Abstract
Klebsiella pneumoniae is a gram-negative, opportunistic pathogen, and a common cause of healthcare-associated infections such as pneumonia, septicemia, and urinary tract infection. The purpose of this study was to survey the occurrence of and characterize K. pneumoniae in different environmental sources in a rural area of Shandong province, China. Two hundred and thirty-one samples from different environmental sources in 12 villages were screened for extended-spectrum β-lactamase-(ESBL)-producing K. pneumoniae, and 14 (6%) samples were positive. All isolates were multidrug-resistant and a few of them belonged to clinically relevant strains which are known to cause hospital outbreaks worldwide. Serotypes, virulence genes, serum survival, and phagocytosis survival were analyzed and the results showed the presence of virulence factors associated with highly virulent clones and a high degree of phagocytosis survivability, indicating the potential virulence of these isolates. These results emphasize the need for further studies designed to elucidate the role of the environment in transmission and dissemination of ESBL-producing K. pneumoniae and the potential risk posed to human and environmental health.
Keywords: Klebsiella pneumoniae, extended-spectrum β-lactamase, environment, feces, water, multilocus sequence typing, pulsed-field gel electrophoresis, whole-genome sequencing
Introduction
The increasing prevalence of antibiotic resistance among bacteria worldwide has become a major issue affecting the global public health (Grundmann, 2014). Extended-spectrum β-lactamases (ESBLs) are a group of enzymes that can hydrolyze penicillins, cephalosporins, and aztreonam, and is one of the main resistance determinants causing resistance to extended-spectrum β-lactam antibiotics in gram-negative bacteria. The main types of ESBLs are SHV, TEM, and CTX-M, the latter of which has become the main epidemic genotype worldwide.
Klebsiella pneumoniae is a gram-negative, opportunistic pathogen commonly associated with health-care associated infections (HAIs) such as pneumonia, septicemia, and urinary tract infections (Lee et al., 2012). K. pneumoniae has a high capacity to acquire mobile genetic elements such as plasmids containing antibiotic resistance genes (Burmolle et al., 2012). Because of the selection pressure exerted by a widespread use of antibiotics, strains of K. pneumoniae endemic at hospitals are often multidrug-resistant (Magiorakos et al., 2012), and treatment of HAIs caused by these strains are often difficult (Hou et al., 2015; Huang et al., 2015). Particularly problematic are strains of K. pneumoniae which are resistant to the last-resort antibiotics carbapenems (Little et al., 2014; Tijet et al., 2014; Cubero et al., 2016). For these reasons, monitoring of K. pneumoniae in the clinics is essential in the management of HAIs.
In recent years, the environment has been highlighted as an important factor in the dissemination of antibiotic-resistant bacteria and antibiotic resistance genes (Machado and Bordalo, 2014). An important factor in facilitating the dissemination of antibiotic resistance is likely selection pressures exerted by antibiotic residues in waste from anthropogenic wastewater and discharge from animal production facilities. The impact and effect of environmental contamination of antibiotic-resistant bacteria on human and environmental health has not yet been fully elucidated. A first step to determining this is to survey, monitor, and characterize the clinically important bacteria with clinically relevant antibiotic resistance determinants occurring in the environment, however, data on the occurrence and characteristics of ESBL-producing K. pneumoniae in environmental sources is scarce. Additionally, the few studies performed usually focus on prevalence in a single environmental matrix such as a body of water or animal manure (Mollenkopf et al., 2013; Zhang et al., 2015), and whole-genome characterization of the isolates are lacking (Zadoks et al., 2011; Ben Said et al., 2015).
In the current study, we screened 231 environmental samples for ESBL-producing K. pneumoniae and performed whole-genome sequencing on the 14 positive isolates detected, in order to survey the occurrence of and characteristics of this bacterial pathogen in different environmental sources in a rural area of Shandong province, China.
Materials and Methods
Sample Collection
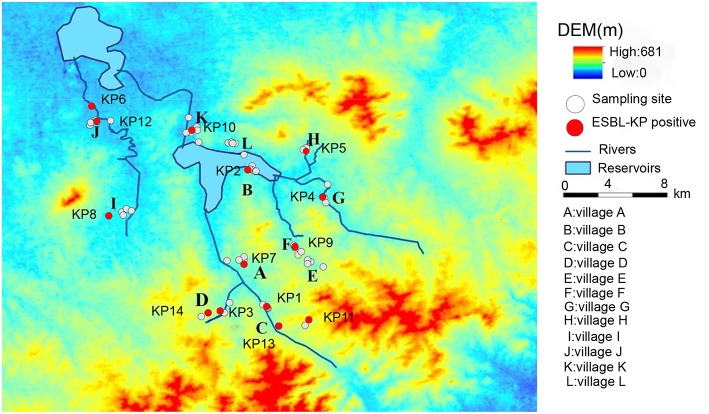
Samples were collected in a rural area of Shandong Province, China, in July 2015 (Figure 1). Details of the sampling procedure have been described elsewhere (Sun et al., 2018). Pig manure, wild bird feces, vegetables (cucumber, beans, and chives), and environmental samples were collected with ESwab tubes (Copan, Brescia, Italy) from the study villages. Sample types included: pig manure (n = 30), wastewater (n = 42), soil (n = 23), vegetables (n = 23), outlet sediment (n = 14), drinking water (n = 44), river water (n = 25), river sediment (n = 24), and feces from wild birds (n = 6). All samples were put in cool-boxes with ice-packs (4–8°C) upon collection and were transported for ~6 h to Shandong Center for Disease Control and Prevention (Jinan). Upon arrival, all samples were put in long-term storage at −80°C until cultivation.
Figure 1.
Map showing village locations (A–L) and the surrounding area in a region of rural Shandong, China, where 231 environmental samples were collected for screening for ESBL-producing K. pneumoniae. White spots denote locations where samples were negative for ESBL-producing K. pneumoniae and red spots denote locations were samples were positive for ESBL-producing K. pneumoniae.
Cultivation of Samples and Screening for ESBL-Producing K. pneumoniae
Prior to cultivation, the samples were collected from the low temperature freezers and allowed to reach room temperature. All samples were screened for ESBL-producing K. pneumoniae by cultivation on ChromID ESBL agar (bioMérieux, Marcy l'Etoile, France). For water, soil, river sediment, and vegetable samples, a pre-enrichment step in 2–3 ml Brain Heart Infusion Broth (BD, Sparks, USA) was performed overnight before being applied on the screening plates. Each sample was cultivated unselectively on a sheep blood agar plate (Babio, Jinan, China) to verify the viability of bacteria in the sample. All samples were applied on agar plates by a primary streak using the brush in the ESwab and then sub-streaked using a sterile 1 μl plastic loop. The agar plates were incubated for 18–24 h at 37°C. After incubation, growth was recorded and suspected K. pneumoniae colonies were identified based on color and morphology, according to the manufacturer's instructions (bioMérieux), sub-cultured on CHROMagar Orientation agar (CHROMagar Company, Paris, France), and incubated overnight at 37°C. From the CHROMagar Orientation agar, suspected K. pneumoniae were transferred to storage tubes with glycerol and stored in −80°C until further analyses. After applying samples on the respective agar plates, the remaining fluid in the ESwab tubes were transferred to 2 ml tubes and mixed with 30% glycerol for long-term storage at −80°C.
Verification of ESBL-Producing K. pneumoniae
Species confirmation of presumptive K. pneumoniae isolates were performed with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) by using a Shimadzu with saramis premium (Shimadzu Corporation, Kyoto, Japan). ESBL-production was verified by using the double disc diffusion method using cefotaxime, ceftazidime, each alone, and in combination with clavulanic acid according to the manufacturer's protocol (Oxoid, Basingstoke, UK). Klebsiella pneumoniae ATCC700603 was used as control strain.
