The liver X receptors α and β (LXRα and LXRβ) are oxysterol-activated transcription factors that coordinately regulate gene expression that is important for cholesterol and fatty acid metabolism. In addition to their roles in lipid metabolism, LXRs participate in the transcriptional regulation of macrophage activation and are considered potent regulators of inflammation.
KEYWORDS: LXR, liver X receptor, gene expression, inflammation, macrophage, nuclear receptor, transcription
ABSTRACT
The liver X receptors α and β (LXRα and LXRβ) are oxysterol-activated transcription factors that coordinately regulate gene expression that is important for cholesterol and fatty acid metabolism. In addition to their roles in lipid metabolism, LXRs participate in the transcriptional regulation of macrophage activation and are considered potent regulators of inflammation. LXRs are highly similar, and despite notable exceptions, most of their reported functions are substantially overlapping. However, their individual genomic distribution and transcriptional capacities have not been characterized. Here, we report a macrophage cellular model expressing equivalent levels of tagged LXRs. Analysis of data from chromatin immunoprecipitation coupled with deep sequencing revealed that LXRα and LXRβ occupy both overlapping and exclusive genomic regulatory sites of target genes and also control the transcription of a receptor-exclusive set of genes. Analysis of genomic H3K27 acetylation and mRNA transcriptional changes in response to synthetic agonist or antagonist treatments revealed a putative mode of pharmacologically independent regulation of transcription. Integration of microarray and sequencing data enabled the description of three possible mechanisms of LXR transcriptional activation. Together, these results contribute to our understanding of the common and differential genomic actions of LXRs and their impact on biological processes in macrophages.
INTRODUCTION
Macrophages are professional phagocytic cells that play crucial roles in immune processes such as pathogen clearance and cytokine secretion, but they also perform other important functions in the regulation of metabolism and maintenance of tissue homeostasis (1). Macrophages exhibit a unique genetic plasticity, particularly at the precursor stage, when myeloid progenitors from different ontogenetic origins (yolk sac, fetal liver, and adult blood monocytes) can give rise to most types of tissue macrophages (2). Transcriptional control of macrophage gene expression is orchestrated by a cross talk between myeloid-specific master regulators, a small set of lineage-determining transcription factors, and chromatin-remodelling enzymes involved in epigenetic modifications, all acting on key enhancer genomic regions (3–5). Recent studies have also demonstrated that adult macrophages from different anatomic locations present a particular transcriptional profile that is decisively determined by their local environment (6).
The liver X receptors α and β (LXRα and LXRβ; encoded by Nr1h3 and Nr1h2, respectively) are transcription factors belonging to the nuclear receptor superfamily that bind to the DNA as obligate heterodimers with the retinoid X receptors (RXR). LXRs are sterol-sensing transcription factors that play essential roles in lipid and cholesterol metabolism and the immune response (7–9). LXRs control the expression of several genes that are pivotal for reverse cholesterol transport as well as fatty acid and phospholipid metabolism. Several studies have demonstrated that LXRα activity is predominant in the control of cholesterol and fatty acid metabolism in the liver (10–12), where its expression is markedly higher than that of LXRβ. LXRα is also expressed in adipose tissue, intestine, kidney, and macrophages (9). On the other hand, LXRβ is expressed ubiquitously (13). Naturally occurring cholesterol derivatives, named oxysterols, have been shown to be potent LXR activators in vitro and in vivo (14–18). In vivo administration of synthetic LXR ligands has shown beneficial effects in several animal models of disease, including atherosclerosis, Alzheimer’s disease, and psoriasis (19; reviewed in reference 20). In addition, however, LXR ligands promote an elevation of plasma triglyceride levels and liver steatosis due to hepatic induction of the master regulator of the lipogenic pathway, SREBP1c (encoded by Srebf1) (21, 22). Ever since these discoveries, the design of LXRβ-specific synthetic agonists to treat metabolic disorders or inflammatory diseases, avoiding de novo lipogenesis, has been a challenging effort (7). Interestingly, recent studies have shown promising therapeutic potential of novel compounds (23, 24) with immunomodulatory and antineoplasic activities (25).
LXRα and LXRβ proteins share 77% sequence homology, and most gene regulatory functions are believed to be performed similarly by LXRα and LXRβ (13). Initial studies, using electrophoretic mobility shift assays (EMSA) and promoter analyses, identified direct repeats of the classic nuclear receptor-binding motif AGGTCA separated by four nucleotides (DR4) as high-affinity binding sites for LXR-RXR heterodimers (9). This sequence binding preference has largely been confirmed by previous genome-wide chromatin immunoprecipitation experiments. However, these initial analyses using chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) were performed in cells expressing unequal levels of LXRα and LXRβ and antibodies that do not discriminate between the two LXRs (18, 26–28).
Receptor-exclusive functions have been described for LXRα, such as transcriptional control of Cd5l expression (29) or the differentiation of the splenic marginal-zone macrophages (30). Transcriptional regulation of target gene expression orchestrated preferentially by LXRβ has also been described (31–33). However, most LXR isoform-specific functions have been ascribed to the prominent expression of a particular receptor in a given cell type. A detailed analysis of specific LXRα and LXRβ transcriptional actions has not been conducted to date.
In this study, we developed a macrophage cellular model that stably expresses FLAG-tagged versions of either LXRα or LXRβ in an LXR-deficient background. Reconstituted cells were used to dissect LXR individual transcriptional actions and binding pattern dynamics to mouse genome upon targeting with commonly used synthetic LXR agonist and antagonist. Using microarray data in combination with ChIP-sequencing data, we identify novel mechanisms of LXR-mediated gene activation, involving LXR pharmacologically dependent and independent activation. This approach will contribute to better characterization of LXRα and LXRβ common and differential genomic actions that further impact biological processes in macrophages.
RESULTS
LXRα and LXRβ expression and activity in macrophage culture in vitro models.
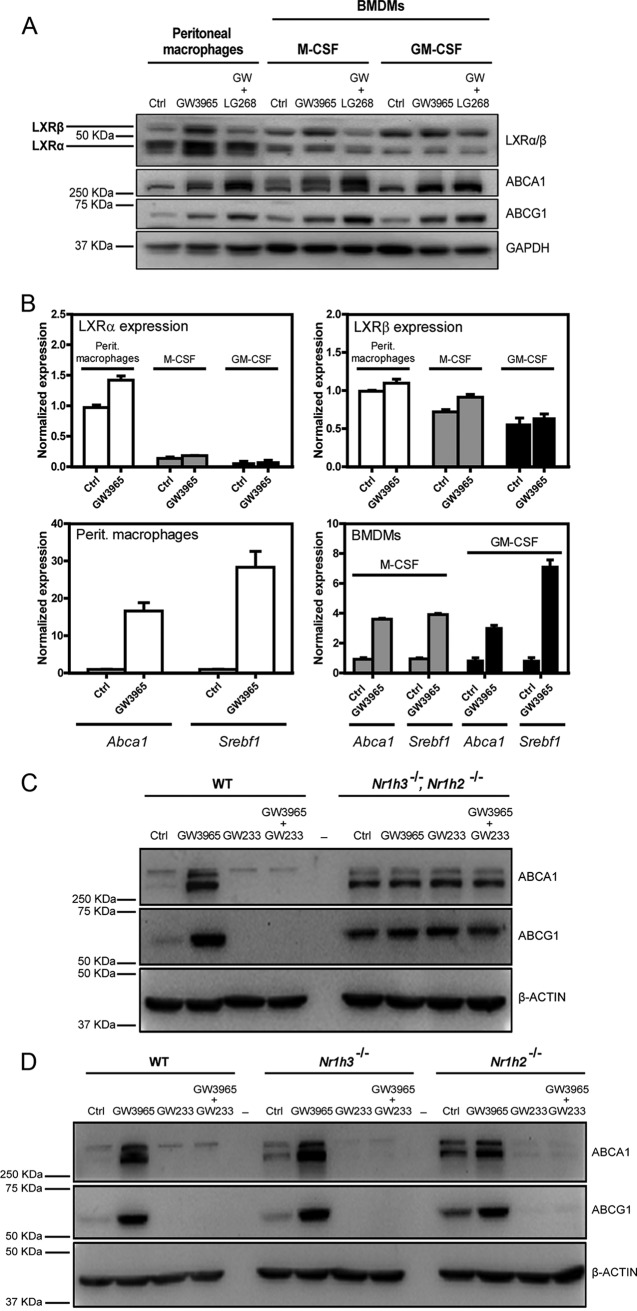
Traditionally, LXR-specific biological functions in the macrophage have been characterized using pharmacological strategies combined with genetic receptor deficiency. However, the relative expression levels of the LXRα and LXRβ proteins in most prior studies were not carefully defined, and in many cases LXRα and LXRβ protein levels were simply assumed to be equivalent across different macrophage populations. We examined the protein expression of LXRα and LXRβ in thioglycolate-elicited peritoneal and bone marrow-derived macrophages, differentiated with macrophage colony-stimulating factor (M-CSF) or granulocyte-macrophage colony-stimulating factor (GM-CSF) (Fig. 1A). The highest expression of LXRα was found in elicited peritoneal macrophages, followed by M-CSF-derived macrophages, whereas the lowest LXRα expression was displayed by GM-CSF-derived cells. In contrast, LXRβ was similarly expressed across all the tested macrophage types. These results were further confirmed by real-time quantitative PCR (RT-qPCR) (Fig. 1B). Additionally, we found that endogenous LXRs displayed posttranscriptional protein stabilization when exposed to the synthetic LXR agonist GW3965, in agreement with previous reports describing these effects in human cells (34). We also found that the degree of target gene induction varied with the type of macrophage (Fig. 1B).
FIG 1.
Protein and RNA levels of LXRα and LXRβ in different in vitro macrophage models. (A and B) Expression levels of LXRα, LXRβ, ABCA1, and ABCG1 from murine thioglycolate-elicited peritoneal macrophages and bone marrow-derived macrophages differentiated with M-CSF and GM-CSF cytokines were tested by Western blotting after cell treatment with LXR and RXR synthetic ligands (A) and by qPCR under GW3965 ligand treatment conditions (B). Drug antagonism mediated by GW233 on LXR target gene expression was investigated on LXR-WT and LXR-DKO peritoneal murine macrophages by Western blotting. (C) Cells were cultured with GW3965 (1 μM) or GW233 (1 μM), alone or in combination, for 24 h. (D) The ability of GW233 to target both LXR nuclear receptors was tested on LXR-WT and LXR single-knockout macrophage cells under culture conditions similar to those described for panel C. One representative experiment out of three is presented in each case, and mean (SD) values from qPCR triplicates are shown in panel B.
