Abstract
Induced pluripotent stem cells derived from normal somatic cells could be utilized to study tumorigenesis through overexpression of specific oncogenes, downregulation of tumor suppressors and dysregulation of other factors thought to promote tumorigenesis. Therefore, effective approaches that provide direct modifications of induced pluripotent stem cell genome are extremely needed. Emerging strategies are expected to provide the ability to more effectively introduce diverse genetic alterations, from as small as single-nucleotide modifications to whole gene amplification or deletion, all with a high degree of target specificity. To date, several techniques have been applied in stem cell studies to directly edit cell genome (ZFNs, TALENs or CRISPR/Cas9). In this review, we summarize specific gene delivery strategies that were applied to stem cell studies together with genome editing techniques, which enable a direct modification of endogenous DNA sequences in the context of cancer studies.
Keywords: Genome editing, Stem cells, Induced pluripotent stem cells, ZFN, TALEN, CRISPR/Cas9
1. Human pluripotent stem cells – an emerging model in basic and clinical research studies
Human pluripotent stem cells, which include embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), are a type of cells that have several crucial characteristics – unlimited self-renewal, high proliferation abilities, and the potential to differentiate into cells from all three germ layers (ectoderm, endoderm and mesoderm).1 Pluripotent stem cells maintain high telomerase activity and exhibit remarkable long-term proliferative potential, while displaying normal karyotype.2 These specific traits give pluripotent stem cells the potential to have far-reaching applications in many different aspects of basic and clinical research studies.3
Human ESCs are derived from the inner cell mass of cultured pre-implantation human blastocysts. When grown on a feeder cell layer (mouse embryonic fibroblasts), these cells can self-renew indefinitely in culture while maintaining the ability to become derivatives of all three germ layers. However, isolation of the inner cell mass cells results in destruction of the pre-implantation blastocyst, which raises ethical issues.4
Fortunately, in 2007 it was shown that human pluripotent stem cells highly resembling ESCs can be generated by the delivery of so-called pluripotency genes (namely OCT-3/4, SOX2, KLF4 and c-MYC) to differentiated somatic cells.5 The enforced expression of these genes reprograms fully differentiated cells to induced pluripotent stem (iPS) cells in vitro, providing the generation of pluripotent stem cells without any manipulations with embryos. Since the first discovery, numerous methods have been developed to generate iPS cells and all emerging strategies were oriented to increase the safety of iPSC derivation.6 Beside reducing the number of pluripotency genes used in reprogramming,7 also the transgene-free delivery strategies to the host cell were applied, greatly enhancing the efficiency of reprogramming.8
Induced pluripotent stem cells provide indisputable advantages as a model system in basic research. To analyze the role of specific genes in iPS cells and their differentiated progeny, two primary strategies might be utilized. In one approach, somatic cells from patients with genetic disorders are reprogrammed to iPS cells. These patient-derived iPS cells as well as their differentiated progeny are then used to elucidate a disease phenotype in vitro or clarify disease-relevant mechanisms.9 Abovementioned approach has been applied successfully to explain the genetic causes of metabolic10 or mental disorders,11 neurodegeneration,12, 13, 14 and heart disease.15, 16, 17, 18
Alternatively, the genetic disease trait can be directly introduced into pluripotent stem cells that basically have an intact genetic background. This approach is supported remarkably by recent developments in gene delivery systems, gene manipulation techniques and genome editing tools, all of which are described below. Modification of pluripotent stem cells can be accomplished both by the enforced expression of exogenous DNA sequence19 or modification of endogenously expressed transcript (at the gene level or the transcript level).20 Gene expression can be modulated by reversibly targeting the endogenous promoter.21, 22 Also, the genetic information of iPS cells can be deleted or inverted23, 24 and modifications as small as single-nucleotide changes can be introduced to mimic disease-associated mutations.25 The emerging genetically modified iPS cells are isogenic with their wild type predecessors – they differ from each other exclusively at the edited locus. Parallel differentiation of such isogenic cell lines into disease-specific cells or tissues can be used to directly assess the contribution of a specific mutation to a cellular pathological state.26, 27, 28, 29
However, to successfully expand potential applications of iPS cells, a rapid, reliable and simple gene delivery technique is needed. Most recently used techniques of genome editing in pluripotent stem cells are presented below.
2. Transgene delivery to pluripotent stem cells
Technologies that allow modification of gene expression in human pluripotent stem cells are numerous and vary in their complexity, efficiency, reliability, and safety. They are frequently categorized by whether the delivered gene remains separate from the host cell genome (transient gene delivery), or whether it is integrated into the chromosome of a target cell (stable gene delivery). Gene transduction options for pluripotent stem cells are most commonly divided into two groups: viral and non-viral gene delivery (further subdivided into chemical or physical gene delivery methods).
