Estrogen-related receptors (ERRs) have emerged as major metabolic regulators in various tissues. However, their expression and function in the vasculature remains unknown.
KEYWORDS: angiogenesis, endothelial cell, estrogen-related receptors, nuclear receptor, regulation of gene expression
ABSTRACT
Estrogen-related receptors (ERRs) have emerged as major metabolic regulators in various tissues. However, their expression and function in the vasculature remains unknown. Here, we report the transcriptional program and cellular function of ERRα in endothelial cells (ECs), a cell type with a multifaceted role in vasculature. Of the three ERR subtypes, ECs exclusively express ERRα. Gene expression profiling of ECs lacking ERRα revealed that ERRα predominantly acts as a transcriptional repressor, targeting genes linked with angiogenesis, cell migration, and cell adhesion. ERRα-deficient ECs exhibit decreased proliferation but increased migration and tube formation. ERRα depletion increased basal as well as vascular endothelial growth factor A (VEGFA)- and ANG1/2-stimulated angiogenic sprouting in endothelial spheroids. Moreover, retinal angiogenesis is enhanced in ERRα knockout mice compared to that in wild-type mice. Surprisingly, ERRα is dispensable for the regulation of its classic targets, such as metabolism, mitochondrial biogenesis, and cellular respiration in the ECs. ERRα is enriched at the promoters of angiogenic, migratory, and cell adhesion genes. Further, VEGFA increased ERRα recruitment to angiogenesis-associated genes and simultaneously decreased their expression. Despite increasing its gene occupancy, proangiogenic stimuli decrease ERRα expression in ECs. Our work shows that endothelial ERRα plays a repressive role in angiogenesis and potentially fine-tunes growth factor-mediated angiogenesis.
INTRODUCTION
Endothelial cells (ECs) are the building blocks of blood vessels, which continue to play an important role in the adult vasculature. These cells are the site of a wide range of cellular activities, including regulation of barrier function, nitric oxide synthesis, and inflammation (1–5). They are directly involved in vascular health, tone, and growth, and endothelial dysfunction plays a causative role in atherosclerosis, hypertension, and vascular complications. Angiogenesis, which involves proliferation, migration, and differentiation of ECs, is a central phenomenon in blood vessel growth. It is important during development of vascular system and tissue vascularization, and its impairment can contribute to various vascular complications, such as myopathies, retinopathies, and peripheral vascular disease. Therefore, there is an urgent need to understand the molecular regulation of angiogenesis, which could ultimately facilitate therapeutic development.
Several signaling pathways (e.g., fibroblast growth factor, transforming growth factor beta, and angiopoietins) have been described in the process of angiogenesis (6–8), most prominent of which is the vascular endothelial growth factor A (VEGFA)-NOTCH signaling pathway (9, 10). This pathway plays an important role in activation of quiescent ECs and commitment of these cells to tip or stalk cell fate. Activation of ECs via the VEGFA-VEGFR2 pathway results in the induction of DLL4, and these cells acquire the tip cell phenotype. Tip cells are characterized by their high migratory capacity, low proliferative capacity, and presence of filopodia and typically form the leading edge of the sprout during vessel morphogenesis. DLL4 expressed on tip cells acts on adjacent ECs to activate Notch signaling, repress VEGFR2 expression, and transform these cells to stalk cells, which have fewer filopodia, are proliferative, and contribute to blood vessel growth. The stalk cells also express Jagged 1, which is another Notch ligand that reciprocally acts on tip cells to antagonize DLL4-NOTCH signaling. Tip versus stalk cell phenotype is dynamic and interchangeable until angiogenesis is complete. In addition to growth factors, transcription regulators are also involved in tight control of endothelial function during both development and adulthood. For example, developmental factors such as Ets, Sox, Hif, Gata, and Coup-tf2 transcriptionally regulate endothelial cell specification (5, 6, 11–14). Despite these key advances in cell surface and transcriptional signaling, endothelial molecular pathways and their interactions with growth factors in the regulation of angiogenesis still remain incompletely understood.
Classical steroid nuclear receptors such as estrogen, progesterone, testosterone, and aldosterone receptors play a critical role in vascular physiology (15, 16). For example, estrogen receptors are involved in angiogenesis, vascular tone, and atheroprotection in endothelial and/or vascular smooth-muscle cells (17–22). Progesterone receptors are negative regulators of angiogenesis, whereas androgen receptors promote ischemic reperfusion and are atheroprotective (23–27). Glucocorticoids and mineralocorticoids regulate vascular tone and negatively regulate angiogenesis by acting on vascular smooth-muscle cells and ECs (28–31). While the steroid receptors have been extensively studied, only a few nonsteroid orphan receptors have known functions in the vasculature (32, 33). For example, peroxisome proliferator activator receptors (PPARs) have been implicated in angiogenesis and vascular development (34). PPARγ inhibits migration and capillary formation in ECs and also contributes to the maintenance of vascular tone via nitric oxide synthesis (35). Nurr/NOR receptors (nuclear receptor-related/neuron-derived orphan receptor) are involved in the atherogenic response and positively modulate angiogenesis in ECs while inhibiting smooth-muscle cell proliferation (36). Coup-Tf2 (chicken ovalbumin upstream promoter transcriptional factor 2) is critical for vascular development and determining venous endothelial cell fate (11, 37). These studies highlight a critical role for nuclear receptors in fine-tuning vascular growth and function, warranting investigations on the other members in this superfamily.
One such subfamily is the estrogen-related receptors (ERRs), which include ERRα, ERRβ, and ERRγ (38, 39). ERRα and ERRγ are enriched in highly metabolic tissues, such as skeletal and cardiac muscle, adipose tissue, and the kidney (40). ERRβ is more selectively expressed in embryonic cells and in adult retina and kidney. ERRs have emerged as master regulators of mitochondrial biogenesis, oxidative metabolism, lipid metabolism, gluconeogenesis, and thermogenesis in the aforementioned tissues (40–50). ERRs are also involved in immune responses by regulating effector T cell maturation and gamma interferon-induced reactive oxygen species production (51, 52). Interestingly, ERRs are involved in paracrine regulation of angiogenesis by promoting Vegfa and other angiogenic factor expression in tissues, such as skeletal muscle and hepatocytes, as well as in breast cancer and monocytic leukemia cells (46, 53–55). The dynamic role of ERRs as regulators of metabolism, angiogenesis, and immune response genes in peripheral tissues raises the likelihood that these receptors serve a critical role in the vasculature.
As a step toward deciphering the role of ERRs in the vasculature, we have investigated ERR subtype expression and transcriptional function in ECs. We report that ECs exclusively express ERRα, where this receptor controls an angiogenesis-associated gene program and acts as a negative regulator of angiogenesis. Our work provides insight into the role of ERRs in vascular cells, warranting broader investigations on ERRs in the vasculature and related diseases.
RESULTS
Endothelial ERRα controls an angiogenesis-linked gene program.
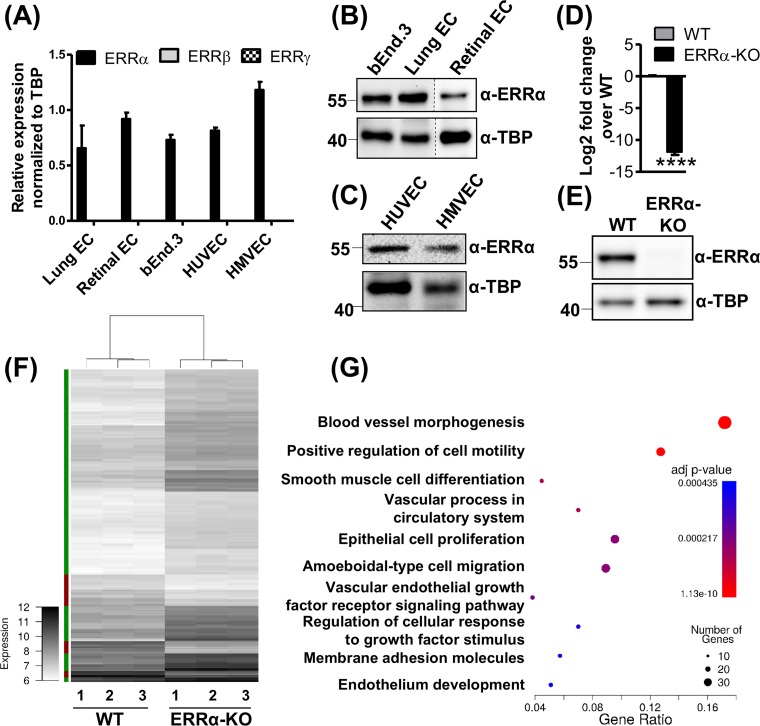
We first measured the expression of the three subtypes of ERRs (ERRα, ERRβ, and ERRγ) in ECs isolated from murine lungs and retinas, as well as in commercially available mouse (bEnd.3) and human (human umbilical vein endothelial cells [HUVEC] and human microvascular endothelial cells [HMVEC]) ECs. We observed that ERRα is robustly expressed in all the ECs tested, whereas the expression of ERRβ and ERRγ are undetectable in these cells (Fig. 1A to C). Therefore, we focused our investigation on the potential transcriptional role of ERRα in the ECs.
FIG 1.
Estrogen-related receptor alpha (ERRα) targets angiogenic genes in endothelial cells. (A) ERRα, ERRβ, and ERRγ mRNA expression in different ECs (n = 3). (B and C) ERRα protein expression in ECs (n = 3). (D and E) ERRα mRNA (D) and protein expression (E) in ERRα-KO compared to WT mouse lung ECs (n = 3). ****, P < 0.00005 by unpaired Student's t test. (F) Heat map representing differentially expressed genes from the microarray analysis in ERRα-KO and WT ECs. Differentially expressed genes were defined as having an absolute fold change of ≥2 and a P value of <0.05 (Bonferroni’s multiple-comparison test). The color bar on the left indicates the direction of differentially expressed genes (green, upregulated; red, downregulated). (G) GO term enrichment was calculated for differentially expressed genes using Cluster Profiler. The 10 most significant categories are shown. Each GO term is represented as a fraction of genes associated with a given GO term that were differentially expressed in ERRα-KO versus WT cells (x axis). The size of the circle represents the number of genes in the GO term, which were differentially expressed. The color of the circles represents the adjusted P value.