We next used an LXR-specific agonist and antagonist (GW3965 and GW233, respectively) to study LXR actions in wild-type (WT), LXR double knockout (Nr1h3−/− Nr1h2−/−) (LXR-DKO), and single knockout (Nr1h3−/− or Nr1h2−/−) primary peritoneal macrophages. As expected, GW3965 was able to effectively induce the expression of Abca1 and Abcg1 in LXR-WT peritoneal macrophages, but its activity was completely abolished in the presence of GW233 (Fig. 1C). In contrast, these drugs were ineffective in LXR-DKO cells, which, conversely, displayed elevated levels of ABCA1 and ABCG1 proteins, in agreement with previous reports (35, 36). Second, the ability of GW233 to effectively target each of the LXR nuclear receptors was tested in cells expressing only LXRα or LXRβ (Fig. 1D). GW233 blocked the expression of Abca1 and Abcg1 to the same extent in LXRα−/− and LXRβ−/− cells. These results establish the pharmacological action of GW233 in primary murine macrophages, showing its ability to effectively target both LXRα and LXRβ nuclear receptors.
Ectopic expression of LXRα and LXRβ in immortalized macrophages (iBMDM).
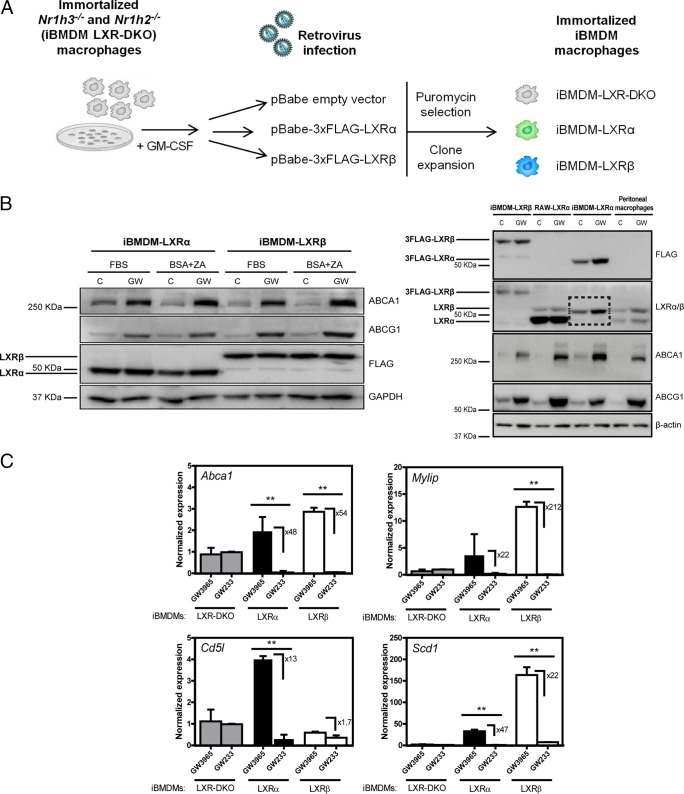
To be able to pinpoint common and LXR-specific transcriptional actions and to gain better insight into the molecular interaction networks underlying LXR biological effects, we developed an immortalized bone marrow-derived macrophage cell model expressing one LXR at a time. Initially, an immortalized LXR-DKO bone marrow macrophage cell line was established as described previously (37–39). The expression of LXRα and LXRβ receptors next was reconstituted separately in this LXR-DKO parental cell line in order to obtain two additional immortalized cell lines (Fig. 2A). The virally expressed LXR proteins were tagged with FLAG (3×FLAG-LXR) to normalize LXR protein recognition in both cell lines using FLAG antibody. We selected FLAG-positive clones exhibiting similar expression of LXRα and LXRβ. Importantly, we also selected for lines in which the level of LXR protein expression was not excessively higher than that in primary peritoneal macrophages (Fig. 2B and C). Because in vitro primary BMDM and many in vivo tissue macrophages present low levels of LXRα expression (6), we used elicited peritoneal macrophages to compare with our clones. For simplicity, these immortalized cell lines will be referred to as iBMDM-LXR-DKO, iBMDM-LXRα, and iBMDM-LXRβ. Reconstituted LXR cells effectively induced target genes such as Abca1, Abcg1, and others upon agonist treatment with GW3965 (Fig. 2B and C). As expected, the induction of LXRα-specific target gene Cd5l (also known as AIM [40]), analyzed by real-time qPCR, was only observed in the iBMDM-LXRα line upon stimulation with GW3965 (Fig. 2C). Thus, we developed and validated model cell lines with defined levels of LXR expression that respond to pharmacological stimulation, inducing target genes in the presence of an agonist and repressing them in the presence of an antagonist.
FIG 2.
Reconstitution of LXRα and LXRβ expression in immortalized macrophages from LXR-DKO bone marrow. (A) Outline of the experimental design for the generation of immortalized macrophage cell lines, iBMDMs, expressing FLAG-tagged LXRα, LXRβ, or no LXRs. (B, left) Whole-protein extracts from iBMDM-LXR macrophages cultured under different serum depletion conditions (see Materials and Methods) were analyzed by Western blotting for the expression of virally transduced LXRα or LXRβ. Induction levels of the LXR target genes ABCA1 and ABCG1 were also examined. GAPDH was used as a loading control. (B, right) LXRα and LXRβ protein expression was tested in whole-protein extracts from iBMDM macrophages, RAW cells virally transduced with LXRα (29), and WT peritoneal macrophages treated with GW3965 (1 μM). β-Actin was used as a loading control. The box in the LXRα/β panel indicates the specific 3FLAG-LXRα protein band, which shows a weight slightly similar to that of endogenous LXRβ protein. (C) Expression of dual and LXRα-specific (Cd5l) target genes upon GW3965 or GW233 (1 μM) treatment was examined by real-time qPCR. Results are represented as mean (±SD) values from three independent experiments. Asterisks indicate statistical significance between treatments: *, P < 0.05; **, P < 0.01.
LXRα- and LXRβ-specific binding and H3K27 acetylation through targeted ChIP.
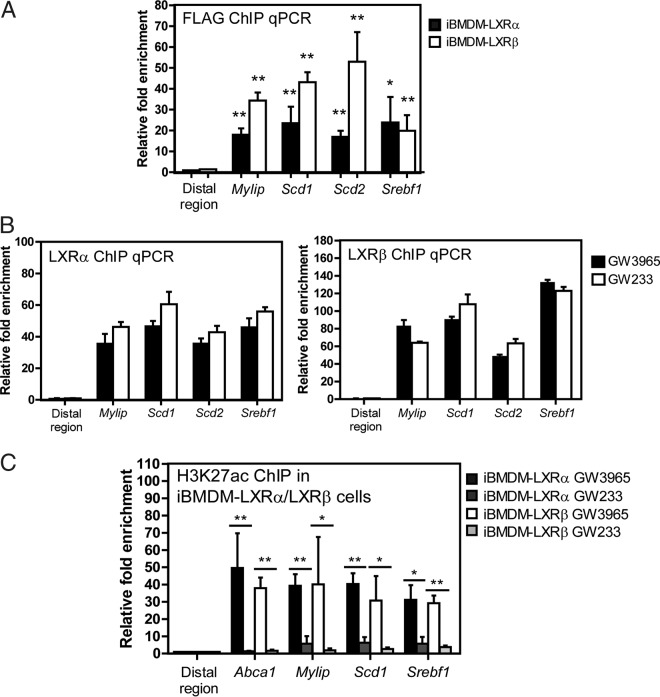
We optimized ChIP conditions for these macrophage lines using the monoclonal FLAG M2 antibody and targeted qPCR amplification of known LXR target gene regulatory sequences (Fig. 3A). To verify the effect of agonistic and antagonistic functions of GW3965 and GW233 on LXR binding ability, iBMDM cell lines were stimulated with GW3965 or GW233 (both 1 μM) for 24 h. LXR binding in the regulatory regions of selected target genes was assessed by ChIP-qPCR (Fig. 3B). We did not find differences in LXR binding to their DNA target sequences when comparing agonist to antagonist treatments. However, since these synthetic molecules promote protein stabilization, it is possible that liganded LXRs exhibit increased LXR-DNA interactions compared to vehicle, nontreated control cells.
FIG 3.
LXRα/β ligand-induced binding and histone H3 acetylation in the cis-acting regulatory regions of known LXR target genes. (A) LXR occupancy was detected in the regulatory sites of known target genes in iBMDM macrophages using anti-FLAG antibody. Data are expressed as mean (±SD) values from three independent experiments. Asterisks indicate statistical significance relative to an irrelevant distal region: *, P < 0.05; **, P < 0.01. (B) LXRα/β binding capacity to LXR regulatory sites was tested in cells cultured with GW3965 and GW233 (24 h, 1 μM). Data are expressed as mean (±SD) values from two independent experiments. (C) Acetylation/deacetylation dynamics of histone H3 (H3K27ac) upon iBMDM treatment with GW3965 and GW233 was examined by ChIP-qPCR. Statistical significance was calculated between treatments in each iBMDM-LXR cell line with unpaired Student´s t test. *, P < 0.05; **, P < 0.01.
We analyzed histone H3 tail acetylation (H3K27ac) at the regulatory genomic regions of known LXR target genes under agonist or antagonist stimulation. The alternating presence and absence of this histone mark under these treatments are indicative of cycles of highly accessible chromatin and compaction, associated with transcriptional activation and repression. Immunoprecipitation of acetylated regions was verified by qPCR in iBMDM LXR-expressing lines (Fig. 3C). Acetylation levels of regulatory regions of LXR targets were strongly dependent on the presence of agonist or antagonist (Fig. 3C).
LXRα and LXRβ display distinctive genome-wide binding signatures.