Viral delivery exploits the backbone of a viral genome with insertion of the gene or genes of interest to be expressed in the stem cell. Generally, viral transduction is highly efficient and results in less damage to pluripotent stem cells among other delivery methods.30 However, viruses can only deliver very small fragments of DNA into the host cell, their production is labor-intensive, and they carry the risk of random insertion sites, cytopathic effects and mutagenesis. To date, retroviral, lentiviral, adenoviral, adeno-associated viral (AAV) or baculoviral transductions have been utilized to modify human pluripotent stem cells.30, 31, 32, 33, 34, 35 Their brief characteristic is presented in Table 1.
Table 1.
Virus-based gene delivery methods applied to modify human pluripotent stem cells.
| Trait | Vector |
||||
|---|---|---|---|---|---|
| Retrovirus | Lentivirus | Adenovirus | Adeno-associated virus (AAV) | Baculovirus | |
| Viral genome | ssRNA | ssRNA | dsDNA | ssDNA | dsDNA |
| Packaging capacity | 8–8.5 kb | 8–9 kb | Up to 38 kb | <5 kb | No known upper limit |
| Genome integration | Yes | Yes | No | Noa | No |
| Infection | Dividing cells only | Dividing and non-dividing cells | Dividing and non-dividing cells | Dividing and non-dividing cells | Dividing and non-dividing cells |
| Transgene expression | Stable | Stable | Transient | Transient/stable | Transient/stable |
| Advantages | + Long term expression | + High transduction efficiency + Long term expression |
+ Very high transduction efficiency + High packaging capacity |
+ Site-specific integration | + High transduction efficiency + Large transgene capacity |
| Disadvantages | - Random integration site - Instability (due to methylation) |
- Random integration site | - Short term gene expression - Requires helper virus for replication |
- Small packaging capacity - Complicated process of vector production |
- Lack of permanent transgene expression - Random integration site |
kb – kilobase; ss – single stranded; ds – double stranded.
AAV vector genomes can persist within cells as episomes, however AAV vector integration has been observed in various experimental settings.30
On the other hand, several non-viral delivery techniques have recently been developed. The most commonly studied and practiced non-viral delivery methods include electroporation, lipofection and nucleofection of plasmid DNA encoding the gene of interest.36, 37, 38 Several other techniques, such as ultrasound, microinjection, and molecular-vibration-mediated delivery may also be tested in human pluripotent stem cell studies.38 Generally, the utilization of non-viral gene delivery systems provides decreased immunogenicity and less insertional mutagenesis when compared to viral vector delivery. However, their main disadvantages are decreased transfection efficiency and potential loss of transgene expression during prolonged cell culture which may limit long-term studies.
3. Genome editing of pluripotent stem cells
Genome engineering of iPS cells represents a powerful tool to study gene function in human development and disease. The comparison between patient-derived and isogenic gene-corrected iPS cell lines allows genotype-phenotype functional correlation studies. Also, the ability to genetically modify human iPS cells holds tremendous clinical promise for generating artificial organs and safer gene therapies. Recently, genome editing strategies that base on custom-engineered endonucleases have been applied in stem cell studies (Fig. 1). These strategies have expanded our possibility to study human biology.
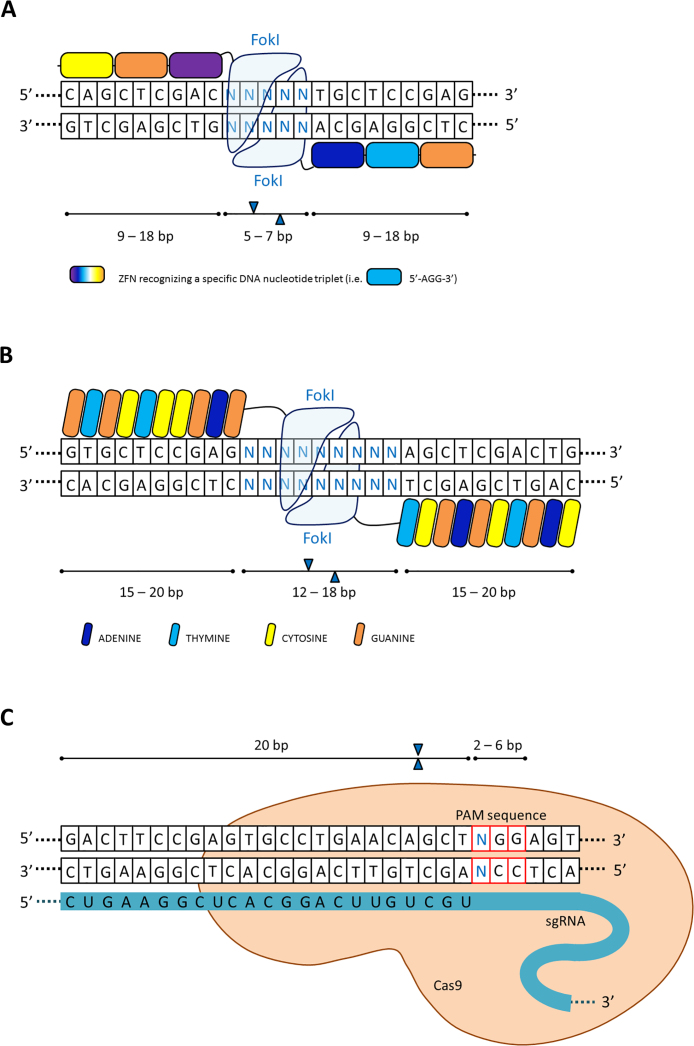
Fig. 1.
