoglucanase 4 were up-regulated by the CCN infection (Table 2), which was consistent with previous reports that the genes encoding expansins, endoglucanases and pectate lyases were up-regulated by nematode infection9. The gene set of the auxin-mediated signalling pathway was up-regulated at 3 dpi and 8 dpi (Fig. 2b,c). Genes involved in auxin biosynthesis such as those encoding flavin-containing monooxygenase YUCCA9, probable indole-3-acetic acid-amido synthetase, aromatic-L-amino-acid decarboxylase25 and auxin response, as well as induced proteins, were up-regulated by the CCN infection at 1 dpi and 3 dpi (Table 3), which was consistent with the previous report that auxin was a key element for syncytium formation22. An auxin influx carrier gene was significantly up-regulated, but a probable auxin efflux carrier component 1c gene was down-regulated (Table 3). A similar result that auxin transporter PIN was important for CCN infection has been reported26.
Metabolic activity and nutrient transport play an important role in CCN infection27,28. GO terms of biological process enrichment showed that the genes involved in the metabolic process, protein phosphorylation, and transport progression were greatly enriched in both up- and down-regulated genes during the CCN infection. It was reported that metabolic and transport progress was very active in the syncytia27,28. We found that in the carbohydrate metabolic process, the genes encoding glucan endo-1,3-beta-glucosidase and beta-glucosidase were up-regulated (Fig. 3; Table S6), which suggested that polysaccharide catabolism was induced. Simultaneously, the genes encoding the sugar transport protein 4 and sugar carrier protein A were up-regulated (Fig. 3; Table S6), indicating a profound role of sugar transportation in the CCN infection. This result is consistent with previous reports that the content of soluble sugars in the stems of tomato has increased after RKN infestation29. Sugar transporters were specifically expressed and active in the syncytia in the previous studies30. In this study, we found that amino acid, peptide metabolic and transporter genes were greatly induced by the CCN (Table S7), which suggests that amino acid metabolism plays a crucial role for nematode infection. Previous metabolic profiling revealed that amino acids and phosphorylated metabolites were increased in the syncytia28. In addition, we found that aquaporin NIP1-1 genes (Table S7), which are associated with stress tolerance31, were significantly induced by CCN.
Nematode invasion may have induced basal host resistance32,33. We found that several genes associated with wounding response were up-regulated at 1 dpi and 3 dpi (Fig. 2a,b), which may be a result of the penetration and migration of CCN. It has been reported that damage-induced defence is controlled by the JA signalling pathway34. In our dataset, the genes that respond to jasmonic acid stimulus were enriched in up-regulated GO terms at 1 dpi (Fig. 2a). In addition, the probable linoleate 9S-lipoxygenase 4 gene was intensely induced at 1 dpi but not in the later infection stages. The allene oxide cyclase 3 gene was induced only at 3 dpi (Table 3), which may indicate that the host induces JA response at the early stage of CCN infection. In the interactions of Arabidopsis thaliana and H. schachtii, JA-triggered early defence has been reported35. In tomato, the JA signalling pathway is required during compatible interaction with RKN36. In addition, we found that the respiratory burst oxidase genes were induced by CCN infection at 3 dpi (Table S3). Early host defence, including ROS induction, was reported during the compatible interactions of Musa acuminata with RKN15. Alternatively, the negative regulation of the defence progress was up-regulated (Fig. 2a), and the response to oxidative stress genes was down-regulated at 1 dpi (Fig. 2d), suggesting that the resistance reaction was under control. The antioxidant enzymes peroxidase, ascorbate and anthocyanidin reductase were induced by CCN. Pioneering research illustrated that the overexpression of anthocyanidin reductase increased ROS scavenging and enhanced tobacco tolerance37. It was reported that the effector MjTTL5 from the RKN can interact with