To study the role of endothelial ERRα, we isolated primary ECs from lungs of wild-type (WT) and ERRα knockout (ERRα-KO) mice (47, 56) and confirmed complete deletion of ERRα mRNA and protein (Fig. 1D and E). We next performed unbiased microarray gene expression analysis in ERRα-KO versus WT murine lung ECs using an Illumina Sentrix Beadchip array mouse WG-6.v2 array. Using a selection criteria of gene expression change of ≥2-fold and significance at a P value of ≤0.05, we found that a total of 157 genes were upregulated, whereas 34 genes were downregulated in ERRα-KO versus WT murine lung ECs. A heat map generated for differentially expressed genes (Fig. 1F) clearly shows that ERRα knockout predominantly increases gene expression, suggesting that endogenous ERRα primarily represses transcription of its target genes in ECs. To determine the functional relevance of the differentially expressed genes, we performed GO pathway enrichment analysis (Fig. 1G). The highly ranked biological categories were linked to angiogenesis, including blood vessel morphogenesis, cell motility and adhesion, and endothelial development (Fig. 1G; see also Tables S1 and S2 in the supplemental material). Consequently, we examined the role of endothelial ERRα in regulating angiogenesis.
Lack of ERRα increases angiogenesis in murine lung ECs.
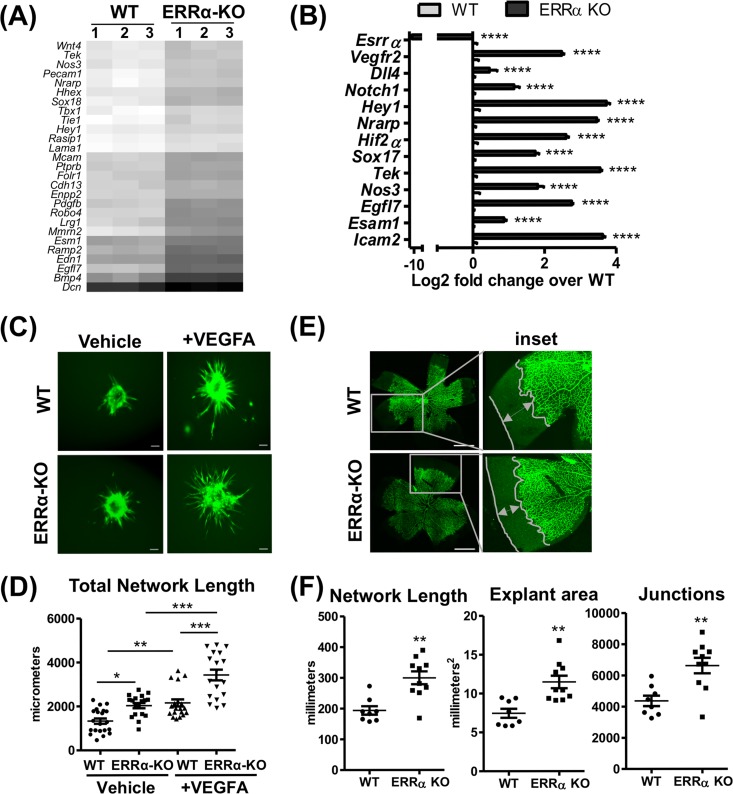
To investigate angiogenesis, we generated a heat map now focused on genes in the “blood vessel morphogenesis” gene ontology (GO) biological processes category. As seen in Fig. 2A, the majority of the genes in this category were upregulated in ERRα-KO murine lung ECs compared to those of the WT. Quantitative reverse transcriptase PCR (RT-qPCR) analysis of candidate angiogenic genes (e.g., Vegfr2, Dll4, Notch1, Nos3, and Hif2α) confirmed that angiogenic gene expression is increased in ERRα-KO compared to WT murine lung ECs (Fig. 2B).
FIG 2.
Loss of ERRα increases angiogenesis in murine lung endothelial cells. (A) Heat map of differentially expressed genes belonging to the GO category “blood vessel morphogenesis” (GO:0048514) from the WT and ERRα-KO cells. (B) Expression of candidate angiogenic genes in ERRα-KO versus WT cells (n = 3). ****, P < 0.00005, unpaired Student's t test. (C) Representative images of calcein AM-stained sprouting angiogenesis in WT and ERRα-KO cells treated with vehicle or VEGFA (30 ng/ml) for 12 h. Scale bars, 100 μm. (D) Quantification of sprouting presented as total network length measured using ImageJ and the Sprout Morphology plug-in (n = 3 experiments, 10 to 20 spheroids per replicate). *, P < 0.05; **, P < 0.005; ***, P = 0.0001, all by Tukey’s multiple-comparison test. (E) Representative images of isolectin B4-stained ERRα-KO P5 mouse retinas and WT littermate controls showing developmental angiogenesis. Scale bars, 1,000 μm. (F) Quantification of explant area, total network area, and number of junctions in retinal vasculature was performed using AngioTool (n = 8 to 10). **, P < 0.005, unpaired Student's t test.
Based on the gene expression patterns, we next asked whether ERRα regulated angiogenesis using the in vitro sprouting assay known to recapitulate key endothelial processes involved in angiogenesis (57, 58). Spheroids prepared from ERRα-KO murine lung ECs exhibited enhanced sprouting compared to that of WT spheroids (Fig. 2C), as depicted in the quantification of the total network length (Fig. 2D). This effect was further enhanced in the VEGFA-treated ERRα-KO spheroids (Fig. 2C and D). We also measured the effect of ERRα knockout on retinal angiogenesis in passage 5 (P5) pups. ERRα deletion enhanced retinal angiogenesis in ERRα-KO versus the WT P5 pups (Fig. 2E), which is quantitatively presented as explant area, total network length, and the number of junctions (Fig. 2F). Therefore, loss of ERRα in murine lung ECs triggers a proangiogenic gene program, which increases the propensity of the mutant ECs to undergo angiogenesis.
ERRα knockdown increases angiogenesis in HUVEC.
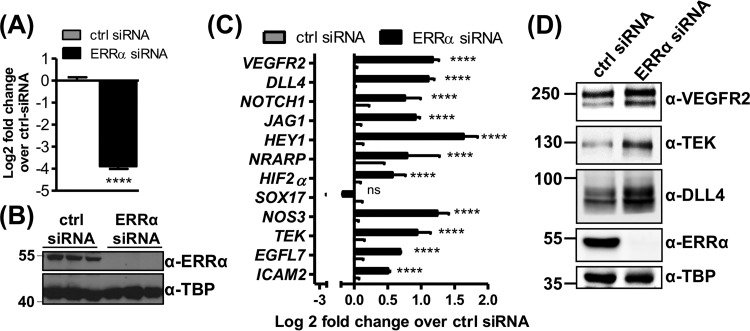
To further characterize the role of ERRα in endothelial angiogenesis, we used transient knockdown of ERRα in HUVEC, a commonly used human endothelial cell line. Efficient knockdown of ERRα mRNA and protein was confirmed by RT-qPCR and Western blotting, respectively (Fig. 3A and B). We measured the expression of some of the same angiogenesis-associated genes that were upregulated in the ERRα-KO mouse ECs, as shown in Fig. 2B. Similar to the case for ERRα-KO murine lung ECs, we found that ERRα knockdown in HUVEC increased the expression of proangiogenic genes (Fig. 3C) and their encoded proteins (Fig. 3D).
FIG 3.
Depletion of ERRα in HUVEC induces expression of angiogenesis-associated genes. (A and B) ERRα knockdown in HUVEC at mRNA (A) and protein (B) levels (n = 3). (C and D) Gene (C) and protein (D) expression of angiogenesis-associated genes in HUVEC (n = 3). ****, P < 0.00005, unpaired Student's t test.
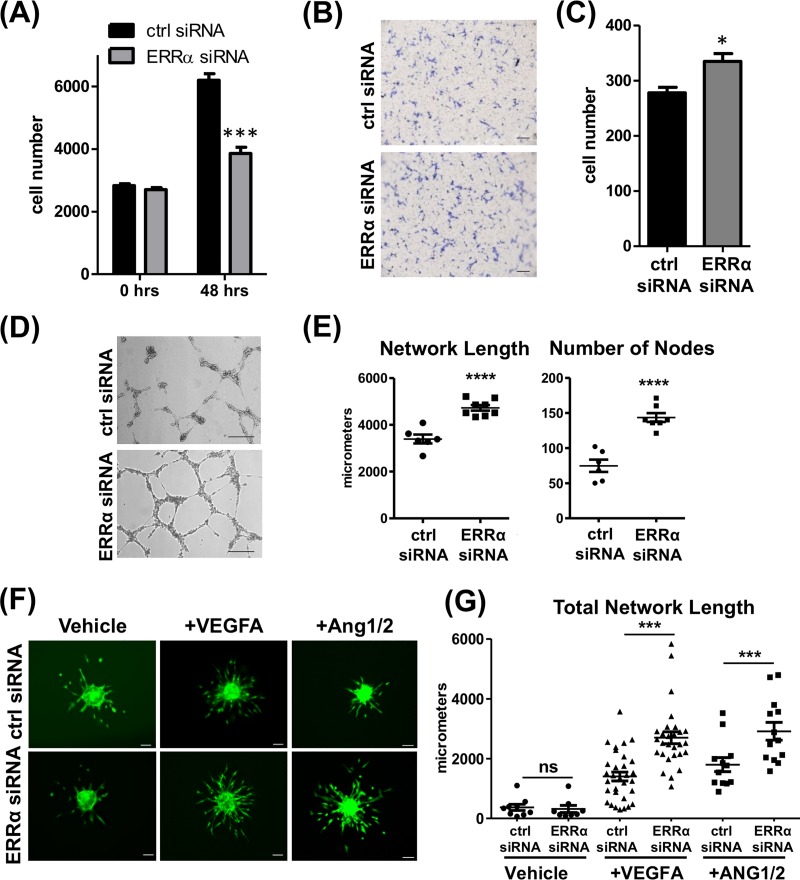
To test whether changes in angiogenic gene expression upon ERRα knockdown impacts angiogenesis, we first measured the effect on EC proliferation, migration, and tube formation. ERRα knockdown in HUVEC resulted in decreased proliferation (Fig. 4A), whereas it increased migration (Fig. 4B and C) and tube formation (Fig. 4D and E). We next performed sprouting angiogenesis assay under vehicle and VEGFA- and Ang1/2-treated conditions in control and knockdown HUVEC spheroids. ERRα knockdown in the ECs resulted in an increase in VEGFA- and Ang1/2-stimulated sprouting of the spheroids, as measured by an increase in total network length (Fig. 4F and G). However, unlike the ERRα-KO murine lung ECs, transient ERRα knockdown did not affect baseline sprout formation. These results show that while ERRα promotes endothelial cell proliferation, it restricts other angiogenic processes, such as migration, tube formation, and sprouting.
