Laboratory-adapted strains of bacteria are used throughout the world for microbiology research. These prototype strains help keep research data consistent and comparable between laboratories. However, we have observed phenotypic variability when using different strains of Pseudomonas aeruginosa PAO1, one of the major laboratory-adopted research strains. Here, we describe the genomic and phenotypic differences among 10 PAO1 strains acquired from independent sources over 15 years to understand how individual maintenance affects strain characteristics. We observed limited genomic changes but variable phenotypic changes, which may have consequences for cross-comparison of data generated using different PAO1 strains. Our research highlights the importance of limiting practices that may promote the microevolution of model strains and calls for researchers to specify the strain origin to ensure reproducibility.
KEYWORDS: Pseudomonas aeruginosa, evolution, genome analysis, variable phenotypes
ABSTRACT
Pseudomonas aeruginosa is an opportunistic pathogen found ubiquitously in the environment and commonly associated with airway infection in patients with cystic fibrosis. P. aeruginosa strain PAO1 is one of the most commonly used laboratory-adapted research strains and is a standard laboratory-adapted strain in multiple laboratories and strain banks worldwide. Due to potential isolate-to-isolate variability, we investigated the genomic and phenotypic diversity among 10 PAO1 strains (henceforth called sublines) obtained from multiple research laboratories and commercial sources. Genomic analysis predicted a total of 5,682 genes, with 5,434 (95.63%) being identical across all 10 strains. Phenotypic analyses revealed comparable growth phenotypes in rich media and biofilm formation profiles. Limited differences were observed in antibiotic susceptibility profiles and immunostimulatory potential, measured using heat-killed whole-cell preparations in four immortalized cell lines followed by quantification of interleukin-6 (IL-6) and IL-1β secretion. However, variability was observed in the profiles of secreted molecular products, most notably, in rhamnolipid, pyoverdine, pyocyanin, Pseudomonas quinolone signal (PQS), extracellular DNA, exopolysaccharide, and outer membrane vesicle production. Many of the observed phenotypic differences did not correlate with subline-specific genetic changes, suggesting alterations in transcriptional and translational regulation. Taken together, these results suggest that individually maintained sublines of PAO1, even when acquired from the same parent subline, are continuously undergoing microevolution during culture and storage that results in alterations in phenotype, potentially affecting the outcomes of in vitro phenotypic analyses and in vivo pathogenesis studies.
IMPORTANCE Laboratory-adapted strains of bacteria are used throughout the world for microbiology research. These prototype strains help keep research data consistent and comparable between laboratories. However, we have observed phenotypic variability when using different strains of Pseudomonas aeruginosa PAO1, one of the major laboratory-adopted research strains. Here, we describe the genomic and phenotypic differences among 10 PAO1 strains acquired from independent sources over 15 years to understand how individual maintenance affects strain characteristics. We observed limited genomic changes but variable phenotypic changes, which may have consequences for cross-comparison of data generated using different PAO1 strains. Our research highlights the importance of limiting practices that may promote the microevolution of model strains and calls for researchers to specify the strain origin to ensure reproducibility.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative rod-shaped bacterium found ubiquitously in the environment. P. aeruginosa has remarkable metabolic versatility and can survive in aerobic, hypoxic, and anaerobic environments, making it an opportunistic pathogen for plants, animals, and humans (1, 2). Although many isolates of P. aeruginosa have been reported and described, strain PAO1 remains the collective laboratory-adapted reference strain and a common strain used for Pseudomonas research in laboratories worldwide. This strain originated from the PAO isolate (previously called “P. aeruginosa strain 1”) in Bruce Holloway’s laboratory (3, 4). PAO was isolated from a wound in Melbourne, Australia, in 1954. The PAO1 strain arose after a spontaneous mutation in the original PAO strain that yielded chloramphenicol resistance and has risen to prominence as the primary reference strain for Pseudomonas genetic and phenotypic analyses (3, 4). PAO1 was the first fully sequenced P. aeruginosa strain, with the complete genome being published in 2000 (5). Additionally, an alternate substrain of PAO1 served as the basis for the P. aeruginosa transposon mutant library developed by the Manoil laboratory at the University of Washington—Seattle (6, 7). PAO1 sublines have been distributed, maintained, and propagated worldwide. One version of PAO1 from Holloway was deposited in the American Type Culture Collection (ATCC) and is available for purchase and use (ATCC 15692; also termed 1C, ATCC 17503, ATCC 25247, ATCC 25375, CIP 104116, PRS 101, Stanier 131) (https://www.atcc.org/Products/All/15692).
Due to the persistence and genetic adaptability of P. aeruginosa, the relatedness of different laboratory-propagated and -maintained PAO1 isolates is of broad interest to the Pseudomonas field. Differences between sublines of PAO1 were observed as early as 1995, when Preston et al. reported variability in the ability of three PAO1 sublines to establish corneal infections in mice (9). More recently, an article from Klockgether et al. in 2010 (10) described that three sublines of PAO1 demonstrated variability in nutrient utilization and in vivo virulence in mice and displayed a minimum of 39 single nucleotide polymorphisms (SNPs) between them. They concluded that “the maintenance and propagation of P. aeruginosa PAO1 in laboratories throughout the world have entertained an ongoing microevolution of genotype and phenotype that jeopardizes the reproducibility of research” (10). To further evaluate this concern, we used 10 PAO1 sublines previously obtained from eight independent research laboratories in the United States and the American Type Culture Collection (ATCC 15692; the genome for this strain has already been sequenced and published) (Table 1) (11). Genomic analysis predicted a total of 5,682 genes, with 5,434 (95.63%) being identical across all 10 sublines. Only 248 genes showed any level of genetic diversity. Phenotypically, several characteristics remained comparable across all 10 sublines, including growth in rich media, biofilm formation, and human and murine Toll-like receptor 4 (TLR4) stimulation. However, differences were observed in motility (swimming and swarming), rhamnolipid and Pseudomonas quinolone signal (PQS) production, excretion of pyoverdine and pyocyanin, and outer membrane vesicle production. Interestingly, the observed phenotypic differences did not often correlate with genomic differences, suggesting that other potential differences in transcription and translation may be responsible for the observed variation.
TABLE 1.
Strain selection and genome characteristicsa
| Strain identifier | GenBank accession no. | Strain name | Source | Date acquired (mo/yr) | Genome size (bp) | % GC content | N50 |
|---|---|---|---|---|---|---|---|
| PAO1-2017-A | QZFW00000000 | H103 PAO-1 AK957 | R. Hancock, University of British Columbia | 02/2000 | 6,218,105 | 66.57 | 277,905 |
| PAO1-2017-B | QZFX00000000 | PAO-1 | J. Burns, University of Washington | 02/2000 | 6,200,122 | 66.56 | 61,263 |
| PAO1-2017-C | QZFY00000000 | PAO-1 (B. Iglewski) | E. P. Greenberg, University of Washington | 10/2000 | 6,219,177 | 66.57 | 277,884 |
| PAO1-2017-D | QZFZ00000000 | PAO-1 (B. Holloway) | D. Ohman, Virginia Commonwealth University | 05/2001 | 6,229,074 | 66.53 | 151,244 |
| PAO1-2017-E | QZGA00000000 | MPAO-1 (C. Manoil) | C. Manoil, University of Washington | 01/2003 | 6,221,774 | 66.57 | 263,062 |
| PAO1-2017-F | QZGB00000000 | PAO-1 V | J. Goldberg, University of Virginia | 07/2003 | 6,219,532 | 66.57 | 160,862 |
| PAO1-2017-G | QZGC00000000 | PAO-1 | H. Nikaido, University of California, Berkeley | 08/2003 | 6,218,750 | 66.57 | 171,105 |
| PAO1-2017-H | QZGD00000000 | PAO-1 | A. Prince, Columbia University | 11/2003 | 6,217,973 | 66.57 | 172,630 |
| PAO1-2017-I | QZGE00000000 | MPAO-1 (C. Manoil) | C. Manoil, University of Washington | 08/2008 | 6,218,510 | 66.57 | 233,461 |
| ATCC 15692 | NZ_CP017149.1 | PAO-1 | ATCC | 12/2017 | 6,276,434 | 66.50 | https://www.atcc.org/Products/All/15692 |
Each PAO1 subline was given a strain identifier based on the date that it was acquired (earliest to latest). The source represents the collaborator from which the strain was received. The names in parentheses represent the lab from which the strain originated. Strain ATCC 15692 genome data were acquired from GenBank (accession number NZ_CP017149.1). All PAO1-2017 subline accession numbers are associated with BioProject accession number PRJNA490649.
RESULTS
Genomic differences between sublines.
