Abstract
Aside from classical loops among G-quadruplexes, the unique leaped V-shape scaffold spans over three G-tetrads, without any intervening residues. This scaffold enables a sharp reversal of two adjacent strand directions and simultaneously participates in forming the G-tetrad core. These features make this scaffold itself distinctive and thus an essentially more accessible target. As an alternative to the conventional antisense method using a complementary chain, forming an intermolecular G-quadruplex from two different oligomers, in which the longer one as the target is captured by a short G-rich fragment, could be helpful for recognizing G-rich sequences and structural motifs. However, such an intermolecular leaped V-shape G-quadruplex consisting of DNA oligomers of quite different lengths has not been evaluated. Here, we present the first nuclear magnetic resonance (NMR) study of an asymmetric intermolecular leaped V-shape G-quadruplex assembled between an Oxytricha nova telomeric sequence d(G2T4G4T4G4) and a single G-tract fragment d(TG4A). Furthermore, we explored the selectivity of this short fragment as a potential probe, examined the kinetic discrimination for probing a specific mutant, and proposed the key sequence motif d(G2NG3NG4) essential for building the leaped V-shape G-quadruplexes.
INTRODUCTION
G-quadruplexes (GQs), which consist of two or more π-π stacked G-quartets (1,2), have been regarded as potential targets for disease diagnosis and drug design because of their important roles in a number of biological processes, including telomere maintenance (3–5), DNA replication (6,7), transcription (8,9), gene recombination (10,11), and translation (12,13).
The structures of GQs are highly diverse (14), and this structural polymorphism confers GQs with various functions and potential applications. Many factors contribute to the polymorphic topologies of GQs, such as strand orientations, connecting loop arrangements, glycosidic torsion angles, numbers of G-quartets, and groove sizes (14–16). Three classic types of loop variants have primarily been observed: edgewise, diagonal and double-chain reversal (15). The diagonal loop links two antiparallel strands across the diagonal, whereas the edgewise loop and the double-chain reversal loop connect adjacent antiparallel and parallel strands, respectively.
Unlike the above classic loops, which often consist of several or at least one single bridging residues, another distinctive type, termed the V-shaped loop or V-shaped scaffold, has also been described (15,17,18). This type of loop not only enables a sharp reversal of strand orientation to directly connect, without any extra intervening residues, the corners of G-tetrads located on adjacent columns, but also simultaneously participates in the hydrogen-bond formation within the G-tetrad core (Figure 1). Among those reported GQs containing the V-shaped scaffold, the two G-residues serving as the linking segment without any intervening residues in between could span two (Figure 1A) or even three adjacent layers of G-tetrads stacked on each other (Figure 1B, C). The latter structure, termed the leaped V-shape scaffold (18–20), represents a unique motif with a surprisingly stable spatial arrangement well adapted to the elongated sugar-phosphate backbone of the two consecutive G-residues as a linking segment.
Figure 1.
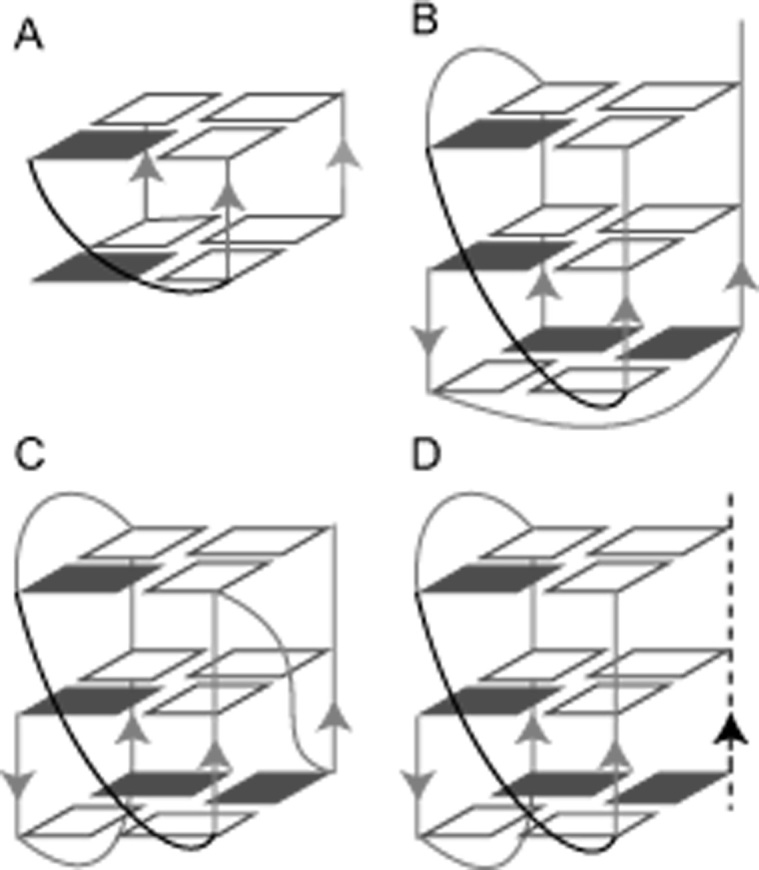
Schematic structure of V-shaped G-quadruplexes. (A) Two-layer V-shaped G-quadruplex, adapted with permission from (17). (B) A self-assembled homodimeric leaped V-shape G-quadruplex (PDB: 1U64). (C) An intramolecular leaped V-shape G-quadruplex (PDB: 2KPR). (D) An intermolecular heterodimeric leaped V-shape G-quadruplex formed by two chains of different lengths. The short chain is indicated by the black dotted line, and the V-shaped loops are colored in black. Other loops and G-columns are colored in light gray. Syn and anti guanines are indicated by solid and hollow rectangles, respectively.
The number of leaped V-shape scaffold-containing GQs showing intramolecular monomeric or self-assembled homodimeric structures has increased from recent studies (17–26). Nevertheless, no intermolecular GQ complexes bearing a leaped V-shape scaffold and having two G-rich oligonucleotides of quite different lengths or sequence compositions have been described in the literature to date (Figure 1D).
Notably, DNA or RNA recognition through the formation of an intermolecular GQ offers unique opportunities for recognition of G-rich sequences and structural motifs (27–30). As previously reported, the association between a three-repeat and another a single-repeat human telomeric DNA sequences, two strands of quite different lengths, leads to the assembly of a heterodimeric GQ (27). Although only conventional edgewise loops were present in this asymmetric intermolecular GQ complex, these findings revealed the capacity for intermolecular recognition of a longer G-rich sequence, and possibly other novel structural motifs, by a homologous short G-rich fragment as a probe.
DNA GQs themselves have some propensity towards forming V-shaped loops (22), and this unique motif may therefore provide an inherently more accessible target. In this regard, one relatively longer sequence of three G-tracts as long as containing the leaped V-shape scaffold, might have an opportunity to be captured by another short single G-tract fragment to form the asymmetric leaped V-shape GQ complex.
Here, we present an NMR analysis of the assembly between the Oxytricha nova telomeric fragment d(G2T4G4T4G4) (termed Otel3Δ2) and another short chain fragment d(TG4A) (termed P6) containing only a single G-tract (Table 1). To the best of our knowledge, this study is the first to report the asymmetric intermolecular leaped V-shape GQ structure. Furthermore, we explored the ability of this short chain as a potential probe to selectively target the sequence containing a leaped V-shape scaffold and proposed the key sequence motif d(G2NG3NG4) that was crucial for building the leaped V-shape GQs.
Table 1.