FIG 4.
Depletion of ERRα increases angiogenesis in HUVEC. (A) Quantification of DAPI-stained nuclei in control and ERRα knockdown HUVEC grown in EGM2. ***, P < 0.005, unpaired Student's t test. (B) Representative images of transwell migration of HUVEC toward EGM2. Scale bars, 1,000 μm. (C) Quantification of migrated cells via crystal violet staining. *, P < 0.05, unpaired Student's t test. (D) Representative images of tube formation in control and ERRα knockdown HUVEC. Scale bars, 100 μm. (E) Tube formation quantification as total network length and number of nodes using ImageJ and the Angiogenesis Analyzer plug-in (n = 3 experiments, three wells were measured in each replicate). ****, P < 0.0001, unpaired Student's t test. (F) Sprouting angiogenesis. Representative images of calcein AM-stained spheroids generated from control and ERRα knockdown HUVEC are shown. Scale bars, 100 μm. (G) Sprouting angiogenesis quantified as total network length using ImageJ and the Sprout Morphology plug-in (n = 3 experiments, 10 to 20 spheroids per replicate). *, P < 0.05; ***, P < 0.001, Tukey’s multiple-comparison test.
ERRα is dispensable for oxidative metabolism in ECs.
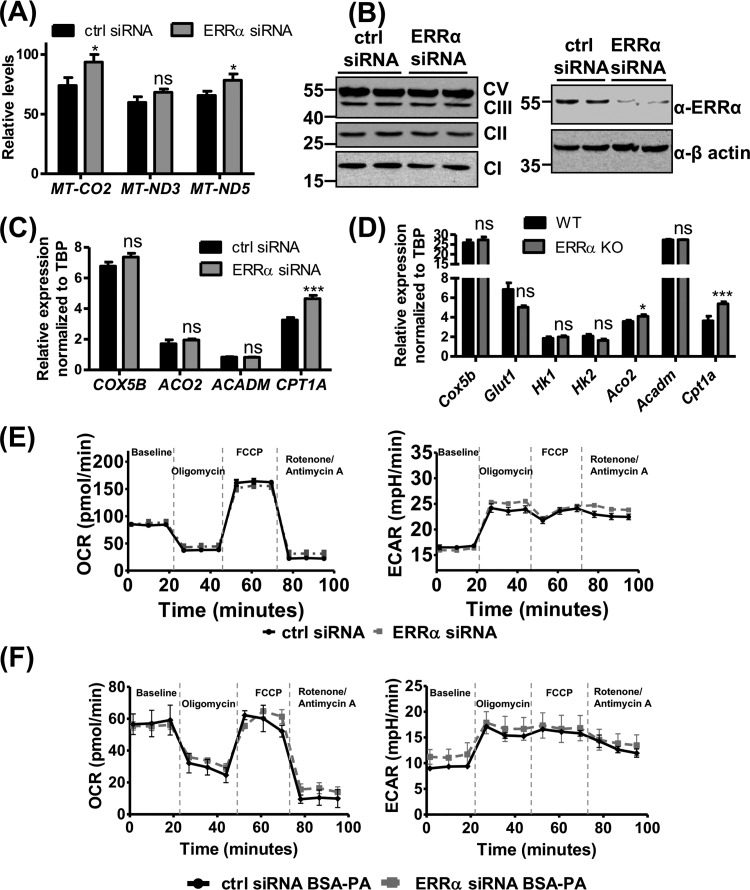
ERRα is a master regulator of oxidative fatty acid metabolism and mitochondrial biogenesis (40, 45, 49, 50, 59–61), raising the possibility that ERRα exerts similar metabolic control in the ECs. Notably, metabolic regulation in ECs can directly impact angiogenesis (62–64). We first determined the mitochondrial content in ERRα-depleted HUVEC by quantifying mitochondrial DNA. Paradoxically, mitochondrial DNA (MT-CO2, MT-ND3, and MT-ND5 genomic loci) was slightly increased in ERRα-KO compared to WT ECs (Fig. 5A). However, ERRα knockdown in HUVEC did not alter the levels of mitochondrial oxidative phosphorylation complex proteins (Fig. 5B). We also measured the expression of COX5B (electron transport chain), hexokinase genes HK1 and HK2, glucose transporter gene GLUT1 (glycolysis), ACO2 (tricarboxylic acid cycle), and ACADM (fatty acid oxidation), none of which changed in ERRα-deficient ECs (murine lung ECs or HUVEC) compared to the control (Fig. 5C and D). One exception was CPT1A, a rate-limiting fatty acid oxidation gene, which was increased upon ERRα knockdown (Fig. 5C and D).
FIG 5.
ERRα does not affect endothelial metabolism under normal growth conditions. (A) Mitochondrial DNA content in control and ERRα knockdown ECs (n = 3). (B) Protein expression levels of ERRα and mitochondrial oxidative phosphorylation complexes in control and ERRα knockdown HUVEC (n = 2). (C and D) Gene expression of known ERRα metabolic target genes in ERRα-KO versus WT murine lung ECs (C) and ERRα knockdown versus control HUVEC (D) (n = 6). (E) Representative graphs of mitochondrial respiration, with glucose-pyruvate as the energy source measured as the oxygen consumption rate (OCR) (left) and glycolysis measured as the extracellular acidification rate (ECAR) (right) in ERRα knockdown versus control HUVEC at baseline and upon sequential drug treatment. Unpaired Student's t test was used (n = 3, each performed in 5 or 6 technical replicates). (F) Representative graphs of mitochondrial respiration with palmitate (BSA-PA) as the energy source measured as the oxygen consumption rate (OCR) (left) and glycolysis measured as the extracellular acidification rate (ECAR) (right) in ERRα knockdown versus control HUVEC at baseline and upon sequential drug treatment (n = 3). *, P < 0.05; ***, P < 0.001, unpaired Student's t test. ns, not significant.
We next measured the rate of mitochondrial respiration in ERRα-depleted and control HUVEC with glucose-pyruvate as the energy source using the Seahorse assay. ERRα knockdown did not alter basal mitochondrial respiration, measured as oxygen consumption rate (OCR). Mitochondrial ATP production (difference in OCR at baseline and post-oligomycin treatment), maximal respiratory capacity (OCR post-FCCP treatment), and spare respiratory capacity (difference between maximal and baseline respiratory capacity) were unaffected by ERRα knockdown in HUVEC (Fig. 5E). We did not observe any difference in the basal glycolytic rate (measured as extracellular acidification rate [ECAR]) in ERRα knockdown cells. Addition of oligomycin inhibits mitochondrial ATP production, resulting in the cells reaching their maximal glycolytic capacity, which was also unchanged in ERRα-depleted versus control ECs (Fig. 5E).
Additionally, we measured fatty acid oxidation in control and ERRα-depleted ECs (Fig. 5F). When palmitate was provided as the energy substrate, we observed no change in OCR at baseline or under oligomycin-, FCCP-, and rotenone/antimycin A-treated conditions between control and ERRα-depleted ECs. No differences were observed in ECAR between ERRα-depleted and control ECs when palmitate was provided as the energy source (Fig. 5F). Therefore, ERRα does not affect fatty acid oxidation or glucose metabolism in ECs, and metabolic remodeling may not be involved in ERRα-mediated regulation of angiogenesis.
ERRα binding is enriched at angiogenesis-associated gene promoters.
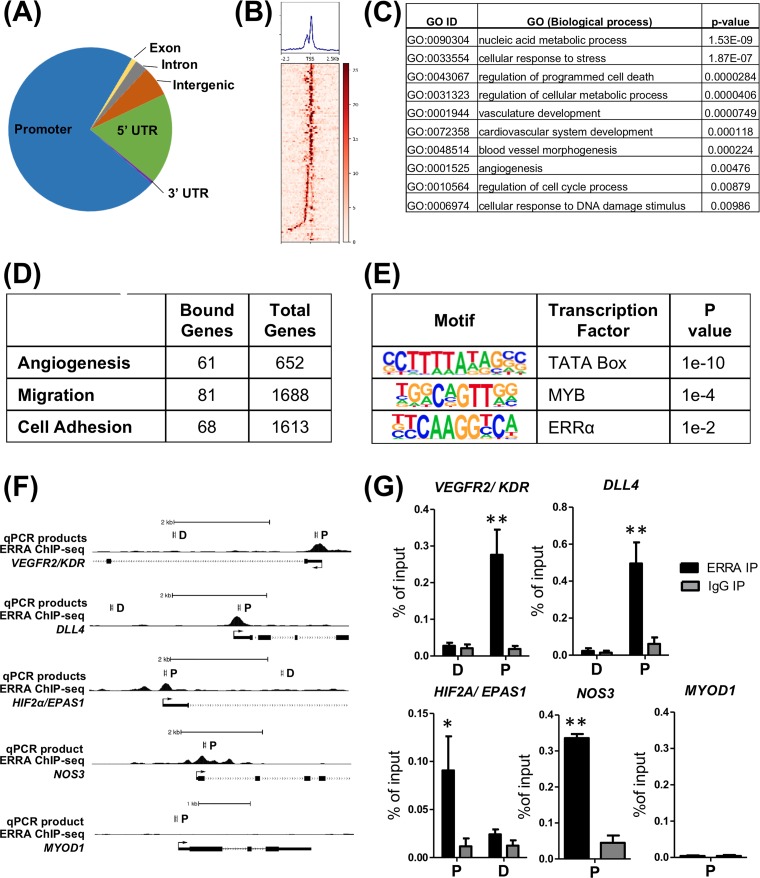
Because ERRα is a DNA binding transcriptional factor and regulates angiogenic gene expression, we used chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP sequencing) to determine genome-wide enrichment sites for ERRα in HUVEC. We found that ERRα was predominantly enriched in gene promoter regions (Fig. 6A and B). However, enrichment in 3′ untranslated regions (UTR), intergenic regions, introns, and exons was also observed (Fig. 6A). Pathway analysis of genes with an ERRα binding site at its transcription start site (TSS) revealed a number of biological processes, including nucleic acid metabolism, stress response, angiogenesis, and DNA damage response (Fig. 6C and Data Set S1). Notably, ERRα was enriched at angiogenic, cell migration, and cell adhesion genes (Fig. 6D and Data Set S2). Using the top 500 scoring peaks, we found three significantly enriched motifs, which are known binding sites for TATA box binding protein, MYB, and ERRα (Fig. 6E). To confirm the results from ChIP sequencing, we performed ChIP-qPCR on candidate angiogenic genes selected from a ChIP sequencing data set. ChIP-qPCR confirmed that ERRα was enriched at the promoters of VEGFR2/KDR, DLL4, EPAS1/HIF2α, and NOS3 and not at sites more distal to these genes’ promoters (Fig. 6F and G). MYOD1, which is a skeletal muscle-specific gene and silent in ECs, was used as a negative control. These data indicate that ERRα regulates angiogenic gene expression by binding in close proximity to the angiogenesis-associated gene promoters.