The construction of site-specific nuclease systems. (A) Zinc Finger Nucleases are hybrid proteins composed of zinc-finger arrays (3–6 ZFNs in each hybrid protein) and the catalytic domain of FokI endonuclease. Each ZFN subunit recognizes and binds to a specific DNA nucleotide triplet in the target sequence. Dimerization of the FokI catalytic domain leads to the formation of double-strand breaks (DSBs). (B) TALEN are hybrid proteins composed of TAL effector DNA-binding domain arrays (15–20 TALE subunits in each TALEN protein) and the catalytic domain of FokI endonuclease. One monomer of TALE recognizes one nucleotide of a target DNA sequence. Dimerized FokI endonuclease introduces a double-strand break into DNA sequence. (C) The CRISPR/Cas9 two-component system is composed of Cas9 endonuclease and the single-guide RNA (sgRNA) molecule (20 base pair long). Cas9 endonuclease domains cleave both strands within the target sequence preceding the protospacer-associated motif (PAM) NGG trinucleotide sequence. bp – base pair; FokI – endonuclease FokI.
3.1. Zinc Finger Nucleases
The first successful genetic modification of human pluripotent stem cells with the custom-engineered, site-specific endonucleases was demonstrated by Lombardo et al.39 in 2007. The authors have inserted an EGFP expression cassette into the CCR5 gene locus of human embryonic stem cells with the 3.5% frequency (in the absence of selection) using Zinc Finger Nucleases (ZFNs). ZFNs are fusion proteins composed of several tandem Zinc-finger DNA binding domains conjugated to the FokI endonuclease catalytic domain. The customized array of zinc-fingers is engineered to bind to a specific DNA sequence. When two ZFNs are tethered to the DNA sequence in the proper orientation, the nuclease domain dimerizes and creates a sequence-specific double-stranded break (DSB). These breaks are then efficiently repaired by either homologous recombination (HR) or error prone non-homologous end-joining (NHEJ). Therefore, this approach can induce a site-specific insertion or deletion at the site of DSB after NHEJ or can insert a defined DNA sequence at the site of the DSB by homologous recombination with an exogenous donor DNA fragment.40
Later, Zou et al.41 applied Zinc Finger Nucleases to perform gene targeting at the endogenous PIG-A gene and at a defective, chromosomally integrated EGFP reporter gene both in human embryonic stem cells and induced pluripotent stem cells. In their work, Zou et al.41 demonstrated that the transient expression of sequence-specific ZFNs significantly enhanced HR in human embryonic stem cells. Importantly, they also showed that these ZFNs enhance gene targeting without detrimental effects on either cell karyotypes or pluripotency. Also, Hockemeyer et al.42 reported the highly efficient targeting of three genes in human pluripotent cells using ZFN-mediated genome editing. Firstly, they inserted a reporter gene into the POU5F1 locus encoding OCT-3/4 pluripotency marker to monitor the pluripotent state of modified cells. Also, they inserted a transgene into the AAVS1 locus to generate a robust drug-inducible overexpression system in human ES cells. Finally, they demonstrated that ZFNs can be used to generate pluripotent stem cells that express exogenous genes specific for differentiated cells. Altogether, Hockemeyer et al. reported efficient targeting of both expressed and silent genes in human pluripotent stem cells using Zinc Finger Nucleases.42
3.2. Transcription activator-like effector nucleases
To date, genome editing aided by ZFNs in human pluripotent stem cells have been described to be highly efficient.42 However, designing ZFNs for a specific target sequence is quite inefficient and laborious, which largely hinders their widespread application. Recently, transcription activator-like effector nucleases (TALENs) have rapidly occurred as an interesting alternative to ZFNs for genome editing. TALENs as ZFNs are restriction enzymes genetically engineered to cut specific sequences of DNA. Comprised of TAL effector DNA-binding domain (multiple units arranged in tandem – TALE repeats) that naturally occurs in Xanthomonas bacteria, and DNA cleavage domain (nuclease domain), TALENs can bind to practically any desired DNA sequence and cut the DNA at specific locations.