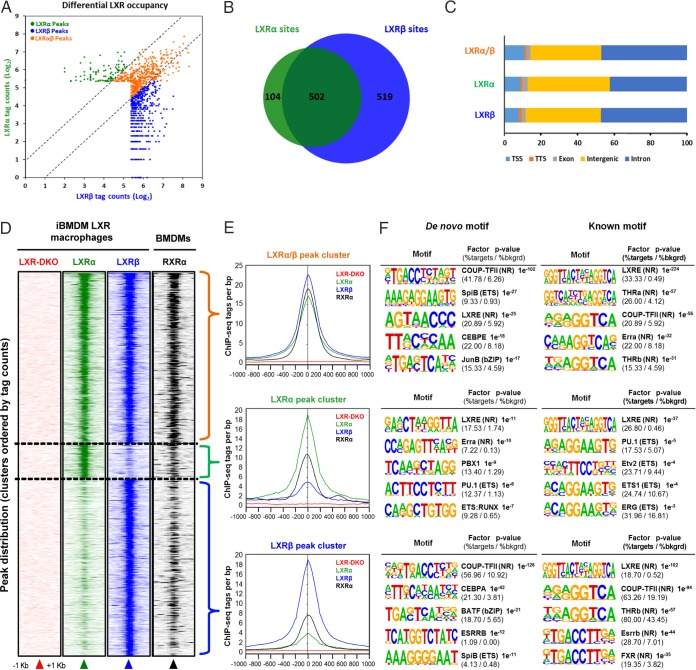
In order to study the individual contribution of LXRα and LXRβ receptors to the LXR genomic landscape, we performed ChIP-seq in iBMDM-LXR-DKO, iBMDM-LXRα, and iBMDM-LXRβ lines in response to GW3965. Details about peak calling, performed to discriminate background signal and false positives from significant LXR binding events, are given in Materials and Methods. Surprisingly, sequencing analysis revealed extensive differences in the number of genomic binding sites of LXRs (Fig. 4A and B). LXRβ was present at a total of 1,021 highly confident genomic locations, whereas LXRα could be detected at 606. Of all the sites, 502 were binding locations common to both LXRs, which represent 49% of LXRβ sites and 83% of LXRα sites. These data indicate that a large proportion of LXRα in cultured macrophages is bound to genomic sites that can be occupied by LXRβ as well (i.e., dual sites). In sharp contrast, almost 50% LXRβ binding was observed at selective sites that were not occupied by LXRα. The genomic distribution of genomic peaks (in reference to transcription start site/transcription termination site/exons/introns/intergenic regions) was similar between receptors (Fig. 4C). A complete list with all genomic locations of LXR peaks and their annotation to proximal genes is enumerated in Table S1 in the supplemental material.
FIG 4.
Genome-wide occupancy of LXRα and LXRβ nuclear receptors in iBMDM cells. (A) Genomic binding locations of LXRα and LXRβ nuclear receptors in iBMDM macrophages are represented in a scatter plot by receptor-normalized ChIP-seq tag counts (log2). (B) Number of unique and shared genomic LXR-bound sites, depicted as a Venn diagram. (C) Distribution of LXRα, LXRβ, and shared LXRα/β binding sites in reference to gene features are shown. TSS, transcription start site; TTS, transcription termination site. (D) Density heatmap of LXRα, LXRβ, and RXRα ChIP-seq peak intensities in a 2-kb window, detected in iBMDM and primary macrophages (accession number GSE63698). Genomic regions are clustered according to shared LXRα/β as well as LXRα- and LXRβ-specific occupancies. (E) LXR and RXR binding (ChIP sequencing tags per bp) in dual, LXRα, and LXRβ peak clusters. (F) Top five de novo and known sequence motif enrichment associated with LXR/RXRα-bound sites in iBMDM macrophages (see Table S3 in the supplemental material for a complete list). bkgrd, background.
We next correlated the binding of LXRs in iBMDM-LXRα and iBMDM-LXRβ immortalized cell lines with that of their heterodimeric partner, retinoid X receptor alpha (RXRα). We compared RXRα ChIP-seq data obtained from the public NCBI GEO database (accession number GSE63698) with our LXRα/β ChIP-seq data (Fig. 4D). As expected, the RXRα receptor mapped to sites that largely overlap those of LXR. RXRα peaks displayed higher tag counts in those genomic locations where LXRα and LXRβ were bound simultaneously (Fig. 4E). Accordingly, LXR/RXRα binding locations could be classified into three clusters, depending on dual or, alternatively, LXRα- or LXRβ-selective peaks.
To further characterize the sequence composition of regions with LXR binding and to predict coexisting transcription factor binding, we performed de novo and known motif sequence analysis with HOMER software on the three LXR/RXRα-bound clusters (Fig. 4F). Interestingly, LXR-binding clusters were associated with common and distinct patterns of transcription factor binding sequences (all motifs are listed in Table S2). The most enriched known motif in all clusters was the DR-4 element (LXR response element, or LXRE), followed by diverse nuclear receptor motifs in the LXRα/β and LXRβ clusters. De novo motif discovery analysis also revealed that LXRE and COUP-TFII motifs were the most robustly enriched sequence elements in all clusters. Strikingly, some sequence motifs, identified by either motif analysis strategy, were only significantly enriched in those peak set sequences bound exclusively by one LXR. This was the case for PBX1, found in the LXRα-specific peak set and BATF or C/EBP, identified in the LXRβ cluster (Fig. 4F and Table S2). The apparent interdependence between certain factors and a specific LXR-bound peak sets suggests a collaborative binding mechanism in either direction. Given the differential motifs associated with the sequences contained in the LXR peak clusters, we performed Gene Ontology (GO) term enrichment analysis of the genes annotated to these sets of peaks (Table S3). Besides the expected functions related to cholesterol and lipid metabolism, other functions associated with leukocyte and nonimmune cell homeostasis were found for the dual-peak cluster. The LXR-specific clusters were enriched for heterogeneous functions, most importantly, cell differentiation for the LXRα-specific cluster and DNA-binding activity and signal transduction for the LXRβ cluster.
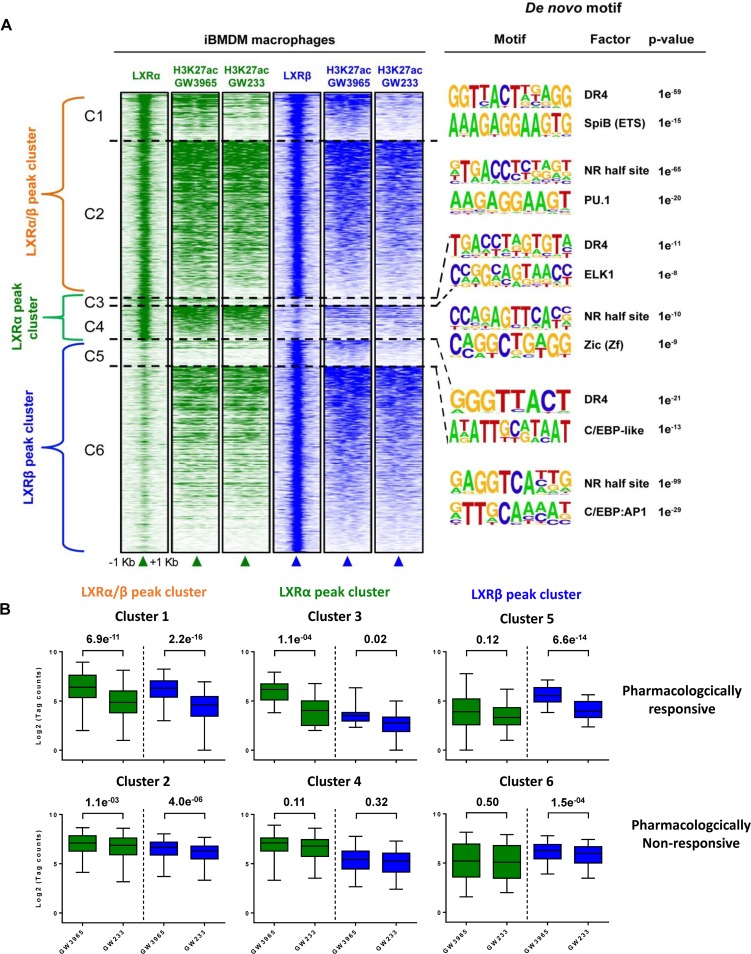
In order to gain a more comprehensive vision of the LXR genomic binding pattern and its relationship to transcriptional control of target gene expression, we performed H3K27ac ChIP-seq upon pharmacological treatment with GW3965 and GW233. As previously indicated, this epigenetic mark is a reliable indicator of active transcription (6, 41, 42). We used the same pharmacological strategy of agonist versus antagonist shown in Fig. 3C (without a baseline, vehicle-treated control) in order to get the most relevant information between maximal activation and repression. Analysis of H3K27ac changes located within LXR-bound regions (representing a 2-kb window around the LXR peak center), after a 24-h pharmacological treatment, is presented as a density heatmap in Fig. 5A. Surprisingly, we could identify two types of H3K27 acetylated regions: those pharmacologically responsive to synthetic compounds and those either poorly responsive or nonresponsive to pharmacological stimulation. Strikingly, the majority of the acetylated regions fell within this last category, in the three LXR-bound peak clusters. Accordingly, LXR peak-associated acetylated genomic regions could be further subdivided into clusters and arranged owing to pharmacological responsiveness (C1 and C2 for dual LXR-bound peaks, C3 and C4 for LXRα-selective peaks, and C5 and C6 LXRβ-selective peaks) (Fig. 5A). De novo motif analysis was performed with HOMER software on clusters C1 to C6, and the most representative predicted factors, with their associated P values, are indicated for each cluster. Importantly, an LXRE site was the most enriched sequence motif found in all C1 to C6 clusters. This finding clearly indicates that a bona fide classic LXR binding site (DR-4), and not indirect, alternative, or degenerate sites, mediates the recruitment of LXRα and LXRβ to their functional genomic locations.
FIG 5.
Genome-wide colocalization of LXRα/β binding peaks and their corresponding H3K27ac marks in iBMDM macrophages. (A) Changes in acetylation marks (H3K27ac) upon agonist and antagonist drug treatment of iBMDM macrophages were examined by ChIP-seq. Acetylated areas are represented as a density heatmap within a 2-kb window of centered LXRα/β, LXRα, and LXRβ peaks as described in the legend to Fig. 4D. LXR peak-associated acetylated genomic regions are subdivided into six clusters (C1 to C6) and arranged depending on pharmacological responsiveness. Clusters C1, C3, and C5, pharmacologically responsive acetylation marks; clusters C2, C4, and C6, weakly or nonresponsive acetylated regions. Top de novo sequence motifs identified in clusters C1 to C6 and their associated P values are indicated. (B) Box plot representation of genomic mean changes in H3K27ac mark intensity, measured as normalized tag counts (log2) in LXR peak subclusters (C1 to C6), after GW3965 and GW233 stimulation and P value changes.
We next focused on acetylated regions that showed a clear correlation with LXR binding, i.e., those where an LXR(s) could be found at the core of the open chromatin region. Box plot representation of mean acetylation tag counts (log2) revealed that H3K27ac changes experienced similar variations with pharmacological treatments at locations where LXRα or LXRβ is present (clusters C1, C3, and C5) (Fig. 5B, upper). These results indicate that analysis of H3K27ac changes, in the vicinity of LXR peak locations, does not distinguish relevant differences between LXRα and LXRβ transactivation power.
LXRα and LXRβ transcriptional profiling.