DNA sequences used in this study
| Type | Name | Sequence (5′-3′)* |
|---|---|---|
| Oxytricha telomeric sequences | Otel3Δ2 | GGTTTTGGGGTTTTGGGG |
| Otel3 | GGGGTTTTGGGGTTTTGGGG | |
| P1 | TTGGGGTT | |
| P2 | TGGGGTT | |
| P3 | TTGGGGT | |
| P4 | TGGGG | |
| P5 | GGGGT | |
| P6 | TGGGGA | |
| Human telomeric sequences | htel3Δ1 | GGTTAGGGTTAGGG |
| htel3Δ1-Ino11 | GGTTAGGGTTIGGG | |
| htel3Δ1-G11 | GGTTAGGGTTGGGG | |
| htel1 | TTAGGG | |
| Inosine substitution | Otel3Δ2-Ino1 | IGTTTTGGGGTTTTGGGG |
| Otel3Δ2-Ino2 | GITTTTGGGGTTTTGGGG | |
| Otel3Δ2-Ino7 | GGTTTTIGGGTTTTGGGG | |
| Otel3Δ2-Ino8 | GGTTTTGIGGTTTTGGGG | |
| Otel3Δ2-Ino9 | GGTTTTGGIGTTTTGGGG | |
| Otel3Δ2-Ino10 | GGTTTTGGGITTTTGGGG | |
| Otel3Δ2-Ino15 | GGTTTTGGGGTTTTIGGG | |
| Otel3Δ2-Ino16 | GGTTTTGGGGTTTTGIGG | |
| Otel3Δ2-Ino17 | GGTTTTGGGGTTTTGGIG | |
| Otel3Δ2-Ino18 | GGTTTTGGGGTTTTGGGI | |
| P6-Ino20 | TIGGGA | |
| P6-Ino21 | TGIGGA | |
| P6-Ino22 | TGGIGA | |
| P6-Ino23 | TGGGIA | |
| Uridine substitution | Otel3Δ2-dU4 | GGTdUTTGGGGTTTTGGGG |
| Otel3Δ2-dU6 | GGTTTdUGGGGTTTTGGGG | |
| Otel3Δ2-dU11 | GGTTTTGGGGdUTTTGGGG | |
| Otel3Δ2-dU13 | GGTTTTGGGGTTdUTGGGG | |
| Mutations on Otel3Δ2 or P6 | Otel3Δ2-T15 | GGTTTTGGGGTTTTTGGG |
| Otel3Δ2-T16 | GGTTTTGGGGTTTTGTGG | |
| Otel3Δ2-A16 | GGTTTTGGGGTTTTGAGG | |
| Otel3Δ2-YA | GGTTTTGGGGTTTTGAGGG | |
| Otel3Δ2-Y2A | GGTTTTGGGGTTTTGAAGGG | |
| Otel3Δ2-YT | GGTTTTGGGGTTTTGTGGG | |
| Otel3Δ2-Y2T | GGTTTTGGGGTTTTGTTGGG | |
| Otel3Δ2-XA | AGGTTTTGGGGTTTTGGGG | |
| Otel3Δ2-XT | TGGTTTTGGGGTTTTGGGG | |
| Otel3Δ2-X2T | TTGGTTTTGGGGTTTTGGGG | |
| Otel3Δ2-X3T | TTTGGTTTTGGGGTTTTGGGG | |
| Otel3Δ2-X4T | TTTTGGTTTTGGGGTTTTGGGG | |
| Otel3Δ2-T7 | GGTTTTTGGGTTTTGGGG | |
| Otel3Δ2-D7 | GGTTTTGGGTTTTGGGG | |
| P6-T20 | TTGGGA | |
| Complementary chains | C3Δ2 | CCCCAAAACCCCAAAACC |
| C3 | CCCCAAAACCCCAAAACCCC |
*I and dU refer to an inosine and a uridine, respectively.
MATERIALS AND METHODS
DNA sample preparation
Unlabeled oligonucleotides were purchased from Sangon Biotech (Shanghai) Co., Ltd (China). The guanine-specific 15N,13C-labeled P6 of d(TGGGGA) was prepared by using the enzymatic synthesis methods (31–34). The designed oligomer template of d(TCCCCAGACTGCATGCAGT)-rC with a 3′ ribose functioned as a template for the following enzymatic reactions, in which the palindrome fragement d(GACTGCATGCAGT)-rC served as a primer, whereas the bold and underlined fragment of d(TCCCCA) served as a coding strand. This designed template was chemically synthesized by Sangon Biotech (Shanghai) Co., Ltd. The enzymatic reaction was performed with 0.045 mM template, Taq polymerase (100 U, Sangon), 0.04 mM unlabeled dATP, 0.04 mM unlabeled dTTP and 0.16 mM 15N, 13C-labeled dGTPs (Cambridge Isotopes, USA) in 1 ml of 10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, (pH 9.0) and 0.1% Triton X-100. The mixtures were heated at 90°C for 5 min and then placed at 72°C for 5 h. The polymerization was stopped by freezing and thawing twice. The labeled DNA d(TGGGGA) was separated from the template through breaking the phosphodiester bond of 3′ ribose rC by an alkaline cleavage. The procedure of the alkaline cleavage step involved adjusting the pH of the mixtures to 12.5 with KOH and incubating at 90°C for 0.5 h. Finally, the products were purified using C18 reversed-phase high-performance liquid chromatography with elution of various combinations of triethylammonium acetate (TEAA) buffer and acetonitrile. The TEAA buffer was prepared in water, the pH was adjusted to 7.0 with acetic acid over several hours with stirring in an ice bath, and the prepared solution was then stored in a refrigerator.
NMR samples were prepared in 100 mM NaCl and 20 mM sodium phosphate buffer at pH 6.8. The strand concentration of the NMR sample was 0.2–2 mM, unless otherwise stated. The samples were heated at 90°C for several minutes and then slowly annealed to room temperature (termed the annealing procedure). Moreover, the samples were heated at 90°C for several minutes and then cooled by quickly placing into an ice-water bath (termed the quench procedure). The NMR samples were prepared under annealing conditions, unless otherwise stated. The DNA concentrations were determined by measuring the UV absorbance at 260 nm.
NMR spectroscopy
NMR data were collected on the 500, 600, and 850 MHz Bruker spectrometers with cryoprobes at 30°C, unless otherwise stated. Two-dimenional homonuclear correlation spectroscopy (2D-COSY), total correlation spectroscopy (TOCSY), 1H–13C heteronuclear multiple bond correlation (HMBC), 1H-15N heteronuclear single quantum coherence (HSQC), 1H-13C HSQC, and nuclear Overhauser effect spectroscopy (NOESY) spectra in H2O and D2O were recorded for resonance assignments and G-tetrad arrangements identification. All data sets were processed and analyzed using Bruker Topspin 3.2, Sparky (UCSF) (35), and CcpNmr Analysis software (version 2.4.1) (36).
NMR structure calculation
Distance restraints for nonexchangeable protons were derived from NOESY spectra with different mixing times (50, 100, 150, 200 and 250 ms) in D2O. The average of all independent thymine H6-CH3 distances was set to 2.99 Å as the distance reference (37). The lower and upper bounds were assigned to ±20–30%. Only a few overlapping resonances were used and given larger bounds (38). The exchangeable proton restraints were deduced from NOESY spectra with two mixing times (80 and 250 ms) in H2O. These distances were restrained to strong 2.7 (±0.9), medium 3.8 (±1.2), or weak 5.0 (±1.5) Å according to the cross peak intensities in the NOESY spectra, corresponding to strong (strong intensity at 80 ms), medium (weak intensity at 80 ms), and weak (observed only at 250 ms), respectively. Within individual G-tetrads, eight hydrogen-bond restraints were added to retain the hydrogen-bonding. The residues of the glycosidic χ torsion angle for syn and anti bases were set to 60° ± 35° and 240° ± 70°, respectively. The ν2 torsion angles of G15 and G16 were deduced from 3JH1′H2′ and 3JH1′H2″ coupling constants, which were achieved from COSY spectra and analyzed by the Matlab Pseudorotation GUI procedure (39).
All structure calculations were performed using the XPLOR-NIH (40) and AMBER (41) programs as described in reported methods (42–44). First, the initial 200 molecules were generated by a distance geometry simulated annealing protocol in the XPLOR-NIH program, which incorporated all restraints, including distance restraints, dihedral angles, hydrogen-bonding restraints, and planarity restraints. Then, the obtained 50 lowest-energy structures were selected to be further refined and optimized using Amber 14 software. The refinements were calculated in 300 ps of NMR restrained simulated annealing simulations using the generalized Born implicit model to account for solvent effects. The system was first minimized over 500 steps of steepest descent minimization followed by 500 steps of conjugated gradient minimization. In the first 6 ps, the system was heated from 0 to 300 K. Molecules were held at constant temperature of 300 K for 54 ps and then cooled to 0 K in the next 90 ps, after which the temperature was kept at 0 K for an additional 150 ps. Force constants were 20 kcal mol−1 A−2 for hydrogen bond restraints, 20 kcal mol−1 A−2 for NOE distances, 200 kcal mol−1 A−2 for torsion angle ν2, and 20 kcal mol−1 A−2 for torsion angle χ. The 10 lowest energy structures were selected as the structure ensemble. The structures were displayed using PyMOL.
Circular dichroism (CD) spectroscopy experiment
CD spectra were obtained at 25°C using a JASCO J-810 (Japan) spectropolarimeter with a 1-mm path length quartz cuvette. To ensure the precise ratio of the different strands in the complex, the samples were prepared in NMR buffer, and the structures were confirmed by NMR first. The NMR samples were then diluted to 20 μM in a 200 μl of 5 mM sodium phosphate buffer containing 100 mM NaCl for CD measurements. An average of three scans was taken for each measurement, and the baseline was corrected with the same buffer. The thermal stability of the complex Otel3Δ2/P6 was evaluated by CD melting experiments measured at 290 nm. Heating and cooling experiments were performed across the temperature range of 25–95°C, as described previously (45,46). The CD spectra were recorded at 5°C intervals. The melting curve was fitted to the Boltzmann function. The melting temperature (Tm) was defined as the temperature at which there were 50% folded and 50% unfolded species (45).