FIG 6.
ERRα is enriched at angiogenesis-associated gene promoters. (A) Occupancy of ERRα across various genomic elements in HUVEC. (B) Heat map of ERRα enrichment at angiogenesis-associated gene promoters in HUVEC. (C) Pathway enrichment of ERRα-bound genes. (D) Number of ERRα-bound genes in angiogenesis-associated gene categories. (E) Motif enrichment in the top 500 ERRα peaks. (F) Representative ChIP sequencing tracks showing ERRα recruitment to angiogenesis-associated genes in HUVEC. P, promoter site; D, distal site. (G) Confirmation of ERRα enrichment at angiogenesis-associated gene promoters by ChIP-qPCR. *, P < 0.05; **, P < 0.005, unpaired Student's t test (n = 4).
Regulation of ERRα expression and gene occupancy.
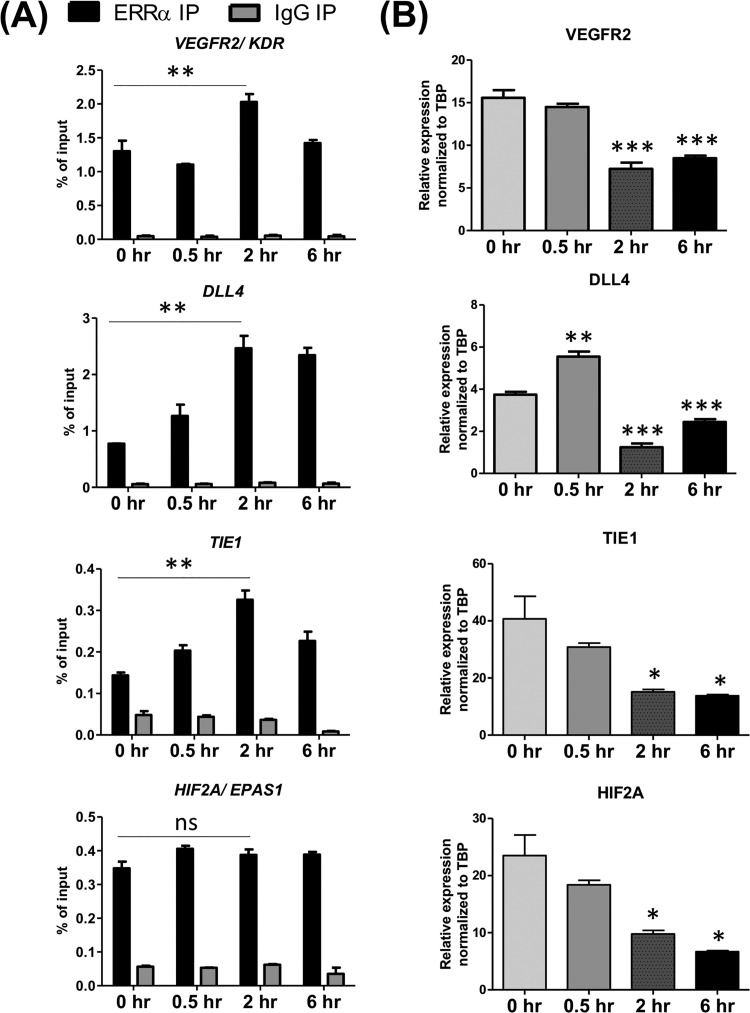
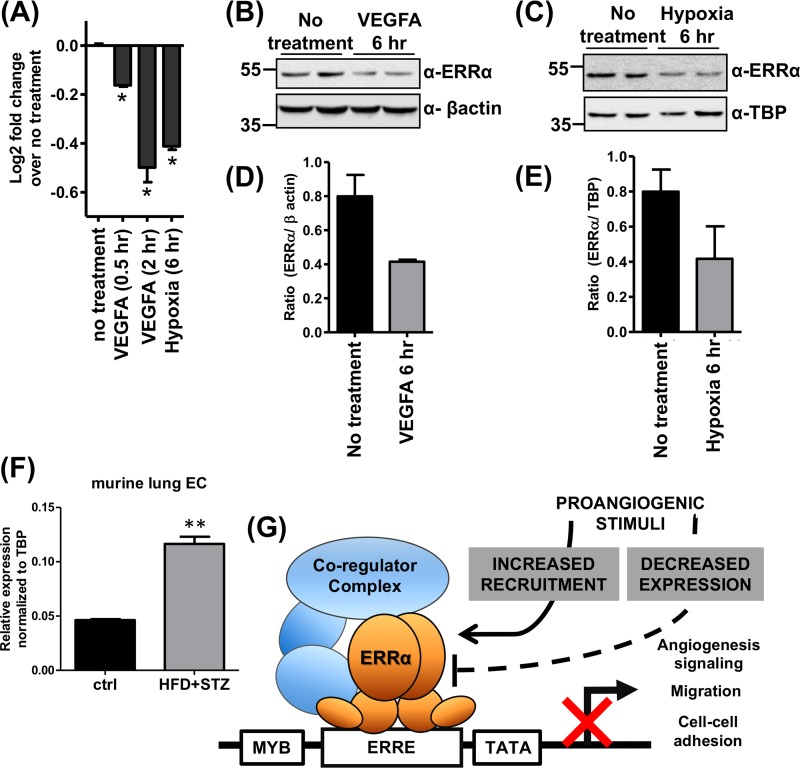
How might proangiogenic stimuli or chronic diseases such as diabetes regulate ERRα? We measured the effect of VEGFA and/or hypoxia on ERRα recruitment to angiogenic genes and ERRα expression in HUVEC. We also measured ERRα expression in lung endothelial cells isolated from control and diabetic mice. Upon 2 h of VEGFA treatment, we observed increased occupancy of ERRα at several candidate angiogenic genes and decreased expression of the same angiogenic genes in a similar time frame (Fig. 7). This is in agreement with our microarray/gene expression data (Fig. 1F, 2A and B, and 3C), reflecting a repressive role of ERRα on angiogenesis-associated genes. Surprisingly, stimulation of ECs with VEGFA or hypoxia decreased ERRα expression at the transcript and protein levels (Fig. 8A to E); however, receptor expression was still detectable. Finally, we found that ERRα expression was increased in lung ECs isolated from diabetic mice (Fig. 8F). Therefore, proangiogenic stimulation of ECs, despite decreasing the expression of ERRα, promotes receptor recruitment to angiogenic genes and decreases the transcription of these genes. Furthermore, changes in ERRα expression in diabetic ECs could play an adaptive role in pathological angiogenesis associated with this chronic condition.
FIG 7.
VEGFA signaling alters ERRα occupancy and gene expression of angiogenesis-associated genes. (A) A time course (0 to 6 h) of ERRα occupancy at angiogenesis-associated genes in response to VEGFA treatment (30 ng/ml) measured by ChIP (n = 3). (B) Angiogenesis-associated gene expression changes upon VEGFA (0 to 6 h) treatment (n = 3). *, P < 0.05; **, P < 0.005; ***, P < 0.0005, all by Tukey’s multiple-comparison test.
FIG 8.
Regulation of ERRα expression and schematic model. (A) ERRα mRNA expression in untreated, VEGFA (30 ng/ml)-treated, and hypoxia-treated HUVEC (n = 3). (B and C) Representative images of ERRα protein expression in VEGFA-treated HUVEC (B) and hypoxia-treated HUVEC (C) (n = 3). (D and E) Densitometry of Western blots quantifying ERRα expression in VEGF-treated (D) and hypoxia-treated (E) HUVEC. (F) ERRα expression in lung endothelial cells isolated from control and diabetic (high-fat diet plus STZ) mice (n = 5, technical replicates from pooled lungs). (G) Schematic model depicting regulation of angiogenesis by ERRα in ECs. ERRα acts as a transcriptional repressor of angiogenic genes and restricts angiogenesis. Proangiogenic stimuli have a dual effect on ERRα, decreasing its expression as well as increasing recruitment to angiogenic genes.
DISCUSSION
In this study, we have found that ECs exclusively express ERRα. Endothelial ERRα predominantly acts as a repressor of gene expression, and genes linked to angiogenesis are its major targets. Consequently, loss of ERRα facilitates angiogenesis in ECs. ERRα expression is downregulated, whereas its recruitment to angiogenic genes is induced by proangiogenic stimuli in ECs, revealing a complex relationship between angiogenic stimulants and ERRα. Surprisingly, ERRα is generally dispensable for the regulation of its known targets from other tissues, such as mitochondrial biogenesis and oxidative metabolism in ECs. Our work identifies a new function of ERRα in restricting angiogenesis and related transcriptional programs in ECs (Fig. 8G).