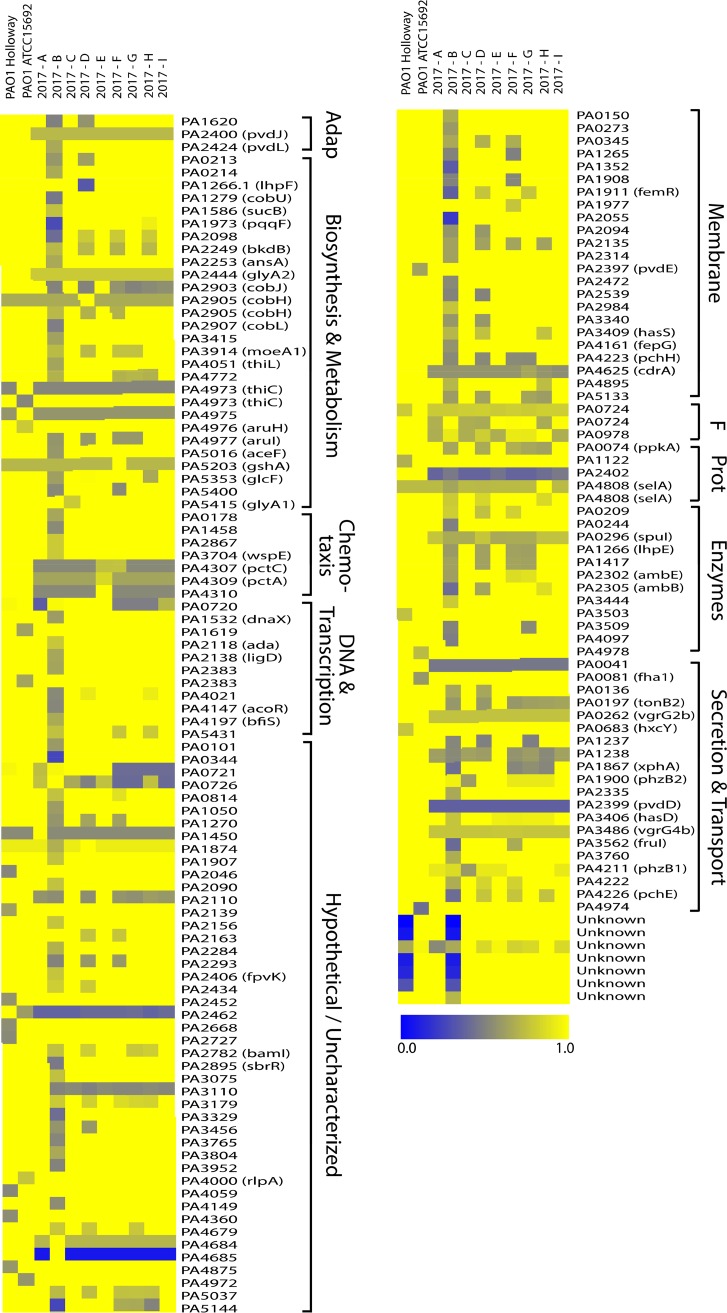
Direct comparison of the 10 PAO1 subline genome sequences revealed an average genome size of 6.2181 Mb (±0.7157 Mb; range, 6.2001 to 6.2291 Mb) and an average GC content of 66.56% (±0.00013%; range, 66.53% to 66.57%) (Table 1). Changes in genome size are likely due to the nature of the draft genomes, which may have repetitive elements that are not resolved using the chosen sequencing technology. To examine the total gene content, a large-scale BLAST score ratio (LS-BSR) analysis was conducted to determine the degree of similarity between the subline genomes (see Table S1 in the supplemental material) (12). A total of 5,682 genes were predicted, and the core genome was determined to consist of 5,434 highly similar genes (LS-BSR score, 1, representing 100% sequence identity), indicating that 95.63% of the genome is highly conserved among these 10 PAO1 sublines. A total of 248 predicted genes were identified as being variable across the sublines (4.37%), with 164 being different by >20% in at least one isolate (2.88%) (Table S2). The genes that shared 80% sequence identity or greater across all sublines were considered to be conserved and excluded from Table S2. Additionally, three gene calls were determined to be contamination and removed from the analyses (data not shown). The divergence of each predicted gene compared to the reference was visualized using a heat map representing the LS-BSR values (Fig. 1). Genes are listed by the locus tag and are loosely categorized by the PseudoCap (13) description (full PseudoCap terms for each gene call are listed in Table S2). The majority of divergent genes were identified as hypothetical genes of unknown function. Genes involved in secretion and transport showed the greatest level of diversity across all sublines. Subline 2017-B had the greatest level of gene divergence and variability among the sublines examined (Fig. 1) and had the greatest number of genes divergent from the ATCC PAO1 reference strain (Table S1). Additionally, 10 genes were not identified from the PAO1 reference strain or during manual BLAST analysis of the published PAO1 genomes (called “unknown” at the bottom of the heat map).
FIG 1.
Unique gene profiles based on large-scale BLAST score ratio (LS-BSR) analysis. All genomes underwent LS-BSR analysis to determine the presence and level of protein sequence identity of gene products. The 164 genes that were not conserved across the genomes are represented here (LS-BSR score, <0.8). Reference protein sequences were taken from the originally sequenced PAO1 strain (GenBank accession number NZ_CP017149.1). Blue represents a lower level of similarity over the length of the queried sequence (full blue indicates 0% identity), and yellow represents a higher level of similarity (full yellow indicates 100% identity). Classifications on the right are loosely based on PseudoCap identifiers. Adap, adaptation; F, foreign (phage, plasmid, transposon); Prot, protein and translation.
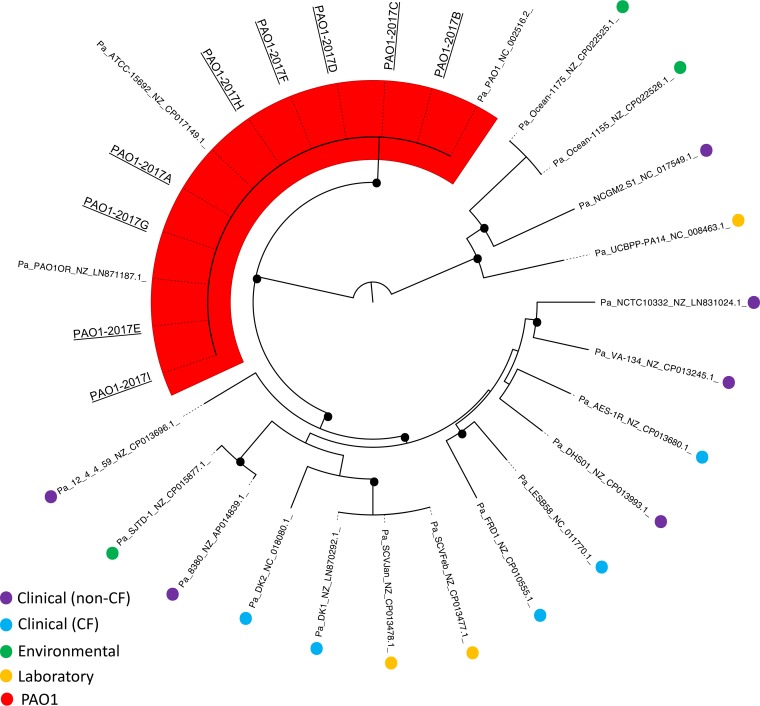
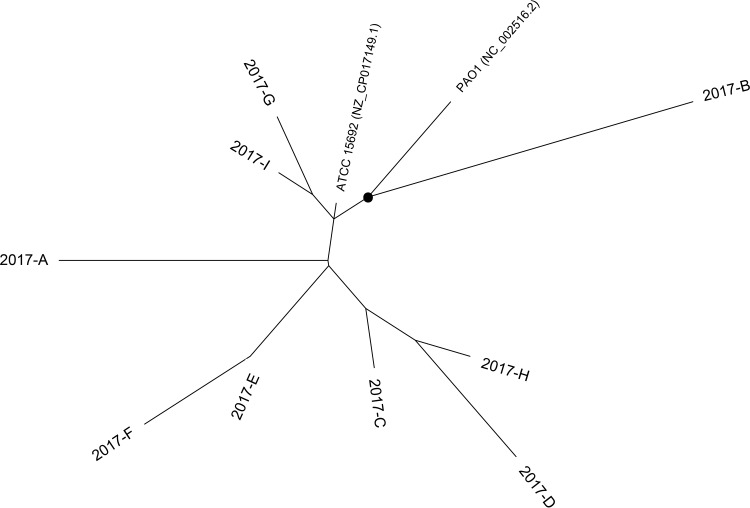
To assess how the individual PAO1 sublines fit within the current P. aeruginosa genomic landscape, we conducted phylogenomic analysis using available PAO1 and non-PAO1 whole-genome sequences (Fig. 2). Currently, there are 12 available PAO1 strain sequences: 9 generated in this study, in addition to the 3 previously published genome sequences from PAO1 Olson (GenBank accession number NC_002516.2) (5), the PAO1 ATCC 15692 strain (GenBank accession number NZ_CP017149.1) (11), and the PAO1 Orsay strain from the University of Paris Sud (GenBank accession number NZ_LN871187.1) (14). Seventeen non-PAO1 representative P. aeruginosa sequences were included in this analysis. These strains were isolated from acute infections (clinical [non-CF]), the airway of cystic fibrosis (CF) patients (clinical [CF]), the environment (environmental), and other laboratory-adapted strains (laboratory) (Fig. 2; Table S3). Within the inferred phylogeny, all PAO1 sublines clustered to a single branch, indicating that they are genomically distinct from other laboratory and nonlaboratory strains of P. aeruginosa. We conducted an additional phylogenetic analysis of just the PAO1 sublines to visualize their level of relatedness (Fig. 3). Subline 2017-B, which was observed to have the highest degree of genetic dissimilarity to the PAO1 reference strain in the LS-BSR analysis, occupied its own branch (indicated with a circle in Fig. 3). Generally, sublines did not appear to group based on the date on which they were acquired. This is not surprising, considering the unknown history of the strain before acquisition by the R. K. Ernst laboratory.