RESULTS AND DISCUSSION
GQ formation by the shortened three-repeat Oxytricha nova telomeric sequence Otel3Δ2 associated with a short fragment of the single G-tract
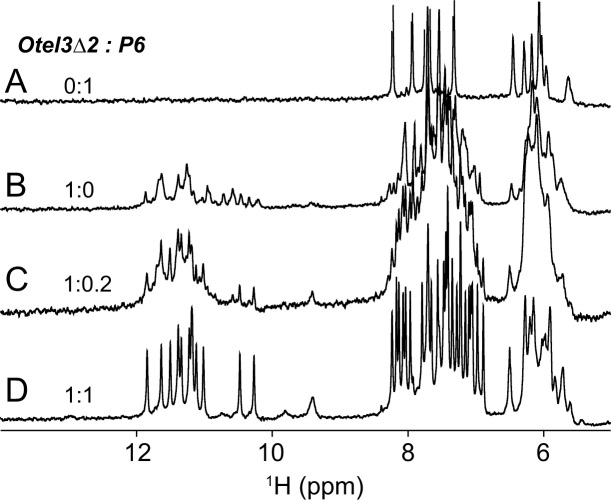
The specific Oxytricha nova telomeric fragment d(G2T4G4T4G4) (termed Otel3Δ2), which was shorten by two guanines at the 5′-end compared with its count partner in the intact three-repeat sequence d(G4T4G4T4G4), was examined to determine its ability to associate with a short chain fragment containing only a single G-tract. Accordingly, increasing amounts of d(TG4A) (designated as P6), an analogue of the single-repeat Oxytricha nova telomeric sequence d(T2G4T2) (termed P1), were titrated stepwise into a solution of the Otel3Δ2, as shown in Figure 2. Under the given experimental conditions, P6 alone was essentially unstructured, and no hydrogen-bonded imino proton signals were observed around 10–12 ppm (Figure 2A), whereas Otel3Δ2 alone self-assembled into a mixture of conformationally heterogeneous GQs, as indicated by the presence of multiple sets of imino peaks at 10–12 ppm (Figure 2B), the characteristic 1H NMR region for GQ formation. Upon increasing the amounts of P6 d(TG4A) during titration, the original NMR signals of Otel3Δ2 alone gradually vanished, and a distinctly new set of guanine imino proton peaks appeared at 10–12 ppm, becoming much more well-resolved and sharper (Figure 2C). Eventually, for an equimolar mixture of Otel3Δ2 and P6, a clean new single set of totally 12 imino proton signals at 10–12 ppm was built up, as observed in Figure 2D, suggesting the formation of an intermolecular GQ complex between Otel3Δ2 and P6, probably containing three layers of G-tetrads. The observation of separated imino proton signals distinct for surplus free Otel3Δ2 alone and the Otel3Δ2/P6 complex (Figure 2C) suggested slow exchange on the NMR time scale between the free and bound states of Otel3Δ2. This phenomenon is usually explained as an indication of tight and specific binding for a given complex. Indeed, the GQ complex Otel3Δ2/P6 exhibited considerable stability, even at an elevated temperature of 40°C (Supplementary Figure S1). This result is consistent with our observation in the CD melting experiments (Supplementary Figure S2). The melting temperature (Tm) of Otel3Δ2/P6 was 51.4 ± 1.0°C. Similarly, a reverse titration by adding increasing amounts of Otel3Δ2 into P6 was consistent with the formation of the same Otel3Δ2/P6 GQ complex (data not shown).
Figure 2.
Strand-titration experiments by solution NMR. One-dimensional 1H spectra of the single G-tract chain P6 alone (A), the Oxytricha nova telomeric fragment Otel3Δ2 alone (B), 20% P6 (C), and 100% P6titrated into Otel3Δ2 (D) showing the formation of an intermolecular G-quadruplex.
A short single-repeat fragment of the natural Oxytricha nova sequence d(T2G4T2) (P1), and other analogues (P2-P6), was also titrated respectively with Otel3Δ2 (Supplementary Figure S3). These NMR spectra were only slightly different from those of the Otel3Δ2/P6 GQ complex, suggesting the formation of heterodimeric GQ complexes with a similar folding topology. Notably, the NMR spectra of the Otel3Δ2/P6 complex (Figures 2 and 3) exhibited the best quality with mostly well-resolved resonances, thus, Otel3Δ2/P6 was chosen for further NMR structure characterization.
Figure 3.
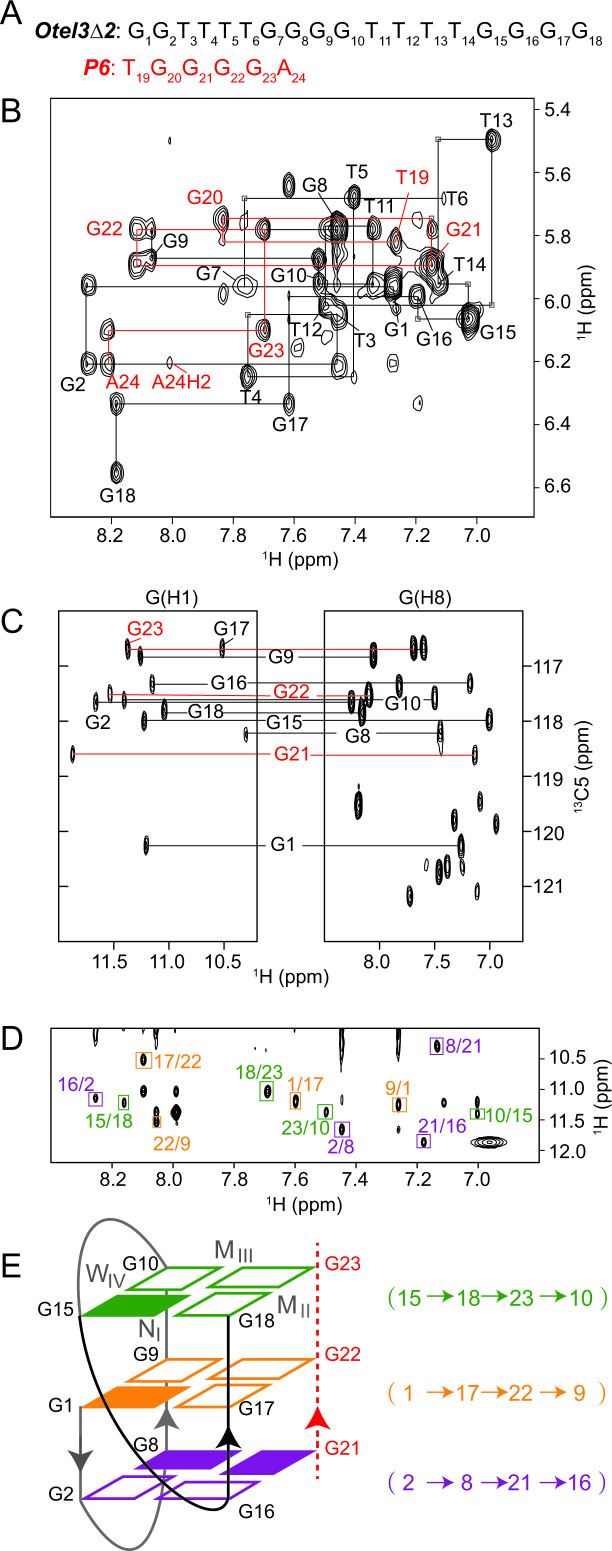
Folding topology of Otel3Δ2/P6 determined by NMR. (A) The DNA sequences of P6 and Otel3Δ2. The short chain P6 is colored in red. (B) Sequential walking in the NOESY spectrum (250 ms mixing time, D2O) showing the H8/H6-H1′ connectivity of Otel3Δ2/P6. Cross peaks are labeled with residue names. The residues from the short chain P6 are colored in red. Weak or missing sequential connectivities are labeled with a light gray frame. (C) 1H-13C HMBC spectrum indicating the correlation of guanine H8 and imino protons via 13C5 at natural abundance. G21, G22, and G23 are colored in red. (D) NOESY spectrum (250 ms mixing time, 10% D2O/90% H2O) showing inter-residue imino-H8 cross peaks for the identification of the arrangement of the three G-tetrads. The guanine H1–H8 cross peaks are framed and labeled with residue numbers of the H1 and H8 protons in the first and second positions, respectively. Residues in the same G-tetrad are shown in the same color. (E) Schematic topology of Otel3Δ2/P6. The backbones of the short chain P6 are shown as red dotted lines, and its residues are colored in red. The backbones of the long chain Otel3Δ2 are shown as solid lines, and the V-shape turn (G15–G18) is colored in black. The top, middle, and bottom G-tetrads are colored in green, orange, and purple, respectively. The hydrogen-bond directionality within each G-tetrad is shown in the same color. Syn and anti guanines are indicated by solid and hollow rectangles, respectively. W, M, and N represent wide, medium, and narrow grooves, respectively. I, II, III, and IV indicate grooves I, II, III, and IV, respectively.
NMR spectral assignments of the GQ complex Otel3Δ2/P6
To gain more insight into the complex Otel3Δ2/P6, we performed a series of through-bond (2D COSY, TOCSY, 1H-15N HSQC, 1H-13C HSQC and 1H-13C HMBC) and through-space (NOESY) NMR experiments at various temperatures. Initially, the 12 exchangeable signals at 10–12 ppm were identified as the guanine imino protons through 1H–15N HSQC in H2O (Supplementary Figure S4), according to the characteristic 15N chemical shifts at approximately 145 ppm for guanines and 155 ppm for thymines, if any.
Nonexchangeable base H8/H6 and sugar H1′ proton assignments were accomplished by tracing the sequential NOE connectivities for Otel3Δ2 of d(G1G2T3T4T5T6G7G8G9G10T11T12T13T14G15G16G17G18) or P6 of d(T19G20G21G22G23A24) respectively, in the NOESY spectrum with a mixing time of 250 ms recorded in D2O (Figure 3A and B). The guanine imino proton assignments for these two individual strands of Otel3Δ2 and P6 were achieved by the 1H–13C HMBC experiment (Figure 3C), which was based on the correlation between guanine base H8 and imino H1 protons through 13C5 at natural abundance (47,48). As a result, nine guanine imino protons were sorted to the Otel3Δ2 strand, whereas the other three were sorted to the P6 strand.