Antibiotic Susceptibility Testing of ESBL-Producing K. pneumoniae
The confirmed ESBL-producing isolates were cultured on blood agar overnight at 37°C and were subsequently tested for susceptibility to 17 different antibiotics listed in Table 1. Antibiotic susceptibility was determined by evaluating minimal inhibitory concentrations (MICs) obtained with the agar dilution method by using the protocol recommended by the Clinical and Laboratory Standards institute (CLSI). Susceptibility was determined in accordance with the European Committee on Antimicrobial Susceptibility Testing, breakpoint tables for interpretation of MICs and zone diameters version 8.1, 2018 (http://www.eucast.org) for all antibiotics except tetracycline and nitrofurantoin, for which CLSI breakpoints version 2017 were used. Since clinical breakpoints of florfenicol are not available for Enterobacteriaceae at EUCAST or CLSI, a resistance breakpoint of >16 mg/L, based on the epidemiological cut-off values for the closely related Escherichia coli and Salmonella spp. available at EUCAST, was used. Isolates were considered to be multidrug-resistant if resistant to antibiotics from more than two different classes of antibiotics (Magiorakos et al., 2012).
Table 1.
Antibiotic susceptibility profiles of ESBL-producing K. pneumoniae of isolates from river water (RW), rivers sediment (RS), pig manure (PM), soil (S), and vegetables (V).
| Isolate | Source | MIC (mg/L)a | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CL | AMC | TZP | CTX | CAZ | MEM | ETP | IPM | GEN | AMK | TE | TGC | CIP | SXT | FOF | F | FFC | ||
| KP1 | RW | 0.125 | 16 | 32 | 2 | 2 | 0.015 | 0.015 | 0.25 | 32 | 4 | 128 | 2 | 16 | >16 | 256 | 256 | >32 |
| KP2 | RW | 0.06 | 16 | 256 | 8 | 0.5 | 0.03 | 0.06 | 0.125 | 32 | 8 | 128 | 2 | 16 | >16 | 256 | 512 | >32 |
| KP3 | RW | 0.125 | 16 | 128 | 8 | 16 | 0.015 | 0.015 | 0.06 | 32 | 2 | 128 | 2 | 0.5 | >16 | 128 | 256 | 4 |
| KP4 | RW | 0.125 | 32 | 256 | 32 | 1 | 0.015 | 0.015 | 0.125 | 128 | 2 | 64 | 2 | 8 | 8 | >256 | 256 | >32 |
| KP5 | RW | 0.125 | 32 | 256 | >32 | 1 | 0.03 | 0.015 | 0.125 | 1 | 2 | 128 | 2 | 1 | 8 | >256 | 256 | 8 |
| KP6 | RW | 0.125 | 32 | 256 | >32 | 1 | 0.015 | 0.03 | 0.125 | 64 | 4 | 128 | 4 | 16 | 8 | 256 | 256 | >32 |
| KP7 | PM | 0125 | 16 | 256 | 8 | 0.5 | 0.015 | 0.06 | 0.125 | 32 | 4 | 128 | 2 | 16 | >16 | 256 | 256 | >32 |
| KP8 | PM | 0.125 | 16 | 4 | 8 | 1 | 0.015 | 0.015 | 0.125 | 16 | 2 | 128 | 1 | 4 | 8 | 256 | 128 | 32 |
| KP9 | S | 0.125 | 32 | 256 | 4 | 1 | 0.015 | 0.03 | 0.125 | 64 | 4 | 128 | 2 | 8 | 8 | 512 | 256 | >32 |
| KP10 | S | 0.06 | 32 | 256 | >32 | 1 | 0.015 | 0.05 | 0.125 | 64 | 2 | 128 | 4 | 0.5 | 8 | >256 | 64 | 2 |
| KP11 | V | 0.125 | 16 | 256 | 8 | 1 | 0.015 | 0.015 | 0.125 | 16 | 2 | 128 | 1 | 1 | >16 | 256 | 64 | >32 |
| KP12 | V | 0.06 | 32 | 256 | >32 | 1 | 0.03 | 0.03 | 0.125 | >128 | 8 | 128 | 2 | 4 | 8 | >256 | 256 | >32 |
| KP13 | RS | 0.125 | 16 | 256 | 8 | 0.5 | 0.015 | 0.06 | 0.125 | 32 | 4 | 128 | 0.5 | 16 | >16 | 256 | 128 | >32 |
| KP14 | WW | 0.125 | 16 | 256 | 8 | 1 | 0.015 | 0.03 | 0.06 | 32 | 4 | 128 | 1 | 16 | >16 | 256 | 512 | >32 |
Resistance is indicated in bold and intermediate is indicated in underlined.
MICs were determined by agar dilution methods for all antibiotics except for colistin, for which broth microdilution was used. Resistance is indicated in bold. CL, colistin; AMC, amoxicillin/clavulanate; TZP, piperacillin/tazobactam; CTX, cefotaxime; CAZ, ceftazidime; MEM, meropenem; GEN, gentamicin; AMK, amikacin; TE, tetracycline; TGC, tigecycline, CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; FOF, fosfomycin; F, nitrofurantoin; FFC, florfenicol. Antimicrobial susceptibility profiles were determined using clinical breakpoints recommended by the EUCAST criteria (http://www.eucast.org/clinical_breakpoints/), except for tetracycline and nitrofurantion, for which the Clinical and Laboratory Standards Institute (CLSI, 2017) breakpoints were used, and florfenicol, for which a resistance breakpoint of >16 mg/L, based on the epidemiological cut-off values for the closely related Escherichia coli and Salmonella spp. available at EUCAST, was used.
Determination of Clonal Relatedness With Pulsed-Field Gel Electrophoresis
The clonal relatedness of the ESBL-positive K. pneumoniae isolates was determined via pulsed-field gel electrophoresis (PFGE) as previously described (Chen et al., 2017). Briefly, DNA plugs were digested using XbaI restriction enzyme (Takara Bio Inc., Kyoto, Japan) for 2 h. PFGE was undertaken on a CHEF-DR III (Bio-Rad, Hercules, CA, USA) using the following parameters: running time 18 h, temperature 14°C, field strength 6 V/cm2, angles 120°, initial pulse time 2.2 s, final pulse time 63.8 s. Two isolates were considered related to each other if the homology of the digested patterns was more than or equal to 80%.
Serum Bactericidal and Phagocytosis Assays
Serum bactericidal and phagocytosis assays were modified from previously described protocols (O'Shaughnessy et al., 2012; Kobayashi et al., 2016). In short, blood was venesected from 10 healthy persons for the serum bactericidal assay and from three healthy persons for the phagocytosis assay. The participants had provided written informed consent prior to participation in the study. Blood was left to clot at 4°C for 8 h, and subsequently centrifuged at 4°C. Inactivated serum was acquired by incubation in a water bath at 56°C for 30 min. Serum bactericidal assays were performed by adding 20 μL of bacterial suspension (108 cfu/mL) to 180 μL of normal or inactivated serum, and incubating the mixture at 37°C for 1 h after which the mixture was quickly placed on ice. From the reaction mixture, 50 μL were diluted 100-fold in PBS and viable count was performed by plating 100 μL on Mueller-Hinton agar and inoculating for 18–24 h. Serum bactericidal assays for each isolate were performed in duplicates at three separate times, and the survival rates were calculated as the ratios between the viable counts in the normal and inactivated serum, based on the means of the replicates.
For the phagocytosis assays; neutrophils were purified from the freshly drawn blood, and enumerated in a Neubauer chamber. Neutrophils were cultured in 24-well plates for 30 min. The phagocytosis assay was performed by adding either 106 neutrophils or an equal volume of autoclaved water (for the control experiments) to a mixture of 200 μL of bacterial suspension (108 cfu/mL), 100 μL of inactivated serum, and 600 μL phosphate-buffered saline, with subsequent incubation of the mixture at 37°C for 1 h, followed by termination of the reaction by adding 100 μL 0.1% Triton X-100 (Shanghai Aladdin Biochemical Technology, Shanghai, China) and placing on ice for 15 min. Bacteria were enumerated by viable count performed by plating 100 μL of the reaction mixture, diluted 1000-fold dilution in PBS, on Mueller-Hinton agar plates, and inoculating for 18–24 h. Phagocytosis assays for each isolate were performed in duplicates at two separate times, and the survival rates were calculated as the ratios between the viable counts in the phagocytosis assay and the controls, based on the means of the replicates.