FIG 2.
Whole-genome phylogeny of 12 PAO1 sublines and 17 non-PAO1 P. aeruginosa strains. Whole-genome sequences of nine PAO1 sublines (sequenced in this study; underlined), three already-sequenced PAO1 strains, and various other non-PAO1 P. aeruginosa strains representing distinct strain types (sequence data were acquired from GenBank; see Table S3 in the supplemental material) were aligned based on regions of conserved sequence. Black dots represent a bootstrap value of >80. Shading highlights the PAO1 strains, which all occupy their own branch.
FIG 3.
Whole-genome phylogeny of P. aeruginosa PAO1 sublines. The whole-genome sequences of the nine PAO1 sublines sequenced in this study, the originally sequenced PAO1 strain (sequence data from GenBank, NC_002516.2), and ATCC 15692 (sequence data are from GenBank accession number NZ_CP017149.1) were aligned based on regions of conserved sequence. The dot represents a bootstrap value of >80.
Phenotypic characterization of PAO1 sublines.
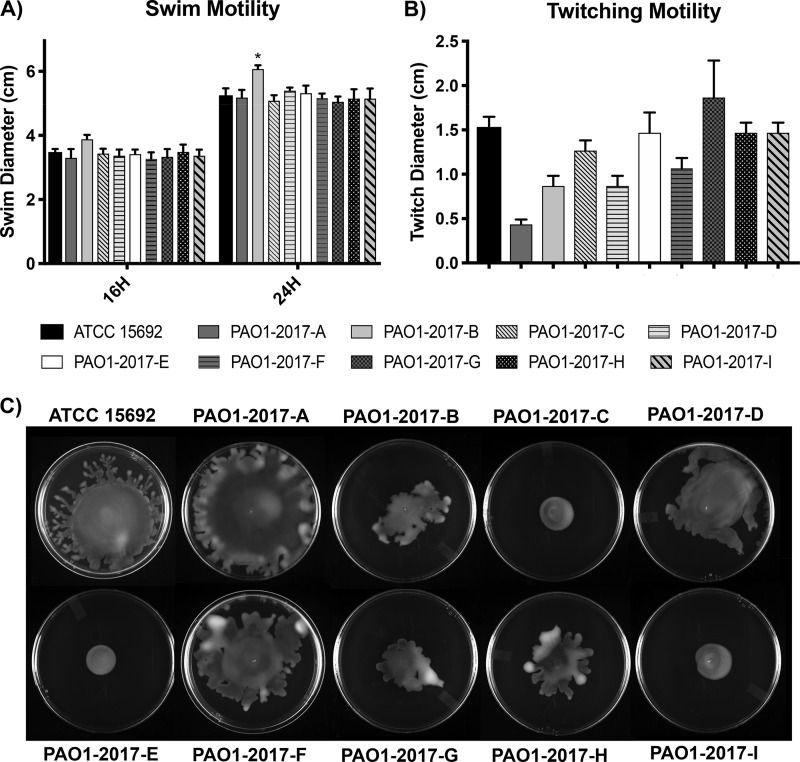
(i) Growth rates, motility, and antibiotic susceptibility. To determine potential differences in replication, the optical density (OD) of the PAO1 sublines was measured during culture in rich medium. No significant differences were observed between the strains when they were grown at both 37°C and 25°C (Fig. S1) in lysogenic broth (LB). Low-viscosity agar plates were used to measure motility (swimming, swarming, and twitching). Flagellum-mediated swimming motility (15) was consistent across the 10 sublines, with an average swim diameter of 3.4 ± 0.2 cm after 16 h (range, 3.0 to 4.0 cm) (Fig. 4A). After 24 h, strains averaged a diameter of 5.2 ± 0.1 cm (range, 4.9 to 6.2 cm), with the exception of subline 2017-B, which had a diameter significantly greater than that of the other sublines (6.1 cm, P < 0.02) (Fig. 4A; Table S8). All sublines displayed comparable profiles of twitching, which is a form of motility dependent on type IV pili (16), with the exception of strain 2017-A. This strain had a significantly decreased twitching diameter compared to all other sublines except 2017-B and 2017-D (P < 0.02) (Fig. 4B).
FIG 4.
Motility characteristics. (A) Swim motility was assayed after 16 and 24 h and is reported as swim diameter (n = 6). (B) Twitching motility was assayed after 24 h (n = 3). (C) Swarm motility was assayed after 16 h. Swarm shape appeared variable between sublines (circular swarm pattern versus tendril pattern). The observed swarming pattern was consistent across all replicates (n = 3).
Unique phenotypes were observed when assaying swarming, which is the movement across a semisolid surface requiring flagellum and biosurfactants (17). Sublines 2017-C, -E, and -I swarmed in a circular pattern with a diameter smaller than that of the other sublines. ATCC 15692 swarmed in a mostly circular pattern, with tendrils protruding from the periphery of the circle. Sublines 2017-A, -B, -D, -F, and -H collectively had more blebs and tendrils than the other strains. Subline 2017-A consistently had the largest swarm diameter, often reaching the plate edges (Fig. 4C).
It has been previously reported that sublines of PAO1 have differences in antibiotic susceptibility patterns (10). Using MIC assays, the susceptibility of the individual sublines to eight antibiotics was determined (Table 2). This analysis revealed similar MICs (<4-fold difference) for ciprofloxacin (fluoroquinolone class), rifampin (rifamycin class), ampicillin (beta-lactam class), polymyxin B (a cationic antimicrobial peptide), amikacin (aminoglycoside class), and ceftazidime (beta-lactam class). Three sublines, ATCC 15692, 2017-G, and 2017-H, showed increased resistance to erythromycin (macrolide class), and as previously shown, variability in the levels of chloramphenicol resistance was observed (10, 18). Sublines 2017-B, -C, -D, -E, -F, -G, and -I all maintained high chloramphenicol resistance, characteristic of the originally described PAO1 strain from Holloway’s lab (3, 4) (MIC, greater than 128 μg/ml), whereas 2017-H showed a decreased MIC (32 μg/ml). Several genes related to antibiotic resistance were identified from the strain genomes, including the mex genes (Table S4) (18). With the exception of mexT, the sequences of all of the identified genes were 100% conserved across the sublines. The mexT sequence had various levels of divergence between the sublines, although the mexT sequences of all strains were 88% identical or greater at the nucleotide level, suggesting that little functional differences may be observed. This is reflected in the MIC data, where little to no phenotypic variability was observed between sublines.
TABLE 2.
Antibiotic susceptibility profiles
| Strain or subline | MICa (μg/ml) of: |
|||||||
|---|---|---|---|---|---|---|---|---|
| ERYTH | AMIK | CHLOR | CIPRO | POLYB | RIF | AMP | CEFT | |
| ATCC 15692 | 128 | 1 | 64 | 0.25 | 0.5 | >128 | >128 | 2 |
| 2017-A | 64 | 0.5 | 128 | 0.25 | 0.25 | >128 | >128 | 1 |
| 2017-B | 64 | 0.5 | >128 | 0.25 | 0.5 | >128 | >128 | 4 |
| 2017-C | 64 | 0.25 | >128 | 0.25 | 1 | >128 | >128 | 1 |
| 2017-D | 64 | 0.25 | >128 | 0.25 | 0.5 | >128 | >128 | 1 |
| 2017-E | 64 | 0.25 | >128 | 0.25 | 2 | >128 | >128 | 1 |
| 2017-F | 64 | 0.5 | >128 | 0.25 | 0.25 | >128 | >128 | 2 |
| 2017-G | 128 | 0.5 | >128 | 0.25 | 0.5 | >128 | >128 | 2 |
| 2017-H | 128 | 0.5 | 32 | 0.25 | 0.5 | >128 | >128 | 0.25 |
| 2017-I | 64 | 0.25 | >128 | 0.25 | 0.25 | >128 | >128 | 1 |
MIC values were determined using a broth microtiter dish assay for erythromycin (ERYTH), amikacin (AMIK), chloramphenicol (CHLOR), ciprofloxacin (CIPRO), polymyxin B (POLYB), rifampin (RIF), ampicillin (AMP), and ceftazidime (CEFT) (n = 4).