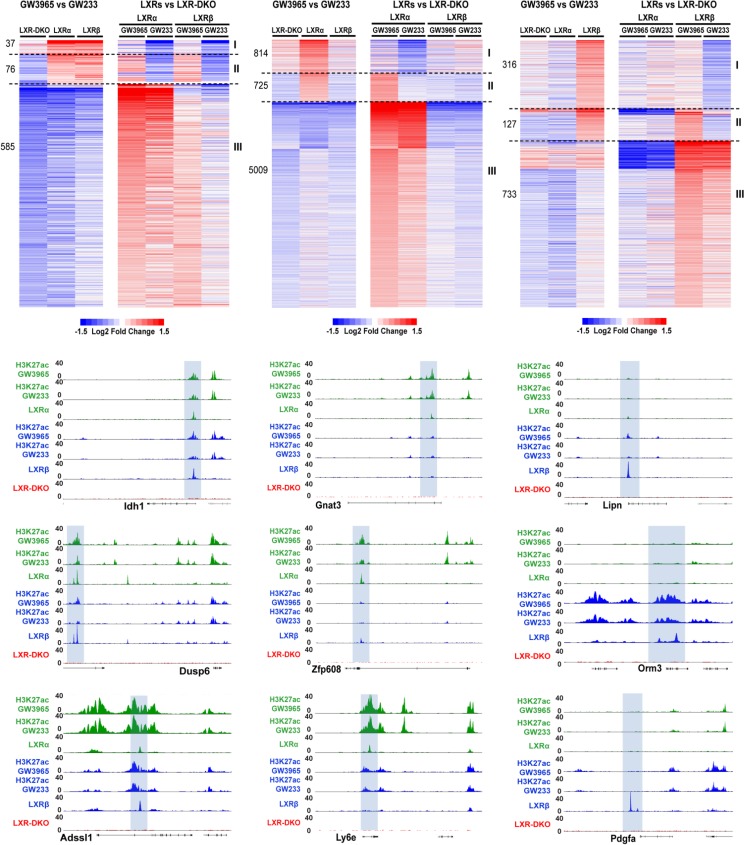
To explore the possibility that differential receptor binding was linked to the selective gene transcription profiles of LXRα and LXRβ in response to ligand, we performed genome-wide gene expression analysis with a mouse gene 2.0 ST Affymetrix microarray, using RNA from cells stimulated with GW3965 (maximal activation) and GW233 (control, maximal repression). Expression data were relativized in two ways: (i) expression values in response to GW3965 were represented relative to GW233 treatment in each iBMDM macrophage cell line, and alternatively, (ii) gene expression under each treatment condition was referenced to expression values obtained in iBMDM-LXR-DKO macrophages. One representation aimed to discover induced/repressed genes by synthetic compounds, and the second analysis focused on the analysis of genes induced/repressed by the ectopic expression of each LXR isoform relative to the LXR-DKO control. Fold changes in transcript levels were depicted in separate heatmaps, depending on LXRα/β-, LXRα-, or LXRβ-mediated (Fig. 6, left, middle, and right, respectively) transcriptional control. The number of transcripts induced in each case is also indicated on the left side of each heatmap.
FIG 6.
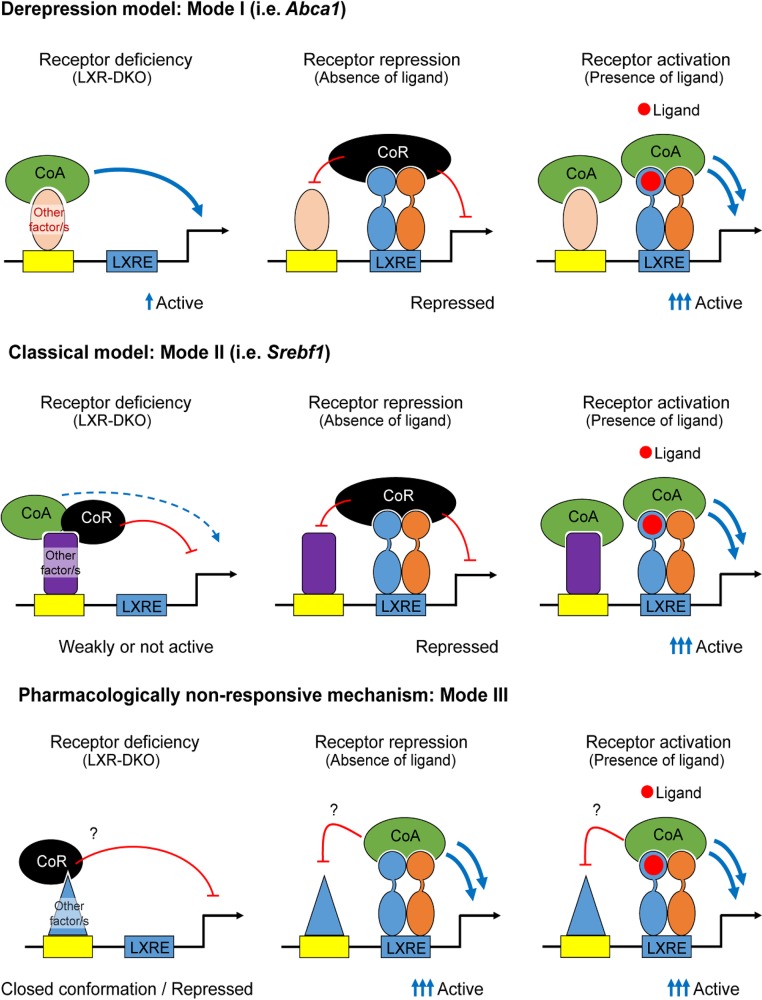
Expression profiling uncovers LXR dual and isoform-specific targets and reveals putative LXR transcriptional modes of action in response to an agonist/antagonist. Microarray analysis in iBMDM macrophages was performed using GW3965 and GW233 culture conditions as described in Materials and Methods. Heatmap panels are presented in sets of two, representing gene expression that depends on LXRα/β (dual targets, top left heatmaps), is LXRα selective (top middle heatmaps), or is LXRβ selective (top right heatmaps). Each pair of heatmaps shows fold changes in response to GW3965 relative to GW233 in each iBMDM cell line (left) or gene expression in response to each drug treatment relative to that of LXR-DKO iBMDMs (right). Relativized data within each category (LXRα/β [dual], LXRα selective, or LXRβ selective) highlight three possible mechanisms mediating gene activation (modes I, II, and III, as indicated at the right of each set of heatmaps). The number of transcripts regulated through each mechanism is indicated on the left. Lower panels show UCSC Genome Browser snapshots of representative genes as examples of each mechanism. Idh1, Dusp6, and Adssl1, gene loci for modes I, II, and III, respectively, of genes dually regulated by LXRα/β; Gnat3, Zfp608, and Ly6e, gene loci for modes I, II, and III, respectively, of genes regulated selectively by LXRα; and Lipn, Orm3, and Pdgfa, gene loci for modes I, II, and III, respectively, of genes regulated selectively by LXRβ.
Our analysis revealed three possible transcriptional activation mechanisms or modes of action, which we designated I, II, and III (heatmaps in Fig. 6). Mode of action I involves transcript induction in a pharmacologically responsive fashion and derepressed expression of the transcript in the absence of an LXR(s). Expression of genes in this class is higher in iBMDM-LXR-DKO cells than that in LXR-expressing iBMDMs under antagonistic conditions (GW233). Mode II represents the canonical model for transcriptional activation, where agonist binding to the LXR/RXR heterodimer triggers a conformational change, displacing the corepressor complex and facilitating the interaction with coactivator complexes. Expression of transcripts is highly dependent on LXR pharmacological activation and concomitant presence of the LXR(s) in the macrophage cell. Consequently, expression of these genes is higher with GW3965 treatment than iBMDM-LXR-DKO cells. Lastly, induction of transcripts in mode III occurred in a pharmacologically nonresponsive manner, but expression values were higher in LXR-expressing lines than in iBMDM-LXR-DKO macrophages under both agonistic and antagonistic treatment conditions, displaying a stark LXR dependence. Genome browser snapshots of representative genes and their associated acetylation modifications are shown below to illustrate each mode of action (Fig. 6, lower).
We found ∼100 transcripts upregulated jointly by LXRα and LXRβ after pharmacological activation (including mode I and mode II) (Fig. 6, top left). In contrast, GW3965-activated LXRα promoted the expression of a strikingly elevated number of genes (∼1,500 transcripts) (Fig. 6, top part of middle heatmap). LXRβ activation induced the expression of ∼450 genes (Fig. 6, top part of right heatmap). Modes of action I and II comprised less upregulated transcripts than mode III in all receptor categories. Surprisingly, the number of transcripts regulated by LXRα was an order of magnitude higher than that by LXRβ. These results highlight that LXRα, despite being present at a reduced number of genomic locations than LXRβ, is able to promote transactivation of a wide collection of genes (Fig. 6, middle). Collectively, these results provide the first indication that LXR nuclear receptors regulate gene expression through three distinct transcriptional modes of action.
We next assessed the global correlation between gene expression and LXRα and LXRβ occupancy. This type of analysis distinguishes putative targets that could be induced directly by the influence of nearby LXR binding or indirectly either by inducing the expression of other proteins or perhaps by a direct but distant regulation. We associated each LXR peak with the nearby upstream and downstream genes that appear within a 50-kb window. This correlation delivered two lists of genes: genes proximal to LXRα binding locations and genes proximal to LXRβ binding locations (within a ±50-kb window). The resulting gene lists were compared to our microarray gene expression profiling performed in iBMDM-3F-LXRα and iBMDM-3F-LXRβ, respectively, using ranked gene set enrichment analysis (GSEA). In both cases, GSEA analysis revealed that genes which present LXR binding in close proximity strongly clustered with genes that were upregulated by GW3965 agonist in the microarray gene expression list (Table S4, represented by red lines under the curve). Gene lists of each analysis (complete lists are in Table S4) show a high core enrichment score associated with those genes intensely regulated in the microarray. LXRα binding positively correlated with 138 genes regulated in the microarray, whereas LXRβ appears to be influencing the positive expression of 266 genes. These results suggest that LXRα modulates the expression of many genes, possibly through indirect mechanisms (only 138/1,500 correlation), whereas LXRβ binding is present in the vicinity of many of its regulated genes (266/450). Globally, these correlation studies strongly support the idea that direct binding of LXR to genomic regions promotes the induction of gene expression and not gene repression.
Bioinformatic analysis of pharmacologically sensitive and insensitive LXR dual or isotype-selective targets.