All of these assignments were further confirmed unambiguously by either guanine-to-inosine or thymine-to-uridine chemical substitutions (Supplementary Figure S5 and Table 1). Upon loss of the N2 amino group in guanine and loss of the C5 methyl group in thymine, guanine and thymine were changed to inosine and uridine, respectively (49). These were among the smallest changes in nucleic acids (50), and inosine substitution for a guanine and uridine substitution for a thymine have been commonly used for unambiguous assignments in NMR structural studies of GQs (49). In addition, the guanine-specific 15N,13C-labeled P6 was prepared and titrated with the unlabeled long chain Otel3Δ2. As expected, the 15N-edited 1D 1H spectrum of Otel3Δ2/P6 displayed 3 imino resonances of G21, G22 and G23 for P6, whose chemical shifts were consistent with the previous assignments for P6 (Supplementary Figure S6), confirming that the left G20 of P6 was not hydrogen-bonded.
In the stacked NOESY spectrum with a short mixing time of 50 ms, four strong H8-H1′ cross peaks were observed for residues G1, G8, G15 and G21, indicating their adoption of a syn glycosidic conformation (Supplementary Figure S7). Furthermore, the hydrogen-bond alignments and directionality within each G-tetrad were determined based on the establishment of imino-H8 connectivities in the NOESY spectrum with a mixing time of 250 ms in H2O (Figure 3D), yielding a total of three G-tetrads, including G15–G18–G23–G10, G1–G17–G22–G9, and G2–G8–G21–G16 (Figure 3E). Accordingly, the folding topology of the complex Otel3Δ2/P6 was established as an asymmetric heterodimeric intermolecular GQ consisting of three G-tetrad layers (Figure 3E). The subsequent hydrogen-deuterium exchange experiment supported this folding topology, and the guanines from the central G-tetrad (G1, G17, G22 and G9) were among the most protected imino protons and exchanged with D2O relatively slower (Supplementary Figure S8).
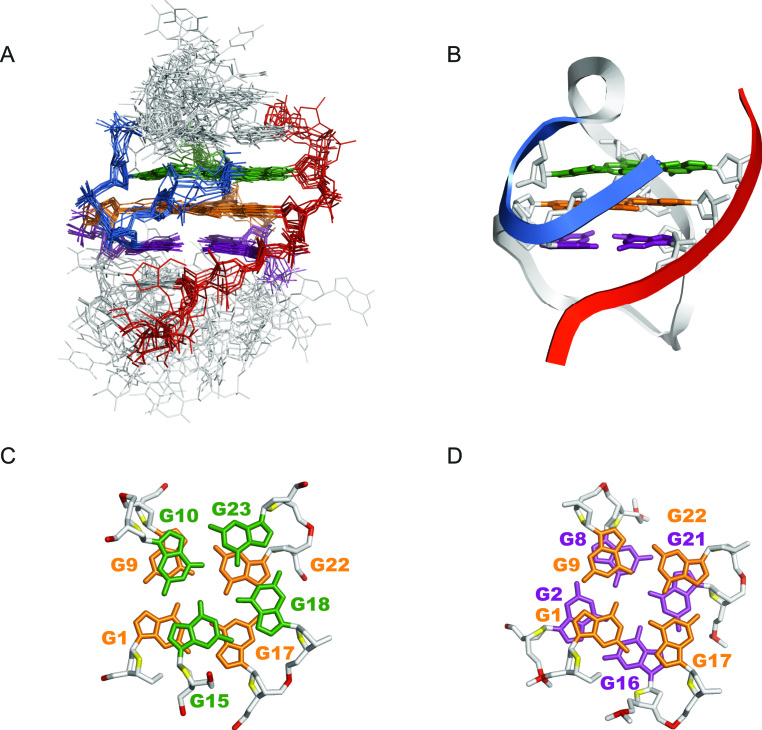
Solution structure of the Otel3Δ2/P6 GQ
The overall solution structure was calculated on the basis of NMR restraints using X-PLOR-NIH and AMBER programs. The NMR restraints and structural statistics are listed in Table 2. Ten superimposed lowest-energy structures are displayed in Figure 4A. The G-tetrad core of Otel3Δ2/P6 was well converged, with a root mean squared deviation (R.M.S.D.) of 1.05 ± 0.14 Å. The edgewise loops were more flexible than the G-tetrad core. A representative refined structure is shown as a ribbon representation in Figure 4B. The structural features of Otel3Δ2/P6 were quite similar to those of other previously reported three-layer leaped V-shape GQs (18–21).
Table 2.
NMR restraints and structure statistics
| A. NMR restraints | ||
| Distance restraints | Exchangeable | Non-exchangeable |
| Intra-residue | 0 | 288 |
| Sequential (i, i+1) | 8 | 66 |
| Long-range(i, >i+1) | 54 | 21 |
| Other restraints | ||
| Hydrogen bond | 48 | |
| Dihedral angle | 26 | |
| B. Structure statistics | ||
| NOE violations | ||
| Numbers (>0.3 Å) | 0.70±0.64 | |
| Mean violations (Å) | 0.34±0.12 | |
| Deviations from ideal covalent geometry | ||
| Bond length (Å) | 0.01±0.00 | |
| Bond angle (deg) | 2.22±0.03 | |
| Pairwise all heavy atom r.m.s.d. values (Å) | ||
| G-tetrad core | 1.05±0.14 | |
| Without T3–G7 & T19-G20 | 2.05±0.38 | |
| Without T11–T14 & A24 | 3.90±0.88 | |
| All heavy atoms | 4.47±0.98 | |
Figure 4.
Solution structure of Otel3Δ2/P6. (A) The 10 lowest energy structures are superimposed. The guanine residues in the top, middle, and bottom G-tetrads are indicated in green, orange, and purple, respectively. The backbone of the V-shaped turn (G15–G18) is indicated in marine. The backbone of the short chain P6 is colored in red. Edgewise loops are colored in light gray. (B) Ribbon representation of a representative refined structure. (C) Stacking of G10-G15-G18-G23 (base in green) over G1–G17–G22–G9 (base in orange). (D) Stacking of G1–G17–G22–G9 (base in orange) over G2–G16–G21–G8 (base in purple). The backbone P is colored in red, and the sugar O4′ is colored in yellow. The other atoms of the backbone are colored in light gray.
The G-tetrad core was established with four G-columns, among which three originated from three contiguous G-tracts (G8–G9–G10, G16–G17–G18, and G21-G22-G23), whereas the fourth was a broken G-column composed of G15 and G1–G2. Overall, there were three parallel and one anti-parallel G-columns in the assembly of Otel3Δ2/P6 GQ. The backbone of the consecutive G15–G16, which functioned as a linker segment that directly connected two adjacent antiparallel G-columns, adopted a V-shaped scaffold. This scaffold leaped over the middle G-tetrad and caused a sharp reversal of the G15-G18 strand direction (Figure 4B), whereas the bases of both G15 and G16 participated in the buildup of G-tetrads. Moreover, the remaining guanine G7 of Otel3Δ2was not hydrogen bonded, and instead functioned as a part of the edgewise loop T3–G7, and the overhanging residue G20 of P6 flanked around the terminal G-tetrad. As expected, the loops and overhanging residues were more flexible (Figure 4A), as evidenced that the broadness of their NMR signals appeared more sensitive to temperature variations (data not shown).
As shown in the surface views, this V-shaped GQ contained four grooves of different widths: two medium (grooves II and III), one wide (groove IV), and one narrow (groove I, Figure 3E and Supplementary Figure S9). In addition, the stacking patterns between G-tetrads are shown in Figure 4C and D. Partial overlaps between the five- and six-membered rings of guanines were observed in the top two G-tetrads, whose hydrogen-bond directionalities were the same, i.e. anticlockwise (Figures 3E and 4C). In contrast, full overlaps through the five-membered rings of guanines were observed in the bottom two G-tetrads, whose hydrogen-bond directionalities were opposite (Figures 3E and 4D). In addition, we also investigated the potassium form of the complex Otel3Δ2/P6. However, poor NMR spectrum quality was observed compared with that of the sodium form, and it was clearly impossible to achieve detailed NMR structural identification (Supplementary Figure S10). Nevertheless, the NMR spectra of Otel3Δ2/P6 in potassium and sodium were still distinct, allowing us to readily monitor the subsequent competition titration. When we titrated the increasing amounts of potassium ions into the sodium form of Otel3Δ2/P6, the sodium form of Otel3Δ2/P6 disappeared, and the potassium form of Otel3Δ2/P6 gradually became the dominant structure, suggesting that this GQ complex structure was more sensitive to the potassium concentration (Supplementary Figure S10).
Effects of mutations on the Otel3Δ2/P6 GQ
To assess the robustness of the V-shaped scaffold in the Otel3Δ2/P6 GQ complex, base substitutions or insertions were introduced into the G15-G16 linker segment. Any single substitution of G15 or G16 to T or A, as well as the insertion of T or A between the G15 and G16 residues, all led to multiple sets of imino protons, or abolished the GQ structure (Supplementary Figure S11). In contrast, the addition of adenine or thymine beyond the 5′ terminus of the G1-G2 segment remained nearly completely unchanged compared with that of the unmodified sequence (Supplementary Figure S12). These results indicated that these two continuous guanine bases of G15–G16 were extremely critical for the formation of the leaped V-shape GQ structure. These findings supported that the contiguous G4-tract within the proposed d(G2NG3NG4) sequence motif (see below), in which N represented 1–5 nucleotides as a linking loop, was critical for the formation of the leaped V-shape GQ.