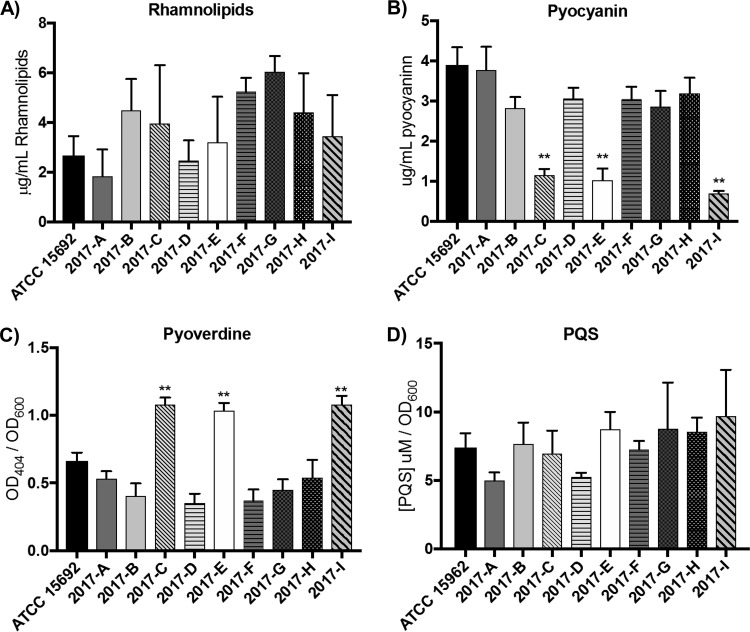
(ii) Extracellular products. P. aeruginosa secretes a number of extracellular products that aid in survival and motility. These include biosurfactants, scavenger molecules, and quorum-sensing molecules. To analyze if the profiles of these secreted substances are altered between sublines, we quantified the production of rhamnolipids, pyoverdine, pyocyanin, and the Pseudomonas quinolone signal (PQS).
Rhamnolipids are glycolipids involved in biofilm growth, antimicrobial defenses, motility, and virulence (17, 19, 20). The level of rhamnolipid production, which is dependent on the rhl operon (19, 21), ranged from 1.02 to 6.77 μg/ml, with 2017-G showing the highest level of production of all sublines (Fig. 5A). However, this increased production was significantly greater than only that of 2017-A (P < 0.05), and the differences in all other subline comparisons were nonsignificant (Table S8).
FIG 5.
Sublines have variable extracellular product profiles. (A) Rhamnolipids were extracted from 72-h cultures and measured using a standard curve (n = 3). (B, C) Pyocyanin (B) and pyoverdine (C) (both n = 6) were extracted from 24-h cultures and measured via determination of the absorbance. (D) Pseudomonas quinolone signal (PQS) production was assayed using an extraction and fluorescence technique. **, P < 0.01.
Pyocyanin is a redox-active secondary metabolite responsible for the blue-green pigmentation often observed in Pseudomonas cultures (22). Quantification of pyocyanin production demonstrated that all sublines, with the exception of subline 2017-A, had decreased levels of pyocyanin production compared to ATCC 15692 (P < 0.02) (Fig. 5B; Table S8). Additionally, sublines 2017-C, -E, and -I had significantly decreased levels of pyocyanin (sublines 2017-C, -E, and -I are all of the MPAO1 background lineage; P < 0.0001) (Table S8). Pyocyanin synthesis is dependent on the synthesis of phenazine-1-carboxylic acid (PCA), which is subsequently converted to pyocyanin (23). 2017-C showed sequence divergence in two of the PCA biosynthesis genes (phzB1 and phzB2) in LS-BSR analysis (Fig. 1), although none of the other biosynthetic genes were divergent between the strains, including pyocyanin-specific synthesis genes phzH, phzM, and phzS (Table S5). The upstream regions of the PCA biosynthesis operons were also probed and revealed no sequence divergence (500 bp upstream of phzA1 and phzA2; data not shown) (23).
Pyoverdine is a siderophore important for iron acquisition and is synthesized and transported by products of the pvd, pvc, and fpv genes (24–26). We observed that MPAO1-derived sublines 2017-C, -E, and -I produced significantly more pyoverdine than the other sublines (P < 0.0001) (Fig. 5C; Table S8). However, when analyzing the sequences of genes involved in pyoverdine synthesis (pvdADEFIJPS, pvcABCD, and fpvAIR) (25, 26), the level of sequence variability was consistent across the sublines and did not correlate with the observed phenotype differences (Table S5). Analysis of the promoter regions upstream of pvdS (the gene for the alternative sigma factor that regulates expression of the other pyoverdine biosynthesis genes) revealed no sequence dissimilarity (data not shown). Similarly, analysis of the upstream regions (500 bp) of genes targeted by pvdS regulation (pvdA, pvdE, and pvdF) showed that they were 100% identical across all sublines (data not shown) (27).
The Pseudomonas quinolone signal (PQS) is an interbacterial quorum-sensing molecule that is important for population dynamics, fitness, and infection (28). We observed variable levels of total PQS in our sublines, ranging from 4.98 μM/OD at 600 nm (OD600) to 9.70 μM/OD600, although statistical analysis revealed no significant differences between the sublines (Fig. 5D; Table S7). Genetic analysis of the PQS synthesis genes (pqsABCDE and pqsR, also called mvfR) (29) revealed 100% identity at these loci in all sublines (Table S6). Therefore, strain-specific variability in PQS induction is independent of the primary sequence of the synthesis genes and suggests that secondary regulation is occurring in these sublines.
Finally, the sequences of selected regulatory genes were analyzed to determine if some of the observed phenotypic changes could be explained by changes in global regulation factors. These included the rhl and las genes, as well as fleQ (a major flagellar regulator) and vsqM (a regulator of quorum sensing and antibiotic resistance) (30, 31). Commonly mutated genes, such as vfr and algR, were also analyzed. Interestingly, no sequence divergence from the PAO1 reference sequence was observed for any of the genes (Table S7). P. aeruginosa has many additional two-component regulatory systems that were not investigated and could be of interest for future queries.
Membrane dynamics are altered in the PAO1 sublines.
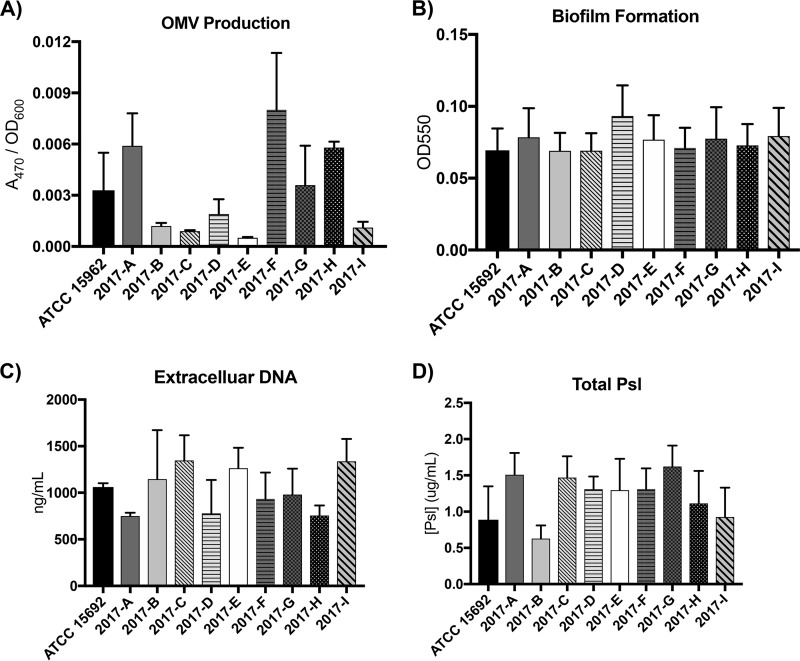
In recent years, the importance of outer membrane vesicles (OMVs) and membrane shedding has become increasingly apparent. In P. aeruginosa, OMVs are involved in quorum sensing and virulence (32, 33). We observed variable OMV production from the 10 PAO1 sublines, with a range of 0.0005 to 0.008 OD470/OD600 (Fig. 6A). Subline 2017-H had significantly less OMV production than sublines 2017-B, -C, -D, and -E; all other differences were not significant (Table S8). Variability in OMV production has been documented between different strains of the same species and between the same species grown under different environmental conditions, and these numbers are in line with previously reported data for P. aeruginosa (32, 34). Additional elements impacting OMV production could account for the variability in OMV production observed here, including the observed variable PQS production, general differences in membrane stress and fluidity, or membrane remodeling (35–38). The data presented here suggest that the intraisolate variability of these factors alters OMV production, which may play an important part in bacterial fitness in natural environments and within the host.
FIG 6.
OMV, eDNA, and Psl production is variable between sublines yet has no effect on biofilm formation. (A) OMV production was assayed and normalized to the cell density (n = 3). (B) Biofilm formation was assayed using a microtiter dish method (n = 6). (C) Extracellular DNA was quantified after extraction from a 5-ml culture (n = 6). Additional extracellular DNA measurements are in Fig. S2 in the supplemental material. (D) Total Psl was quantified as the sum of cell-free and surface-associated Psl (n = 3).
P. aeruginosa is known to grow in a biofilm both in the environment and in infectious niches, such as the airways of patients with cystic fibrosis (39, 40). High levels of extracellular DNA (eDNA) production have been linked to biofilm development and propagation (41, 42). Biofilm growth of the 10 PAO1 sublines was assayed using the O’Toole microtiter plate biofilm assay (Fig. 6B) (43). This analysis showed no significant differences in biofilm density between the sublines (Table S8). However, differences in eDNA production after 24 h of culture in rich media were observed. eDNA was assayed using two methods, due to the sample-to-sample variability observed with one method alone (an extraction and absorbance method [Fig. 6C] and a gel-based method [Fig. S2]). Sublines 2017-C, -E, and -I had increased levels of eDNA production. Sublines 2017-A and 2017-H had decreased levels, and the levels in the remaining sublines were relatively comparable to each other and to the level for ATCC 15692. While the level of variability between sublines was different between the two assay methods, the general trends held true (Fig. 6C; Fig. S2).