In stem cell studies, TALENs were used for the first time in 2011 by Hockemeyer et al.43 The Authors assessed the utility of TALENs to insert specific gene modifications in 5 distinct loci using human ES cells and iPS cells. At all five genomic sites tested, they obtained clones carrying transgenes solely at the TALEN-specified locus with different frequencies (ranging from 1% to 100% of correctly targeted clones depending on loci). Although the efficiencies of gene targeting were similar to those observed with ZFNs, this approach is much more easy and convenient to design and construct. In 2013, Sakuma et al.44 demonstrated that using TALENs, the complete process of genome editing (from initiating construction of TALENs to completing evaluation) of induced pluripotent stem cells requires only one week. In their work, Sakuma et al. modified the native TALEN protein to enhance the nuclease activity by replacing the final destination vector with a mammalian expression/in vitro transcription vector bearing both CMV and T7 promoters.44 Also, it was demonstrated that TALENs exhibited lower off target effects and reduced nuclease-associated cytotoxicities compared with ZFNs.45, 46, 47
Later, Ma et al.48 reported a robust process combining efficient generation of integration-free β-Thalasemia iPS cells from the amniotic fluid cells of patients harboring two distinct mutations in HBB (hemoglobin subunit B). They further used TALEN-encoding plasmids electroporation for HBB gene correction and showed that these iPS cells are pluripotent and have normal karyotype. Moreover, the correction process did not generate TALEN-induced off targeting mutations and obtained gene-corrected iPS cells could be induced to differentiate into hematopoietic progenitor cells and then further to erythroblasts expressing normal β-globin.
Moreover, TALENs were also used to repair the mitochondrial DNA of iPS cells. In 2017 Yahata et al.49 generated iPS cells from a patient with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) with the m.13513G>A mutation. Mitochondrial defects can lead to a broad spectrum of clinical manifestations based on the ratio of mutant to wild-type mtDNA, known as heteroplasmy. Therefore, Yahata et al.49 have tried to decrease mutant mtDNA level in iPS cells using TALENs. TALEN has multiple options of DNA recognition motif (TALE) for the target sequence and can be transported into mitochondria by addition of the mitochondrial targeting sequence (MTS) to the TALEN encoding plasmid. They reported significant decrease in the relative mutated mtDNA level and proposed TALEN-modified disease-specific iPS cells as a powerful tool for investigation of specific phenotypes caused by mitochondrial DNA (mtDNA) mutations. Accordingly, similar results were further obtained by Yang et al.50 who successfully eliminated the mitochondrial m.3243A>G mutation in MELAS patient-specific iPS cells and reported no off-target mutagenesis in the TALEN-edited iPS cell colonies.
3.3. CRISPR/Cas9 platform
Although ZFNs and TALENs are powerful tools for genome editing, they must be reengineered for each editing site and it is difficult to use them for the introduction of multiplex edits.
Fortunately, recently discovered CRISPR-associated systems have overcome this drawback and significantly improved the efficiency and simplify the genome editing,51, 52, 53 which enormously expanded our ability to introduce genetic alterations in human cells. The Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-Associated Protein 9 nuclease (Cas9) form an adaptive immunity mechanism of bacteria and archaea, enabling the organisms to respond to and eliminate invading genetic material. Among three types of CRISPR mechanisms identified, type II is the most studied. This type functions through a RNA-guided mechanism, conjugated to associated effector Cas9 nuclease. Firstly, extraneous viral or plasmid DNA is cut into fragments that are further inserted into the CRISPR locus among a series of short repeats. Then, the CRISPR locus is transcribed and transcripts are processed to generate CRISPR RNA (crRNA) complexes together with trans-activating crRNAs sequence (collectively known as single guide RNA, sgRNA) and Cas9 nuclease. The sgRNA guides the effector nucleases that target the invading DNA based on sequence complementarity. As a genome-editing tool, CRISPR/Cas9 system is broadly used to develop gene knock-outs or knock-ins through substitution of a target DNA sequence with a desired donor sequence.51, 52, 53
In pluripotent stem cells studies, the CRISPR/Cas9 system was for the first time used in 2013 and since then it has rapidly become the platform of choice to genetically modify human pluripotent stem cells. Mali et al.54 have reported a successful modification of native AAVS1 locus in induced pluripotent stem cells with the targeting rate reaching 2–4% with CRISPR-mediated gene targeting. Interestingly, Hou et al.55 achieved ∼60% targeting efficiency with two human embryonic stem cell lines and one human induced pluripotent stem cell line when targeting endogenous POU5F1 locus with CRISPR-Cas system from Neisseria meningitidis (to date, only two CRISPR-Cas systems – from Streptococcus pyogenes and Streptococcus thermophilus, each with their own distinct targeting requirements and limitations, have been developed for genome editing). The establishment of CRISPR/Cas9 systems from different organisms has increased the ease and scope of Cas9-mediated genome engineering in pluripotent stem cells. Also, several optimized protocols for CRISPR/Cas9-mediated genome editing of pluripotent stem cells were published recently, further demonstrating the tremendous impact it has on stem cells research and human disease modeling.56, 57, 58, 59, 60
The main advantage of the CRISPR/Cas9 system over the site-specific nucleases (ZFNs or TALENs) comes from the fact that DNA-binding specificity is encoded solely by the single guide RNA and so, unlike previous machineries, does not require laborious engineering of DNA binding proteins. Therefore, it is not surprising that CRISPR/Cas9-based editing has largely replaced previous site-specific nucleases. However, when first discovered, CRISPR/Cas9 system has faced problems with off-target nuclease activity,55 which had also been seen in ZNF or TALEN platforms. Fortunately, further adaptations and improvement have been made to extend the specificity of Cas9.61
4. Modeling cancer progression with genome edited human pluripotent stem cells
As soon as reprogramming of somatic cells to induced pluripotent stem cells was developed, the discovery attracted the attention of scientists, offering new perspectives for personalized medicine and providing a powerful platform for drug testing. The iPSC technology was almost immediately applied to cancer studies62 and has robustly progressed upon invention of genome editing approaches, especially CRISPR/Cas9 system.