ferredoxin to dramatically activate ROS scavenging, resulting in defence suppression38. In this study, the genes encoding UDP-glycosyltransferase and glutathione S-transferases (GSTs) were highly expressed after CCN infection, suggesting that they may play an important role in the wheat-CCN interaction. GSTs play conserved roles in the detoxification of toxic compounds from pathogen attack and oxidative stress39. The RKN and pine wood nematode can secrete GSTs into their hosts to counteract plant resistance40,41. In compatible soybean, GST was induced in the syncytia and whole roots by the soybean cyst nematode42. In this study, 22 of 24 GST genes were significantly up-regulated after nematode infection at 3 dpi. We also found that the UDP-glycosyltransferase genes were significantly up-regulated after CCN infection at 1 dpi and 3 dpi. The overexpression of the UDP-glucosyltransferase from A. thaliana can improve plant tolerance43. The UDP-glucosyltransferase from Brachypodium distachyon can detoxify deoxynivalenol from Fusarium graminearum44. Antioxidant enzymes, such as GSTs and UDP-glycosyltransferase, may be crucial in scavenging ROS and detoxifying toxins in wheat after nematode attack, as well as keeping the plant defence under control. We also found that most NBS-LRR genes were down-regulated, illustrating the critical role of resistance suppression in successful CCN infection. The suppression of tomato defense responce genes upon potato cyst nematode infection indicates a key regulatory role of miRNAs45. Successful nematode parasitism appears to involve recognition by the host, the induction of early resistance reactions, the elimination of ROS and control of resistance46. Cloning the enzyme-encoding genes and verification of their functions will further improve our understanding of the CCN infection and could help to the design of effective management strategies to reduce CCN damage.
In conclusion, from an analysis of compatible wheat-cereal cyst nematode interaction analyses, a large number of genes and pathways associated with CCN infection were revealed. In this study, we found that the metabolic and transporter processes were regulated by CCN. This process may facilitate the development of the nematodes. Plant cell wall hydrolysis, modifying proteins and auxin biosynthesis, signalling and transporter genes were regulated by the nematode. This may facilitate the penetration, migration and syncytium establishment. It seems that CCN infection can induce host defense reactions but can also hijack the antioxidant and detoxification processes of the plant to eliminate ROS and toxic components and suppress the NBS-LRR resistance gene expression.
Materials and Methods
Plant materials and CCN treatments
A compatible wheat variety Triticum aestivum L. cv. Wen 19 was used for transcriptomic profiling in this study. The seeds were germinated in Petri dishes on wet gauze at 25 °C in the dark for 2 days. Seedlings were planted in 50 mL tubes filled with sterilized sands and grown at 16 °C under a 16 h light/8 h dark photoperiod for 7 days6. Cereal cyst nematodes were propagated in the greenhouse under non-sterile conditions as previously described47. Fully developed cysts were incubated in 3 mM ZnCl2, and the J2 larvae were harvested and counted with a microscope. Each seedling was inoculated with 500 J2 larvae and the negative control was not inoculated. At 24 hours past inoculation, the roots were washed three times with sterilized H2O to remove CCN adhering to the roots, transplanted into pots containing 500 mL of sterilized sands and grown as described by Kong et al. and Xu et al.6,48.
RNA extraction and sequencing
Root materials were sampled at 1, 3 and 8 days past inoculation, and immediately frozen in liquid nitrogen. Total RNA was extracted using the TRIzol reagent according to the manufacturer’s instructions and treated with DNase (Invitrogen, USA). The concentration and quality of the RNA was determined using a NanoDrop-2000 (ThermoFisher Scientific Inc., USA). RNA extracted at 1 dpi, 3 dpi and 8 dpi were quantified to an equal concentration by adding RNase-free water and subsequently sequenced (paired-end, 2 × 100 bp) using an Illumina HiSeq™ 2000 (Illumina Inc., USA).