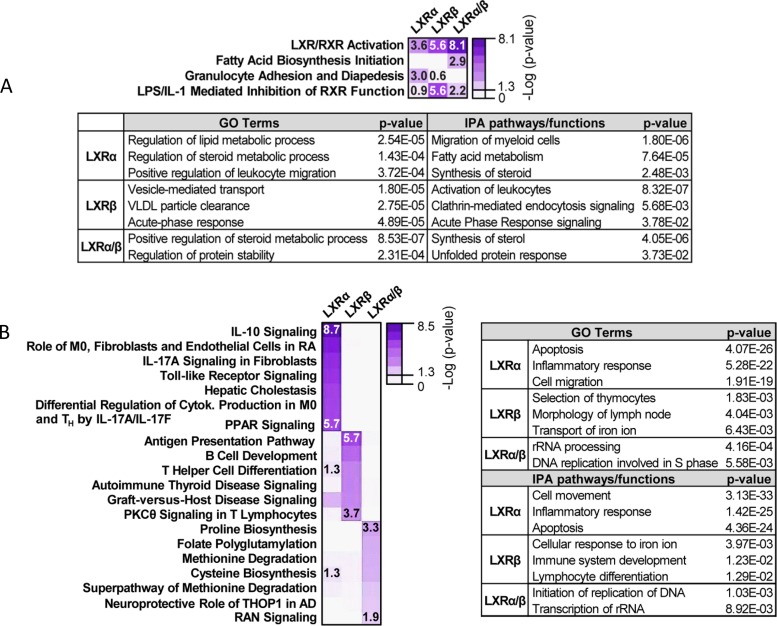
We further assessed the contribution of LXR-regulated (either dual or receptor-specific) genes to biological pathways in macrophages, focusing on the two main categories of mechanistically related LXR transcriptional activation: pharmacologically responsive (modes I and II) and weakly/nonresponsive (mode III). We performed Ingenuity Pathway Analysis (IPA) and Gene Ontology biological process enrichment bioinformatic analysis. Among the enriched functions for the pharmacologically responsive category, we found that LXRα/β and LXRα, in addition to be implicated in lipid metabolism, were also linked with pathways such as the unfolded protein response and leukocyte migration, respectively. LXRβ was strongly associated with the acute-phase response and vesicle-mediated transport (Fig. 7A and Table S5). Analysis on the second category (mode III) yielded a totally different array of functions for each LXR. This analysis linked the genes regulated by both LXRα and LXRβ (dually) to DNA replication and rRNA processing, LXRα-dependent genes to inflammatory responses, and LXRβ-regulated genes to lymphocyte differentiation, among other functions (Fig. 7B and Table S5).
FIG 7.
Gene Ontology analysis and IPA pathway annotation for microarray gene clusters. Biological pathway analysis was performed on genes that belong to pharmacologically responsive (up in GW3965/GW233 ratio, modes I and II) and nonresponsive (up when referred to iBMDM-DKO, mode III) clusters. (A) Most relevant IPA biological pathways associated with modes I and II (pharmacologically responsive) are depicted as a heatmap. Below, additional relevant GO terms and functions identified by IPA are shown. (B) Most relevant IPA biological pathways associated with mode III (pharmacologically nonresponsive) are depicted as a heatmap. Pathways were arranged by receptor dependence. The table on the right shows additional relevant GO terms and IPA functions. Right-tailed Fisher’s exact test P values for each case are shown. The highest, lowest, and borderline statistically significant P values are shown for each category.
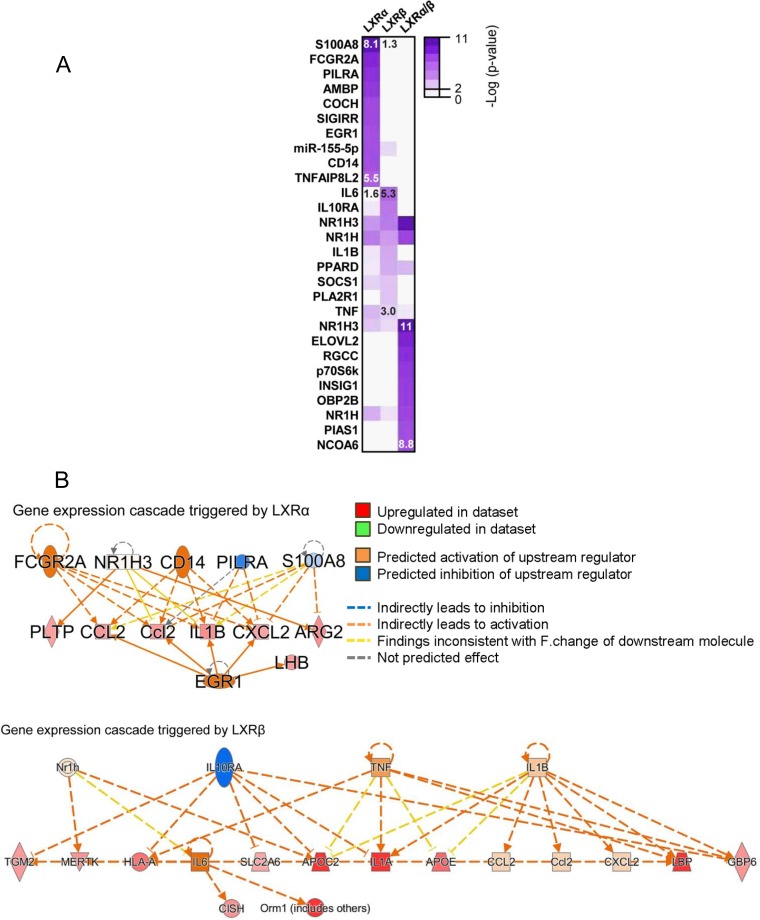
We next sought to identify upstream signaling pathways and factors that had been implicated in the regulation of these biological processes, acting as signaling hubs and regulating transcription of these clusters of genes. This type of bioinformatic analysis would predict possible ways in which LXR activity is connected to signaling factors that control the expression of these clusters of genes. These regulator pathways may amplify LXR activation or trigger parallel actions that activate biological pathways linked to LXRs. Within the pharmacologically responsive category, we found diverse molecules, such as the lipopolysaccharide coreceptor CD14, the transcription factor EGR1, and the chemotactic protein S100A8, associated with the LXRα-specific signaling cascade. On the other hand, molecules connecting gene expression regulated by LXRβ-specific activation were principally cytokines or cytokine-related genes, such as those for interleukin-6 (IL-6), IL-1β, tumor necrosis factor (TNF), or IL-10RA (Fig. 8). Thus, these data predict that LXRα (ligand-dependent responses) are involved in Toll-like receptor (TLR)-dependent immune responses and LXRβ participates in IL-1 and TNF signaling.
FIG 8.
Upstream signaling pathways connecting gene expression cascades triggered by LXR activity in a pharmacologically dependent manner (modes I and II). (A) Molecular regulators of gene expression networks associated with transcriptional modes I and II, identified with IPA. Heatmap color intensities correlate with significance of right-tailed Fisher’s exact test. (B) Diagrams showing molecular interaction networks between signaling regulators and pharmacologically active LXRα and LXRβ, leading to gene expression cascades. Predicted relationships among molecules yielded by IPA are indicated. The highest and lowest statistically significant P values are shown for each category.
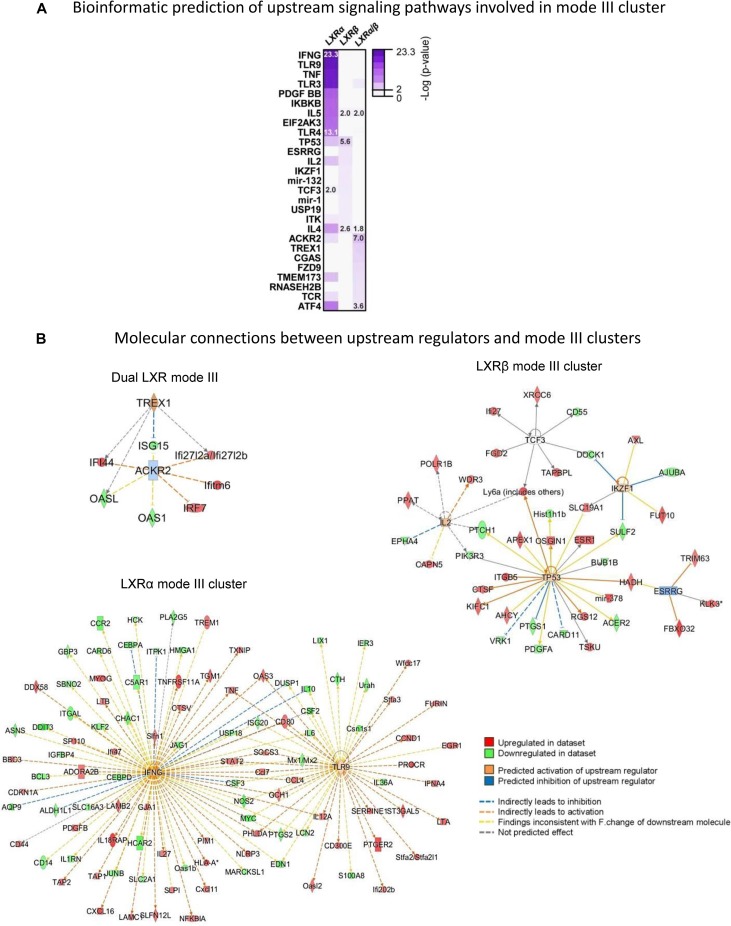
Interestingly, when we examined the computer prediction of molecular regulators of pharmacologically nonresponsive signaling cascades, we found that LXRα-dependent mode III genes relied on few molecular regulators, particularly on gamma interferon and TLR9, which displayed an extensive influence over a manifold of additional molecules, magnifying the initial activating event and triggering several signaling pathways (Fig. 9). On the other hand, LXRβ-dependent genes (insensitive to ligand stimulation) are connected through a wide variety of upstream molecules that display a more limited range of action, exemplified by TP53, ESRRγ, TCF3, and IL-2 (Fig. 9B, right). A similar situation was found for both LXRα/β receptors, which appear to influence signaling pathways through ACKR2 and TREX1. Biological pathway activation by these molecules was also examined by IPA (Table S6).
FIG 9.
Upstream signaling pathways connecting gene expression cascades triggered by LXR activity in a pharmacologically independent manner (mode III). (A) Molecular regulators of gene expression networks associated with transcriptional mode III, identified with IPA. Heatmap color intensities correlate with significance of right-tailed Fisher’s exact test. (B) Diagrams showing molecular interaction networks between signaling regulators and LXRs. Predicted relationships among molecules yielded by IPA are indicated. The highest and lowest statistically significant P values are shown for each category.