In 2017 the induced reprogramming of somatic cells was successfully utilized to establish iPS cell-based experimental platforms that recapitulate disease stages and the clonal architecture of myeloid malignancies.63, 64 The iPSC methodology allowed for the isolation of subclones with different mutational background from the same patient and gave the opportunity to easily introduce additional mutations, or correct mutations, through CRIPSR/Cas9 genome editing. Using this model, Kotini et al.63 were able to capture a range of distinct disease stages encompassing preleukemia, low-risk myelodysplastic syndrome (MDS), high-risk MDS, and secondary acute myeloid leukemia through CRISPR/Cas9-mediated genetic correction or introduction of site-specific mutations to induced pluripotent stem cells. They also demonstrated that iPS cell-based approach can be used to uncover disease-stage-specific responses to drugs.63, 65
Also, Chang et al.65 used a similar platform to determine individual participations of two specific genetic lesions, the splicing factor SRSF2 P95L mutation and the chromosome 7q deletion, to the development of myeloid malignancy. Using a panel of isogenic iPS cells – with none, one, or both genetic lesions – Chang et al.65 characterized the relative phenotypic contributions and identified drug sensitivities specific to each of them through a candidate drug approach and an unbiased large-scale small-molecule screen. CRISPR/Cas9 genome edited iPS cells were also exploited to model the early steps of childhood acute B-lymphoblastic leukemia (cALL) development.66 Introduction of the ETV6-RUNX1, a first-hit mutation that initiates a clinically silent pre-leukemia in utero, into the endogenous ETV6 locus of iPS cells which were further subjected to haemato-endothelial differentiation led to the acquisition of several specific features (collectively) consistent with a pre-leukemic state.
Recently, Sommer et al.67 established an isogenic iPS-based platform (using TALEN-mediated editing of APC gene) to evaluate the effect of APC mutation before and after differentiation into intestinal organoids. Utilization of this model provided novel insights into the early events of APC-mediated tumorigenesis, demonstrating surprising defects in cell identity and chromosomal integrity upon APC heterozygosity.
Induced pluripotent stem cell reprogramming was frequently utilized to generate high-fidelity models to elucidate the pathological mechanism of carcinogenesis in patients harboring genetic diseases that predispose to cancer68, 69, 70; however, these models lack proper experimental controls. Only recently the methodology was enriched in genome editing techniques to generate isogenic cell line models that enable a better understanding of the molecular basis of cancer initiation. The iPS-based platform has been harnessed to unravel the molecular mechanisms underlying the development of the multiple endocrine neoplasia type 2A (MEN2A) hereditary cancer predisposing syndrome that affects patients with germline RET mutations. Hadoux et al.71, 72 generated iPS cells from a patient with RET mutation (who developed medullary thyroid carcinoma and pheochromocytoma) and utilized the CRISPR/Cas9 technology to obtain an isogenic RETWT iPS control cell line in order to specifically study their transcriptional landscape in the context of an oncogenic mutated RET. Interestingly, they identified Early Growth Response 1 (EGR1) as a putative effector of the development of MEN2A features at the embryonic stage independent of any differentiation program (explaining why the tumors develop in tissues of distinct embryonic origins in MEN2A patients).71, 72
5. Reprogramming of cancer cells to induced pluripotency
Reprogramming of cancer cells to induced pluripotency (i) could be used to model carcinogenesis and elucidate the mechanisms which underlie the stages of cancer development, (ii) offers a platform for studying tumor heterogeneity and origin of cancer stem cells and (iii) serves as a source for cancer type-specific drug discovery studies.73, 74, 75 Specific cancer cell reprogramming methods and potential application of induced cancer stem cells (iCSCs) in basic studies was previously summarized.62 Briefly, the reprogramming efficiency of cancer cells is extremely low, suggesting that some properties of these cells impede induced dedifferentiation. The difficulties in cancer cell reprogramming are thought to stem (at least partially) from the accumulation of DNA damage and cancer-specific mutations, epigenetic modifications and reprogramming-triggered cellular senescence. Therefore, the development of more effective reprogramming approaches that would overcome current technological and biological challenges is essential to fully exploit the potential of induced pluripotent stem cell technology in cancer research.76 Also, the observation that induced reprogramming of cancer cells could be achieved with enforced expression of reprogramming factors reveals the possibility that stem cell-like phenotypes could be gained and maintained by cancer cells during tumor progression in response to other exogenous stimuli in vitro or in vivo (i.e. hypoxia, therapeutic agents, heterogenous stromal cells), increasing the complexity of cancer and uncovering the need for developing more complex cancer models in vitro.77, 78, 79 Therefore, reprogramming of cancer cells together with genome editing techniques in vitro should result in generation of more accurate models for investigation of cancer development and progression, directly mirroring the heterogeneity of cancer cells and cancer microenvironment.