Bioinformatic analysis
The data analysis was performed as previously described6. During the reads trimming, low quality reads (Phred score <Q30), reads containing more than two ambiguous bases (‘N’) and reads shorter than 50 nucleotides were filtered out. The CCN transcripts were removed as previously described6. The clean reads from each sample were de novo assembled together using the Trinity program package49. Gene annotation was performed via BLASTx alignments to protein databases (Nr, Swiss-Prot, KEGG and COG) and BLASTn alignments to the NCBI nucleotide databases with a cut-off E-value of 1e−5. We performed the GO annotations of the transcripts by Blast2GO program50. The expressions of unigenes were calculated by Fragments Per kb per Million Fragments (FPKM). The false discovery rate (FDR) was used to determine the P value threshold. A gene was judged to be a DEG if the P value (FDR) ≤0.001 and the fold change (FC) between CCN infection and control samples at each time point ≥251.
Supplementary information
Acknowledgements
The authors thank Dr. Nicholas Clarke at the Division of Environment and Natural Resources of the NIBIO-Norwegian Institute of Bioeconomy Research for linguistic correction. We are grateful to Dr. Houxiang Kang at the Institute of Plant Protection of Chinese Academy of Agricultural Sciences for the RNA-seq data analyses and Xuehong Peng for technical assistance in the lab and greenhouse. This study was supported financially by the National Nature Science Foundation of China (Grant number 31571988) and the Special Fund for Agro-Scientific Research in the Public Interest (Grant number 201503114 and 200903040).
Author Contributions
D.L.P. designed the experiments. L.A.K. and D.Q.W. performed the RNA sequencing. F.Q. visualized the data and wrote the original draft. J.L.C., L.A.K., H.P., S.M.L. revised the manuscript, W.K.H. and D.W.Q. contributed reagents and materials. All authors reviewed and commented on the manuscript.
Data Availability
Sequences of differentially expressed transcripts were submitted to GenBank and GenBank accession numbers are provided in Tables 2, 3 and 4.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fen Qiao and Ling-An Kong contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37824-9.
References
- 1.Shewry PR, Hey SJ. The contribution of wheat to human diet and health. Food Energy Security. 2015;4:178–202. doi: 10.1002/fes3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar M, et al. De novo transcriptome sequencing and analysis of the cereal cyst nematode. Heterodera avenae. PLoS ONE. 2014;9:e96311. doi: 10.1371/journal.pone.0061550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng, D. L. et al. Current knowledge of cereal cyst nematode (Heterodera avenae) on wheat in China In ‘Cereal cyst nematodes: status, research and outlook.’ Editered by Riley, I. T., Nicol J. M. & Dababat A. A., 29–34 (CIMMYT: Ankara, Turkey) (2009).
- 4.Long HB, Peng DL, Huang WK, Peng H, Wang GF. Molecular characterization and functional analysis of two new β-1,4-endoglucanase genes (Ha-eng-2, Ha-eng-3) from the cereal cyst nematode Heterodera avenae. Plant Pathol. 2013;62:953–960. doi: 10.1111/ppa.12000. [DOI] [Google Scholar]
- 5.Williams KJ, Fisher JM. Development of Heterodera avenae Woll. and host cellular responses in susceptible and resistant wheat. Fund. Appl. nematol. 1993;16:417–423. [Google Scholar]
- 6.Kong LA, et al. Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics. 2015;16:801. doi: 10.1186/s12864-015-2037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grymaszewska G, Golinowski W. Structure of syncytia induced by Heterodera avenae Woll. in roots of susceptible and resistant wheat (Triticum aestivum L.) J Phytopathol. 1991;133:307–319. doi: 10.1111/j.1439-0434.1991.tb00166.x. [DOI] [Google Scholar]
- 8.Lasserre F, et al. Genetic variation in natural populations of the cereal cyst nematode (Heterodera avenae Woll.) submitted to resistant and susceptible cultivars of cereals. Theor. Appl. Genet. 1996;93:1–8. doi: 10.1007/BF00225719. [DOI] [PubMed] [Google Scholar]