An analysis of the structure of Otel3Δ2/P6 revealed that residue G7 from the Otel3Δ2 strand and residue G20 from the P6 strand did not participated in the formation of the G-tetrad core (Figure 3E). These findings were consistent with the lack of observation of hydrogen-bonded inosine imino proton signals, which are often characteristically most downfield, in the NMR spectra of Otel3Δ2-Ino7/P6 and Otel3Δ2/P6-Ino20 (Supplementary Figure S5). Further G7-to-T and G20-to-T mutation assays yielded similar NMR spectra and thus once again confirmed the topology of the Otel3Δ2/P6 complex structure (Supplementary Figure S13).
Notably, only three guanines, G8, G9, and G10, from the second G-tract G7–G10 participated in forming the G-tetrad core, and G7 remained a part of the T3–G7 loop. Considering the possible strand slippage, we then investigated whether the GQ complex could still form when G10 rather than G7 looped out. Accordingly, the sequence Otel3Δ2-Ino10, which contained a G10-to-inosine10 substitution (Supplementary Figure S14A), was selected to titrate with P6-Ino20. The resulting complex displayed 12 imino peaks at 10–12 ppm for guanines only, without any observation of inosine imino peaks, which are typically more downfield shifted above 14 ppm (Supplementary Figure S14B). In particular, there was no observation of a 15N chemical shift around 175 ppm for the characteristic inosine N1, further confirming that the imino proton of inosine10 was indeed not involved in the hydrogen-bond formation in the G-tetrad core but rather served as a part of the loop. In contrast, the observed 15N1 chemical shift at 142–145 ppm indicated that the remaining 12 guanines, including G7, were all hydrogen-bonded (Supplementary Figure S14C). Further CD measurements showed that Otel3Δ2-Ino10/P6-Ino20 had a CD profile similar to that of the previously described Otel3Δ2/P6 (Supplementary Figure S14D), suggesting that Otel3Δ2-Ino10/P6-Ino20 still exhibited a similar leaped V-shape topology, but accepted G7, G8, and G9 rather than G8, G9, and inosine10 as a G-column (Supplementary Figure S14E). Further analysis of 2D NOESY and 1H,13C-HMBC spectra of Otel3Δ2-ino10/P6-ino20 also supported our schematic structure (Supplementary Figures S14F–H). As explained, the lack of an amino NH2 group on C2 of inosine caused the G-tetrad core to become less stable in terms of hydrogen-bonding capability, thus shifting the equilibrium of strand slippage and leading inosine10 to become switchable as a part of the loop. Therefore, regardless of how the G-tract of G7–G8–G9–G10 slipped, the GQ complex could still assemble as long as three continuous guanines were selected to serve as a G-column in the G3-tract required by the d(G2NG3NG4) sequence motif. Further analysis of the deletion mutant Otel3Δ2-D7 of G2T4G3T4G4 also confirmed this conclusion (Supplementary Figure S13B). These results provided additional supports that the d(G2NG3NG4) sequence motif formed a leaped V-shape scaffold (see below).
The short chain P6 as a potential probe to distinguish the two guanines shortened Otel3Δ2 from the intact sequence Otel3
Sequence-specific targeting of a given nucleic acid fragment can usually be achieved quite well by the antisense probe of a complementary chain based on Watson-Crick base pairings. However, this approach may not be sufficient for repetitive sequences. Indeed, for Otel3Δ2 and Otel3, for which both fragments shared the common repetitive octamer unit of d(GGGGTTTT), neither corresponding complementary segments d(C4A4C4A4C2) nor d(C4A4C4A4C4) exhibited sufficient specificity for simultaneously distinguishing the targets Otel3Δ2 and Otel3, as shown in Supplementary Figure S15.
Collectively, quite a few examples of intramolecular or self-dimeric GQs bearing the leaped V-shape scaffold have been reported, and all exhibit considerable stability (18–21). Further inspired by our finding in this work regarding the formation of the GQ complex Otel3Δ2/P6, in which two quite different fragments assembled together through the novel leaped V-shape scaffold inherently responsible for the sequence specificity, we anticipated that the short chain P6 may have the potential to serve as a probe to distinguish Otel3Δ2 from other analogous sequences. Therefore, rather than the conventional antisense method using a complementary chain, we used an alternative approach with a short homologous G-rich sequence, to specifically probe the highly repetitive sequences.
To verify our hypothesis, we examined whether the short probe P6 could distinguish Otel3Δ2 from Otel3. Either Otel3Δ2 or Otel3 alone was folded into multiple GQ structures in a broad envelope of multiple imino resonances at 10–12 ppm (Supplementary Figures S16A, and S16C). Upon the addition of P6 to Otel3 at a molar ratio of 1:1, only a minor noticeable change was observed, implying weak Otel3/P6 GQ formation (Supplementary Figure S16B). However, the apparent formation of the Otel3Δ2/P6 complex was clearly detected when an equimolar amount of P6 was titrated into Otel3Δ2 (Supplementary Figure S16D), thus suggesting the preferential association between P6 and Otel3Δ2. Indeed, even in the presence of an equimolar mixture of Otel3Δ2 and Otel3 (Supplementary Figure S16E), the subsequently added P6 still preferentially recognized Otel3Δ2 to form the Otel3Δ2/P6 complex, as evidenced by the higher intensity of the characteristic peaks at 10.25 ppm and 10.46 ppm representative of Otel3Δ2/P6 (marked by asterisks) and the less intense signals at 12.25 ppm for the Otel3/P6 complex (marked by hash signs; Supplementary Figure S16F).
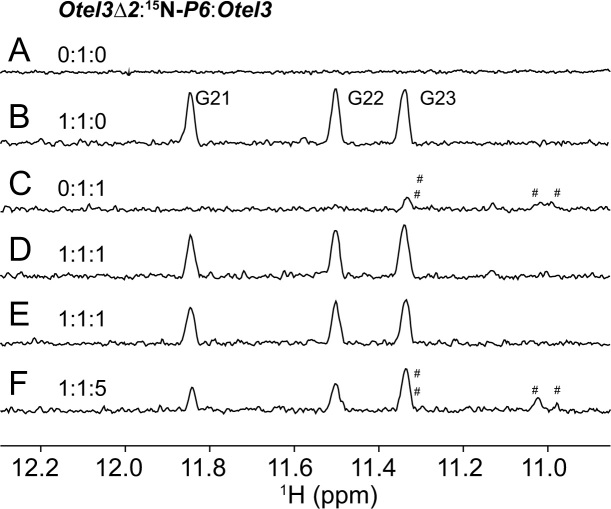
In order to avoid heavily overlapped 1H spectra of the imino proton region as shown in Supplementary Figure S16, the above competition experiments were repeated again using the guanine-specific 15N,13C-labeled P6. Using 15N-edited experiments, it was much more convenient and straightforward to distinguish the complexes Otel3Δ2/P6 and Otel3/P6. Considering the cost and possible self-assembly of the labeled P6 into a GQ at an otherwise high sample concentration, the experimental concentration of the labeled P6 was kept constantly at 0.04 mM to ensure the existence of labeled P6 alone as an unstructured short chain (Figure 5A). As expected, the 15N-edited 1D 1H spectrum of Otel3Δ2/P6 became greatly simplified, with only three sharp imino resonances whose chemical shifts were consistent with the previous assignments (Figure 5B and Supplementary Figure S6). In contrast, only a small portion of Otel3/P6 formed at a molar ratio of 1:1 (Figure 5C). When an equimolar amount of Otel3Δ2 was added to the mixture of Otel3 and P6, however, Otel3Δ2/P6 was observed as the major structure (Figure 5D and E). In particular, even when the Otel3 concentration was further elevated to reach an [Otel3Δ2]/[P6]/[Otel3] molar ratio of 1:1:5, the signal intensity of Otel3Δ2/P6 was still stronger than that of Otel3/P6 (Figure 5F). Therefore, the use of labeled P6 once again verified that the short chain P6 preferred to bind with Otel3Δ2 rather than the intact sequence Otel3.
Figure 5.
Imino protons in one-dimensional 15N-edited spectra displaying the competition experiments among Otel3Δ2, Otel3, and guanine-specific 15N,13C-labeled P6 at the indicated ratio. Peaks representing Otel3/P6 are labeled with hash signs. Three peaks representing Otel3Δ2/P6 are assigned in (B). The concentration of 15N,13C-labeled P6 was 0.04 mM. The concentrations of other strands corresponded to the indicated ratios. The sample in (E) was prepared under quench condition.
Notably, the same results were obtained under both slow annealing and fast quench conditions, indicating that the formation of the complex Otel3Δ2/P6 was both thermodynamically and kinetically favorable (Figure 5D and E). Overall, these competition experiments demonstrated that the short chain P6 had considerable selectivity for distinguishing Otel3Δ2 and the intact sequence Otel3. Our results provided an alternative structure-guided approach to probe the highly repeated sequence. Thus, it is expected that this approach will improve the probing specificity and play an important role in recognizing G-rich sequences.