ERRs have various expression levels in different tissues with ERRα ubiquitously expressed, and most tissues express one or more subtypes of ERRs. These receptors are known to regulate overlapping sets of genes based on studies in the skeletal and cardiac muscle (40, 50). Interestingly, we found that a variety of ECs, including murine lung, retina, and brain, as well as human lung and umbilical vein ECs, exclusively express ERRα. Considering that ECs in different tissues may have unique gene signatures, ERRβ or ERRγ could be expressed in ECs from tissues we have not examined. However, ERRα knockdown/deletion did not result in ERRβ or ERRγ induction, suggesting that these isoforms do not have a compensatory role in the ECs (data not shown). Loss of ERRα changed PPARα (increase in HUVEC, decrease in mouse lung ECs) expression, increased PPARγ (both mouse lung ECs and HUVEC) expression, and did not change PPARδ expression (data not shown). The expression of the coactivator PGC1α was barely detectable, and its expression was unaffected by ERRα in endothelial cells (data not shown). Therefore, it is possible that changes in expression of these other regulators could contribute to ERRα regulation of angiogenesis.
Gene profiling data suggest that ERRα predominantly acts as a transcriptional repressor in ECs. A majority of the differentially expressed genes were upregulated in ERRα-null ECs. In particular, genes linked to blood vessel morphogenesis, cell motility, and cell adhesion were major targets of ERRα in the ECs. Of the differentially expressed genes, 33 genes were classified as blood vessel morphogenesis and angiogenic factor signaling-associated genes, 34 genes were linked to cell motility/migration, and 9 genes were linked to cell adhesion (see Table S2 in the supplemental material). Several prominent genes (e.g., Nos3, Sox 17/18, Robo4, and Bmp4) that regulate angiogenesis (65–68) were upregulated by ERRα deletion in ECs. In addition, several migratory and cell adhesion genes, such as Pecam, Mmp3, Cdh3, and Esam1, that were upregulated upon ERRα deletion are known to be involved in angiogenic regulation (69–72).
ERRs have been shown to regulate gene expression by preferentially binding to conserved estrogen-related receptor response elements (ERREs) in the proximal promoter regions of genes (40). Indeed, ChIP sequencing revealed that even in ECs ERRα predominantly binds to promoter regions. In concert with gene expression data, ERRα was localized to genes linked to angiogenesis, migration, and cell adhesion. Top-ranking motifs found at the receptor enrichment sites are known motifs for TBP (TATA box binding protein), MYB, and ERRα, suggesting that ERRα regulates endothelial gene expression directly or via interaction with other transcriptional regulators. Notably, endothelial MYB is implicated in regulation of angiogenesis in gastric cancer (73). ERRα previously has been found to bind genes such as VEGFR2, DLL4, and HIF2α/EPAS1 in different cell lines (e.g., A549, BT474, K562, and GM12878), but it did so in nonpromoter regions (74, 75). We previously showed that muscle ERRγ promotes angiogenesis by paracrine secretion of angiogenic growth factors such as Vegfa (46, 53). However, reexamination of our published microarray data from muscle-specific ERRγ-overexpressing mice (46) suggests that ERRγ does not regulate any of the other effector signaling angiogenic genes (e.g., Dll4, Hif2α/Epas1, and VegfR2) targeted by endothelial ERRα. Also, to our knowledge genomic recruitment of ERRβ/γ isoforms to angiogenesis pathway genes has not been reported. Therefore, while in tissues such as skeletal muscle ERRγ may play a paracrine proangiogenic role, in ECs ERRα may have a contrasting function to limit angiogenesis.
Transcriptional remodeling of ECs upon ERRα deletion leads to increased angiogenesis. ERRα depletion in murine lung ECs as well as in HUVEC leads to enhanced sprouting angiogenesis. In this assay, we observed an increase in total network length in ERRα-null ECs. The increase in total network length is a combination of a change in the number of sprouts and/or sprout length. This indicates that ERRα could be affecting the balance between the tip and stalk cells in the ECs. The tip cells are migratory and form the leading edge that follows the angiokine gradient cues during angiogenesis. On the other hand, stalk cells are more proliferative, follow the tip cells, and are involved as building blocks in the elongation of the vascular sprout. Depletion of ERRα in HUVEC decreased proliferation and increased migration and tube formation, suggesting effects on tip and stalk cell dynamics. Loss of ERRα increased expression of genes (e.g., VEGFR2 and DLL4) that are the markers of tip cells, which coincides with increased sprouting potential of the ERRα-depleted ECs. The effect on migration is also in agreement with the upregulation of the migratory genes in the ERRα-null ECs. Our findings that loss of ERRα advances the angiogenic front, which in the P5 retinas is primarily comprised of tip cells, also supports the potential effect of ERRα on tip versus stalk cell balance. Furthermore, because lack of ERRα sensitizes the mutant ECs to both VEGFA- or ANG1/2-mediated sprouting, ERRα may not be exclusively downstream of a single angiogenic growth factor. Rather, ERRα-mediated gene programing may generally fine-tune EC sprouting or angiogenic responsiveness to growth factors.
Recent studies have highlighted a major role for metabolism in the regulation of angiogenesis, particularly in tip versus stalk cell fate of ECs during angiogenesis (76). While ECs are generally glycolytic, the tip cells become even more glycolytic, whereas the stalk cells rely on fatty acid oxidation to meet the biosynthetic demands of these cells. Tip cells increase the expression of phosphofructokinase 2/fructose-2,6-bisphosphatase (PFKFB3), an enzyme that catalyzes the rate-limiting step in glycolysis (63). Loss of PFKFB3 decreases tip cell formation and directional cell migration (63, 77). Meanwhile, stalk cells upregulate CPT1a, an enzyme that catalyzes the rate-limiting step of fatty acid oxidation (78). Endothelium-specific deletion of CPT1a results in decreased proliferation and subsequently impaired sprouting (78). Furthermore, inhibition of CPT1a can rescue pathological ocular angiogenesis. CPT1a-driven fatty acid oxidation seems to be necessary for de novo synthesis of nucleotide synthesis and DNA replication in ECs (78). On the other hand, CPT1a and fatty acid oxidation in quiescent endothelial cells is required for redox homeostasis as well as for prevention of EC dysfunction, oxidative stress, and susceptibility to inflammatory diseases (79). As mentioned above, ERRs are major regulators of oxidative metabolism, mitochondrial biogenesis, and fatty acid oxidation in organs such as skeletal muscle, heart, and adipose tissues (50). However, ERRα was found to be dispensable for the regulation of mitochondrial biogenesis and oxidative metabolic genes in the ECs. Cellular oxygen consumption rates as well as extracellular acidification rates measured using Seahorse with glucose-pyruvate or palmitate as the energy source were found to be comparable between control and ERRα-depleted ECs, suggesting that the lack of ERRα does not affect the metabolic preference in these cells and does not contribute to angiogenesis via this mechanism. However, this possibility currently cannot be ruled out for conditions such as hyperglycemia, inflammation, and proangiogenic stimuli.
The relationship between angiogenic stimuli and ERRα was found to be complex. VEGFA treatment of ECs resulted in increased occupancy of ERRα at most angiogenesis-associated genes tested. VEGFA treatment within the same time frame repressed angiogenic gene expression. We speculate that ERRα represents a counterfeedback mechanism to limit excessive angiogenesis in response to growth factors such as VEGFA. This idea is supported by the aforementioned results that both VEGFA- and Ang1/2-mediated sprouting angiogenesis are enhanced in ERRα-depleted ECs. Paradoxically, we found that proangiogenic stimulation of ECs with VEGFA or hypoxia resulted in the downregulation of ERRα gene expression. However, there still remains residual ERRα expression, which potentially gets recruited to angiogenic genes in response to VEGFA treatment. How VEGFA can downregulate ERRα while simultaneously increasing its recruitment to angiogenesis-associated genes remains unclear. We speculate that proangiogenic stimuli induce posttranslational modifications of ERRα, subsequently increasing its recruitment. Nuclear trafficking of ERRα and modulation of interaction with corepressors could also contribute to regulation of ERRα by proangiogenic signaling. Overall, our findings are suggestive of a model where proangiogenic stimulation of ECs represses ERRα expression while increasing its recruitment to angiogenic genes, thereby potentially fine-tuning angiogenesis (Fig. 8G). The physiological and pathological significance of this model remains to be explored. It is possible that dynamic regulation of ERRα is critical for developmental, physiological, or pathological angiogenesis. Our finding of increased ERRα expression in primary lung ECs isolated from diabetic mice suggests that endothelial ERRα play an adaptive role in pathological angiogenesis.
In summary, ERRα is a regulator of EC gene expression potentially via transcriptional repression. Genes linked with angiogenesis form a substantial cohort of ERRα-regulated targets in ECs. Consequently, endothelial ERRα acts as a repressor of angiogenesis. While we have primarily used in vitro and ex vivo experiments to delineate the role of endothelial ERRα in cellular angiogenesis, it opens up potentially interesting future directions. What is the impact of endothelium-specific ERRα deletion on physiological and pathological angiogenesis? What are the components of ERRα transcriptional repressor complexes in ECs? What are additional mechanisms of ERRα regulation in ECs? Finally, what other roles does ERRα play beyond angiogenesis in ECs? Future studies will answer some of these critical questions to establish ERRα as a major player in endothelial function.
MATERIALS AND METHODS
Mouse strains and mouse treatment conditions.
ERRα-null mice (ERRα-KO) have been previously described (47) and were backcrossed to the C57BL/6J background strain. Mice were housed in a temperature-controlled room (20 to 22°C) with ad libitum access to water and food (Pico Lab rodent diet, 13.2% fat) under a 12-h/12-h light-dark cycle. Animals were housed and treated in accordance with the U.S. National Institutes of Health Guide for the Care and Use of Laboratory Animals (80). The Animal Welfare Committee at The University of Texas McGovern Medical School in Houston approved the procedures.