- 9.Gheysen G, Mitchum MG. How nematodes manipulate plant development pathways for infection. Current opinion in plant biology. 2011;14:415–421. doi: 10.1016/j.pbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim HM, et al. Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics. 2011;12:220. doi: 10.1186/1471-2164-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puthoff DP, Nettleton D, Rodermel SR, Baum TJ. Arabidopsis gene expression changes during cyst nematode parasitism revealed by statistical analyses of microarray expression profiles. Plant J. 2003;33:911–921. doi: 10.1046/j.1365-313X.2003.01677.x. [DOI] [PubMed] [Google Scholar]
- 12.Alkharouf NW, et al. Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode) Planta. 2006;224:838–852. doi: 10.1007/s00425-006-0270-8. [DOI] [PubMed] [Google Scholar]
- 13.Villeth GR, et al. Cowpea-Meloidogyne incognita interaction: Root proteomic analysis during early stages of nematode infection. Proteomics. 2015;15:1746–1759. doi: 10.1002/pmic.201400561. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, et al. Transcriptional responses of wheat and the cereal cyst nematode Heterodera avenae during their early contact stage. Sci. Rep. 2017;7:14471. doi: 10.1038/s41598-017-14047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castañeda NEN, et al. Gene expression analysis in Musa acuminata during compatible interactions with Meloidogyne incognita. Annals of Botany. 2017;119:915–930. doi: 10.1093/aob/mcw272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santini L, et al. Host transcriptional profiling at early and later stages of the compatible interaction between Phaseolus vulgaris and Meloidogyne incognita. Phytopathol. 2016;106:282–294. doi: 10.1094/PHYTO-07-15-0160-R. [DOI] [PubMed] [Google Scholar]
- 17.Kyndt T, et al. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New phytol. 2012;196:887–900. doi: 10.1111/j.1469-8137.2012.04311.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Li W, Jorgensen HJ. Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS ONE. 2013;8:e81606. doi: 10.1371/journal.pone.0081606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuluaga AP, et al. Analysis of the tomato leaf transcriptome during successive hemibiotrophic stages of a compatible interaction with the oomycete pathogen Phytophthora infestans. Mol. Plant Pathol. 2016;17:42–54. doi: 10.1111/mpp.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan J, et al. Whole-genome gene expression profiling revealed genes and pathways potentially involved in regulating interactions of soybean with cyst nematode (Heterodera glycines Ichinohe) BMC Genomics. 2015;16:148. doi: 10.1186/s12864-015-1316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, et al. Comparative RNA-Seq analysis uncovers a complex regulatory network for soybean cyst nematode resistance in wild soybean (Glycine soja) Sci. Rep. 2017;7:9699. doi: 10.1038/s41598-017-09945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C, et al. The novel cyst nematode effector protein 19C07 interacts with the Arabidopsis auxin influx transporter LAX3 to control feeding site development. Plant Physiol. 2011;155:866–880. doi: 10.1104/pp.110.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Lilley CJ, Imren M, Knox JP, Urwin PE. The complex cell wall composition of syncytia induced by plant parasitic cyst nematodes reflects both function and host plant. Front. Plant Sci. 2017;8:1087. doi: 10.3389/fpls.2017.01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohlmann H, Sobczak M. The plant cell wall in the feeding sites of cyst nematodes. Front Plant Sci. 2014;5:89. doi: 10.3389/fpls.2014.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. J. Exp. Biol. 2012;63:2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- 26.Grunewald W, Cannoot B, Friml J, Gheysen G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS pathogens. 2009;5:e1000266. doi: 10.1371/journal.ppat.1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammes UZ, et al. Nematode-induced changes of transporter gene expression in Arabidopsis roots. Mol. Plant- Microbe In. 2005;18:1247–1257. doi: 10.1094/MPMI-18-1247. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann J, et al. Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. Plant J. 2010;62:1058–1071. doi: 10.1111/j.1365-313X.2010.04217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eloh K, Sasanelli N, Maxia A, Caboni P. Untargeted metabolomics of tomato plants after root-knot nematode infestation. J. Agric. Food Chem. 2016;64:5963–5968. doi: 10.1021/acs.jafc.6b02181. [DOI] [PubMed] [Google Scholar]