Whole-Genome Sequencing of ESBL-Producing K. pneumoniae
DNA of all ESBL-positive isolates was extracted from pure cultures of K. pneumoniae using a Gentra Puregene Yeast/Bact. Kit (QIAGEN, Hilden, Germany). Whole-genome sequencing was performed on the extracted DNA by Novogene (Beijing, China) using the Illumina HiSeq sequencing platform. Raw sequences were assembled by using SPAdes 3.11 (Anton et al., 2012) (Table S1) and annotated via RAST. All draft genomes were deposited at NCBI (accession number: RCGE00000000-RCGR00000000). Occurrence of antibiotic resistance genes and multilocus sequence typing (MLST) was performed by querying the databases at the Center for Genomic Epidemiology (www.genomicepidemiology.org). Lipopolysaccharide-based serotyping (O-type) and capsule polysaccharide-based serotyping (K-type) were performed by using Kaptive Web (Wick et al., 2018). Presence of virulence genes were determined by using the Institut Pasteur MLST and whole genome MLST databases (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). In addition, the operons encoding type 1 fimbriae (fimABCDEFGHIK) and type 3 fimbriae (mrkABCDFHIJ) were evaluated, as were the gene sequences for the outer membrane porins (OMPs) OmpK35 and OmpK36. ompK36 allele group was determined based on genetic polymorphisms between bp 500 and 1,000, as previously described (Du et al., 2018). The genetic environment surrounding the CTX-M-type ESBL-genes were annotated and investigated using Easyfig 2.2.3. All genomes in the study dataset were annotated using the Prokka prokaryotic (Seemann, 2014) annotation pipeline, and using Roary: the Pan Genome Pipeline (Page et al., 2015) to obtain a system of linked qualified single nucleotide polymorphisms (SNPs) identified by mapping raw reads to core genes development.
Statistical Analysis
Mann-Whitney tests were used to determine differences in MICs for cefoxitin and ceftazidime between isolates carrying genes encoding CTX-M ESBLs of different groups. Viable counts from the serum bactericidal and phagocytosis assays were compared with their respective controls and differences were determined using repeated measures ANOVA with Bonferroni multiple comparison tests performed post-hoc. All statistical tests were carried out using Prism 5 for Windows.
Results
Screening for ESBL-Producing K. pneumoniae
A total of 14 (6%) isolates of ESBL-producing K. pneumoniae were isolated from 231 environmental samples. The positive rate of ESBL-producing K. pneumoniae isolates in different environmental matrices were: river water 24% (6/25), soil 9% (2/23), vegetables 9% (2/23), pig manure 7% (2/30), river sediment 4% (1/24), and wastewater 2% (1/42). No isolates were detected in the sewage outlet sediment, drinking water, or wild bird fecal samples.
Antibiotic Susceptibility Testing
The antibiotic susceptibility testing (Table 1) showed that all isolates were susceptible to amikacin, colistin and the carbapenems meropenem, imipenem, and ertapenem. Susceptibility to ceftazidime was high (86%). None of the isolates were susceptible to cefotaxime, amoxicillin-clavulanic acid, ciprofloxacin, trimethoprim-sulfamethoxazole, tetracycline, nitrofurantoin or fosfomycin. Susceptibility rates to gentamicin (7%), piperacillin-tazobactam (7%), and florfenicol (21%) were low. The isolates showed a low (29%) susceptibility rate to tigecycline. All isolates were multidrug-resistant (i.e., resistant to antibiotics of more than two classes of antibiotics).
Antibiotic Resistance Genes
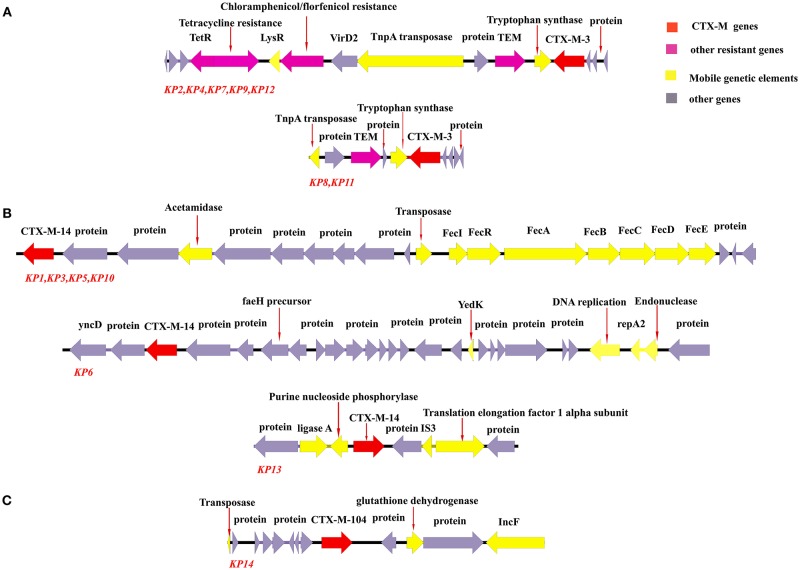
Data on antibiotic resistance genes are presented in Table 2. All isolates carried CTX-M-type ESBL-genes. Seven isolates (50%) carried blaCTX−M−3, six isolates (43%) carried blaCTX−M−14, and only one isolate carried blaCTX−M−104. Additionally, the ESBL-gene blaSHV−27 was detected in two isolates. The genetic environment surrounding the CTX-M genes are shown in Figure 2. Two distinct genetic environments and three distinct genetic environments were determined among the isolates carrying blaCTX−M−3 and blaCTX−M−14, respectively.
Table 2.
Antibiotic resistance genes and STs of ESBL-producing K. pneumoniae isolates from environmental samples.
| Isolate | Source | Antibiotic resistance genes | MLST |
|---|---|---|---|
| KP1 | RW | ARR-3, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, qnrS1, oqxA, oqxB, qnrB52, fosA, mph(A), sul1, sul2, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaTEM−1B, blaSHV−27, blaCTX−M−14 | ST967 |
| KP2 | RW | ARR-3, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, qnrS1, oqxA, oqxB, qnrB49, fosA, mph(A), sul1, sul2, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaCTX−M−3, blaSHV−28, blaTEM−1B | ST15 |
| KP3 | RW | fosA, qnrS1, oqxA, oqxB, aac(3)-IId, strA, strB, sul1, sul2, dfrA1, tet(A), blaTEM−1B, blaCTX−M−14, blaSHV−1 | ST101 |
| KP4 | RW | ARR-3, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, qnrS1, oqxA, oqxB, fosA, mph(A), sul2, sul1, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaCTX−M−3, blaSHV−1, blaTEM−1B | ST3003 |
| KP5 | RW | fosA, qnrS1, oqxA, oqxB, sul1, dfrA1, tet(A), blaSHV−11, blaCTX−M−14 | ST659 |
| KP6 | RW | ARR-3, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, sul1, sul2, fosA, mph(A), qnrS1, oqxB, oqxA, qnrB49, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaSHV−1, blaCTX−M−14 | ST314 |
| KP7 | PM | ARR-3, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, oqxA, oqxB, qnrS1, qnrB49, fosA, mph(A), sul1, sul2, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaCTX−M−3, blaSHV−26, blaTEM−1B | ST1967 |
| KP8 | PM | ARR-3, fosA, qnrS1, qnrB49, oqxA, oqxB, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, mph(A), sul1, sul2, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaTEM−1B, blaSHV−11, blaCTX−M−3 | New |
| KP9 | S | ARR-3, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, qnrS1, oqxA, oqxB, fosA, mph(A), sul2, sul1, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaCTX−M−3, blaSHV−1, blaTEM−1B | ST999 |
| KP10 | S | fosA, sul1, aac(3)-IId, oqxA, oqxB, qnrS1, dfrA1, tet(A), blaCTX−M−14, blaSHV−82 | ST1738 |
| KP11 | V | ARR-3, fosA, oqxA, oqxB, qnrB49, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, mph(A), sul1, sul2, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaTEM−1B, blaSHV−11, blaCTX−M−3 | New |
| KP12 | V | ARR-3, fosA, oqxA, oqxB, strA, strB, aph(3′)-Ia, aadA16, aac(3)-IId, mph(A), sul2, sul1, dfrA27, tet(A), floR, aac(6′)Ib-cr, blaCTX−M−3, blaSHV−27, blaTEM−1B | ST661 |
| KP13 | RS | ARR-3, fosA, oqxA, oqxB, aph(3′)-Ia, aadA16, mph(A), sul1, dfrA27, tet(D), floR, catA2, aac(6′)Ib-cr, blaSHV−11, blaCTX−M−14 | ST11 |
| KP14 | WW | ARR-3, strB, aadA16, aadA2, aac(3)-IId, aph(3′)-Ia, qnrB6, oqxA, oqxB, fosA, erm(B), sul1, sul2, dfrA27, tet(A), tet(D), floR, aac(6′)Ib-cr, cfr, blaSHV−11, blaTEM−1A, blaCTX−M−104 | ST258 |
The samples were collected from river water (RW), pig manure (PM), soil (S), vegetable (V), river sediment (RS), and wastewater (WW).