Cell culture, primary cell isolation, and treatment conditions.
HUVEC (Lonza) were grown in EGM2 medium (PromoCell) in a humidified incubator at 37°C and 5% CO2. HMVEC (Lonza) and bEnd.3 cells (ATCC) were grown on 2% gelatin-coated plates in EGM2 MV medium (PromoCell) in a humidified incubator at 37°C and 5% CO2. Cells were passaged by trypsinization followed by neutralization of trypsin with Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. Cells used for experiments were between passage 3 and passage 6. Mouse lung and retinal primary ECs were isolated from pups (between P10 and P18) by adopting a previously described protocol (56). For diabetic ECs, C57BL/6J mice were fed on a 60 kcal% fat diet (Research Diets Inc.) for 16 weeks along with injection of STZ at 40 mg/kg of body weight for 3 days at 8 weeks. Lungs were harvested at 16 weeks, followed by EC isolation and immediate RNA extraction. For angiogenic stimulus, cells were treated with 30 ng/ml VEGFA (R&D Biosystems) or ANG1 (200 ng/ml) and ANG2 (650 ng/ml) (R&D Biosystems). Hypoxia treatment of cells was carried out in a modular incubator chamber (Billups, Rothenberg) by flushing the chamber containing cells with a preanalyzed gas mixture of 5% CO2 and 95% N2, followed by incubation for 6 h at 37°C. HUVEC were transfected with 30 nM short interfering RNA (siRNA; L-003403-00-0005 [ESSRA] and 001810-10-05 [control]; GE Healthcare, Dharmacon) using Lipofectamine RNAiMax (Thermo Fisher Scientific). Cells were harvested or used for experiments 48 to 60 h posttransfection.
Western blotting.
Cells were harvested and lysed in radioimmunoprecipitation assay buffer (Boston BioProducts). The Pierce bicinchoninic acid protein assay kit (Thermo Scientific) was used to quantify protein content in the lysate. Twenty-five to 40 μg of total protein was used for Western blotting. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, stained using Ponceau S, blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline with 0.1% Tween 20 (PBST), and incubated overnight at 4°C with primary antibody (ERRα [13826; Cell Signaling], TBP [28175; Abcam], VEGFR2 [PA5-16487; Thermo Fisher Scientific], TEK [SC-324; Santa Cruz Biotechnology], DLL4 [2589T; Cell Signaling], β-actin [612656; BD Biosciences], and Oxphos [110413; Abcam]). Membranes were subsequently washed with PBST and incubated with the appropriate secondary antibodies (Cell Signaling) for 1 h at room temperature, followed by washes with PBST. Blots were visualized using chemiluminescence Western blotting detection reagents (Thermo Fisher Scientific) and imaged on the ChemiDoc MP imaging system (Bio-Rad).
RNA isolation and RT-qPCR.
Total RNA was isolated from cells using TRIzol and the PureLink kit (Ambion). Reverse transcription was carried out with 500 to 2,000 ng of total RNA using SuperScript III reverse transcriptase (Thermo Fisher Scientific) by following the manufacturer’s protocol. Quantitative reverse transcriptase PCR (RT-qPCR) analysis was performed using Power Up SYBR PCR master mix (Applied Biosystems) with an ABI-7900 cycler (Applied Biosystems) or CFX 96-Touch (Bio-Rad). Gene expression was normalized using the housekeeping TATA binding protein (TBP) gene and represented as ΔCT or ΔΔCT values (where CT is threshold cycle). The primer sequences are provided in Tables S3 (mouse) and S4 (human) in the supplemental material.
Microarray gene expression.
Genome-wide gene expression analysis was performed on RNA samples from lung ECs (P4) of WT and ERRα KO mice. Total RNA (300 ng/sample) was amplified and purified using an Illumina TotalPrep RNA amplification kit (Ambion) by following the manufacturer’s instructions. In vitro transcription was performed and biotinylated cRNA was synthesized by 14 h of amplification with deoxynucleoside triphosphate (dNTP) mix containing biotin-dUTP and T7 RNA polymerase. An aliquot of 1.5 μg of amplified products was loaded onto Illumina Sentrix BeadChip array mouse WG-6.v2 arrays. Data were analyzed using Genome Studio software (Illumina). Heat maps were generated in R (v3.4.4) using the average signal from the raw array data for each gene/probe that was found to be differentially expressed (absolute fold change of ≥2 and adjusted P value of ≤0.05). Differentially expressed genes (DEG) (fold change of ≥2 and P value of ≤0.05) were tested for statistically enriched GO terms using Cluster Profiler (v3.2.4) in R (v3.4.4), which uses the hypergeometric model to test for overrepresentation of a GO category in the DEG list. Gene identifiers from the array annotation file were used to map DEGs to GO terms. The results from the GO enrichment analysis were simplified by removing enriched GO terms whose associated genes were completely represented by another more significant GO term (81).
In vitro angiogenesis assays.
An in vitro sprouting assay was performed as described previously (58), with slight modifications. Briefly, cells were treated with siRNA for 24 h, followed by spheroid formation. Spheroids were then collected and plated in a 2-mg/ml collagen matrix at a density of 1 spheroid per well in a 96-well plate. After 12 h of incubation (with or without VEGFA or ANG1/2 treatment) in the collagen matrix, spheroids were stained with calcein AM (Life Technologies) and imaged on a Nikon Eclipse TE2000E widefield fluorescence microscope. When specified, spheroids were treated with 30 ng/ml VEGFA or Ang 1 (200 ng/ml) plus Ang 2 (650 ng/ml) (R&D Biosystems) in EBM2 (Lonza). Quantification of sprout network length was carried out using the Sprout Morphology plug-in for ImageJ (82). For tube formation assay, a 96-well plate was prepared by coating it with 50 μl of Matrigel (Gibco), followed by incubating the plate at 37°C for 30 min. Forty-eight hours after transfection, cells were trypsinized and plated at a density of 2 × 104 cells per well in EBM2 (Lonza). Tube formation was imaged 4 h postplating using phase contrast imaging on a Nikon Eclipse TE2000E widefield fluorescence microscope. Tube formation was quantified using the angiogenesis analyzer for ImageJ (83). Endothelial cell proliferation was measured by plating equal numbers of ERRα siRNA or control siRNA-transfected HUVEC in 96-well format, followed by counting cell numbers (4′,6-diamidino-2-phenylindole [DAPI] staining of nuclei) at the stated time points. Endothelial cell migration was measured by loading ERRα siRNA- or control siRNA-transfected HUVEC in minimal medium in the upper chamber of a transwell (8-μm pore size) (Costar) with complete growth medium (EGM2) in the lower chamber. Migrated cells were fixed with paraformaldehyde and visualized using crystal violet.
Mouse retina staining.
Retinas were dissected from P5 neonatal mice as described previously (84). The isolated retinas were fixed with methanol and incubated with biotinylated isolectin B4 (Vector Laboratories) at a dilution of 1:2,000, followed by incubation with DyLight 488-conjugated streptavidin (Vector Laboratories). The retinas were mounted with Prolong Gold (Life Technologies) and imaged using a Leica TCS SP5 confocal microscope. The retinal vasculature was quantitatively analyzed using AngioTool (85).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed using the ChIP-IT Express enzymatic kit (Active Motif). Two 150-mm plates of HUVEC grown to confluence were used per immunoprecipitation reaction (∼107 cells). Chromatin immunoprecipitation was carried out using antibodies (ERRα [13826; Cell Signaling] and rabbit IgG [011-000-003; Jackson Immuno Research]) according to the manufacturers’ protocol. Cells were cross-linked with 1% formaldehyde for 10 min. MNase digestion was performed at 37°C for 20 min. The immunoprecipitated DNA was cleaned up using the chromatin IP DNA purification kit (Active Motif). The purified DNA was quantified using qPCR. Data are represented as a percentage of input DNA. The primer sequences are provided in Table S4.
The ChIP-sequencing experiment was performed in duplicate with the above-mentioned immunoprecipitation conditions and cell numbers. Sequence reads were aligned to the human genome (version hg19) using bowtie2 (86). Greater than 75% of reads aligned to the genome exactly 1 time. ERRα-enriched peaks were identified for each ChIP replicate using Macs2 (87) with default parameters. A consensus set of peaks was created by using the idr procedure in R, with a threshold of 0.05 (88). Peaks were annotated using Homer’s annotate peaks function (89). To find enriched motifs, we performed a de novo search using Homer’s findMotifs.pl using the hypergeometric background model and discovered motifs were matched to known motifs from Encode’s factorbook (90). Signal tracks and heat maps were generated using deeptools (91) with 25-bp bins across the genome. Pathway analysis was performed on genes which had a peak upstream of its TSS within 1 kb or in which a peak overlapped its TSS or the closest gene to every peak using Panther and/or R/Bioconductor (92).
Mitochondrial content measurement.
HUVEC were transfected with control or ERRα siRNA. Forty-eight hours posttransfection, cells were harvested and DNA was isolated using a NucleoSpin DNA isolation kit (Macherey Nagel). In brief, cells were lysed and incubated with proteinase K overnight at 65°C. DNA was then purified using the supplied silica spin columns. The purified DNA was quantified, diluted to a concentration of 50 ng/μl, and used as a template for qPCR. CT values for mitochondrial genes were normalized to CT values of three genomic DNA loci on chromosomes 10, 11, and 14. Data are represented as ΔCT values.
Seahorse assay.