- 30.Hofmann J, et al. Diversity and activity of sugar transporters in nematode-induced root syncytia. J. Exp. Bot. 2009;60:3085–3095. doi: 10.1093/jxb/erp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hove RM, Ziemann M, Bhave M. Identification and expression analysis of the barley (Hordeum vulgare L.) aquaporin gene family. PLoS ONE. 2015;10:e0128025. doi: 10.1371/journal.pone.0128025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clevenger J, et al. Gene expression profiling describes the genetic regulation of Meloidogyne arenaria resistance in Arachis hypogaea and reveals a candidate gene for resistance. Sci. Rep. 2017;7:1317. doi: 10.1038/s41598-017-00971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature. 2012;492:256–260. doi: 10.1038/nature11651. [DOI] [PubMed] [Google Scholar]
- 34.Koo AJ. Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytoch. Reviews. 2017;17:51–80. doi: 10.1007/s11101-017-9510-8. [DOI] [Google Scholar]
- 35.Kammerhofer N, et al. Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New phytol. 2015;207:778–789. doi: 10.1111/nph.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattarai KK, et al. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol. Plant-Microbe In. 2008;21:1205–1214. doi: 10.1094/MPMI-21-9-1205. [DOI] [PubMed] [Google Scholar]
- 37.Luo P, et al. Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016;245:35–49. doi: 10.1016/j.plantsci.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Lin B, et al. A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 2016;209:1159–1173. doi: 10.1111/nph.13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annual review of plant biology. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- 40.Dubreuil G, Magliano M, Deleury E, Abad P, Rosso MN. Transcriptome analysis of root-knot nematode functions induced in the early stages of parasitism. New phytol. 2007;176:426–436. doi: 10.1111/j.1469-8137.2007.02181.x. [DOI] [PubMed] [Google Scholar]
- 41.Mota M, Espada M, Jones JT. Characterization of glutathione S-transferases from the pine wood nematode. Bursaphelenchus xylophilus. Nematology. 2016;18:697–709. doi: 10.1163/15685411-00002985. [DOI] [Google Scholar]
- 42.Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF. Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines) Planta. 2007;226:1389–1409. doi: 10.1007/s00425-007-0578-z. [DOI] [PubMed] [Google Scholar]
- 43.Poppenberger B, et al. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. of Biol.Chem. 2003;278:47905–47914. doi: 10.1074/jbc.M307552200. [DOI] [PubMed] [Google Scholar]
- 44.Schweiger W, et al. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol. Plant-Microbe In. 2013;26:781–792. doi: 10.1094/MPMI-08-12-0205-R. [DOI] [PubMed] [Google Scholar]
- 45.Swiecicka M, et al. The suppression of tomato defence response genes upon potato cyst nematode infection indicates a key regulatory role of miRNAs. Plant Physiol. and Bioch. 2017;113:51–55. doi: 10.1016/j.plaphy.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 46.Melillo MT, Leonetti P, Leone A, Veronico P, Bleve-Zacheo T. ROS and NO production in compatible and incompatible tomato-Meloidogyne incognita interactions. Eur. J. Plant Pathol. 2011;130:489–502. doi: 10.1007/s10658-011-9768-4. [DOI] [Google Scholar]
- 47.Qiao F, et al. Characterization of three novel fatty acid- and retinoid-binding protein genes (Ha-far-1, Ha-far-2 and Hf-far-1) from the cereal cyst nematodes Heterodera avenae and H. filipjevi. PLoS ONE. 2016;11:e0160003. doi: 10.1371/journal.pone.0160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu D-L, et al. De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genomics. 2012;13:33. doi: 10.1186/1471-2164-13-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 51.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of differentially expressed transcripts were submitted to GenBank and GenBank accession numbers are provided in Tables 2, 3 and 4.