In addition to eDNA, P. aeruginosa biofilm formation relies on the production of other matrix components, including abundant exopolysaccharides to provide scaffolding and cohesion of the biofilm community. The exopolysaccharide Psl is critical for initial adherence and structural stability during biofilm development (44–46), as well as serves a protective function against innate immune receptors, neutrophils (47), and antimicrobial agents (48). Previous studies have shown that there are two primary forms of Psl: a high-molecular-weight form associated with the bacterial surface and a low-molecular-weight form that is released from the bacterial cells (45). Additional studies have shown that Psl is the dominant exopolysaccharide produced from the PAO1 strain of P. aeruginosa (49), with Psl production reported to be variable between strains of P. aeruginosa (50). Here, we quantified both the cell-free and surface-associated forms of Psl to determine changes to total Psl across the PAO1 sublines (Fig. 6D). Total Psl was variable across the sublines, with subline 2017-B producing the smallest amount of Psl. However, 2017-B had a significantly reduced amount of Psl only compared to that of subline 2017-G; all other differences were not significant (P < 0.05; Table S8). Interestingly, the trends in variability observed with Psl did not mirror those of eDNA or OMV production, suggesting that Psl level fluctuations are not sufficient to predict the capacity for biofilm formation.
The immunostimulatory profiles of sublines suggest similar virulence potential.
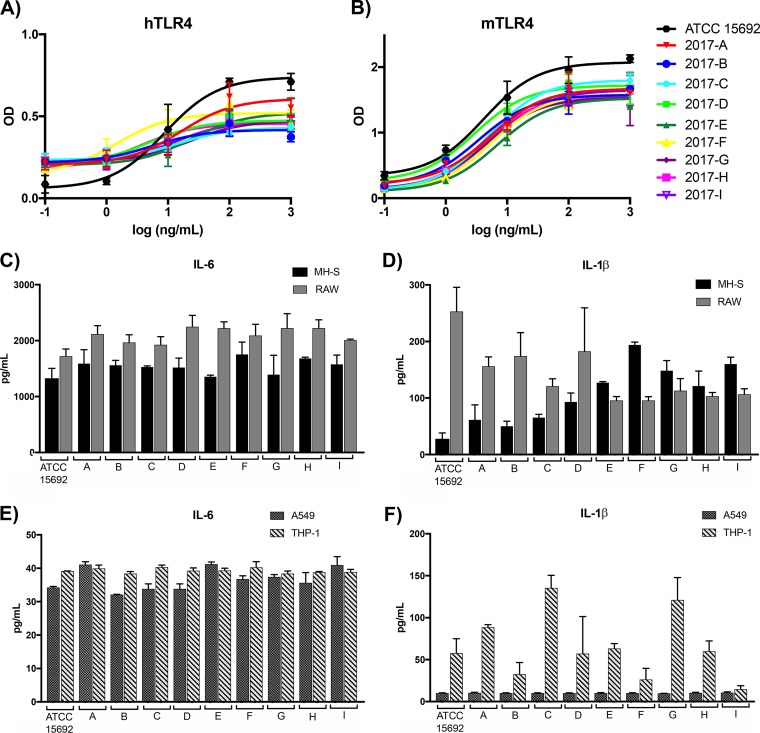
To assess virulence potential, various measures of immunostimulation were assessed. First, Toll-like receptor 4 (TLR4) complex stimulation was evaluated. TLR4 is a pattern recognition receptor (PRR) present on the surface of innate immune cells that is responsible for recognition of the lipid A portion of lipopolysaccharide (LPS), in coordination with its coreceptor, MD-2. Upon recognition, a signaling cascade is initiated that converges on the transcription factor NF-κB and subsequent upregulation of inflammatory cytokines and other immunomodulators (51). TLR4 stimulation was assessed using HEK reporter cells transfected with human or murine TLR4, with NF-κB activation as the readout. No significant differences in stimulation between the sublines were observed over a 5-log range of concentrations with either mouse or human TLR4 (Fig. 7A and B), with the exception of that with the highest concentration tested (1 μg/ml), in which the ATCC strain was significantly more stimulatory than the other sublines (P < 0.05). Unsurprisingly, the major lipid A structures, as analyzed using mass spectrometry of the sublines, were comparable (Fig. S3).
FIG 7.
Immunostimulatory potential of PAO1 sublines. (A and B) HEK reporter cells expressing human TLR4 (hTLR4) (A) or murine TLR4 (mTLR4) (B) were stimulated with a 5-log range of LPS concentrations, and NF-κB activation was recorded. (C to F) Murine cell lines MH-S and RAW 246.7 (C and D) and human cell lines A549 and THP-1 (E and F) were stimulated with 106 heat-killed whole-cell preparations of each strain. IL-6 and IL-1β were quantified from the cell supernatant using ELISA after 16 h of stimulation.
To further analyze the potential immune response to each subline, the cytokines interleukin-6 (IL-6) and IL-1β were analyzed from the supernatants of two murine and two human cell lines after stimulation with heat-killed whole cells for 16 h. In the murine cell lines MH-S (alveolar macrophages) and RAW 264.7 (macrophages), IL-6 production was comparable across sublines (Fig. 7C). IL-1β production in the same cell lines was much more variable, with ATCC 15692 inducing high levels in RAW 264.7 cells compared to the other sublines (Fig. 7D). In the human cell lines A549 (alveolar basal epithelium-derived cells) and THP-1 (monocyte-derived cells), the levels of IL-6 induction were comparable across sublines and between cell types (Fig. 7E). IL-1β production was consistent across the sublines in A549 cells but variable in THP-1 cells. Sublines 2017-C and 2017-G induced greater levels of IL-1β production, with subline 2017-I inducing the lowest level (Fig. 7F). Interestingly, subline trends of high- versus low-level inducers of IL-1β were not consistent between the murine and human cell lines.
DISCUSSION
The genetic and phenotypic diversity between PAO1 sublines has become increasingly apparent in recent years. Despite this, PAO1 remains one of the most common reference strains for P. aeruginosa research worldwide. The analysis by Klockgether et al. of three PAO1 sublines in 2010 revealed genomic and phenotypic differences (10). However, only three sublines were included, which highlights the need for a larger-scale analysis of potential genetic drift and phenotypic variability. Our analyses of 10 PAO1 sublines probed the level of baseline genomic evolution that occurs in individually maintained PAO1 strains in laboratories throughout the world. Despite observing only ∼5% of the genome being different between our PAO1 sublines, significant variability in phenotypes was observed, highlighting the variation beyond the primary genome sequence, including, but not limited to, transcriptional and translational events.
Many of the observed phenotypic differences may have effects on virulence and infection. Swarming, or the coordinated movement of bacteria across a semisolid surface, is predicted to promote infection of tissues covered with mucosal layers, such as in the lungs, which is the major site of infection in patients with cystic fibrosis. Therefore, the differential swarming phenotypes observed between our sublines could result in a differential ability to infect and colonize the lungs of model organisms used in infection studies. Pyoverdine is a siderophore that is important for scavenging iron during infection. Our data demonstrate that PAO1 sublines have variable levels of pyoverdine production in vitro, which could theoretically give certain sublines an advantage during in vivo infection. Additionally, we observed variable levels of pyocyanin production, with the 2017-C, -E, and -I sublines having significantly reduced levels of pyocyanin. Pyocyanin has been implicated in impairing host neutrophil defenses in vivo (52). Therefore, in vivo infection studies may be directly affected by the specific PAO1 strain being used. PQS production was variable between strains but, on average, was not significantly different. Differences in quorum sensing could have consequences for infection, particularly in coinfection models, although recent reports suggest that quorum sensing may be less important in human infection than under in vitro conditions (53, 54).
Membrane dynamics were observed to be variable between sublines. Variability in PQS production was observed between sublines, although the differences did not correlate with variability in OMV production. This is consistent with recent reports demonstrating that, although it is a major driver of OMV biogenesis, total PQS production is a poor predictor of OMV production between strains of the same species (32, 38). Previous studies have reported that pyocyanin promotes extracellular DNA release (55); however, in our studies, sublines that had greater levels of extracellular DNA production were somewhat correlated with lower levels of pyocyanin production. Most notably, the 2017-C, -E, and -I strains had greater levels of extracellular DNA release but significantly reduced levels of pyocyanin production. Additionally, extracellular DNA release did not correlate with biofilm formation, and all sublines had a comparable ability to form biofilms. This was unexpected, as extracellular DNA is thought to be a key component of the biofilm matrix (56). Outer membrane vesicle production likewise did not correlate with biofilm formation, extracellular DNA release, or the levels of Psl, again highlighting the complex relationships between phenotypes. Importantly, the immunostimulatory potentials of the strains seemed to be comparable, based on the TLR4 and cell line stimulation studies, although variability in IL-1β production was observed. Together, these data suggest that the inflammatory potentials of the sublines are comparable, although variance in some of the underlying mechanisms that lead to inflammation may be observed.
There were some observed genetic differences that were not followed up with phenotypic analysis. Most notably, there were several genes related to pyoverdine production and chemotaxis that were variable compared to those in the reference strain. Analysis of chemotactic potential is an interesting avenue for future studies. Chemotaxis has important roles in nutrient acquisition and metabolism and, consequently, can affect biofilm formation, pilus biosynthesis, and other features that may be important in an infectious niche (57).