DISCUSSION
LXRα and LXRβ are nuclear receptors that play a crucial role in the control of whole-body cholesterol metabolism (43). Previous work from our laboratory and others has demonstrated that LXRs also participate in diverse aspects of macrophage transcriptional machinery, including inflammation and host defense (7, 44). Both LXRα and LXRβ proteins are present in macrophages, but their individual functions in macrophage models have not been conclusively addressed. Despite the fact that LXR activity has been extensively studied using synthetic agonists in culture systems, an absence of specific tools to isolate and manipulate LXRα and LXRβ proteins individually has limited our understanding of their specific roles in macrophage biological processes (45). Thus, the main objective of the present work was to characterize the distinctive transcriptional properties of LXRα and LXRβ in murine macrophages. Two main concepts arise from our study. The first is the striking difference in LXRα versus LXRβ genomic binding landscapes, despite the similarity of their direct DNA-binding motifs. The second is the different biological consequences of specific LXR DNA-binding events that reflect distinct modes of transcriptional regulation.
Since naturally occurring macrophage models express various levels of LXRα versus LXRβ (Fig. 1A and B) (6), we generated an immortalized macrophage cellular model (iBMDM) (37, 39) that expresses equivalent levels of each LXR separately. This iBMDM model has proven to be an effective way to interrogate macrophage functions in vitro, as these cells display expression markers and consistent characteristics of functional macrophages (37). Our iBMDM system expressing FLAG-tagged LXRs, reconstituted on an LXR-DKO genetic background, allowed us to unambiguously define characteristic receptor functions. Moreover, we clarified the specific actions of potent, commercially available pharmacological tools in this LXR reconstituted system: the nonsteroidal LXR agonist GW3965, widely accepted as potent stimulator of LXR activity (46), and the synthetic LXR antagonist GW233 (47). We also tested the ability of GW233 to inhibit GW3965-dependent induction of target genes in LXR single-knockout macrophages, which confirmed its potent antagonistic effect on both LXRα and LXRβ. Our iBMDM system appropriately reproduces LXR responses shared by LXRα and LXRβ, such as induction of classical dual target genes (48), as well as individual LXRα-specific transcription of Cd5l (29, 49).
Our genome-wide ChIP-seq analysis of LXRα versus LXRβ binding revealed a common group of DNA regions that can be occupied by both LXRα and LXRβ and a large set of distinctive LXRβ-specific peaks. The frequency of LXRα-exclusive binding regions was surprisingly lower. Thus, despite the high degree of similarity between both receptors, LXRβ is able to bind to a larger number of sites than LXRα in the macrophage genome. We initially expected to find many sites selectively bound by LXRα, as exclusive actions linked to this receptor have been previously described in vitro and in vivo (29, 30, 49). Remarkably, our peak filtering strategy used LXR-null cells and input DNA as negative controls that resulted in a robust set of curated peaks that exhibit DR-4 LXR binding motifs as the most enriched sequence found in all binding sites. Because previous studies used cell lines with uneven levels of LXRs or antibodies that do not discriminate between LXRα and LXRβ, we believe that our data sets represent the most accurate LXR binding repertoire described for macrophages (4, 26, 27). Recently, it was reported that LXR genes arose through a gene duplication event (50). However, it is unclear whether LXRα has acquired specialized functions related to lipid metabolism and immunity, losing its ability to regulate other genes, or whether an expansion of LXRβ-associated functions has occurred, resulting in LXRα occupying a reduced number of genomic sites in comparison.
It is noteworthy that our motif analysis identified accompanying sequences that were selectively associated with the binding of one LXR in particular. For example, C/EBP-like sites were not found in the LXRα-specific peak set, whereas PBX1 sequences were absent from the LXRβ peak cluster. Interestingly, a signaling cascade dependent on an LXRα-C/EBPβ interaction has recently been described as important for insulin induction of SREBP1c in the liver (51). In addition, a cooperative mechanism of action has been previously described for complexes containing the homeodomain protein PBX1. Interaction of Hox transcription factors with PBX1 complexes was demonstrated to be necessary to modulate their binding specificity in Drosophila (52). More recently, PBX1 pioneer binding ability for nonpermissive chromatin was identified in myoblasts, and this activity is believed to facilitate the targeting of MyoD for muscle lineage gene activation (53). It is possible that similar or distinctive interactions are also operating in macrophages. However, to prove the function of these factors in LXR-specific binding capacities, genetic manipulation of neighboring sequences and/or elimination of these factors will be necessary. Nevertheless, it is therefore plausible that, despite a canonical LXRE sequence being present in all of these LXR-specific clusters, the mutually exclusive binding of each LXR could be facilitated by interactions with a cohort of accessory factors that provide a permissive binding environment at these genomic locations.
We also used changes in H3K27 acetylation marks in response to agonist/antagonist as readout of transcriptional activation/repression differences between LXRs. Strikingly, we found that a remarkable number of the enhancer regions flanking LXR peaks displayed weak or no H3K27ac changes in response to pharmacological agonist/antagonist exposure. We hypothesize that this behavior is explained by one or both of the following possibilities: (i) the presence of LXR on these locations is important to confer a certain level of H3K27ac mark but does not promote acetylation modifications in response to ligand, and (ii) LXR binding could be playing a mere bystander role in these pharmacologically insensitive enhancer regions, and other factors could be critically contributing to the appearance of these acetylation marks, including pioneer factors or lineage-determining transcription factors (4).
Analysis of the microarray data set broadened and complemented our ChIP-seq data interpretation. We defined three different groups of genes that were associated with distinct putative mechanisms of transcriptional regulation (referred to as I, II, and III). These three activation modes were found to be employed by both LXRα and LXRβ. Mode I is the derepression mode, which is exemplified by the gold standard LXR target, Abca1. Mode I gene expression is higher under LXR-DKO control conditions than for the iBMDM-LXR lines, but their expression increases upon GW3965 stimulation (20). Mode II represents the canonical transcriptional activation mechanism that has been previously characterized in depth (35, 36), in which pharmacological responsiveness is accompanied by higher expression in LXRα- or LXRβ-expressing macrophages than with the LXR-DKO line. Interestingly, a large set of genes found by expression comparison does not fall in these two classic modes, and we propose here a distinct mode of action, called mode III, which represents pharmacologically nonresponsive transcriptional activation. Within this category of mode III, we observe that expression values of most genes do not respond to pharmacological antagonism with GW233 more so than with GW3965, but their expression is still significantly higher than that observed in LXR-DKO cells. It is possible that ectopic overexpression of LXRα or LXRβ, even if liganded by a potent antagonist, promotes the recruitment of coactivator complexes that results in higher RNA expression levels of a large set of targets than do similar conditions in LXR-DKO cells. However, future experiments are needed to directly test the differential requirement of coregulators in modes I and II versus mode III LXR-regulated gene expression. As mentioned above, this mode III comprises groups of transcripts regulated dually by both LXRα and LXRβ or exclusively by LXRα or LXRβ.
Our bioinformatics analyses suggested that LXRα and LXRβ participate in specific biological functions beyond fatty acid and steroid metabolism. For example, LXRα-selective gene regulation was found to be linked to apoptosis and leukocyte migration. Interestingly, we have recently demonstrated that LXRs regulate leukocyte chemotaxis (54). It will be interesting to validate whether LXRα has a prominent role over LXRβ in leukocyte migration. On the other hand, specific functions for LXRβ identified by Gene Ontology and IPA analysis were linked to selection of thymocytes and lymphocyte differentiation. Remarkably, binding sites for BATF, which is important for lymphoid progenitor and Th differentiation (55, 56), were enriched in the LXRβ-selective cluster. Thus, it is possible that LXRβ cooperates with BATF in pathways related to lymphocyte activation. Processes controlled by LXRβ in lymphocytes that regulate proliferation and the acquired immune response have previously been reported (31).
In conclusion, our data provide compelling evidence that LXRα and LXRβ bind to both common and distinct regulatory sequences in the genome and exert transcriptional control over a wide range of macrophage pathways. Importantly, these studies highlight the importance of LXRs in direct transcriptional regulation of immune-related functions. Moreover, the integration of our DNA binding and RNA expression data reveal three distinct modes of transcriptional regulation by LXRs (depicted as models in Fig. 10). Particularly important is the recognition that most LXR target genes are not responsive to ligand (mode III). In the future it will be important to link the specific LXRα and LXRβ regulatory actions uncovered here to biological functions in different tissue-resident macrophage populations, especially in the context of steady-state homeostasis or disease.
FIG 10.
Proposed mechanisms for LXR nuclear receptor transcriptional activation. Through integration of gene expression and genome binding data, three possible transcriptional LXR-mediated mechanisms or modes (namely, I, II, and III) are proposed.
MATERIALS AND METHODS
Mice.
WT, LXRα-deficient (Nr1h3−/−), LXRβ-deficient (Nr1h2−/−), and LXRα/β-deficient (Nr1h3−/− Nr1h2−/−) (denoted LXR-DKO) mice on a mixed Sv129/C57BL/6 background were originally provided by David Mangelsdorf (UTSW) (11). All mice were maintained under pathogen-free conditions in a temperature-controlled room and a 12-h light-dark cycle in the animal facilities of Universidad de Las Palmas de Gran Canaria (ULPGC). All animal studies were conducted in accordance with institutional participants’ animal ethics research committees (protocol CEEA-ULPGC 2015-002 and resolution 414-2015-ULPGC).
Cell culture and macrophage differentiation.
Thioglycolate-elicited peritoneal macrophages were obtained through injection of 3 ml of 3% sterile thioglycolate (BD Difco), pH 7.0, and after 3 days macrophages were collected after washing the peritoneal cavity 3 times with cold phosphate-buffered saline (PBS). All cells were cultured in Dulbecco´s modified Eagle´s medium (DMEM; Lonza) supplemented with 10% fetal bovine serum (Gibco), penicillin (100 U/ml) (Sigma), and streptomycin (100 μg/ml) (Sigma). For BMDM cell differentiation, bone marrow from femur and tibia of 5- to 7-week-old WT or LXR-DKO mice were isolated and cultured for 7 days in DMEM supplemented with 10% conditioned medium containing M-CSF or GM-CSF and 1% antibiotics (penicillin and streptomycin) (Sigma).
Immortalization of murine macrophages from bone marrow and expression of FLAG-tagged LXRα and LXRβ receptors.
Bone marrow-derived macrophages were immortalized using J2 retrovirus as previously described (37, 38, 57). Ectopic expression of LXRα or LXRβ was performed with a pBabe-based retroviral expression system. Briefly, Phoenix A cells at 90% confluence were transfected with the pBabe-3FLAG-LXRα or pBabe-3FLAG-LXRβ vector (58), expressing either LXRα or LXRβ nuclear receptors and carrying antibiotic resistance to ampicillin and puromycin. For transfection, 10 μg of plasmid and Lipofectamine 2000 (Thermo Fisher Scientific), 1:1.5, were used. After 6 h, the culture medium was replaced with complete DMEM, and after 48 h the medium containing viral particles was collected. Before exposing iBMDM-LXR-DKO cell culture to the viral supernatant, it was filtered through a 45-μm-pore-size filter and mixed with 10 μg/ml Polybrene (Sigma). Cells were cultured with puromycin (2 to 10 μg/ml gradually; Sigma-Aldrich) for 2 weeks. Several clones expressing LXRα or LXRβ were isolated and tested for similar expression using anti-FLAG M2 antibody. A detailed protocol of this procedure is available through a recent review (59).