Figure 2.
Genetic environment of CTX-M genes among K. pneumoniae isolated from environmental sources. Arrows represent direction of transcription. Red open reading frames (ORFs) denote the CTX-M genes, purple ORFs denote other antibiotic resistance genes, yellow ORFs denote genes related to mobile genetic elements, and gray ORFs denote other genes or genes of unknown function. (A) The genetic environments of blaCTX−M−3 were similar in isolates KP2, KP4, KP7, KP9, and KP12. The two isolates KP8 and KP11 also shared a similar genetic environment surrounding blaCTX−M−3. (B) Isolates KP1, KP3, KP5, and KP10 all carried blaCTX−M−14 and shared the same genetic environment surrounding the gene, whereas isolates KP6 and KP13 had unique genetic environments surrounding blaCTX−M−14. (C) The genetic environment of blaCTX−M−104 in the one isolate in the study which carried the gene.
All isolates carried fosA, oqx- sul-, dfr-, and tet-genes, encoding fosfomycin-, quinolone-, sulphonamide-, trimethoprim-, and tetracycline resistance, respectively. Aminoglycosides resistance genes were also common (93% of isolates), as were the florfenicol resistance gene floR (86% of isolates).
Serotyping and Virulence-Associated Genes
Serotyping was performed based on both lipopolysaccharide (O-serotype) and capsular polysaccharide (K-serotype) (Table 3). The O-serotypes were distributed as follows; O2 (n = 5), O1 (n = 4), O3 (n = 2), OL101 (n = 1), and OL103 (n = 1). One isolate (KP4) had poor match confidence against any known O-type (<90% coverage and two truncated/missing genes) and was determined to be non-typeable. The K-serotype-associated K loci among the isolates were diverse; two isolates had KL34, whereas KL3, KL18, KL23, KL28, KL31, KL38, KL49, KL57, KL106, KL107, and KL127 were found in one isolate each. One isolate (KP4) had poor match confidence against any known K-serotype and was determined to be non-typeable.
Table 3.
Serotypes, virulence characteristics, and virulence-associated genes of ESBL-producing K. pneumoniae from environmental samples.
| KP1 | KP2 | KP3 | KP4 | KP5 | KP6 | KP7 | KP8 | KP9 | KP10 | KP11 | KP12 | KP13 | KP14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLST | ST967 | ST15 | ST101 | ST3003 | ST659 | ST314 | ST1967 | New | ST999 | ST1738 | New | ST661 | ST11 | ST258 |
| O serotype | O1 | OL103 | O1 | N.D. | O2 | O2 | O2 | O1 | OL101 | O3 | O1 | O2 | O3 | O2 |
| K locus | KL18 | KL49 | KL106 | N.D. | KL3 | KL23 | KL57 | KL34 | KL127 | KL38 | KL34 | KL28 | KL31 | KL107 |
| Serum survival | 74% | 2% | 82% | 34% | 51% | 30% | 53% | 111% | 47% | 1% | 122% | 68% | 67% | 39% |
| P value | <0.05 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.05 | <0.01 | ||||
| Phagocytosis survival | 74% | 104% | 94% | 75% | 75% | 78% | 81% | 104% | 67% | 74% | 55% | 97% | 108% | 75% |
| P value | <0.01 | |||||||||||||
| OmpK35 | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | Q68Stop | WT |
| ompK36 allele group | C | D | C | A | D | D | N.D. | A | D | D | A | A | A | A |
| Type 1 fimbriae | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Type 3 fimbriae | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| kfu | – | + | + | – | – | – | – | + | – | – | + | – | – | – |
| kvg | – | – | – | – | – | – | – | + | – | – | + | – | – | – |
| Yersiniabactin | – | – | + | – | – | – | – | – | – | + | – | – | – | – |
P-values are indicated for tests comparing viable counts for serum bactericidal and phagocytosis assays to their respective controls. P > 0.05 are omitted for clarity. WT, wild type; N.D., non-determinable.
Querying of virulence-associated genes and serotyping was performed in silico by using the whole-genome sequencing data (Table 3). All isolates carried the fim and mrk operons which encode type 1 and type 3 fimbriae, respectively. The kfu and the kvg operons were detected in four isolates and two isolates, respectively. Additionally, two isolates carried the operon encoding the yersiniabactin siderophore (ybtSXQPA-irp2-irp1-ybtUTE-fyuA) (Table 3). The genes encoding the OMPs OmpK35 and OmpK36 were also evaluated in all isolates. None of the isolates carried an ompK35 with any mutations clearly affecting the protein function except isolate K13, which carried an ompK35 with a nucleotide substitution (c202t) engendering a premature stop codon (Q68Stop) causing a 81% truncation of the protein (shortening it from 359 to 67 aa). Based on ompK36 polymorphisms, the isolates could be divided in three different allele groups; group A (n = 7), group D (n = 5), and group C (n = 2).
Serum Bactericidal and Phagocytosis Assays
The survivability of the isolates in serum bactericidal and phagocytosis assays were evaluated as the differences in viable counts between the assay and control experiments (Table 3). For the serum bactericidal assay, 10 isolates (71%) had significantly lower survivability compared to the controls (P < 0.05); eight of these isolates had survival rates ranging from 30 to 74%. Two isolates (KP2 and KP10) had survival rates of 1 and 2%, respectively, which was significantly lower compared to the other isolates (P < 0.01). Four isolates (KP3, KP8, KP11, and KP12) did not differ significantly from the controls and had survival rates ranging from 68 to 122%. In the phagocytosis assay, only one isolate (KP11) had a significantly lower survival rate (55%) compared to the control (P < 0.01); the remaining isolates showed survival rates ranging from 67 to 108%.
Phylogenetic Analysis
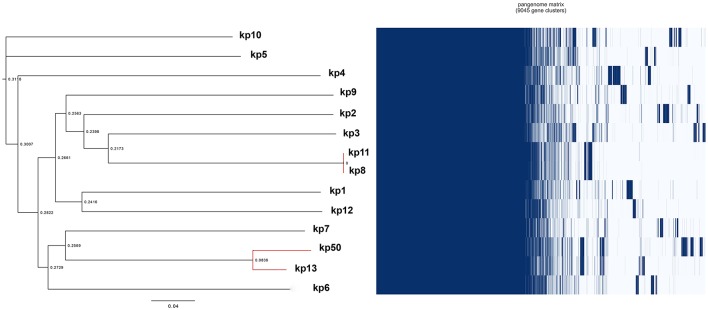
A total of 13 different STs were found among the 14 ESBL-producing K. pneumoniae isolates in this study (Table 2), including ST11, ST15, ST101, ST258, ST659, ST661, ST967, ST999, ST11967, ST3003, and ST1738. Two isolates shared 100% nucleotide sequences (KP8, isolated from vegetables, and KP11, isolated from pig manure) in the MLST genes, but were of a previously undescribed type. These two isolates also presented high similarity in the PFGE-analysis (Figure S1). In addition, the results of the PFGE indicated a close genetic relationship between three other isolates, KP9 (from soil), KP13 (from river sediment), and KP14 (from wastewater). A phylogenetic tree based on SNPs was computed from 9,045 gene clusters obtained from the 14 isolates (Figure 3). The analysis revealed a close genetic relationship between isolates KP8 and KP11, among which no SNP-differences could be discerned. Isolates KP13 and KP14 also showed high genetic similarity.