6. Future perspectives
To date, several technologies have been developed for genome editing of human iPS cells. These methods differ in their complexity, efficiency of gene targeting and the frequency of off-target effects. The incorporation of so-far discovered approaches in patient-derived iPS cells is a promising step toward personalized anti-cancer therapy.
When using isogenic pairs of iPS cells, this technology is also an indispensable tool for assessment of the molecular and cellular phenotypes resulting from single genetic lesions. It gives us the ability to directly uncover the cellular consequences of disease mutations, which is often impossible to determine from human genetic studies alone. Moreover, these isogenic induced pluripotent stem cell lines can be used to perform high throughput genetic and pharmacological screens to both understand the underlying pathological mechanisms and to develop novel therapeutic agents. However, in order to obtain perfect isogenic iPS cells, several factors need to be considered including the type of delivery vector, selection markers, gene correction strategy, off target effects, and genomic integrity.
Hopefully, further optimization of genome editing technologies (to increase their targeting efficiency and reduce off target rates) may aid in improving iPS cell-based studies to ultimately elucidate biological mechanisms of cancer development and progression and provide novel clinical therapies.
Authors’ contributions
PC was a major contributor in writing the manuscript. MW coordinated the manuscript preparation. All authors read and approved the final manuscript.
Conflict of interest
None declared.
Financial disclosure
This work was supported by Greater Poland Cancer Centre intramural grants (Nos. 17/2016 (132) and 22/2016 (137)) and Poznań University of Medical Sciences intramural grant (No. 502-14-02233381-11055) to Patrycja Czerwinska.
Contributor Information
Patrycja Czerwińska, Email: czerwinska.patr@gmail.com.
Sylwia Mazurek, Email: syl.mazurek@gmail.com.
Iga Kołodziejczak, Email: kolodziejczak.iga@gmail.com.
Maciej Wiznerowicz, Email: maciej.wiznerowicz@gmail.com.
References
- 1.Klimczak M. Oncogenesis and induced pluripotency – commonalities of signalling pathways. Contemp Oncol. 2015;19(1A):A16–A21. doi: 10.5114/wo.2014.47133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiyama E., Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96(7):1020–1024. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kavyasudha C., Macrin D., ArulJothi K.N. Clinical applications of induced pluripotent stem cells – Stato Attuale. Adv Exp Med Biol. 2018;1079:127–149. doi: 10.1007/5584_2018_173. [DOI] [PubMed] [Google Scholar]
- 4.Volarevic V., Markovic B.S., Gazdic M. Ethical and safety issues of stem cell-based therapy. Int J Med Sci. 2018;15(1):36–45. doi: 10.7150/ijms.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Okano H., Nakamura M., Yoshida K. Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res. 2013;112(3):523–533. doi: 10.1161/CIRCRESAHA.111.256149. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa M., Koyanagi M., Tanabe K. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 8.Okita K., Matsumura Y., Sato Y. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 9.Cao L., Tan L., Jiang T., Zhu X.-C., Yu J.-T. Induced pluripotent stem cells for disease modeling and drug discovery in neurodegenerative diseases. Mol Neurobiol. 2015;52(1):244–255. doi: 10.1007/s12035-014-8867-6. [DOI] [PubMed] [Google Scholar]
- 10.Rashid S.T., Corbineau S., Hannan N. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Investig. 2010;120(9):3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennand K.J., Simone A., Jou J. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimos J.T., Rodolfa K.T., Niakan K.K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 13.Park I.-H., Arora N., Huo H. Disease-specific induced pluripotent stem cells. Cell. 2008;134(5):877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagi T., Ito D., Okada Y. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20(23):4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 15.Carvajal-Vergara X., Sevilla A., D'Souza S.L. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465(7299):808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paşca S.P., Portmann T., Voineagu I. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17(November):1657. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itzhaki I., Maizels L., Huber I. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471(7337):225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 18.Moretti A., Bellin M., Welling A. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363(15):1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 19.Merkle F.T., Neuhausser W.M., Santos D. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11(6):875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaspar J.A., Srinivasan S.P., Sureshkumar P. Depletion of Mageb16 induces differentiation of pluripotent stem cells predominantly into mesodermal derivatives. Sci Rep. 2017;7(1):14285. doi: 10.1038/s41598-017-14561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K.-H., Oghamian S., Park J.-A., Kang L., Laird P.W. The REMOTE-control system: a system for reversible and tunable control of endogenous gene expression in mice. Nucleic Acids Res. 2017;45(21):12256–12269. doi: 10.1093/nar/gkx829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun S.M.G., Kirkland J.G., Chory E.J., Husmann D., Calarco J.P., Crabtree G.R. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat Commun. 2017;8(1):560. doi: 10.1038/s41467-017-00644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Y., Wang D., Lan F. An episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci Rep. 2017;7(1):1–11. doi: 10.1038/s41598-017-02456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceasar S.A., Rajan V., Prykhozhij S.V., Berman J.N., Ignacimuthu S. Insert, remove or replace: a highly advanced genome editing system using CRISPR/Cas9. Biochim Biophys Acta – Mol Cell Res. 2016;1863(9):2333–2344. doi: 10.1016/j.bbamcr.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Ochiai H. Single-base pair genome editing in human cells by using site-specific endonucleases. Int J Mol Sci. 2015;16(9):21128–21137. doi: 10.3390/ijms160921128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung C.Y., Khurana V., Auluck P.K. Identification and rescue of α-synuclein toxicity in parkinson patient-derived neurons. Science. 2013;342(6161):983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan S.D., Dolatabadi N., Chan S.F. Isogenic human iPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155(6):1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Liang P., Lan F. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64(5):451–459. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yusa K., Rashid S.T., Strick-Marchand H. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapti K., Stillitano F., Karakikes I. Effectiveness of gene delivery systems for pluripotent and differentiated cells. Mol Ther Methods Clin Dev. 2015;2:14067. doi: 10.1038/mtm.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann D., Schott J.W., Geis F.K. Detailed comparison of retroviral vectors and promoter configurations for stable and high transgene expression in human induced pluripotent stem cells. Gene Ther. 2017;24(5):298–307. doi: 10.1038/gt.2017.20. [DOI] [PubMed] [Google Scholar]
- 32.Bajpai R. Lentivirus-mediated modification of pluripotent stem cells. Methods Mol Biol. 2011;767:315–331. doi: 10.1007/978-1-61779-201-4_23. [DOI] [PubMed] [Google Scholar]
- 33.Naujok O., Diekmann U., Elsner M. Gene transfer into pluripotent stem cells via lentiviral transduction. Methods Mol Biol. 2016;1341:67–85. doi: 10.1007/7651_2015_221. [DOI] [PubMed] [Google Scholar]
- 34.Brokhman I., Pomp O., Fishman L. Genetic modification of human embryonic stem cells with adenoviral vectors: differences of infectability between lines and correlation of infectability with expression of the coxsackie and adenovirus receptor. Stem Cells Dev. 2009;18(3):447–456. doi: 10.1089/scd.2008.0127. [DOI] [PubMed] [Google Scholar]
- 35.Sung L.-Y., Chen C.-L., Lin S.-Y. Efficient gene delivery into cell lines and stem cells using baculovirus. Nat Protoc. 2014;9(8):1882–1899. doi: 10.1038/nprot.2014.130. [DOI] [PubMed] [Google Scholar]
- 36.Hohenstein K.A., Pyle A.D., Chern J.Y., Lock L.F., Donovan P.J. Nucleofection mediates high-efficiency stable gene knockdown and transgene expression in human embryonic stem cells. Stem Cells. 2008;26(6):1436–1443. doi: 10.1634/stemcells.2007-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao F., Xie X., Gollan T. Comparison of gene-transfer efficiency in human embryonic stem cells. Mol Imaging Biol. 2010;12(1):15–24. doi: 10.1007/s11307-009-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellott A.J., Forrest M.L., Detamore M.S. Physical non-viral gene delivery methods for tissue engineering. Ann Biomed Eng. 2013;41(3):446–468. doi: 10.1007/s10439-012-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombardo A., Genovese P., Beausejour C.M. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 40.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou J., Maeder M.L., Mali P. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5(1):97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hockemeyer D., Soldner F., Beard C. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hockemeyer D., Wang H., Kiani S. Genetic engineering of human ES and iPS cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakuma T., Hosoi S., Woltjen K. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013;18(4):315–326. doi: 10.1111/gtc.12037. [DOI] [PubMed] [Google Scholar]
- 45.Boch J. TALEs of genome targeting. Nat Biotechnol. 2011;29(2):135–136. doi: 10.1038/nbt.1767. [DOI] [PubMed] [Google Scholar]
- 46.Mussolino C., Morbitzer R., Lutge F., Dannemann N., Lahaye T., Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sander J.D., Cade L., Khayter C. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29(8):697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma N., Liao B., Zhang H. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free beta-thalassemia induced pluripotent stem cells. J Biol Chem. 2013;288(48):34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yahata N., Matsumoto Y., Omi M., Yamamoto N., Hata R. TALEN-mediated shift of mitochondrial DNA heteroplasmy in MELAS-iPSCs with m.13513G>A mutation. Sci Rep. 2017;7(1):1–11. doi: 10.1038/s41598-017-15871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y., Wu H., Kang X. Targeted elimination of mutant mitochondrial DNA in MELAS-iPSCs by mitoTALENs. Protein Cell. 2018;9(3):283–297. doi: 10.1007/s13238-017-0499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doudna J.A., Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213) doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 52.Savić N., Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mali P., Yang L., Esvelt K.M. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou Z., Zhang Y., Propson N.E. Efficient genome engineering in human pluripotent stem cells using Cas9. Proc Natl Acad Sci U S A. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiba K., Hockemeyer D. Genome editing in human pluripotent stem cells using site-specific nucleases. Methods Mol Biol. 2015;1239:267–280. doi: 10.1007/978-1-4939-1862-1_15. [DOI] [PubMed] [Google Scholar]