Treatment with LXR synthetic ligands.
The following pharmacological treatments were used: 1 μM synthetic LXR ligand GW3965 (46) and synthetic LXR antagonist GSK1440233A (here denoted GW233) were both from GlaxoSmithKline (47) at 1 μM in dimethyl sulfoxide (Sigma) stock solution. Additionally, cells were subjected to cholesterol biosynthesis inhibitor culture conditions: serum-free DMEM, supplemented with 0.2% bovine serum albumin (BSA; Sigma), and 2 μM zaragozic acid (squalene synthase inhibitor; Sigma) for 4 h prior to exposure to the synthetic treatments.
Western blotting.
Whole-cell protein extracts were obtained with radioimmunoprecipitation assay buffer (RIPA; 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor; Complete; Roche). Protein extracts were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Primary antibodies that recognize ABCA1 (NB400-105; Novus), ABCG1 (NB400-132; Novus), FLAG M2 (F3165; Sigma), LXRα/β (kindly provided by Knut R. Steffensen, Karolinska Institute [27]), β-actin (sc-47778, C4; Santa Cruz), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; G9545; Sigma) were used. Secondary antibodies were horseradish peroxidase (HRP)-coupled anti-mouse and anti-rabbit antibodies (sc-2005 and sc-2004; Santa Cruz). Reactive bands were detected by Clarity Western ECL substrate (Bio-Rad). One representative Western blot from three independent experiments is shown in each case.
ChIP assay.
ChIP assay for the study of the LXR cistrome and variations in the acetylation status of lysine 27 on histone H3 (H3K27ac) was performed as follows. Cell fixation and cross-linking were performed. First, 2.5 × 107 macrophages were fixed with 2 μM disuccinimidyl glutarate (ThermoFisher Scientific) in PBS for 30 min. Cells next were washed with PBS and fixed for another 10 min with 1% methanol-free formaldehyde (ThermoFisher Scientific). The cross-linking reaction was quenched by adding glycine to a final concentration of 200 mM (Sigma). Chromatin extraction was performed in a two-step lysis reaction. First, a hypotonic buffer was used for nucleus extraction (50 mM Tris-HCl, pH 8, 85 mM KCl, 0.5% NP-40, supplemented with Complete [Roche] protease inhibitor). Second, chromatin was resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, Complete) and stored at −80°C. Chromatin then was sonicated with a Diagenode Bioruptor sonication device for 60 min (30 s on/30 s off) to generate 200- to 400-bp fragments, and 10% of the total volume was set aside to test fragment size (input material). Immunoprecipitation was performed with 4 μg of anti-FLAG M2 antibody or anti-H3K27ac (ab4729; Abcam) on 2 ml of previously diluted chromatin with dilution buffer (10 mM Tris-HCl, pH 8, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl, and 5% glycerol). Protein-bound immune complexes were captured with 100 μg of magnetic Dynabeads protein A (Thermo-Fisher Scientific). Unbound complexes were washed out with 3 buffers of increasing ionic strength: 20 mM Tris-HCl, pH 8, 2 mM EDTA, pH 8, 1% Triton X-100, 0.1% SDS, and either 150 mM (first buffer) or 500 mM NaCl (second buffer), and 10 mM Tris-HCl, 1% sodium deoxycholate, 1 mM EDTA, pH 8, 1% NP-40, 250 mM LiCl (third buffer). These washes were followed by 2 washes with TE buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, pH 8). DNA fragments were reverse cross-linked for 30 min at 37°C in 1% SDS, 0.1 M NaHCO3, 10 μl of 5 M NaCl, 6 μg/ml RNase A for 1 h at 55°C with 400 μg/ml proteinase K (TaKaRa). Column purification was performed with a Qiagen QIAquick PCR purification kit, and DNA was eluted in a final volume of 50 μl, 5 μl of which was used for qPCR amplification. Primers used for ChIP-qPCR analysis are listed in Table S7 in the supplemental material.
High-throughput sequencing.
ChIP DNA was quantified using a Qubit 2.0 fluorometer. To prepare libraries, a minimum of 2 ng of anti-FLAG-immunoprecipitated DNA and 1 to 2 ng of anti-H3K27Ac-immunoprecipitated DNA was pooled from 5 biological replicates per condition. Libraries were prepared by the Genomics Unit of the Centre de Regulacion Genomica (CRG; Barcelona, Spain) using the NEBNext Ultra DNA library preparation kit for Illumina (number 7370) by following the manufacturer’s instructions. Twelve cycles of PCR were done for the final library amplification for all samples. Sequencing was performed using Illumina HiSeq2000 equipment. For ChIP-seq, sequencing data (single-end 50-bp reads) obtained from Illumina HiSeq2000 were aligned to the UCSC mm10 genome using bowtie2 aligner (v2.2.9) (60). Each ChIP-seq experiment was normalized to a total number of 107 uniquely mapped tags. Aligned read files were visualized with IGV (61) genome browser and analyzed with HOMER software (http://homer.ucsd.edu/homer/) (v4.9). LXR peaks in each experiment were identified using HOMER and compared to data obtained from LXR-DKO samples as a negative control (4). The peak list was further filtered using a tag count cutoff of 40. This number was chosen by comparing the average tag count found in LXR peaks in each experiment. LXR peaks and H3K27Ac regions were clustered and represented as tag densities on a heatmap within a window of 4 kb around the LXR peaks using SEQminer (62). Ontology analysis of each LXR peak cluster was performed with the DAVID bioinformatics resource (63). Details of all applied bioinformatic analytical tools are available in a recent review protocol on this particular data processing method (64). GSEA was used to correlate gene expression data with LXR ChIP-seq data (65). GSEA analysis was performed using two lists: a preranked gene expression list obtained from microarray analysis and a list of genes obtained from the annotation of ChIP-seq peaks to the neighboring genes found on a window of ±50-kb using BedTools (66) and the mouse genome annotation of UCSC mm10.
RNA extraction, cDNA synthesis, and real-time qPCR.
Total RNA was extracted from iBMDM-LXR-DKO or iBMDM-LXRα/-LXRβ, using TRI Reagent (MRC) by following product specifications. The RNA pellet was resuspended with diethyl pyrocarbonate-treated water, and 1 μg was used for retrotranscription with an iScript cDNA synthesis kit (Bio-Rad). For RT-qPCR assay, 5 μl of cDNA was mixed with 15 μl of 2× PCR MasterMix (Diagenode) and 0.4 μM qPCR primer mix. Primers used for qPCR analysis are listed in Table S7. Fluorescence emission was captured with a CFX Connect thermal cycler (Bio-Rad). The relative levels of RNA were calculated according to the ΔΔCT method (where CT is threshold cycle), and individual expression data were normalized to 36B4 expression.
Microarray analysis and biological pathway analysis.
Changes in RNA expression promoted by ligand treatment (GW3965 and GW233) in immortalized macrophages were analyzed using a GeneChip mouse gene 2.0 ST array (Affymetrix). Raw expression values, obtained as log2 values and normalized to reference genes, were processed by the Genomic Unit of the Complutense University of Madrid. Heatmap representations were performed according to log-transformed values (log2) of fold changes in expression and arranged in decreasing order of magnitude. Gene Ontology biological process analysis (GO BP terms) and IPA were performed on transcripts classified in the three heatmap categories under program default settings. Only significant terms (P value of <10−2) are shown.
Statistical analysis.
Real-time quantitative PCR expression measurements and immunoprecipitated fragment amplification are presented as means (standard deviations [SD]) and were calculated from three biological replicates. Statistical differences from reference conditions were analyzed with unpaired t test.
Data availability.
Data sets are available under NCBI GEO database accession series GSE104027.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Mangelsdorf (University of Texas, Southwestern) for the LXR null mice and Knut R. Steffensen (Karolinska Institutet, Sweden) for the LXRα/β antibody. We are grateful to Luis del Peso (IIBM-UAM) and Prashant Rajbhandari (UCLA) for their suggestions on bioinformatic analysis. We also thank Jon Collins (GlaxoSmithKline SA, NC) for the LXR agonist and antagonist.
This work was supported by grants to the laboratory of A.C. from the Spanish Ministry of Economy and Competitiveness (MINECO) (SAF2011-29244 and SAF2014-56819-R), a Grant for Networks of Excellence from MINECO (Nuclear Receptors in Cancer, Metabolism and Inflammation [NuRCaMeIn]; SAF2015-71878-REDT and SAF2017-90604-REDT), and a grant from MINECO (SAF2017-82463R) to L.B. A.R-V. received a fellowship from MINECO (reference number BES-2012-058574).
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00376-18.