Figure 3.
Phylogeny of ESBL-producing K. pneumoniae isolated from environmental sources based on differences in SNPs. Red branches indicate isolates which were found related in the PFGE-analysis. The value represents the proportion of the scale of SNP differences.
Discussion
In the current study, 231 environmental samples were collected from a rural farming area in Shandong province, China, and screened for ESBL-producing K. pneumoniae. The results showed that the positive rate among the samples was 6%. Among the different environmental sources sampled, the detection rate was highest in river water (24%), followed by soil (9%), vegetables (9%), pig manure (7%), river sediment (4%), and wastewater (2%). The prevalence in river water was higher than the prevalence in the wastewaters connected to pig discharge. This could reflect the comparatively lower prevalence in pig manure (7%). The river water on the other hand, was mainly influenced by anthropogenic discharge originating from the surrounding community and directly from inhabitants using the water. The higher prevalence in anthropogenically affected water compared to pig discharge wastewater may reflect that human rather than animal sources are more important for the environmental presence of ESBL-producing K. pneumoniae in the local area. In China, ESBL-producing K. pneumoniae has been reported in spring water adjacent to restaurants and hotels at a mountain resort close to Tai'an, Shandong province (Li et al., 2015) and in water from the Pearl River in Guangzhou (Ye et al., 2017). A fairly high prevalence rate of 36% was also reported in the Yangtze River in Chongqing (Chen et al., 2010). Overall, the findings in the different environmental sources in the study area indicate a need to clarify the role and importance of the environment in the potential for transmission and dissemination of ESBL-producing K. pneumoniae and subsequent risk for transmission.
All isolates were susceptible to colistin and carbapenems. The susceptible rate to ceftazidime was high (86%); only one isolate was resistant. Resistance to ceftazidime in K. pneumoniae has been shown to be engendered by loss of the OMP OmpK35 (Etsuko et al., 2016). The single resistant isolate in the current study (KP3) carried an ompK35 with a nonsense mutation which translated into an OmpK35 truncated by 81%, likely rendering the protein nonfunctional. For the antibiotics cefotaxime, amoxicillin-clavulanic acid, piperacillin-tazobactam, ciprofloxacin, trimethoprim-sulfamethoxazole, tetracycline, fosfomycin, gentamicin, and nitrofurantoin, susceptible rates were low, between 0 and 7%. The high MIC-values to these antibiotics correlated to a correspondingly high carriage rate of antibiotic resistance genes. All isolates carried genes conferring resistance to trimethoprim, sulphonamides, quinolones, tetracycline, fosfomycin, and aminoglycosides. The susceptible rate to florfenicol was low (21%), and all of the non-susceptible isolates carried the florfenicol resistance gene floR. Additionally, MICs were high for tigecycline, and only a few isolates were susceptible (29%). Notably, all isolates collected in this study were multidrug-resistant.
CTX-M-genes were carried by all isolates in this study. The most prevalent was blaCTX−M−3, a gene of the CTX-M-1-group, which was detected in seven isolates (50%), followed by blaCTX−M−14, a gene of the CTX-M-9-group, which was detected in five isolates (43%). MICs of the tested cephalosporins ceftazidime and cefoxitin were not significantly different for isolates with CTX-M-1 group enzymes as compared to isolates with CTX-M-9 group enzymes. CTX-M-genes are the most common genes encoding ESBLs worldwide, and are prevalent among Enterobacteriaceae (Naseer and Sundsfjord, 2011; Bevan et al., 2017). Regarding the epidemiology of CTX-M-genes, most studies performed have focused on E. coli. Although there are regional differences pertaining to the distribution of specific CTX-M-genes, blaCTX−M−14 and blaCTX−M−15 are predominant in clinical isolates worldwide, which also holds true in China (Shin et al., 2011; An et al., 2012; Xia et al., 2014; Bevan et al., 2017). For K. pneumoniae in China, the predominance of certain CTX-M-types differ depending on study. For instance, one study on isolates collected from seven hospital in Beijing reported a predominance of CTX-M-14 (An et al., 2012), whereas another study from a single hospital in Beijing reported CTX-M-10 to be the most common (Li et al., 2012). Another study in a hospital in Guangdong reported CTX-M-9 to be the most common CTX-M-type (Du et al., 2014) The most common gene in the current study, blaCTX−M−3, is rarely reported in China, although this is likely due to a larger volume of data being available on ESBLs among E. coli as compared to K. pneumoniae. For instance, a study on clinical isolates of K. pneumoniae from hospitals in Jilin, China, showed that 48% of CTM-X-producing isolates carried blaCTX−M−3 (Li et al., 2014) and another study on K. pneumoniae isolates collected at a hospital in Henan, China, showed that blaCTX−M−3 was carried by 38% of the CTX-M-producers, which was second in prevalence only by carriers of blaCTX−M−14 (40%) (Guo et al., 2017).
Additionally, the genetic environments surrounding the CTX-M-genes were analyzed (Figure 2). blaCTX−M−3 (n = 7) was located on two distinct genetic fragments among the isolates, whereas blaCTX−M−14 (n = 6) was located on three distinct genetic fragments. These results show that several mobile genetic elements are likely responsible for the dissemination of CTX-M-genes in the local environment. Notably, five isolates (KP2, KP4, KP7, KP9, and KP12) sharing the same genetic environment surrounding blaCTX−M−3, also carried the florfenicol resistance gene floR on the same genetic element, indicating the potential of co-selection of ESBL-producing K. pneumoniae via florfenicol exposure. The high variety of ESBL-genes and other antibiotic resistance genes among K. pneumoniae isolated from environmental sources indicate the potential for transfer of these genes to other bacteria present in the same sources. Reversely, the K. pneumoniae isolates may potentially acquire additional antibiotic resistance determinants from environmentally occurring bacteria.
Isolates of K. pneumoniae expressing ESBLs have been linked to higher virulence compared to K. pneumoniae isolates lacking ESBLs. In one study, isolates collected from hospital-acquired bacteremia showed that ESBL-producing K. pneumoniae were significantly more resistant to serum killing compared to isolates lacking ESBLs (Lin et al., 2016). Another study on isolates from clinical specimens from hospitalized patients found that ESBL-producing K. pneumoniae to a higher degree carried both type 1 and type 3 fimbriae and were more capable of invasion of ileocecal and bladder epithelial cells (Sahly et al., 2008). ESBL-production has also been strongly linked to human isolates of K. pneumoniae, and it has further been indicated that community-acquired K. pneumoniae tend to carry fewer acquired antibiotic resistance genes (Holt et al., 2015). In general, the difference between clinical and environmental isolates of K. pneumoniae is not as well-studied.
In the current study, virulence among the environmental isolates of K. pneumoniae was evaluated based on serum bactericidal and phagocytosis assays, serotypes, and virulence-associated genes. Ten isolates had significantly lower survivability in serum compared to the controls (P < 0.05), two of which (KP2 and KP10) had considerably lower survival rates (1 and 2%, respectively) compared to the other isolates (P < 0.01); indicating a difference in degree of survivability in serum for the isolates in this study. Four isolates, KP3, KP8, KP11, and KP12, did not significantly differ in survivability compared to the controls, and had survival rates ranging from 68 to 122%, indicating that these were potentially serum-resistant. For the phagocytosis assay, only one isolate (KP11) had significantly lower survivability compared to the control, implying a high degree of phagocytosis resistance among the isolates. Notably, three isolates (KP3, KP8, and KP12) had high survival rates in both the serum bactericidal and phagocytosis assay, indicating the potential virulence of these isolates.