- 57.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kleinstiver B.P., Pattanayak V., Prew M.S. High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Byrne S.M., Church G.M. Crispr-mediated gene targeting of human induced pluripotent stem cells. Curr Protoc Stem Cell Biol. 2015;35 doi: 10.1002/9780470151808.sc05a08s35. 5A.8.1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soh C.-L., Huangfu D. CRISPR/Cas9-mediated mutagenesis of human pluripotent stem cells in defined xeno-free E8 medium. Methods Mol Biol. 2017;1498:57–78. doi: 10.1007/978-1-4939-6472-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kleinstiver B.P., Prew M.S., Tsai S.Q. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czerwińska P., Mazurek S., Wiznerowicz M. Application of induced pluripotency in cancer studies. Rep Pract Oncol Radiother. 2018;23(3):207–214. doi: 10.1016/j.rpor.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kotini A.G., Chang C.-J., Chow A. Stage-specific human induced pluripotent stem cells map the progression of myeloid transformation to transplantable leukemia. Cell Stem Cell. 2017;20(3) doi: 10.1016/j.stem.2017.01.009. 315–28.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chao M.P., Gentles A.J., Chatterjee S. Human AML-iPSCs reacquire leukemic properties after differentiation and model clonal variation of disease. Cell Stem Cell. 2017;20(3) doi: 10.1016/j.stem.2016.11.018. 329–44.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang C.-J., Kotini A.G., Olszewska M. Dissecting the contributions of cooperating gene mutations to cancer phenotypes and drug responses with patient-derived iPSCs. Stem Cell Rep. 2018;10(5):1610–1624. doi: 10.1016/j.stemcr.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boiers C., Richardson S.E., Laycock E. A human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6-RUNX1. Dev Cell. 2018;44(3) doi: 10.1016/j.devcel.2017.12.005. 362–77.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sommer C.A., Capilla A., Molina-Estevez F.J. Modeling APC mutagenesis and familial adenomatous polyposis using human iPS cells. PLOS ONE. 2018;13(7):e0200657. doi: 10.1371/journal.pone.0200657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soyombo A.A., Wu Y., Kolski L. Analysis of induced pluripotent stem cells from a BRCA1 mutant family. Stem Cell Rep. 2013;1(4):336–349. doi: 10.1016/j.stemcr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee D.F., Su J., Kim H.S. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–254. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mulero-Navarro S., Sevilla A., Roman A.C. Myeloid dysregulation in a human induced pluripotent stem cell model of PTPN11-associated juvenile myelomonocytic leukemia. Cell Rep. 2015;13:504–515. doi: 10.1016/j.celrep.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hadoux J., Féraud O., Griscelli F. Generation of an induced pluripotent stem cell line from a patient with hereditary multiple endocrine neoplasia 2A (MEN2A) syndrome with RET mutation. Stem Cell Res. 2016;17(1):154–157. doi: 10.1016/j.scr.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 72.Hadoux J., Desterke C., Féraud O. Transcriptional landscape of a RETC634Y-mutated iPSC and its CRISPR-corrected isogenic control reveals the putative role of EGR1 transcriptional program in the development of multiple endocrine neoplasia type 2A-associated cancers. Stem Cell Res. 2018;26:8–16. doi: 10.1016/j.scr.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 73.Kim J., Hoffman J.P., Alpaugh R.K. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3(6):2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernhardt M., Novak D., Assenov Y. Melanoma-derived iPCCs show differential tumorigenicity and therapy response. Stem Cell Rep. 2017;8(5):1379–1391. doi: 10.1016/j.stemcr.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X., Cruz F.D., Terry M., Remotti F., Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32(18):2249–2260. doi: 10.1038/onc.2012.237. 2260.e1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao H., Davies T.J., Ning J. A highly optimized protocol for reprogramming cancer cells to pluripotency using nonviral plasmid vectors. Cell Reprogram. 2015;17(1):7–18. doi: 10.1089/cell.2014.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L., Huang X., Zheng X. Enrichment of prostate cancer stem-like cells from human prostate cancer cell lines by culture in serum-free medium and chemoradiotherapy. Int J Biol Sci. 2013;9:472–479. doi: 10.7150/ijbs.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y., Li W., Patel S.S. Blocking the formation of radiation-induced breast cancer stem cells. Oncotarget. 2014;5:3743–3755. doi: 10.18632/oncotarget.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Debeb B.G., Lacerda L., Xu W. Histone deacetylase inhibitors stimulate dedifferentiation of human breast cancer cells through WNT/beta-catenin signaling. Stem Cells. 2012;30:2366–2377. doi: 10.1002/stem.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]