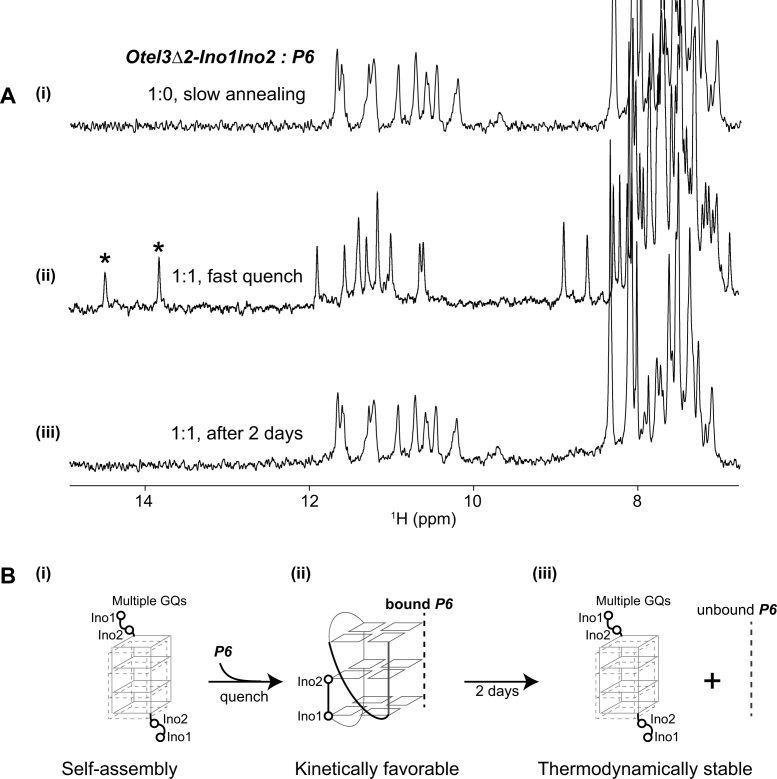
Kinetically favored probing of dual inosine-substituted Otel3Δ2-Ino1Ino2 via the concomitant leaped V-shape scaffold
As described for the resonance assignments of the Otel3Δ2/P6 complex, several single substitutions by chemically modified bases on the sequence of Otel3Δ2, including G1-to-Ino1 and G2-to-Ino2 respectively, were successfully used to tightly associate with P6, resulting in a stable intermolecular GQ complex and further confirming the assignments (Supplementary Figure S5). For the dual inosine-substituted Otel3Δ2-Ino1Ino2, harboring two inosine substitutions simultaneously at the 5′ terminal G1 and G2 residues of Otel3Δ2 (Supplementary Figure S17A), Otel3Δ2-Ino1Ino2 could be probed by P6 however only in a kinetically favorable manner.
In the absence of the short chain probe P6, the majority of Otel3Δ2-Ino1Ino2 alone was the unfolding structure, and another small proportion of ∼10% of Otel3Δ2-Ino1Ino2 alone was the self-assembled GQs. There were multiple guanine imino proton peaks at 10–12 ppm; however, no characteristic inosine imino proton signals, which were typically downfield shifted larger than 14 ppm, were observed in the 1H spectrum (Figure 6A–i). This finding was consistent with the lack of 15N1 peaks at 175 ppm in the 1H–15N HSQC spectrum (Supplementary Figure S17B). In addition, more than one set of methyl peaks of thymines at 1–2 ppm was observed in the 1H–13C HSQC spectrum of Otel3Δ2-Ino1Ino2 alone (Supplementary Figure S17C). These findings suggested that these two inosines, without hydrogen bonding, did not participate into the formation of the G-tetrad cores. Instead, these two residues likely functioned as a part of the protruded loops, as shown in the schematic Figure 6B–i, whereas the remaining two G4-tracts of Otel3Δ2-Ino1Ino2 self-assembled into multiple GQs, potentially similar to cases of self-association of other two-repeat telomeric DNA sequences reported previously (51–53). This result was not surprising because the reduced hydrogen-bonding capability of inosine due to its lack of an amino functional group would weaken the overall stability of a given GQ only if an inosine was forced to be included in the G-tetrad core.
Figure 6.
The dual inosine-substitution mutant Otel3Δ2-Ino1Ino2 probed by the short chain P6. (A) (i) The one-dimensional 1H spectrum of Otel3Δ2-Ino1Ino2 alone showed its self-assembly into multiple G-quadruplexes under annealing condition. (ii) The one-dimensional 1H spectrum of the Otel3Δ2-Ino1Ino2/P6 complex at an equimolar ratio under quench conditions showed the formation of a new G-quadruplex containing two hydrogen-bonded inosine imino peaks (marked by asterisks). (iii) The one-dimensional 1H spectrum of the sample used in panel (ii) after incubation at room temperature for 2 days. The concentration of the DNA strands was 0.1 mM. The possible process of conformational changes under the conditions shown in panel (A) was schematically indicated using the structural model in panel (B), without showing the unfolding proportions of single chain P6 and single chain Otel3Δ2-Ino1Ino2 for clarity. (B) (i) and (iii) These two inosines did not participate in the formation of the G-tetrad core. In the schematic structure, the short chain P6 is indicated by the gray dotted line. The hollow circles represent the 5′ terminal inosines of Otel3Δ2-Ino1Ino2.
Upon addition of an equimolar amount of P6 intoOtel3Δ2-Ino1Ino2, a new minor set of imino peaks immediately appeared (Supplementary Figure S18) and then became the dominant imino peaks (Figure 6A-ii) upon additional rapid quenching, the conditions typically accepted as optimal for forming a kinetically controlled structure. Notably, two rather sharp imino proton signals of inosine were observed at 13–14 ppm, along with another 10 signals representing guanines at 10–12 ppm, indicating the participations of inosines in the G-tetrad core of the newly formed complex (termed InoG4), and tentatively adopting the same topology of non-inosine substituted Otel3Δ2/P6 with a leaped V-shape scaffold (Figure 6B-ii), on the basis of comparable patterns of NMR spectra. Nevertheless, the imino peaks of InoG4 then gradually faded away after a long incubation for 2 days at room temperature, and eventually, the self-assembled Otel3Δ2-Ino1Ino2 reappeared as the major GQ structure once again (Figure 6A-iii and B-iii). These results revealed that the formation of InoG4 was kinetically favorable but thermodynamically less stable. Importantly, based on assessment of the relative intensity ratio between the imino and base H8/H6 protons in the 1H NMR spectrum, the proportion of InoG4 that was newly formed immediately after a rapid quench of an equimolar mixture of P6 andOtel3Δ2-Ino1Ino2 was as low as approximately 10%, whereas the majority of either P6 orOtel3Δ2-Ino1Ino2 remained unfolded, with InoG4 presumably functioning more or less like a kinetically favorable intermediate structure and eventually disassociating away.
Based on the appearance or absence of characteristic inosine imino proton signals, the observed structural switch of Otel3Δ2-Ino1Ino2 upon the addition of P6 could be conveniently monitored in a straightforward manner. However, it was still unclear whether this short chain P6 indeed directly participated in the recognition of Otel3Δ2-Ino1Ino2 to form InoG4 or whether P6 triggered a conformational change in Otel3Δ2-Ino1Ino2. To answer the above question, the preceding binding titrations were repeated again using the guanine-specific 15N,13C-labeled P6 to titrate with unlabeled Otel3Δ2-Ino1Ino2. The 15N isotopic-labeled guanines in the short chain enabled us to specifically trace the imino resonances of P6 itself whether or not is involved in Hoogsteen base paring within the G-tetrad. Consistent with our previous results using unlabeled P6, a total of three imino resonances belonging to P6 itself, in the 15N-edited spectra, appeared under quench conditions but disappeared after incubation at room temperature for 2 days (Supplementary Figure S19). These results clearly demonstrated that the short chain P6 directly participated in the kinetically favored formation of InoG4 when quenched and that only three out of the four guanines from the short chain P6 contributed to the observed imino proton signals, similar to the case of the three-layer leaped V-shape GQ Otel3Δ2/P6.
Although less stable thermodynamically, the short chain P6 was still capable of quickly capturing the mutant Otel3Δ2-Ino1Ino2 into a leaped V-shape GQ whose formation appeared to be kinetically favored. The readily accessible feature of the leaped V-shape scaffold may be responsible for this kinetic discrimination. In general, many biological processes, including those involved in gene expression, are mostly regulated by kinetic control (54). Therefore, the kinetically favored structures appeared to be important transient regulators in vivo and undoubtedly play key roles in the early stage of ligand-target recognition. Accordingly, the potential GQ formation during these processes may be dominated by kinetic rather than thermodynamic control. In this work, the achieved kinetic capture of the mutant Otel3Δ2-Ino1Ino2 into the leaped V-shape GQ provided novel insights into these important mechanisms which has been paid attentions far more than enough.