Serotyping was performed in silico by using the whole-genome sequencing data (Table 3). O-serotypes O1, O2, and O3 are the serotypes most commonly isolated from human hosts, and O1 in particular is the most common serotype associated with disease in humans (Rainer et al., 2016) Interestingly, these serotypes predominated among the environmental isolates collected in this study; five isolates were O2, four isolates were O1 and two isolates were O3. Among K-serotypes, K1 and K2 are associated to hypervirulence in K. pneumoniae (Rainer et al., 2016), however, none of the isolates in this study carried the KL1 or KL2 loci associated to either of these K-serotypes.
Virulence gene analysis revealed that all isolates carried the fim and mrk operons which encodes type 1 and type 3 fimbriae, virulence factors ubiquitous in K. pneumoniae (Wu et al., 2012; Paczosa and Mecsas, 2016). Both fimbriae mediate adhesion and biofilm formation. Type 1 fimbriae are expressed in the urinary bladder and have been shown to contribute to urinary tract infections (Struve et al., 2009; Paczosa and Mecsas, 2016). Type 3 fimbriae mediate biofilm formation and are involved in adhesion to medical devices, and may be an important factor for K. pneumoniae in biofilm-associated infections as well as gaining entry into the host and persistence in the clinical environment rather than facilitating infections in the host (Huang et al., 2009; Struve et al., 2009).
The OMP OmpK36 appears to play a role in K. pneumoniae for both antibiotic resistance and virulence, as deficiencies in the protein has been shown to lead to elevated MICs for some antibiotics (likely due to decreased membrane permeability) and lowered virulence (Paczosa and Mecsas, 2016). Genetic polymorphisms in ompK36 can be used as a basis for classification into four different ompK36 alleles, each of which is associated with altered protein function. Interestingly, isolates carrying a group C allele have been associated with STs, K-serotypes, and virulence genes associated with hypervirulent K. pneumoniae lineages, and have additionally been shown to display higher virulence in a mouse lethality model (Yan et al., 2015; Du et al., 2018). In the current study, two isolates (KP1 and KP3) were found to carry group C alleles of ompK36.
Four isolates carried the kfu operon, which encodes components of an ABC transport system which mediates uptake of ferric iron. Kfu is associated with hypervirulent strains of K. pneumoniae, and its expression has been shown to be important for virulence in mouse models (Luo et al., 2014; Paczosa and Mecsas, 2016). In the current study, two isolates (KP3 and KP10) containing the yersiniabactin virulence operon were found, and where from river water and soil, respectively. Yersiniabactin is a siderophore which helps K. pneumoniae survive during respiratory tract infections (Bachman et al., 2011), and has been shown to be strongly associated with infection in humans (Holt et al., 2015). Yersiniabactin is also overrepresented among hypervirulent strains (Paczosa and Mecsas, 2016). One of these isolates (KP3) also carried the kfu operon.
The genetic relationship of the isolates was analyzed using MLST, PFGE, and SNPs, and the isolates were found to exhibit a high diversity. For the 14 isolates, 13 different STs were determined (Figure 2, Table 2). The two isolates (KP8, KP11) which shared ST (i.e., identical sequence in MLST genes) belonged to a previously undescribed ST, and showed a close relation in the PFGE analysis. This was confirmed by the SNP-analysis, in which the two isolates had identical SNPs (Figure 3). These isolates also shared identical genetic environments surrounding blaCTX−M−3 (Figure 2). Other STs detected in this study included clinically relevant STs such as ST11, ST15, ST101, and ST258. ST11 and ST258, both members of clonal group (CG) 258, are the most common carbapenem-resistant K. pneumoniae isolated from clinical samples worldwide (Lee et al., 2016). ST258 is commonly reported to cause hospital outbreaks in North America, Latin America, and Europe (Ruiz-Garbajosa et al., 2013; Geraci et al., 2015; Gomez et al., 2016; Lee et al., 2016). ST11 is more commonly reported from hospital outbreaks in Asian countries, including China (Qi et al., 2011; Lee et al., 2016). Recently, a clone of carbapenem-resistant ST11 K. pneumoniae having acquired a hypervirulence-plasmid was reported to cause an outbreak at a hospital in Hangzhou, China (Gu et al., 2018). ST15 and ST101 are two other globally occurring strains of K. pneumoniae associated with hospital outbreaks. For example, ESBL-producing ST15 has been reported in hospital outbreaks in the Netherlands (Zhou et al., 2016) and Hungary (Damjanova et al., 2008), and carbapenem-resistant ST15 has been reported in outbreaks in Bulgaria (Markovska et al., 2015), Portugal (Vubil et al., 2017), Vietnam (Tada et al., 2017), and Nepal (Chung The et al., 2015). For ST101, hospital outbreaks of carbapenem-resistant K. pneumoniae have been reported in Algeria (Loucif et al., 2016) and Greece (Avgoulea et al., 2018). A study in Turkey (Can et al., 2018) on colistin-resistant K. pneumoniae isolated from patients found that colistin-resistant ST101 were prevalently carbapenem-resistant, and occurrence of ST101 in the patient was found to be a significant independent predictor of patient mortality. The ST101 isolate in the current study (KP3) had several factors associated with virulence and high-risk clones; it belonged to the commonly disease-associated O-serotype O1, carried a group C ompK36 allele and carried the hypervirulence-associated kfu and yersiniabactin operons. Additionally, the isolate showed high survivability in the serum bactericidal and phagocytosis assays (82 and 94%, respectively) indicating that this is potentially a highly virulent clone.
In our study, the ST11, ST15, ST101, and ST258 isolates were detected in river sediment, river water, and wastewater. The presence of clinically relevant strains of K. pneumoniae in aquatic environments in association to human settlements indicates a potential for the environment as a transmission route of these strains. These isolates may have originated from human or animal guts colonized by the strains, and could have disseminated via feces and untreated wastewater. Although the data collected in this study is not sufficient to conclusively demonstrate transmission of K. pneumoniae strains between different environmental media, some interesting observations could nevertheless be made. For instance, KP8, isolated from pig manure, and KP11, isolated from vegetables, shared STs and were highly similar in the PFGE- and SNP-analysis, which may indicate the possibility of transmission from animals to humans via consumption of produce (Figure S1). The isolates were from the geographically separate villages C and I. Although of different STs, KP9, KP13, and KP14 also showed similar PFGE- and SNP-profiles, indicating that there may exist possible transmission routes between wastewater (KP14), river sediment (KP13), and soil (KP9). Bacteria in these environmental matrices could further potentially spread to humans, via for example surface and ground water, and irrigation of fields used for vegetable cultivation and finally consumption by humans. KP9, KP13, and KP14 were isolated from villages F, C, and D, respectively. Although villages C and D are located adjacent to each other, connected by a river, village F is separated from the aforementioned villages. Although the number of isolates in this study were too few to make a robust spatial analysis, the occurrence of genetically related isolates from different environmental matrices in geographically separate locations indicate the potential existence of a transmission mechanism.
Conclusion
There is a scarcity of studies investigating occurrence of K. pneumoniae in the environment and the role of environmental transmission routes for dissemination of the pathogen. We sampled different environmental sources in a rural region of Shandong province, China, and screened the samples for ESBL-producing K. pneumoniae, and 6% of the samples were determined to be positive. All of the isolates were multidrug-resistant and several belonged to clinically relevant strains which are known to cause hospital outbreaks worldwide. Serotypes, virulence genes, serum survival, and phagocytosis survival were analyzed and showed the presence of virulence factors associated with highly virulent clones and a high degree of phagocytosis survivability, indicating the potential virulence of these isolates. The occurrence of ESBL-producing K. pneumoniae in the environment indicates inadequate treatment of domestic sewage and waste in the rural area. In addition, these results emphasize the need for further studies designed to elucidate the role of the environment in transmission and dissemination of ESBL-producing K. pneumoniae and the potential risk posed to human and environmental health.