Interestingly, the genomic variation observed between sublines did not consistently match with the observed phenotypic variability. Therefore, additional analysis of selected regulatory genes was performed but did not reveal any genetic changes that correlated with the observed variability in phenotype (see Table S7 in the supplemental material), suggesting that additional factors may exert a level of control over protein activity that may include gene induction, transcription, small RNA regulation, or translation. Future studies using the 10 sublines should include large-scale transcriptomic or proteomic studies to further interrogate these differences. Specifically, reverse transcription-PCR analysis of genes involved in phenotypes that had significant differences between strains, such as pyoverdine and pyocyanin production, could be used to better identify the source of the observed divergence. Regulation by small RNAs has also emerged as an important level of control for antibiotic resistance and virulence genes and could serve as another line for future investigation (58).
In conclusion, our analyses support the idea that individually maintained P. aeruginosa PAO1 sublines are undergoing a continuous microevolution that has consequences for both in vitro and in vivo genotypic and phenotypic analysis. The specifics and magnitude of these changes are variable between sublines and are not completely predictable based on sequencing alone. This highlights the need for researchers to specify the lineage of the subline used to increase transparency and reproducibility, as individual culturing and storage practices may promote microevolution. As such, for all of the studies conducted here, cultures were inoculated from the single glycerol stock that was prepared from the culture used for sequencing and were never continuously subcultured. Therefore, stringent quality controls and subline assessments, including sequencing and the use of low-passage cultures, will help ensure the reproducibility and consistency of P. aeruginosa pathogenesis research.
MATERIALS AND METHODS
Bacterial strains.
For a complete list of the strains/sublines used in this study, see Table 1. All sublines were maintained in 25% glycerol, 75% lysogenic broth (LB) at −80°C. For all experiments, cultures were inoculated directly from the stock used for sequencing without subculturing.
Preparation of genomic DNA.
Genomic DNA was isolated using a GenElute bacterial genomic DNA kit (catalog number NA2110; Sigma-Aldrich, St. Louis, MO) following the kit instructions with the following exception: all DNA was eluted in 400 μl of ultrapure diethylpyrocarbonate-treated water (Thermo Fisher Scientific, Waltham, MA). The concentration of the DNA preparations was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). All preparations were stored at −20°C.
Genome sequences.
The genomes of the nine new PAO1 sublines analyzed in this study were sequenced as previously described (59).
Phylogenomic analysis.
The genomes of the nine PAO1 sublines analyzed in this study were compared with 20 previously sequenced P. aeruginosa genomes (see Table S3 in the supplemental material) using an in silico genotyper (ISG) (60, 61). Single nucleotide polymorphisms (SNPs) were detected relative to the completed genome sequence of the reference P. aeruginosa PAO1 strain (GenBank accession number NC_002516.2) using the ISG (61), which uses the NUCmer (v.3.22) program (62) for SNP detection. The SNP sites that were identified in all genomes analyzed were concatenated and used to construct a maximum likelihood phylogeny using the RAxML (v7.2.8) program (63). The phylogeny was constructed using the GTR model of nucleotide substitution with the GAMMA model of rate heterogeneity and 100 bootstrap replicates. The phylogeny was then visualized using the FigTree (v1.4.2) program (http://tree.bio.ed.ac.uk/software/figtree/).
LS-BSR analysis.
The genomes of the 10 sublines were compared using the large-scale BLAST score ratio (LS-BSR) as previously described (Table 1) (12, 59). The predicted protein-encoding genes of each genome that had ≥80% nucleotide sequence identity to each other were assigned to gene clusters using the uclust algorithm (64). Representative sequences of each gene cluster were then compared to the sequence of each genome using the TBLASTN program (65) with the composition-based adjustment turned off, and the TBLASTN scores were used to generate a BLAST score ratio (BSR) value indicating the detection of each gene cluster in each of the genomes (Table S1). The BSR value was determined by dividing the score of a gene compared to a genome sequence by the score of the gene compared to its own sequence. The LS-BSR values and the nucleotide sequences of each gene cluster for the strains are included in the supplemental material (Tables S1 and S4 to S7). Heat maps of the LS-BSR values of the predicted genes were generated using the multiple experiment viewer software MeV (Table S2) (66).
Motility analysis.
Swimming was assessed using 0.3% agar plates (per 1 liter, 3 g Bacto agar, 5 g proteose peptone, 3 g yeast extract). Overnight cultures (37°C; LB, 1 mM MgCl2; n = 6 per subline) were used to inoculate swim plates by depositing 2.5 μl of culture directly into the agar in the center of the plate. Plates were incubated face up at 37°C, and the swim diameter (in centimeters) was recorded at 16 and 24 h. Twitching was assayed using the macroscopic method described by Turnbull and Whitchurch (16). Briefly, a sterile toothpick was used to inoculate 1% Lennox LB plates (n = 3 per subline). The plates were inverted and kept in a humidified chamber at 37°C. Twitching (in centimeters) was measured after 24 h. Swarming was assessed using 0.5% agar plates (per 1 liter, 5 g Bacto agar, 5 g proteose peptone, 3 g yeast extract). Plates were poured at 30 ml/plate and allowed to solidify at room temperature for 5 h. Overnight cultures of each subline (37°C; LB, 1 mM MgCl2; n = 3 per subline) were used to inoculate the swarm plates by spotting 2.5 μl of culture directly in the center of the plate. The plates were incubated face up at 37°C for 16 h and imaged (FluorChem 8900; Alpha InnoTech, San Leandro, CA).
Pyoverdine quantification.
Pyoverdine production was quantified as previously described (67). Briefly, an individual colony was used to inoculate 5-ml cultures in King’s B medium (per 1 liter, 10 g proteose peptone and 15 ml glycerol; after sterilization, 1.5 g K2HPO4 and 5 ml 1 M MgSO4) (68). Cultures were incubated at 37°C for 24 h. The OD600 was recorded using a DU730 UV/visible spectrophotometer (Beckman Coulter, Brea, CA). Cultures were centrifuged (5,000 × g, 10 min), and the OD404 of the cell-free supernatant was recorded using King’s B medium as a blank. Data are represented as the relative pyoverdine concentration (OD404/OD600) and as the average for six independent cultures per subline.
Pyocyanin quantification.
Pyocyanin production was quantified as previously described (69). Briefly, an individual colony was used to inoculate 5-ml cultures in King’s A medium (per 1 liter, 20 g proteose peptone; after sterilization, 1.4 g MgCl2 and 10 g K2SO4) (68). Cultures were incubated at 37°C for 24 h. The OD600 was recorded using a DU730 UV/visible spectrophotometer (Beckman Coulter, Brea, CA). The cultures were centrifuged (5,000 × g, 10 min), and the supernatant was transferred to a glass tube with 3 ml chloroform. The chloroform mixture was incubated at room temperature with rocking for 1 h and then centrifuged at 1,000 × g for 5 min to allow for phase separation. The organic phase was extracted to a new glass tube, and 1 ml of 0.2 N HCl was added. Samples were vortexed for 10 s, and the aqueous phase was transferred to a microcuvette. The OD520 was recorded using 0.2 N HCl as a blank. All data are represented as the relative pyocyanin concentration (OD520/OD600) and as the average for six independent cultures per subline.
Rhamnolipid quantification.
The sublines were inoculated into 30-ml cultures of autoinducer bioassay (AB) medium supplemented with 2.5 mg/liter thiamine and 0.2% glucose and statically incubated for 72 h at 37°C (n = 3). Rhamnolipids from cell-free supernatants were assayed as previously described (70). A rhamnolipid standard was purchased from Agae Technologies (catalog number R90; Corvallis, OR).
PQS extraction and quantification.
PQS was extracted from a whole culture with a 1:1, 1:2, or 1:3 addition of acidified ethyl acetate (0.1 ml/liter acetic acid). The organic phase was removed and dried under nitrogen gas. Dried samples were resuspended in 100% methanol (Optima grade; Fisher Scientific, Hampton, NH), and 20-µl specimens were spotted onto a straight-phase phosphate-impregnated thin-layer chromatography plate (EMD MilliporeSigma, Burlington, MA) which had been activated for 1 h at 100°C. The mobile phase was 95:5 dichloromethane-methanol. PQS was visualized by intrinsic fluorescence after excitation under long-wave UV light. Digital images were captured and analyzed using the UVP, Inc., Gel Doc-It2 imaging system and its densitometry software. The concentrations of PQS were calculated by comparison to known standard-concentration preparations that were run concurrently with whole-culture extracts.
Culture growth conditions and isolation of outer membrane vesicles.
Cultures were inoculated into 25 ml fresh defined growth medium from overnight cultures at an OD600 of 0.01 and grown to early stationary phase. The cells were pelleted at 15,000 × g for 15 min at 4°C using a Sorvall Legend XTR centrifuge F15 rotor (Thermo Fisher Scientific, Waltham, MA). The supernatant was collected and filtered through a 0.45-μm-pore-size filter. To isolate OMVs, 20 ml of supernatant was subjected to ultracentrifugation (Thermo Scientific S50-A rotor; Thermo Fisher Scientific, Waltham, MA) at 209,438 × g for 1.5 h at 4°C. Pelleted vesicles were resuspended in 500 μl of phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 2 mM KH2PO4) and kept at 4°C until analysis.