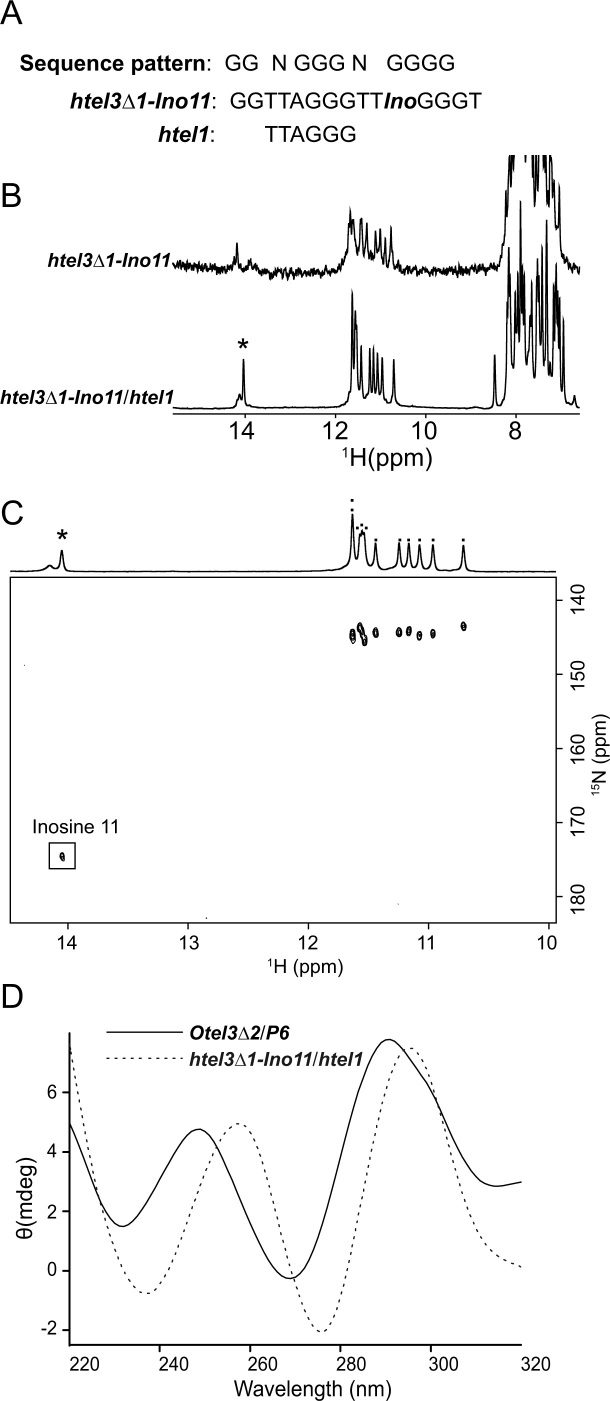
The mutated human telomeric sequence d(G2T2AG3T2-I-G3T) was recognized by a short chain G-rich probe
Taking advantage of the novel leaped V-shape scaffold, several fragments of Oxytricha nova telomeric DNA long enough to fulfill the d(G2NG3NG4) sequence motif could be trapped by a short chain probe containing a single G-tract (Figures 3E and 6B-ii). Compared with the other two G-tracts (G2 and G3) within the d(G2NG3NG4) sequence motif, the last consensus G4-tract at the 3′-end was responsible for the formation of the leaped V-shape scaffold, which required four contiguous guanines, and was essential for overall stability and selectivity.
Unlike Oxytricha nova telomeric sequences, whose repeat unit of d(TTTTGGGG) generated a G4-tract naturally, the repeating unit of human telomeric sequences d(TTAGGG) was only composed of three contiguous guanines as a single G-tract of G3 at maximum, thus hardly satisfying the demand for the critical G4-tract within the d(G2NG3NG4) sequence motif. Indeed, no apparent association with the probe was detected (Supplementary Figures S20 and S21) for a given fragment of natural human telomeric DNA d(G2T2AG3T2AG3) (termed htel3Δ1), despite such a minor deviation, with only one fewer guanine in the last G4-tract of d(G2NG3NG4).
As an analog of guanine, inosine substitution for guanine has been commonly used for unambiguous assignments in NMR structural studies of GQs. In addition, adenine-to-inosine mutation actually occurs in DNA damage in nature, with a noticeable occurrence within human telomeric DNA (55). Given the availability of the d(G2T2AG3T2-I-G3T) sequence (termed htel3Δ1-Ino11), regarded as an adenine-to-inosine-mutated human telomeric sequence of d(GGTTAGGGTTAGGG), we could create a pseudo G4-tract containing one inosine as an analog of guanine along with other three naturally canonical guanines together to fulfill the d(G2NG3NG4) sequence motif (Figure 7A).
Figure 7.
Interaction of htel3Δ1-Ino11 and htel1. (A) Sequences of htel3Δ1-Ino11 and htel1. The sequence motif d(G2NG3NG4) is shown on top. The residue inosine11 is coded as Ino. (B) One-dimensional 1H spectra of htel3Δ1-Ino11 in the absence and presence of htel1. The inosine imino peak is marked by an asterisk. (C) Two-dimensional 1H-15N HSQC spectrum of htel3Δ1-Ino11/htel1. A one-dimensional imino proton projection is shown. The inosine imino peak is marked by an asterisk in the one-dimensional projection and framed in the two-dimensional spectrum. Other guanine imino peaks are marked by dots in the one-dimensional projection. (D) CD spectra of Otel3Δ2/P6 (indicated by the black solid line) and htel3Δ1-Ino11/htel1 (indicated by the gray dotted line).
As expected, htel3Δ1-Ino11 was able to associate with the short chain htel1 of d(TTAGGG). The one-dimensional 1H NMR spectrum of the complex htel3Δ1-Ino11/htel1 displayed 11 peaks at 10–12 ppm and one downfield imino peak at 14 ppm for inosine 11 (Figure 7B and C), indicating the participation of this inosine into the G-tetrad core of a possible three-layer GQ. The two dimensional 1H-15N HSQC spectrum was used to confirm that the most downfield imino signal around 14 ppm was indeed from inosine (Figure 7C), based on the observed distinctive 15N1 chemical shift of 175 ppm. Accordingly, upon further replacement of this inosine with a canonical guanine, the given htel3Δ1-G11 of d(GGTTAGGGTTGGGG), as expected, enabled association with the short chain htel1, and the resulting complex htel3Δ1-G11/htel1 displayed an NMR spectral pattern almost identical to that of htel3Δ1-Ino11/htel1 (Supplementary Figure S22). In addition, the similarly comparable CD spectra of the newly formed complexes htel3Δ1-Ino11/htel1 and the previous complex Otel3Δ2/P6, implied their sharing of the same folding topology of a leaped V-shape GQ (Figure 7D). Further analysis of 2D NOESY and 1H,13C-HMBC spectra of htel3Δ1-ino11/htel1 also confirmed that htel3Δ1-Ino11/htel1 folded into a three-layer leaped V-shape GQ (Supplementary Figure S23).
Furthermore, the short chain probes of htel1 and P6 were found to preferentially bind to htel3Δ1-Ino11 rather than htel3Δ1, enabling htel3Δ1-Ino11 to be distinguished from htel3Δ1 (Supplementary Figures S20 and S21). However, the selectivity of P6 was not as good as the human telomeric sequence htel1. Therefore, htel1 was more appropriate for targeting the mutant htel3Δ1-Ino11. These results suggested a new avenue to detect specific mutants based on the d(G2NG3NG4) sequence motif.
Our results confirmed that the d(G2NG3NG4) sequence motif, particularly the last G4-tract, was essential for building the three-layer V-shaped GQ. This sequence motif would enhance the accuracy of GQ prediction in bioinformatic tools and was expected to provide more information for subsequent drug design.
CONCLUSION
In this work, we described the first NMR structure of intermolecular assembly of a leaped V-shape GQ complex consisting of two strands of quite different lengths, in which the longer one was the target, and the shorter one, with only a single G-tract, could be used as a potential probe. Taking advantage of the novel leaped V-shape scaffold structure, the tested G-rich probes exhibited advantageous and considerable selectivity for capture of the d(G2NG3NG4) sequence motif proposed by us. More interestingly, even for less thermodynamically favorable cases of inosine-mutated sequences, kinetic discrimination could still be achieved through the kinetically favored formation of an intermolecular GQ complex as long as the structure contained the leaped V-shape scaffold. Overall, our findings in this work will be helpful for improving our understanding of the nature of this novel V-shaped scaffold and will enrich GQ prediction algorithms. Our results will also provide insights into the exploration of other nucleic acid variants, such as the use of engineered PNA as a fluorescence in situ hybridization probe with higher stability and improved dye brightness in vivo (56), enabling the specific targeting of distinct V-shaped scaffolds in cells via the formation of a PNA/DNA heteroquadruplex (29). Furthermore, the sequence motif d(G2NG3NG4) is not necessarily limited to telomeric sequences but may also be applicable to other potential GQ sequences, such as the promoter region. This structure-guided approach would be helpful for identifying more V-shaped GQ sequences in genes.