The Seahorse assay was performed using the Agilent Seahorse XFe24 extracellular flux assay kit (Agilent Technologies) according to the manufacturer’s protocol. Briefly, ERRα was knocked down in HUVEC using siRNA. Thirty hours posttransfection, cells were plated in the assay plate at a density of 40,000 cells/well. The assay was performed 48 to 60 h posttransfection. Cells were incubated in the assay medium containing 25 mM glucose, 2 mM glutamine, and 2 mM sodium pyruvate at 37°C for 1 h prior to measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). After measurement of baseline OCR and ECAR, cells were treated sequentially with oligomycin (1 μM), FCCP (2 μM), and antimycin A-rotenone (0.5 μM), followed by measurement of OCR and ECAR after every measurement. Fatty acid oxidation was measured using the XF palmitate-BSA FAO substrate (Agilent Technologies) by adhering to the manufacturer’s protocol. Cells were incubated with 150 μM palmitate conjugated to 250 μM BSA. OCR and ECAR were measured at baseline, followed by measurement after each sequential drug treatment of oligomycin (2 μM), FCCP (3 μM), and antimycin A (2 μM)-rotenone (4 μM).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism. Experiments with two test groups were analyzed using unpaired Student's t test. Experiments with more than two groups were analyzed using one-way analysis of variance with Tukey’s multiple-comparison post hoc test. Microarray data were analyzed using Genome Studio using Bonferroni’s multiple-comparison test. A P value of ≤0.05 was considered statistically significant. Bioinformatics analysis of microarray data are described in Results.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from The Welch Foundation (no. L-AU-0002) and NIH/NHLBI (R01 HL129191-01) to V.N., as well as by UTHealth intramural funds. We have no financial disclosures to report.
We have no conflicts of interest to declare.
We thank Zhengmei Mao and the IMM Microscopy Core for assistance with imaging. We also thank Zhanguo Gao and Misha Kolonin for help with the Seahorse assay.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00411-18.
REFERENCES
- 1.Rodrigues SF, Granger DN. 2015. Blood cells and endothelial barrier function. Tissue Barriers 3:e978720. doi: 10.4161/21688370.2014.978720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Förstermann U, Sessa WC. 2012. Nitric oxide synthases: regulation and function. Eur Heart J 33:829–837. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pober JS, Sessa WC. 2007. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol 7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 4.Risau W. 1997. Mechanisms of angiogenesis. Nature 386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 5.Blanco R, Gerhardt H. 2013. VEGF and Notch in tip and stalk cell selection. Cold Spring Harb Perspect Med 3:a006569. doi: 10.1101/cshperspect.a006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcelo KL, Goldie LC, Hirschi KK. 2013. Regulation of endothelial cell differentiation and specification. Circ Res 112:1272–1287. doi: 10.1161/CIRCRESAHA.113.300506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel-Hett S, Damore PA. 2011. Signal transduction in vasculogenesis and developmental angiogenesis. Int J Dev Biol 55:353–363. doi: 10.1387/ijdb.103213sp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ucuzian AA, Gassman AA, East AT, Greisler HP. 2010. Molecular mediators of angiogenesis. J Burn Care Res 31:158–175. doi: 10.1097/BCR.0b013e3181c7ed82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potente M, Gerhardt H, Carmeliet P. 2011. Basic and therapeutic aspects of angiogenesis. Cell 146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 10.Jakobsson L, Bentley K, Gerhardt H. 2009. VEGFRs and Notch: a dynamic collaboration in vascular patterning. Biochem Soc Trans 37:1233–1236. doi: 10.1042/BST0371233. [DOI] [PubMed] [Google Scholar]
- 11.Park C, Kim TM, Malik AB. 2013. Transcriptional regulation of endothelial cell and vascular development. Circ Res 112:1380–1400. doi: 10.1161/CIRCRESAHA.113.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Val S, Black BL. 2009. Transcriptional control of endothelial cell development. Dev Cell 16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bautch VL. 2012. VEGF-directed blood vessel patterning: from cells to organism. Cold Spring Harb Perspect Med 2:a006452. doi: 10.1101/cshperspect.a006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeltsch M, Leppanen VM, Saharinen P, Alitalo K. 2013. Receptor tyrosine kinase-mediated angiogenesis. Cold Spring Harb Perspect Biol 5:a009183. doi: 10.1101/cshperspect.a009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839. doi: 10.1016/0092-8674(95)90199-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T, Nakamura Y, Moriya T, Sasano H. 2003. Effects of steroid hormones on vascular functions. Microsc Res Tech 60:76–84. doi: 10.1002/jemt.10246. [DOI] [PubMed] [Google Scholar]
- 17.Brouchet L, Krust A, Dupont S, Chambon P, Bayard F, Arnal JF. 2001. Estradiol accelerates reendothelialization in mouse carotid artery through estrogen receptor-alpha but not estrogen receptor-beta. Circulation 103:423–428. doi: 10.1161/01.CIR.103.3.423. [DOI] [PubMed] [Google Scholar]
- 18.Caulin-Glaser T, Farrell WJ, Pfau SE, Zaret B, Bunger K, Setaro JF, Brennan JJ, Bender JR, Cleman MW, Cabin HS, Remetz MS. 1998. Modulation of circulating cellular adhesion molecules in postmenopausal women with coronary artery disease. J Am Coll Cardiol 31:1555–1560. doi: 10.1016/S0735-1097(98)00145-4. [DOI] [PubMed] [Google Scholar]
- 19.Jun SS, Chen Z, Pace MC, Shaul PW. 1998. Estrogen upregulates cyclooxygenase-1 gene expression in ovine fetal pulmonary artery endothelium. J Clin Investig 102:176–183. doi: 10.1172/JCI2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losordo DW, Isner JM. 2001. Estrogen and angiogenesis: a review. Arterioscler Thromb Vasc Biol 21:6–12. doi: 10.1161/01.ATV.21.1.6. [DOI] [PubMed] [Google Scholar]
- 21.Morales DE, McGowan KA, Grant DS, Maheshwari S, Bhartiya D, Cid MC, Kleinman HK, Schnaper HW. 1995. Estrogen promotes angiogenic activity in human umbilical vein endothelial cells in vitro and in a murine model. Circulation 91:755–763. doi: 10.1161/01.CIR.91.3.755. [DOI] [PubMed] [Google Scholar]
- 22.Prabhushankar R, Krueger C, Manrique C. 2014. Membrane estrogen receptors: their role in blood pressure regulation and cardiovascular disease. Curr Hypertens Rep 16:013–0408. [DOI] [PubMed] [Google Scholar]
- 23.Ingegno MD, Money SR, Thelmo W, Greene GL, Davidian M, Jaffe BM, Pertschuk LP. 1988. Progesterone receptors in the human heart and great vessels. Lab Investig 59:353–356. [PubMed] [Google Scholar]
- 24.Schnaper HW, McGuire J, Runyan C, Hubchak SC. 2000. Sex steroids and the endothelium. Curr Med Chem 7:519–531. doi: 10.2174/0929867003374949. [DOI] [PubMed] [Google Scholar]
- 25.Lee WS, Liu CW, Juan SH, Liang YC, Ho PY, Lee YH. 2003. Molecular mechanism of progesterone-induced antiproliferation in rat aortic smooth muscle cells. Endocrinology 144:2785–2790. doi: 10.1210/en.2003-0045. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Hadoke PWF, Takov K, Korczak A, Denvir MA, Smith LB. 2016. Influence of androgen receptor in vascular cells on reperfusion following hindlimb ischaemia. PLoS One 11:e0154987. doi: 10.1371/journal.pone.0154987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takov K, Wu J, Denvir MA, Smith LB, Hadoke PWF. 2018. The role of androgen receptors in atherosclerosis. Mol Cell Endocrinol 465:82–91. doi: 10.1016/j.mce.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Ullian ME. 1999. The role of corticosteriods in the regulation of vascular tone. Cardiovasc Res 41:55–64. doi: 10.1016/S0008-6363(98)00230-2. [DOI] [PubMed] [Google Scholar]
- 29.Logie JJ, Ali S, Marshall KM, Heck MMS, Walker BR, Hadoke PWF. 2010. Glucocorticoid-mediated inhibition of angiogenic changes in human endothelial cells is not caused by reductions in cell proliferation or migration. PLoS One 5:e14476. doi: 10.1371/journal.pone.0014476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yano A, Fujii Y, Iwai A, Kageyama Y, Kihara K. 2006. Glucocorticoids suppress tumor angiogenesis and in vivo growth of prostate cancer cells. Clin Cancer Res 12:3003–3009. doi: 10.1158/1078-0432.CCR-05-2085. [DOI] [PubMed] [Google Scholar]
- 31.Lother A, Moser M, Bode C, Feldman RD, Hein L. 2015. Mineralocorticoids in the heart and vasculature: new insights for old hormones. Annu Rev Pharmacol Toxicol 55:289–312. doi: 10.1146/annurev-pharmtox-010814-124302. [DOI] [PubMed] [Google Scholar]
- 32.Giguere V. 1999. Orphan nuclear receptors: from gene to function. Endocr Rev 20:689–725. doi: 10.1210/er.20.5.689. [DOI] [PubMed] [Google Scholar]
- 33.Mullican SE, DiSpirito JR, Lazar MA. 2013. The orphan nuclear receptors at their 25th year reunion. J Mol Endocrinol 51:T115–T140. doi: 10.1530/JME-13-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotlinowski J, Jozkowicz A. 2016. PPAR gamma and angiogenesis: endothelial cells perspective. J Diabetes Res 2016:8492353. doi: 10.1155/2016/8492353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong HK, Cho YM, Park KH, Lee CT, Lee HK, Park KS. 2003. Peroxisome proliferator-activated receptor gamma mediated inhibition of plasminogen activator inhibitor type 1 production and proliferation of human umbilical vein endothelial cells. Diabetes Res Clin Pract 62:1–8. doi: 10.1016/S0168-8227(03)00142-6. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Bruemmer D. 2010. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol 30:1535–1541. doi: 10.1161/ATVBAHA.109.191163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. 1999. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev 13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giguere V. 2008. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev 29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 39.Giguere V, Yang N, Segui P, Evans RM. 1988. Identification of a new class of steroid hormone receptors. Nature 331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 40.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. 2007. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab 5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Audet-Walsh É, Giguére V. 2015. The multiple universes of estrogen-related receptor alpha and gamma in metabolic control and related diseases. Acta Pharmacol Sin 36:51–61. doi: 10.1038/aps.