Lipid assay for OMV quantification.
Lipid quantification was determined via extrapolation of the method developed by Stewart (71). OMV preparations were lipid extracted with 1:1, 1:2, or 1:3 chloroform. The chloroform layer was retained and exposed to 1:1 aqueous ammonium ferrothiocyanate to stain the lipid. Two hundred microliters of the resulting chloroform layer was aliquoted into a quartz cuvette, and the absorbance of the solution at an λ of 470 nm was measured.
Extracellular DNA quantification.
Extracellular DNA was assayed from cell-free supernatant (n = 6) using NaCl-ethanol precipitation as previously described (72). DNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Biofilm assay.
Biofilm formation was assayed as previously described (43). Briefly, overnight cultures (LB, 1 mM MgCl2; 37°C; n = 2 per subline) were diluted 1:100 in 1× M63 minimal medium. One hundred microliters of the dilution was plated in technical triplicate in a round-bottom, untreated, sterile 96-well plate (CellTreat Scientific Products, Pepperell, MA) and statically incubated at 37°C for 16 h. The plate was washed with endotoxin-free water and dyed with 125 μl 0.1% crystal violet for 15 min. The plate was washed again, and the remaining dye was solubilized with 125 μl 30% acetic acid in water. One hundred microliters of the crystal violet-acetic acid solution was transferred to a new 96-well plate, and the absorbance at 550 nm was measured using a multimode detector (catalog number DTX-880; Beckman Coulter, Brea, CA). The results for a blank control consisting of wells with medium only were subtracted and represent the average for all biological and technical replicates.
Psl polysaccharide extraction and immunoblotting.
Strains were inoculated into lysogenic broth without salt (LBNS) and grown overnight at 37°C and 200 rpm. Cultures were normalized to an OD A600 of 5.0, and polysaccharides were extracted as described by Chiba et al. (73). Briefly, bacterial cells were centrifuged at 8,000 × g for 10 min at room temperature. The supernatants were collected, lyophilized, and resuspended in 1 ml of sterile water. This material is the cell-free Psl extract. The bacterial pellets were suspended in 1 ml of 1.5 M NaCl, vortexed vigorously, and placed on an orbital rocker for 15 min at room temperature. The samples were centrifuged at 5,000 × g for 10 min. The supernatants were harvested; this material is the surface-associated (SA) Psl. For immunoblotting, 2 μl of each polysaccharide extract was spotted onto a 0.45-μm-pore-size nitrocellulose membrane (Bio-Rad, Hercules, CA) and allowed to air dry. The membrane was blocked with 5% nonfat dry milk (NFDM) in Tris-buffered saline–Tween 20 (TBST; 20 mM Tris, 137 mM NaCl, 0.1% Tween 20, pH 7.6), followed by probing with an anti-Psl polyclonal antibody mixture diluted 1:3,000 in NFDM-TBST (kindly provided by MedImmune, LLC) (74). After the blot was washed with TBST, the secondary antibody, horseradish peroxidase-conjugated goat anti-human whole IgG monoclonal antibody (Abcam, Cambridge, MA), was added at a dilution of 1:5,000 in NFDM-TBST. The membrane was washed with TBST, and the Amersham ECL Prime Western blotting detection agent (GE Healthcare Biosciences, Pittsburgh, PA) was used for detection of the polysaccharides. Images were visualized using a ChemiDoc XRS imaging system (Bio-Rad, Hercules, CA), and densitometry was performed using open-source Fiji ImageJ software. The densitometry values for the cell-free and SA extracts were combined to give the total Psl quantity for each subline.
LPS purification, TLR4 stimulation, and lipid A analysis.
LPS was prepared from 1 liter of culture grown at 37°C in LB (with 1 mM MgCl2) using a phenol-water extraction followed by removal of contaminating phospholipids, as previously described (75–77). TLR4 stimulation was conducted as previously described (76). Lipid A was prepared from the LPS preparations using mild acid hydrolysis as previously described (78). Lipid A was analyzed on a Bruker microFlex mass spectrometer (Bruker, Billerica, MA) using 10 mg/ml Norharmane matrix (Sigma-Aldrich, St. Louis, MO) in 2:1 (vol/vol) chloroform-methanol in the negative-ion mode. Data were processed in Bruker flexAnalysis software.
Cell culture stimulation.
Cell cultures were maintained at 37°C with 5% CO2, and all cultures were passaged less than 10 times. The RAW 264.7 and MH-S cell lines were maintained in Dulbecco modified Eagle medium (with 5% penicillin-streptomycin [PenStrep], 10% fetal bovine serum, 4.5 g/liter d-glucose and l-glutamine; Gibco, Gaithersburg, MD). The A549 and THP-1 cell lines were maintained in RPMI medium (5% PenStrep, 10% fetal bovine serum, 4.5 g/liter d-glucose and l-glutamine; Corning, Corning, NY). Cells were seeded to a 96-well tissue culture-treated flat-bottom plate (100,000 cells/well) and allowed to adhere for 48 h. To prepare the stimulating dose, overnight cultures of each subline (grown in LB with 1 mM MgCl2) were subcultured 1:50 and incubated for 3 h (two biological duplicates per strain). Doses were diluted so that the final volume contained 106 bacteria in sterile phosphate-buffered saline (PBS) using a linear calibration curve based on optical density (OD600). The doses were heat inactivated in 1 ml PBS at 65°C for 1 h, and heat inactivation was confirmed by plating 20 μl of each dose. The doses were centrifuged (8,000 × g, 10 min), PBS was aspirated, and the doses were resuspended in the appropriate medium (260 μl total; each tube was prepared with 13 times the required dose to ensure that the same dose could be used for all cell lines). For stimulation, the medium was aspirated and 180 μl of fresh medium was added. Twenty microliters of each strain dose (in biological duplicate and technical triplicate) was added. An equivalent number of unstimulated control cells were also included. The cells were centrifuged at 1,000 × g for 5 min to ensure bacterial deposition and incubated at 37°C with 5% CO2 for 16 h. The plates were promptly frozen until analysis via enzyme-linked immunosorbent assay (ELISA). ELISA analysis was conducted using a DuoSet ELISA development system (R&D Systems, Minneapolis, MN) of the following types: mouse IL-6, mouse IL-1β, human IL-6, and human IL-1β. The developed plates were read at 450 nm on a multimode detector (model DTX-880; Beckman Coulter, Brea, CA). All data are reported as averages.
Data analysis.
Data are expressed as means ± standard deviations. Significance was determined using one-way analysis of variance with multiple comparisons in GraphPad Prism (version 7.03) software. A P value of ≤0.05 was considered significant. The results of all statistical analyses are available in Table S8.
Accession number(s).
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession numbers listed in Table 1. The BioProject accession number for all PAO1 2017 samples is PRJNA490649.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge and thank all of the researchers that have sent us PAO1 strains through the years: Robert Hancock (University of British Columbia), Jane Burns (University of Washington), E. Pete Greenberg (University of Washington), Dennis Ohman (Virginia Commonwealth University), Collin Manoil (University of Washington), Joanna Goldberg (University of Virginia), Hiroshi Nikaido (University of California, Berkeley), and Alice Prince (Columbia University). We thank Tracy Hazen for her assistance with the sequencing and genomic analyses.
The genomic data for the Pseudomonas aeruginosa isolates were generated by National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services, grant number U19 AI110820.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00595-18.