REFERENCES
- 1.Okabe Y, Medzhitov R. 2016. Tissue biology perspective on macrophages. Nat Immunol 17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 2.van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, Hoffmann E, Beyaert R, Saeys Y, Lambrecht BN, Guilliams M. 2016. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44:755–768. doi: 10.1016/j.immuni.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Glass CK, Natoli G. 2016. Molecular control of activation and priming in macrophages. Nat Immunol 17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smale ST, Tarakhovsky A, Natoli G. 2014. Chromatin contributions to the regulation of innate immunity. Annu Rev Immunol 32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 6.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulman IG. 2017. Liver X receptors link lipid metabolism and inflammation. FEBS Lett 591:2978–2991. doi: 10.1002/1873-3468.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B, Tontonoz P. 14 June 2018. Liver X receptors in lipid signalling and membrane homeostasis. Nat Rev Endocrinol. doi: 10.1038/s41574-018-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev 9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 10.Hong C, Bradley MN, Rong X, Wang X, Wagner A, Grijalva V, Castellani LW, Salazar J, Realegeno S, Boyadjian R, Fogelman AM, Van Lenten BJ, Reddy ST, Lusis AJ, Tangirala RK, Tontonoz P. 2012. LXRalpha is uniquely required for maximal reverse cholesterol transport and atheroprotection in ApoE-deficient mice. J Lipid Res 53:1126–1133. doi: 10.1194/jlr.M022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693–704. doi: 10.1016/S0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Breevoort SR, Angdisen J, Fu M, Schmidt DR, Holmstrom SR, Kliewer SA, Mangelsdorf DJ, Schulman IG. 2012. Liver LXRalpha expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J Clin Investig 122:1688–1699. doi: 10.1172/JCI59817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Repa JJ, Mangelsdorf DJ. 2000. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol 16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. 2007. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab 5:73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. 1999. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci U S A 96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature 383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, Winegar DA, Blanchard DE, Spencer TA, Willson TM. 1997. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem 272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 18.Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, Raetz CR, Wang EW, Kelly SL, Sullards MC, Murphy RC, Merrill AH Jr, Brown HA, Dennis EA, Li AC, Ley K, Tsimikas S, Fahy E, Subramaniam S, Quehenberger O, Russell DW, Glass CK. 2012. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. 2003. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med 9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 20.Hong C, Tontonoz P. 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov 13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 21.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev 14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikawa T, Shimano H, Amemiya-Kudo M, Yahagi N, Hasty AH, Matsuzaka T, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Osuga J, Harada K, Gotoda T, Kimura S, Ishibashi S, Yamada N. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol 21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kick E, Martin R, Xie Y, Flatt B, Schweiger E, Wang TL, Busch B, Nyman M, Gu XH, Yan G, Wagner B, Nanao M, Nguyen L, Stout T, Plonowski A, Schulman I, Ostrowski J, Kirchgessner T, Wexler R, Mohan R. 2015. Liver X receptor (LXR) partial agonists: biaryl pyrazoles and imidazoles displaying a preference for LXRbeta. Bioorg Med Chem Lett 25:372–377. doi: 10.1016/j.bmcl.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 24.Kick EK, Busch BB, Martin R, Stevens WC, Bollu V, Xie Y, Boren BC, Nyman MC, Nanao MH, Nguyen L, Plonowski A, Schulman IG, Yan G, Zhang H, Hou X, Valente MN, Narayanan R, Behnia K, Rodrigues AD, Brock B, Smalley J, Cantor GH, Lupisella J, Sleph P, Grimm D, Ostrowski J, Wexler RR, Kirchgessner T, Mohan R. 2016. Discovery of highly potent liver X receptor beta agonists. ACS Med Chem Lett 7:1207–1212. doi: 10.1021/acsmedchemlett.6b00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavazoie MF, Pollack I, Tanqueco R, Ostendorf BN, Reis BS, Gonsalves FC, Kurth I, Andreu-Agullo C, Derbyshire ML, Posada J, Takeda S, Tafreshian KN, Rowinsky E, Szarek M, Waltzman RJ, McMillan EA, Zhao C, Mita M, Mita A, Chmielowski B, Postow MA, Ribas A, Mucida D, Tavazoie SF. 2018. LXR/ApoE activation restricts innate immune suppression in cancer. Cell 172:825–840. e18. doi: 10.1016/j.cell.2017.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B, Mandrup S. 2012. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pehkonen P, Welter-Stahl L, Diwo J, Ryynänen J, Wienecke-Baldacchino A, Heikkinen S, Treuter E, Steffensen KR, Carlberg C. 2012. Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics 13:50. doi: 10.1186/1471-2164-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson LM, Parini P, Janne OA, Gustafsson JA, Steffensen KR, Treuter E. 2010. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev 24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O'Connell RM, Cheng G, Saez E, Miller JF, Tontonoz P. 2004. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell 119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 30.A-Gonzalez N, Guillen JA, Gallardo G, Diaz M, de la Rosa JV, Hernandez IH, Casanova-Acebes M, Lopez F, Tabraue C, Beceiro S, Hong C, Lara PC, Andujar M, Arai S, Miyazaki T, Li S, Corbi AL, Tontonoz P, Hidalgo A, Castrillo A. 2013. The nuclear receptor LXRalpha controls the functional specialization of splenic macrophages. Nat Immunol 14:831–839. doi: 10.1038/ni.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, Tontonoz P. 2008. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell 134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mak PA, Laffitte BA, Desrumaux C, Joseph SB, Curtiss LK, Mangelsdorf DJ, Tontonoz P, Edwards PA. 2002. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors alpha and beta. J Biol Chem 277:31900–31908. doi: 10.1074/jbc.M202993200. [DOI] [PubMed] [Google Scholar]
- 33.Walczak R, Joseph SB, Laffitte BA, Castrillo A, Pei L, Tontonoz P. 2004. Transcription of the vascular endothelial growth factor gene in macrophages is regulated by liver X receptors. J Biol Chem 279:9905. doi: 10.1074/jbc.M310587200. [DOI] [PubMed] [Google Scholar]
- 34.Ignatova ID, Angdisen J, Moran E, Schulman IG. 2013. Differential regulation of gene expression by LXRs in response to macrophage cholesterol loading. Mol Endocrinol 27:1036–1047. doi: 10.1210/me.2013-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ. 2000. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 36.Wagner BL, Valledor AF, Shao G, Daige CL, Bischoff ED, Petrowski M, Jepsen K, Baek SH, Heyman RA, Rosenfeld MG, Schulman IG, Glass CK. 2003. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol 23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. 1985. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature 318:667–670. doi: 10.1038/318667a0. [DOI] [PubMed] [Google Scholar]
- 38.Ito A, Hong C, Rong X, Zhu X, Tarling EJ, Hedde PN, Gratton E, Parks J, Tontonoz P. 2015. LXRs link metabolism to inflammation through Abca1-dependent regulation of membrane composition and TLR signaling. Elife 4:e08009. doi: 10.7554/eLife.08009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapp UR, Cleveland JL, Fredrickson TN, Holmes KL, Morse HC III, Jansen HW, Patschinsky T, Bister K. 1985. Rapid induction of hemopoietic neoplasms in newborn mice by a raf(mil)/myc recombinant murine retrovirus. J Virol 55:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazaki T, Hirokami Y, Matsuhashi N, Takatsuka H, Naito M. 1999. Increased susceptibility of thymocytes to apoptosis in mice lacking AIM, a novel murine macrophage-derived soluble factor belonging to the scavenger receptor cysteine-rich domain superfamily. J Exp Med 189:413–422. doi: 10.1084/jem.189.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SD, Tontonoz P. 2015. Liver X receptors at the intersection of lipid metabolism and atherogenesis. Atherosclerosis 242:29–36. doi: 10.1016/j.atherosclerosis.2015.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A-Gonzalez N, Castrillo A. 2011. Liver X receptors as regulators of macrophage inflammatory and metabolic pathways. Biochim Biophys Acta 1812:982–994. doi: 10.1016/j.bbadis.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 45.Bischoff ED, Daige CL, Petrowski M, Dedman H, Pattison J, Juliano J, Li AC, Schulman IG. 2010. Non-redundant roles for LXRalpha and LXRbeta in atherosclerosis susceptibility in low density lipoprotein receptor knockout mice. J Lipid Res 51:900–906. doi: 10.1194/jlr.M900096-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis M, Moore L, Parks D, Wilson J, Tippin TK, Binz JG, Plunket KD, Morgan DG, Beaudet EJ, Whitney KD, Kliewer SA, Willson TM. 2002. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem 45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 47.Zuercher WJ, Buckholz RG, Campobasso N, Collins JL, Galardi CM, Gampe RT, Hyatt SM, Merrihew SL, Moore JT, Oplinger JA, Reid PR, Spearing PK, Stanley TB, Stewart EL, Willson TM. 2010. Discovery of tertiary sulfonamides as potent liver X receptor antagonists. J Med Chem 53:3412–3416. doi: 10.1021/jm901797p. [DOI] [PubMed] [Google Scholar]
- 48.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc Natl Acad Sci U S A 97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valledor AF, Hsu LC, Ogawa S, Sawka-Verhelle D, Karin M, Glass CK. 2004. Activation of liver X receptors and retinoid X receptors prevents bacterial-induced macrophage apoptosis. Proc Natl Acad Sci U S A 101:17813–17818. doi: 10.1073/pnas.0407749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fonseca E, Ruivo R, Lopes-Marques M, Zhang H, Santos MM, Venkatesh B, Castro LFC. 2017. LXRα and LXRβ nuclear receptors evolved in the common ancestor of gnathostomes. Genome Biol Evol 9:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian J, Goldstein JL, Brown MS. 2016. Insulin induction of SREBP-1c in rodent liver requires LXRalpha-C/EBPbeta complex. Proc Natl Acad Sci U S A 113:8182–8187. doi: 10.1073/pnas.1608987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mann RS, Affolter M. 1998. Hox proteins meet more partners. Curr Opin Genet Dev 8:423–429. doi: 10.1016/S0959-437X(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 53.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14:465–477. doi: 10.1016/S1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 54.Beceiro SA, Pap A, Czimmerer Z, Sallam T, Guillen JA, Gallardo G, Hong C, A-Gonzalez N, Tabraue C, Diaz M, Lopez F, Matalonga J, Valledor AF, Dominguez P, Ardavin C, Delgado-Martin C, Partida-Sanchez S, Rodriguez-Fernandez JL, Nagy L, Tontonoz P, Castrillo A. 2018. LXR nuclear receptors are transcriptional regulators of dendritic cell chemotaxis. Mol Cell Biol doi: 10.1128/MCB.00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, Wherry EJ, Haining WN. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Sun Q, Morita Y, Jiang H, Groß A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, Schambach A, Wuestefeld T, Dauch D, Schrezenmeier H, Hofmann W-K, Nakauchi H, Ju Z, Kestler HA, Zender L, Rudolph KL. 2012. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 57.Blasi E, Radzioch D, Durum SK, Varesio L. 1987. A murine macrophage cell line, immortalized by v-raf and v-myc oncogenes, exhibits normal macrophage functions. Eur J Immunol 17:1491–1498. doi: 10.1002/eji.1830171016. [DOI] [PubMed] [Google Scholar]
- 58.Chen M, Beaven S, Tontonoz P. 2005. Identification and characterization of two alternatively spliced transcript variants of human liver X receptor alpha. J Lipid Res 46:2570–2579. doi: 10.1194/jlr.M500157-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Ramón-Vázquez A, de la Rosa J, Tabraue C, Castrillo A. Bone marrow-derived macrophage immortalization of LXR nuclear receptor-deficient cells. Methods Mol Biol, in press. [DOI] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye T, Ravens S, Krebs AR, Tora L. 2014. Interpreting and visualizing ChIP-seq data with the seqMINER software. Methods Mol Biol 1150:141–152. doi: 10.1007/978-1-4939-0512-6_8. [DOI] [PubMed] [Google Scholar]
- 63.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. 2007. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de la Rosa JA, Ramón-Vázquez C, Tabraue C, Castrillo A. Analysis of LXR nuclear receptor cistrome through ChIP-seq data bioinformatics. Methods Mol Biol, in press. [DOI] [PubMed] [Google Scholar]
- 65.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets are available under NCBI GEO database accession series GSE104027.