Ethics Statement
Ethical permission was granted by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University, reference number 2018#1031.
Author Contributions
XL, LN, CSL, JO, SB, BZ and BB conceived of and designed the study. XC, HZ, XJ and XL performed the sampling. XC and XJ performed the experiments. XC, BB and HZ analyzed the data. XC completed the first draft of the manuscript. BB, BZ, SB, JO, CSL, XL and LN critically revised the manuscript. All co-authors approved of the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the participants, co-ordinators, and administrators for their support during the study. Thanks to the IMPACT Consortium for its efforts in this project.
Footnotes
Funding. This project was supported by the National Natural Science Foundation of China (41771499, 81361138021), the Fundamental Research Funds of Shandong University (2018JC102), the Swedish Research Council (D0879801) and the Swedish Research Council Formas (2016-00640).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00211/full#supplementary-material
References
- An S., Chen J., Wang Z., Wang X., Yan X., Li J., et al. (2012). Predominant characteristics of CTX-M-producing Klebsiella pneumoniae isolates from patients with lower respiratory tract infection in multiple medical centers in China. FEMS Microbiol. Lett. 332, 137–145. 10.1111/j.1574-6968.2012.02586.x [DOI] [PubMed] [Google Scholar]
- Anton B., Sergey N., Dmitry A., Alexey A., Gurevich A., Dvorkin M., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgoulea K., Di Pilato V., Zarkotou O., Sennati S., Politi L., Cannatelli A., et al. (2018). Characterization of extensively- or pandrug-resistant ST147 and ST101 OXA-48-producing Klebsiella pneumoniae isolates causing bloodstream infections in ICU patients. Antimicrob Agents Chemother. 62, AAC.02457–17. 10.1128/AAC.02457-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman M. A., Oyler J. E., Burns S. H., Caza M., Lepine F., Dozois C. M., et al. (2011). Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79, 3309–3316. 10.1128/IAI.05114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Said L., Jouini A., Klibi N., Dziri R., Alonso C. A., Boudabous A., et al. (2015). Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 203, 86–92. 10.1016/j.ijfoodmicro.2015.02.023 [DOI] [PubMed] [Google Scholar]
- Bevan E. R., Jones A. M., Hawkey P. M. (2017). Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 72, 2145–2155. 10.1093/jac/dkx146 [DOI] [PubMed] [Google Scholar]
- Burmolle M., Norman A., Sorensen S. J., Hansen L. H. (2012). Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae. PLoS ONE 7:e41259. 10.1371/journal.pone.0041259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can F., Menekse S., Ispir P., Atac N., Albayrak O., Demir T., et al. (2018). Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 73, 1235–41. 10.1093/jac/dkx532 [DOI] [PubMed] [Google Scholar]
- Chen H., Shu W., Chang X., Chen J., Guo A. Y., Tan Y. (2010). The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ. Pollut. 158, 2459–2464. 10.1016/j.envpol.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Chen K., Chan E. W., Xie M., Ye L., Dong N., Chen S. (2017). Widespread distribution of mcr-1-bearing bacteria in the ecosystem, 2015 to 2016. Eurosurveillance 22:17-00206. 10.2807/1560-7917.ES.2017.22.39.17-00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung The H., Karkey A., Pham Thanh D., Boinett C. J., Cain A. K., Ellington M., et al. (2015). A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol. Med. 7, 227–239. 10.15252/emmm.201404767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2017). M100 Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. Wayne, PA: CLSI. [Google Scholar]
- Cubero M., Grau I., Tubau F., Pallares R., Dominguez M. A., Liñares J., et al. (2016). Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin. Microbiol. Infect. 22, 154–160. 10.1016/j.cmi.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Damjanova I., Toth A., Paszti J., Hajbel-Vekony G., Jakab M., Berta J., et al. (2008). Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new ‘MRSAs’? J. Antimicrob. Chemother. 62, 978–985. 10.1093/jac/dkn287 [DOI] [PubMed] [Google Scholar]
- Du F., Wei D. D., Wan L., Cao X. W., Zhang W., Liu Y. (2018). Evaluation of ompK36 allele groups on clinical characteristics and virulence features of Klebsiella pneumoniae from bacteremia. J. Microbiol. Immunol. Infect. [Epub ahead of print]. 10.1016/j.jmii.2018.08.018 [DOI] [PubMed] [Google Scholar]
- Du J., Li P., Liu H., Lu D., Liang H., Dou Y. (2014). Phenotypic and molecular characterization of multidrug resistant Klebsiella pneumoniae isolated from a university teaching hospital, China. PLoS ONE 9:e95181. 10.1371/journal.pone.0095181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etsuko S., Seiji K., Hiroshi N. (2016). Klebsiella pneumoniae major porins OmpK35 and OmpK36 allow more efficient diffusion of β-Lactams than their Escherichia coli homologs OmpF and OmpC. J. Bacteriol. 198, 3200–3208. 10.1128/JB.00590-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraci D. M., Bonura C., Giuffre M., Saporito L., Graziano G., Aleo A., et al. (2015). Is the monoclonal spread of the ST258, KPC-3-producing clone being replaced in southern Italy by the dissemination of multiple clones of carbapenem-nonsusceptible, KPC-3-producing Klebsiella pneumoniae? Clin. Microbiol. Infect. 21, e15–e17. 10.1016/j.cmi.2014.08.022 [DOI] [PubMed] [Google Scholar]
- Gomez S. A., Rapoport M., Piergrossi N., Faccone D., Pasteran F., De Belder D., et al. (2016). Performance of a PCR assay for the rapid identification of the Klebsiella pneumoniae ST258 epidemic clone in Latin American clinical isolates. Infect. Genet. Evol. 44, 145–146. 10.1016/j.meegid.2016.06.018 [DOI] [PubMed] [Google Scholar]
- Grundmann H. (2014). Towards a global antibiotic resistance surveillance system: a primer for a roadmap. Ups J. Med. Sci. 119, 87–95. 10.3109/03009734.2014.904458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- Guo X., Cao Z., Dai Z., Li Y., He X., Hu X., et al. (2017). Antimicrobial susceptibility and molecular epidemiology of multidrug-resistant Klebsiella pneumoniae in Central China. Jpn. J. Infect. Dis. 70, 229–234. 10.7883/yoken.JJID.2016.049 [DOI] [PubMed] [Google Scholar]
- Holt K. E., Wertheim H., Zadoks R. N., Baker S., Whitehouse C. A., Dance D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. 112, E3574–E3581. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X. H., Song X. Y., Ma X. B., Zhang S. Y., Zhang J. Q. (2015). Molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates. Braz. J. Microbiol. 46, 759–768. 10.1590/S1517-838246320140138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Y., Pan K. Y., Liu X. Q., Xie X. Y., Dai X. L., Chen B. J., et al. (2015). Analysis of the drug-resistant characteristics of Klebsiella pneumoniae isolated from the respiratory tract and CTX-M ESBL genes. Genet. Mol. Res. 14, 12043–12048. 10.4238/2015.October.5.17 [DOI] [PubMed] [Google Scholar]
- Huang Y. J., Liao H. W., Wu C. C., Peng H. L. (2009). MrkF is a component of type 3 fimbriae in Klebsiella pneumoniae. Res. Microbiol. 160, 71–79. 10.1016/j.resmic.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Kobayashi S. D., Porter A. R., Dorward D. W., Brinkworth A. J., Chen L., Kreiswirth B. N., et al. (2016). Phagocytosis and killing of carbapenem-resistant ST258 Klebsiella pneumoniae by human neutrophils. J. Infect. Dis. 213, 1615–1622. 10.1093/infdis/jiw001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park K. S., Kim Y. B., Jeong B. C., Lee S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C., Lee E. J., Lee J. H., Jun S. H., Choi C. W., Kim S. I., et al. (2012). Klebsiella pneumoniae secretes outer membrane vesicles that induce the innate immune response. FEMS Microbiol. Lett. 331, 17–24. 10.1111/j.1574-6968.2012.02549.x [DOI] [PubMed] [Google Scholar]