DATA AVAILABILITY
The coordinates of 10 structures of the Otel3Δ2/P6 GQ have been deposited in the Protein Data Bank (accession code 6A7Y). The chemical shifts have been deposited in the BioMagResBank under accession code 36199.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Bo Wu, Dr Lei Zhu and Xiaojuan Xu for proofreading the manuscript, as well as Dr Pengzhi Wu for assistance with NMR structure refinement. All the NMR experiments were performed on the Steady High Magnetic Field Laboratory, Chinese Academy of Sciences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31600621 to C.J.W., 21372223 and U1232145 to N.Z.]; National Key Research and Development Program of China [2016YFA0400900 to N.Z.]; Hefei Science Center CAS [2012FXCX001 to N.Z.]; Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology [2018ZYFX004 to N.Z.]; Anhui Province Grant [1308085MC41 to N.Z.]. Funding for open access charge: National Natural Science Foundation of China [31600621 to C.J.W.]; National Key Research and Development Program of China [2016YFA0400900 to N.Z.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 2. Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002; 2:188–200. [DOI] [PubMed] [Google Scholar]
- 3. Blackburn E.H. Switching and signaling at the telomere. Cell. 2001; 106:661–673. [DOI] [PubMed] [Google Scholar]
- 4. de Lange T. Protection of mammalian telomeres. Oncogene. 2002; 21:532–540. [DOI] [PubMed] [Google Scholar]
- 5. Lei M., Podell E.R., Cech T.R.. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004; 11:1223–1229. [DOI] [PubMed] [Google Scholar]
- 6. Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A.. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011; 30:4033–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paeschke K., Capra J.A., Zakian V.A.. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011; 145:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kerkour A., Marquevielle J., Ivashchenko S., Yatsunyk L.A., Mergny J.L., Salgado G.F.. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J. Biol. Chem. 2017; 292:8082–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cahoon L.A., Seifert H.S.. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009; 325:764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mani P., Yadav V.K., Das S.K., Chowdhury S.. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009; 4:e4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumari S., Bugaut A., Huppert J.L., Balasubramanian S.. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eddy J., Maizels N.. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008; 36:1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010; 277:1107–1117. [DOI] [PubMed] [Google Scholar]
- 15. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adrian M., Heddi B., Phan A.T.. NMR spectroscopy of G-quadruplexes. Methods. 2012; 57:11–24. [DOI] [PubMed] [Google Scholar]
- 17. Zhang N., Gorin A., Majumdar A., Kettani A., Chernichenko N., Skripkin E., Patel D.J.. V-shaped scaffold: a new architectural motif identified in an A x (G x G x G x G) pentad-containing dimeric DNA quadruplex involving stacked G(anti) x G(anti) x G(anti) x G(syn) tetrads. J. Mol. Biol. 2001; 311:1063–1079. [DOI] [PubMed] [Google Scholar]
- 18. Crnugelj M., Sket P., Plavec J.. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003; 125:7866–7871. [DOI] [PubMed] [Google Scholar]
- 19. Kuryavyi V., Patel D.J.. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010; 18:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y., Lan W., Wang C., Cao C.. A putative G-quadruplex structure in the proximal promoter of VEGFR-2 has implications for drug design to inhibit tumor angiogenesis. J. Biol. Chem. 2018; 293:8947–8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sket P., Crnugelj M., Plavec J.. d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions. Bioorg. Med. Chem. 2004; 12:5735–5744. [DOI] [PubMed] [Google Scholar]
- 22. Nielsen J.T., Arar K., Petersen M.. Solution structure of a locked nucleic acid modified quadruplex: introducing the V4 folding topology. Angew. Chem. Int. Ed. Engl. 2009; 48:3099–3103. [DOI] [PubMed] [Google Scholar]
- 23. Marusic M., Veedu R.N., Wengel J., Plavec J.. G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold. Nucleic Acids Res. 2013; 41:9524–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adrian M., Ang D.J., Lech C.J., Heddi B., Nicolas A., Phan A.T.. Structure and conformational dynamics of a stacked dimeric G-quadruplex formed by the human CEB1 minisatellite. J. Am. Chem. Soc. 2014; 136:6297–6305. [DOI] [PubMed] [Google Scholar]
- 25. Piazza A., Cui X., Adrian M., Samazan F., Heddi B., Phan A.T., Nicolas A.G.. Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae. Elife. 2017; 6:e26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marusic M., Plavec J.. The effect of DNA sequence directionality on G-Quadruplex folding. Angew. Chem. Int. Ed. Engl. 2015; 54:11716–11719. [DOI] [PubMed] [Google Scholar]
- 27. Zhang N., Phan A.T., Patel D.J.. (3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005; 127:17277–17285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu Y., Kimura T., Komiyama M.. Human telomere RNA and DNA form an intermolecular G-quadruplex. Nucleic Acids Symp. Ser. (Oxf.). 2008; 169–170. [DOI] [PubMed] [Google Scholar]
- 29. Kormuth K.A., Woolford J.L. Jr, Armitage B.A.. Homologous PNA Hybridization to Noncanonical DNA G-Quadruplexes. Biochemistry. 2016; 55:1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Datta B., Schmitt C., Armitage B.A.. Formation of a PNA2-DNA2 hybrid quadruplex. J. Am. Chem. Soc. 2003; 125:4111–4118. [DOI] [PubMed] [Google Scholar]
- 31. Masse J.E., Bortmann P., Dieckmann T., Feigon J.. Simple, efficient protocol for enzymatic synthesis of uniformly 13C, 15N-labeled DNA for heteronuclear NMR studies. Nucleic Acids Res. 1998; 26:2618–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zimmer D.P., Crothers D.M.. NMR of enzymatically synthesized uniformly 13C15N-labeled DNA oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3091–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith D.E., Su J.Y., Jucker F.M.. Efficient enzymatic synthesis of 13C,15N-labeled DNA for NMR studies. J. Biomol. NMR. 1997; 10:245–253. [DOI] [PubMed] [Google Scholar]
- 34. Nelissen F.H.T., Tessari M., Wijmenga S.S., Heus H.A.. Stable isotope labeling methods for DNA. Prog. Nucl. Magn. Reson. Spectrosc. 2016; 96:89–108. [DOI] [PubMed] [Google Scholar]
- 35. Goddard T.D., Kneller D.G.. San Francisco: University of California. [Google Scholar]
- 36. Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D.. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005; 59:687–696. [DOI] [PubMed] [Google Scholar]
- 37. Webba da Silva M. NMR methods for studying quadruplex nucleic acids. Methods. 2007; 43:264–277. [DOI] [PubMed] [Google Scholar]
- 38. Wang Y., Patel D.J.. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 1995; 251:76–94. [DOI] [PubMed] [Google Scholar]
- 39. Hendrickx P.M., Martins J.C.. A user-friendly Matlab program and GUI for the pseudorotation analysis of saturated five-membered ring systems based on scalar coupling constants. Chem. Cent. J. 2008; 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M.. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003; 160:65–73. [DOI] [PubMed] [Google Scholar]
- 41. Case D.A., Cheatham T.E. 3rd, Darden T., Gohlke H., Luo R., Merz K.M. Jr, Onufriev A., Simmerling C., Wang B., Woods R.J.. The Amber biomolecular simulation programs. J. Comput. Chem. 2005; 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Do N.Q., Chung W.J., Truong T.H.A., Heddi B., Phan A.T.. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017; 45:7487–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brcic J., Plavec J.. Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 2015; 43:8590–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chung W.J., Heddi B., Schmitt E., Lim K.W., Mechulam Y., Phan A.T.. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad Sci. U.S.A. 2015; 112:2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim K.W., Khong Z.J., Phan A.T.. Thermal stability of DNA quadruplex-duplex hybrids. Biochemistry. 2014; 53:247–257. [DOI] [PubMed] [Google Scholar]
- 46. Lim K.W., Lacroix L., Yue D.J., Lim J.K., Lim J.M., Phan A.T.. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010; 132:12331–12342. [DOI] [PubMed] [Google Scholar]
- 47. Phan A.T. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000; 16:175–178. [DOI] [PubMed] [Google Scholar]
- 48. Phan A.T., Patel D.J.. A site-specific low-enrichment (15)N,(13)C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002; 124:1160–1161. [DOI] [PubMed] [Google Scholar]
- 49. Phan A.T., Gueron M., Leroy J.L.. Investigation of unusual DNA motifs. Methods Enzymol. 2001; 338:341–371. [DOI] [PubMed] [Google Scholar]
- 50. Krepl M., Otyepka M., Banas P., Sponer J.. Effect of guanine to inosine substitution on stability of canonical DNA and RNA duplexes: molecular dynamics thermodynamics integration study. J. Phys. Chem. B. 2013; 117:1872–1879. [DOI] [PubMed] [Google Scholar]
- 51. Smith F.W., Feigon J.. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992; 356:164–168. [DOI] [PubMed] [Google Scholar]
- 52. Schultze P., Smith F.W., Feigon J.. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG). Structure. 1994; 2:221–233. [DOI] [PubMed] [Google Scholar]
- 53. Haider S., Parkinson G.N., Neidle S.. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002; 320:189–200. [DOI] [PubMed] [Google Scholar]
- 54. Xue Y., Liu J.Q., Zheng K.W., Kan Z.Y., Hao Y.H., Tan Z.. Kinetic and thermodynamic control of G-quadruplex folding. Angew. Chem. Int. Ed. Engl. 2011; 50:8046–8050. [DOI] [PubMed] [Google Scholar]
- 55. Kuraoka I. Diversity of endonuclease V: from DNA repair to RNA editing. Biomolecules. 2015; 5:2194–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stender H. PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2003; 3:649–655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of 10 structures of the Otel3Δ2/P6 GQ have been deposited in the Protein Data Bank (accession code 6A7Y). The chemical shifts have been deposited in the BioMagResBank under accession code 36199.