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huss JM, Kopp RP, Kelly DP. 2002. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem 277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 43.Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, Takahashi N, Kawada T, Miyoshi M, Ezaki O, Kakizuka A. 2003. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A 100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. 2004. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A 101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry MC, Dufour CR, Tam IS, B'Chir W, Giguere V. 2014. Estrogen-related receptor-alpha coordinates transcriptional programs essential for exercise tolerance and muscle fitness. Mol Endocrinol 28:2060–2071. doi: 10.1210/me.2014-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narkar VA, Fan W, Downes M, Yu RT, Jonker JW, Alaynick WA, Banayo E, Karunasiri MS, Lorca S, Evans RM. 2011. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metab 13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. 2003. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol Cell Biol 23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, Tang X, Straubhaar J, Cherniack AD, Parker MG, Czech MP. 2006. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Investig 116:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A. 2007. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A 104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huss JM, Garbacz WG, Xie W. 2015. Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim Biophys Acta 9:24. [DOI] [PubMed] [Google Scholar]
- 51.Sonoda J, Laganiere J, Mehl IR, Barish GD, Chong LW, Li X, Scheffler IE, Mock DC, Bataille AR, Robert F, Lee CH, Giguere V, Evans RM. 2007. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev 21:1909–1920. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michalek RD, Gerriets VA, Nichols AG, Inoue M, Kazmin D, Chang CY, Dwyer MA, Nelson ER, Pollizzi KN, Ilkayeva O, Giguere V, Zuercher WJ, Powell JD, Shinohara ML, McDonnell DP, Rathmell JC. 2011. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proc Natl Acad Sci U S A 108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsakas A, Yadav V, Lorca S, Evans RM, Narkar VA. 2012. Revascularization of ischemic skeletal muscle by estrogen-related receptor-gamma. Circ Res 110:1087–1096. doi: 10.1161/CIRCRESAHA.112.266478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein RA, Gaillard S, McDonnell DP. 2009. Estrogen-related receptor alpha induces the expression of vascular endothelial growth factor in breast cancer cells. J Steroid Biochem Mol Biol 114:106–112. doi: 10.1016/j.jsbmb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang J, Han F, Chen Y. 2013. Transcriptional regulation of VEGF expression by estrogen-related receptor γ. Acta Pharmaceutica Sinica B 3:373–380. doi: 10.1016/j.apsb.2013.09.001. [DOI] [Google Scholar]
- 56.Sobczak M, Dargatz J, Chrzanowska-Wodnicka M. 2010. Isolation and culture of pulmonary endothelial cells from neonatal mice. J Vis Exp 14:2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korff T, Augustin HG. 1999. Tensional forces in fibrillar extracellular matrices control directional capillary sprouting. J Cell Sci 112:3249–3258. [DOI] [PubMed] [Google Scholar]
- 58.Pfisterer L, Korff T. 2016. Spheroid-based in vitro angiogenesis model, p 167–177. In Martin SG, Hewett PW (ed), Angiogenesis protocols. Springer New York, New York, NY. doi: 10.1007/978-1-4939-3628-1_11. [DOI] [PubMed] [Google Scholar]
- 59.Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, Beaulieu V, Gao J, Turner G, Markovits J. 2010. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem 285:22619–22629. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, Evans RM. 2007. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab 6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 61.Vega RB, Kelly DP. 1997. A role for estrogen-related receptor alpha in the control of mitochondrial fatty acid beta-oxidation during brown adipocyte differentiation. J Biol Chem 272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- 62.Cruys B, Wong BW, Kuchnio A, Verdegem D, Cantelmo AR, Conradi LC, Vandekeere S, Bouche A, Cornelissen I, Vinckier S, Merks RM, Dejana E, Gerhardt H, Dewerchin M, Bentley K, Carmeliet P. 2016. Glycolytic regulation of cell rearrangement in angiogenesis. Nat Commun 7:12240. doi: 10.1038/ncomms12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Bock K, Georgiadou M, Carmeliet P. 2013. Role of endothelial cell metabolism in vessel sprouting. Cell Metab 18:634–647. doi: 10.1016/j.cmet.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 64.Treps L, Conradi LC, Harjes U, Carmeliet P. 2016. Manipulating angiogenesis by targeting endothelial metabolism: hitting the engine rather than the drivers-a new perspective? Pharmacol Rev 68:872–887. doi: 10.1124/pr.116.012492. [DOI] [PubMed] [Google Scholar]
- 65.Rothhammer T, Bataille F, Spruss T, Eissner G, Bosserhoff AK. 2007. Functional implication of BMP4 expression on angiogenesis in malignant melanoma. Oncogene 26:4158–4170. doi: 10.1038/sj.onc.1210182. [DOI] [PubMed] [Google Scholar]
- 66.Yadav SS, Narayan G. 2014. Role of ROBO4 signalling in developmental and pathological angiogenesis. Biomed Res Int 2014:683025. doi: 10.1155/2014/683025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Y, Williams J, Smallwood PM, Nathans J. 2015. Sox7, Sox17, and Sox18 cooperatively regulate vascular development in the mouse retina. PLoS One 10:e0143650. doi: 10.1371/journal.pone.0143650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukumura D, Gohongi T, Kadambi A, Izumi Y, Ang J, Yun CO, Buerk DG, Huang PL, Jain RK. 2001. Predominant role of endothelial nitric oxide synthase in vascular endothelial growth factor-induced angiogenesis and vascular permeability. Proc Natl Acad Sci U S A 98:2604–2609. doi: 10.1073/pnas.041359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamalice L, Le Boeuf F, Huot J. 2007. Endothelial cell migration during angiogenesis. Circ Res 100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 70.Cao G, O'Brien CD, Zhou Z, Sanders SM, Greenbaum JN, Makrigiannakis A, DeLisser HM. 2002. Involvement of human PECAM-1 in angiogenesis and in vitro endothelial cell migration. Am J Physiol Cell Physiol 282:C1181–C1190. doi: 10.1152/ajpcell.00524.2001. [DOI] [PubMed] [Google Scholar]
- 71.van Hinsbergh VW, Koolwijk P. 2008. Endothelial sprouting and angiogenesis: matrix metalloproteinases in the lead. Cardiovasc Res 78:203–212. doi: 10.1093/cvr/cvm102. [DOI] [PubMed] [Google Scholar]
- 72.Ishida T, Kundu RK, Yang E, Hirata K, Ho YD, Quertermous T. 2003. Targeted disruption of endothelial cell-selective adhesion molecule inhibits angiogenic processes in vitro and in vivo. J Biol Chem 278:34598–34604. doi: 10.1074/jbc.M304890200. [DOI] [PubMed] [Google Scholar]
- 73.Yang H, Zhang H, Ge S, Ning T, Bai M, Li J, Li S, Sun W, Deng T, Zhang L, Ying G, Ba Y. 2018. Exosome-derived miR-130a activates angiogenesis in gastric cancer by targeting C-MYB in vascular endothelial cells. Mol Ther 26:2466–2475. doi: 10.1016/j.ymthe.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Audet-Walsh É, Papadopoli DJ, Gravel S-P, Yee T, Bridon G, Caron M, Bourque G, Giguère V, St-Pierre J. 2016. The PGC-1alpha/ERRalpha axis represses one-carbon metabolism and promotes sensitivity to anti-folate therapy in breast cancer. Cell Rep 14:920–931. doi: 10.1016/j.celrep.2015.12.086. [DOI] [PubMed] [Google Scholar]
- 76.Potente M, Carmeliet P. 2017. The link between angiogenesis and endothelial metabolism. Annu Rev Physiol 79:43–66. doi: 10.1146/annurev-physiol-021115-105134. [DOI] [PubMed] [Google Scholar]
- 77.Xu Y, An X, Guo X, Habtetsion TG, Wang Y, Xu X, Kandala S, Li Q, Li H, Zhang C, Caldwell RB, Fulton DJ, Su Y, Hoda MN, Zhou G, Wu C, Huo Y. 2014. Endothelial PFKFB3 plays a critical role in angiogenesis. Arterioscler Thromb Vasc Biol 34:1231–1239. doi: 10.1161/ATVBAHA.113.303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, Heggermont W, Godde L, Vinckier S, Van Veldhoven PP, Eelen G, Schoonjans L, Gerhardt H, Dewerchin M, Baes M, De Bock K, Ghesquiere B, Lunt SY, Fendt SM, Carmeliet P. 2015. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 520:192–197. doi: 10.1038/nature14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalucka J, Bierhansl L, Conchinha NV, Missiaen R, Elia I, Bruning U, Scheinok S, Treps L, Cantelmo AR, Dubois C, de Zeeuw P, Goveia J, Zecchin A, Taverna F, Morales-Rodriguez F, Brajic A, Conradi LC, Schoors S, Harjes U, Vriens K, Pilz GA, Chen R, Cubbon R, Thienpont B, Cruys B, Wong BW, Ghesquiere B, Dewerchin M, De Bock K, Sagaert X, Jessberger S, Jones EAV, Gallez B, Lambrechts D, Mazzone M, Eelen G, Li X, Fendt SM, Carmeliet P. 2018. Quiescent endothelial cells upregulate fatty acid beta-oxidation for vasculoprotection via redox homeostasis. Cell Metab 28:881–894. doi: 10.1016/j.cmet.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 80.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 81.Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eglinger J, Karsjens H, Lammert E. 2017. Quantitative assessment of angiogenesis and pericyte coverage in human cell-derived vascular sprouts. Inflamm Regen 37:016–0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carpentier GMM, Courty J, Cascone I. 2012. Angiogenesis analyzer for ImageJ, p 198–201. 4th ImageJ User and Developer Conference Proceedings. Centre de Recherche Henri Tudor, Luxembourg, Belgium. [Google Scholar]
- 84.Tual-Chalot S, Allinson KR, Fruttiger M, Arthur HM. 2013. Whole mount immunofluorescent staining of the neonatal mouse retina to investigate angiogenesis in vivo. J Vis Exp 9:50546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zudaire E, Gambardella L, Kurcz C, Vermeren S. 2011. A computational tool for quantitative analysis of vascular networks. PLoS One 6:e27385. doi: 10.1371/journal.pone.0027385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:2008–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li QB, James B, Huang H, Bickel PJ. 2011. Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5:1752–1779. doi: 10.1214/11-AOAS466. [DOI] [Google Scholar]
- 89.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, Rando OJ, Birney E, Myers RM, Noble WS, Snyder M, Weng Z. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.