REFERENCES
- 1.Savoia D. 2014. New perspectives in the management of Pseudomonas aeruginosa infections. Future Microbiol 9:917–928. doi: 10.2217/fmb.14.42. [DOI] [PubMed] [Google Scholar]
- 2.Gellatly SL, Hancock REW. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 3.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. doi: 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 4.Holloway BW, Morgan AF. 1986. Genome organization in Pseudomonas. Annu Rev Microbiol 40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- 5.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FSL, Hancock REW. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res 15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Preston MJ, Fleiszig SM, Zaidi TS, Goldberg JB, Shortridge VD, Vasil ML, Pier GB. 1995. Rapid and sensitive method for evaluating Pseudomonas aeruginosa virulence factors during corneal infections in mice. Infect Immun 63:3497–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schöck U, Pohl TM, Wiehlmann L, Tümmler B. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahl JW, Caporaso JG, Rasko DA, Keim P. 2014. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winsor GL, Lo R, Ho Sui SJ, Ung KSE, Huang S, Cheng D, Ching W-KH, Hancock REW, Brinkman FSL. 2004. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res 33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergnaud G, Hauck Y, Midoux C, Pourcel C, Latino L. 2016. Pseudolysogeny and sequential mutations build multiresistance to virulent bacteriophages in Pseudomonas aeruginosa. Microbiology 162:748–763. doi: 10.1099/mic.0.000263. [DOI] [PubMed] [Google Scholar]
- 15.Ha D-G, Kuchma SL, O'Toole GA. 2014. Plate-based assay for swimming motility in Pseudomonas aeruginosa. Methods Mol Biol 1149:59–65. doi: 10.1007/978-1-4939-0473-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbull L, Whitchurch CB. 2014. Motility assay: twitching motility. Methods Mol Biol 1149:73–86. doi: 10.1007/978-1-4939-0473-0_9. [DOI] [PubMed] [Google Scholar]
- 17.Caiazza NC, Shanks RMQ, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morita Y, Kimura N, Mima T, Mizushima T, Tsuchiya T. 2001. Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J Gen Appl Microbiol 47:27–32. [DOI] [PubMed] [Google Scholar]
- 19.Ochsner UA, Reiser J, Fiechter A, Witholt B. 1995. Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl Environ Microbiol 61:3503–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soberón-Chávez G, Lépine F, Déziel E. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 21.Reis RS, Pereira AG, Neves BC, Freire DMG. 2011. Gene regulation of rhamnolipid production in Pseudomonas aeruginosa—a review. Bioresour Technol 102:6377–6384. doi: 10.1016/j.biortech.2011.03.074. [DOI] [PubMed] [Google Scholar]
- 22.Lau GW, Hassett DJ, Ran H, Kong F. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schalk IJ, Guillon L. 2013. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ Microbiol 15:1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 25.Stintzi A, Cornelis P, Hohnadel D, Meyer JM, Dean C, Poole K, Kourambas S, Krishnapillai V. 1996. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology 142:1181–1190. doi: 10.1099/13500872-142-5-1181. [DOI] [PubMed] [Google Scholar]
- 26.Lamont IL, Martin LW. 2003. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833–842. doi: 10.1099/mic.0.26085-0. [DOI] [PubMed] [Google Scholar]
- 27.Loubens I, Peng W-T, Storey DG, Hunt TA. 2002. The Pseudomonas aeruginosa alternative sigma factor PvdS controls exotoxin A expression and is expressed in lung infections associated with cystic fibrosis. Microbiology 148:3183–3193. doi: 10.1099/00221287-148-10-3183. [DOI] [PubMed] [Google Scholar]
- 28.Häussler S, Becker T. 2008. The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4:e1000166. doi: 10.1371/journal.ppat.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol 187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H, Deng X, Li X, Ye Y, Wu M. 2014. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res 42:10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dasgupta N, Ferrell EP, Kanack KJ, West SEH, Ramphal R. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J Bacteriol 184:5240–5250. doi: 10.1128/JB.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florez C, Raab JE, Cooke AC, Schertzer JW. 2017. Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 8:e01034-17. doi: 10.1128/mBio.01034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis TN, Leiman SA, Kuehn MJ. 2010. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect Immun 78:3822–3831. doi: 10.1128/IAI.00433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chuanchuen R, Murata T, Gotoh N, Schweizer HP. 2005. Substrate-dependent utilization of OprM or OpmH by the Pseudomonas aeruginosa MexJK efflux pump. Antimicrob Agents Chemother 49:2133–2136. doi: 10.1128/AAC.49.5.2133-2136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 36.Schertzer JW, Whiteley M. 2012. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3:e00297-11. doi: 10.1128/mBio.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO, Klug L, Gadermaier B, Weinzerl K, Prassl R, Lass A, Daum G, Reidl J, Feldman MF, Schild S. 2016. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. doi: 10.1038/ncomms10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horspool AM, Schertzer JW. 2018. Reciprocal cross-species induction of outer membrane vesicle biogenesis via secreted factors. Sci Rep 8:9873. doi: 10.1038/s41598-018-28042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee K, Yoon SS. 2017. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J Microbiol Biotechnol 27:1053–1064. doi: 10.4014/jmb.1611.11056. [DOI] [PubMed] [Google Scholar]
- 40.Høiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 41.Tang L, Schramm A, Neu TR, Revsbech NP, Meyer RL. 2013. Extracellular DNA in adhesion and biofilm formation of four environmental isolates: a quantitative study. FEMS Microbiol Ecol 86:394–403. doi: 10.1111/1574-6941.12168. [DOI] [PubMed] [Google Scholar]
- 42.Das T, Ibugo AI, Klare W, Manefield M. 2016. Role of pyocyanin and extracellular DNA in facilitating Pseudomonas aeruginosa biofilm formation, p 1072–1078. In Dhanasekaran D, Thajuddin N (ed), Microbial biofilms—importance and applications. InTechOpen, London, United Kingdom. doi: 10.5772/63497. [DOI] [Google Scholar]
- 43.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp 2011:2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. 2012. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol 14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. 2013. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. 2012. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman L, Kolter R. 2003. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 51.Chandler CE, Ernst RK. 2017. Bacterial lipids: powerful modifiers of the innate immune response. F1000Res 6:1334. doi: 10.12688/f1000research.11388.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen L, Dockrell DH, Pattery T, Lee DG, Cornelis P, Hellewell PG, Whyte MKB. 2005. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J Immunol 174:3643–3649. doi: 10.4049/jimmunol.174.6.3643. [DOI] [PubMed] [Google Scholar]
- 53.Cornforth DM, Dees JL, Ibberson CB, Huse HK, Mathiesen IH, Kirketerp-Møller K, Wolcott RD, Rumbaugh KP, Bjarnsholt T, Whiteley M. 2018. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A 11:E5125–E5134. doi: 10.1073/pnas.1717525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y-C, Chan K-G, Chang C-Y. 2015. Modulation of host biology by Pseudomonas aeruginosa quorum sensing signal molecules: messengers or traitors. Front Microbiol 6:1226. doi: 10.3389/fmicb.2015.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das T, Manefield M. 2012. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One 7:e46718. doi: 10.1371/journal.pone.0046718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuxman Bass JI, Russo DM, Gabelloni ML, Geffner JR, Giordano M, Catalano M, Zorreguieta A, Trevani AS. 2010. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. J Immunol 184:6386–6395. doi: 10.4049/jimmunol.0901640. [DOI] [PubMed] [Google Scholar]
- 57.Kato J, Kim H-E, Takiguchi N, Kuroda A, Ohtake H. 2008. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y-F, Han K, Chandler CE, Tjaden B, Ernst RK, Lory S. 2017. Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol Microbiol 106:919–937. doi: 10.1111/mmi.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hazen TH, Donnenberg MS, Panchalingam S, Antonio M, Hossain A, Mandomando I, Ochieng JB, Ramamurthy T, Tamboura B, Qureshi S, Quadri F, Zaidi A, Kotloff KL, Levine MM, Barry EM, Kaper JB, Rasko DA, Nataro JP. 2016. Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat Microbiol 1:15014. doi: 10.1038/nmicrobiol.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hazen TH, Kaper JB, Nataro JP, Rasko DA. 2015. Comparative genomics provides insight into the diversity of the attaching and effacing Escherichia coli virulence plasmids. Infect Immun 83:4103–4117. doi: 10.1128/IAI.00769-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahl JW, Beckstrom-Sternberg SM, Babic-Sternberg J, Gillece JD, Hepp CM, Auerbach RK, Tembe W, Wagner DM, Keim PS, Pearson T. 2015. The In Silico Genotyper (ISG): an open-source pipeline to rapidly identify and annotate nucleotide variants for comparative genomics applications. bioRxiv 015578. doi: 10.1101/015578. [DOI]
- 62.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets Curr Protoc Bioinformatics Chapter 10:Unit 10.3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 63.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 64.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 65.Gertz EM, Yu Y-K, Agarwala R, Schäffer AA, Altschul SF. 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol 4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Methods Enzymol 411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 67.Gallagher LA, Manoil C. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol 183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 69.El-Fouly MZ, Sharaf AM, Shahin AAM, El-Bialy HA, Omara AMA. 2015. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J Radiat Res Appl Sci 8:36–48. doi: 10.1016/j.jrras.2014.10.007. [DOI] [Google Scholar]
- 70.Pinzon NM, Ju L-K. 2009. Analysis of rhamnolipid biosurfactants by methylene blue complexation. Appl Microbiol Biotechnol 82:975–981. doi: 10.1007/s00253-009-1896-9. [DOI] [PubMed] [Google Scholar]
- 71.Stewart JC. 1980. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem 104:10–14. [DOI] [PubMed] [Google Scholar]
- 72.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 73.Chiba A, Sugimoto S, Sato F, Hori S, Mizunoe Y. 2015. A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb Biotechnol 8:392–403. doi: 10.1111/1751-7915.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DiGiandomenico A, Warrener P, Hamilton M, Guillard S, Ravn P, Minter R, Camara MM, Venkatraman V, MacGill RS, Lin J, Wang Q, Keller AE, Bonnell JC, Tomich M, Jermutus L, McCarthy MP, Melnick DA, Suzich JA, Stover CK. 2012. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med 209:1273–1287. doi: 10.1084/jem.20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Apicella MA. 2008. Isolation and characterization of lipopolysaccharides. Methods Mol Biol 431:3–13. [DOI] [PubMed] [Google Scholar]
- 76.Gregg KA, Harberts E, Gardner FM, Pelletier MR, Cayatte C, Yu L, McCarthy MP, Marshall JD, Ernst RK. 2017. Rationally designed TLR4 ligands for vaccine adjuvant discovery. mBio 8:e00492-17. doi: 10.1128/mBio.00492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J Immunol 165:618–622. [DOI] [PubMed] [Google Scholar]
- 78.Rosner MR, Tang J, Barzilay I, Khorana HG. 1979. Structure of the lipopolysaccharide from an Escherichia coli heptose-less mutant. I. Chemical degradations and identification of products. J Biol Chem 254:5906–5917. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.