- Li B., Li M., Qu L., Wang M., Guo J. (2014). Prevalence and characteristics of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated from pediatric inpatients with respiratory tract infections at a teaching hospital in China. Scand. J. Infect. Dis. 46, 200–203. 10.3109/00365548.2013.859393 [DOI] [PubMed] [Google Scholar]
- Li B., Yi Y., Wang Q., Woo P. C., Tan L., Jing H., et al. (2012). Analysis of drug resistance determinants in Klebsiella pneumoniae isolates from a tertiary-care hospital in Beijing, China. PLoS ONE 7:e42280. 10.1371/journal.pone.0042280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhu Z. C., Wang L., Zhou Y. F., Tang Y. J., Miao Z. M. (2015). Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae in spring waters. Lett. Appl. Microbiol. 61, 544–548. 10.1111/lam.12489 [DOI] [PubMed] [Google Scholar]
- Lin H. A., Huang Y. L., Yeh K. M., Siu L. K., Lin J. C., Chang F. Y. (2016). Regulator of the mucoid phenotype A gene increases the virulent ability of extended-spectrum beta-lactamase-producing serotype non-K1/K2 Klebsiella pneumonia. Sci. Direct 49, 494–501. 10.1016/j.jmii.2014.08.023 [DOI] [PubMed] [Google Scholar]
- Little M. L., Qin X., Zerr D. M., Weissman S. J. (2014). Molecular epidemiology of colonizing and disease-causing Klebsiella pneumoniae in paediatric patients. J. Med. Microbiol. 63, 610–616. 10.1099/jmm.0.063354-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loucif L., Kassah-Laouar A., Saidi M., Messala A., Chelaghma W., Rolain J. M. (2016). Outbreak of OXA-48-producing Klebsiella pneumoniae involving a sequence type 101 clone in Batna University hospital, algeria. Antimicrob. Agents Chemother. 60, 7494–7497. 10.1128/AAC.00525-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Wang Y., Ye L., Yang J. (2014). Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 20, O818–O824. 10.1111/1469-0691.12664 [DOI] [PubMed] [Google Scholar]
- Machado A., Bordalo A. A. (2014). Prevalence of antibiotic resistance in bacteria isolated from drinking well water available in Guinea-Bissau (West Africa). Ecotoxicol. Environ Saf. 106, 188–194. 10.1016/j.ecoenv.2014.04.037 [DOI] [PubMed] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Markovska R., Stoeva T., Schneider I., Boyanova L., Popova V., Dacheva D., et al. (2015). Clonal dissemination of multilocus sequence type ST15 KPC-2-producing Klebsiella pneumoniae in Bulgaria. APMIS 123, 887–894. 10.1111/apm.12433 [DOI] [PubMed] [Google Scholar]
- Mollenkopf D. F., Mirecki J. M., Daniels J. B., Funk J. A., Henry S. C., Hansen G. E., et al. (2013). Escherichia coli and Klebsiella pneumoniae producing CTX-M cephalosporinase from swine finishing barns and their association with antimicrobial use. Appl. Environ. Microbiol. 79, 1052–1054. 10.1128/AEM.03169-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer U., Sundsfjord A. (2011). The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17, 83–97. 10.1089/mdr.2010.0132 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy C. M., Cunningham A. F., MacLennan A. C. (2012). The stability of complement-mediated bactericidal activity in human serum against salmonella. PLoS One. 7:e49147. 10.1371/journal.pone.0049147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczosa M. K., Mecsas J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K, Reuter S., Holden M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Wei Z., Ji S., Du X., Shen P., Yu Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- Rainer F., Eva H., Kelly Wyres L., Matthew Ellington J., Kowarik M., Holt K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genomics 2:e000073 10.1099/mgen.0.000073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Garbajosa P., Curiao T., Tato M., Gijon D., Pintado V., Valverde A., et al. (2013). Multiclonal dispersal of KPC genes following the emergence of non-ST258 KPC-producing Klebsiella pneumoniae clones in Madrid, Spain. J. Antimicrob. Chemother. 68, 2487–2492. 10.1093/jac/dkt237 [DOI] [PubMed] [Google Scholar]
- Sahly H., Navon-Venezia S., Roesler L., Hay A., Carmeli Y., Podschun R., et al. (2008). Extended-spectrum beta-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesins in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52, 3029–3034. 10.1128/AAC.00010-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Shin J., Kim D. H., Ko K. S. (2011). Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 63, 39–47. 10.1016/j.jinf.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Struve C., Bojer M., Krogfelt K. A. (2009). Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect. Immun. 77, 5016–5024. 10.1128/IAI.00585-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Wang Y., Li X. W., Zheng B. W. (2018). Study protocol for One Health data collections, analyses and intervention of the Sino-Swedish integrated multisectoral partnership for antibiotic resistance containment (IMPACT). BMJ Open 8:e017832. 10.1136/bmjopen-2017-017832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Tsuchiya M., Shimada K., Nga T. T. T., Thu L. T. A, Phu T. T., et al. (2017). Dissemination of Carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of Carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA Methylases (RmtB and RmtC) in Vietnam. BMC Infect. Dis. 17:467. 10.1186/s12879-017-2570-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N., Sheth P. M., Lastovetska O., Chung C., Patel S. N., Melano R. G. (2014). Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008-2011. PLoS ONE 9:e116421. 10.1371/journal.pone.0116421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vubil D., Figueiredo R., Reis T., Canha C., Boaventura L., DA Silva G. J. (2017). Outbreak of KPC-3-producing ST15 and ST348 Klebsiella pneumoniae in a Portuguese hospital. Epidemiol. Infect. 145, 595–599. 10.1017/S0950268816002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R. R., Heinz E., Holt K. E., Wyres K. L. (2018). Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56, e00197–e00118. 10.1128/JCM.00197-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. C., Lin C. T., Cheng W. Y., Huang C. J., Wang Z. C., Peng H. L. (2012). Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology 158, 1045–1056. 10.1099/mic.0.053801-0 [DOI] [PubMed] [Google Scholar]
- Xia S., Fan X., Huang Z., Xia L., Xiao M., Chen R., et al. (2014). Dominance of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS ONE 9:e100707. 10.1371/journal.pone.0100707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. J., Wang M. C., Zheng P. X., Tsai L. H., Wu J. J. (2015). Associations of the major international high-risk resistant clones and virulent clones with specific ompK36 allele groups in Klebsiella pneumoniae in Taiwan. N. Microb. N. Infect. 5, 1–4. 10.1016/j.nmni.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Wu Q., Zhang S., Zhang J., Yang G., Wang H., et al. (2017). Antibiotic-resistant extended spectrum ss-lactamase- and plasmid-mediated AmpC-producing enterobacteriaceae isolated from retail food products and the pearl river in Guangzhou, China. Front. Microbiol. 8:96. 10.3389/fmicb.2017.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks R. N., Griffiths H. M., Munoz M. A., Ahlstrom C., Bennett G. J., Thomas E., et al. (2011). Sources of Klebsiella and Raoultella species on dairy farms: be careful where you walk. J. Dairy Sci. 94, 1045–1051. 10.3168/jds.2010-3603 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhou Y., Guo S., Chang W. (2015). Prevalence and characteristics of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae isolated from rural well water in Taian, China, 2014. Environ. Sci. Pollut. Res. Int. 22, 11488–11492. 10.1007/s11356-015-4387-9 [DOI] [PubMed] [Google Scholar]
- Zhou K., Lokate M., Deurenberg R. H., Tepper M., Arends J. P., Raangs E. G. C., et al. (2016). Use of whole-genome sequencing to trace, control and characterize the regional expansion of extended-spectrum beta-lactamase producing ST15 Klebsiella pneumoniae. Sci. Rep. 6:20840. 10.1038/srep20840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.