Abstract
Nucleic acid triplexes may regulate many important biological processes. Persistent accumulation of the oncogenic 7-kb long noncoding RNA MALAT1 is dependent on an unusually long intramolecular triple helix. This triplex structure is positioned within a conserved ENE (element for nuclear expression) motif at the lncRNA 3′ terminus and protects the entire transcript from degradation in a polyA-independent manner. A requisite 3′ maturation step leads to triplex formation though the precise mechanism of triplex folding remains unclear. Furthermore, the contributions of several peripheral structural elements to triplex formation and protective function have not been determined. We evaluated the stability, conformational fluctuations, and function of this MALAT1 ENE triple helix (M1TH) protective element using in vitro mutational analyses coupled with biochemical and biophysical characterizations. Using fluorescence and UV melts, FRET, and an exonucleolytic decay assay we define a concerted mechanism for triplex formation and uncover a metastable, dynamic triplex population under near-physiological conditions. Structural elements surrounding the triplex regulate the dynamic M1TH conformational variability, but increased triplex dynamics lead to M1TH degradation. Taken together, we suggest that finely tuned dynamics may be a general mechanism regulating triplex-mediated functions.
INTRODUCTION
Nucleic acid triplex formation using RNA, DNA and RNA–DNA hybrids are thought to be an essential element of several biological regulatory processes (1,2). Recent discoveries have elucidated evidence for diverse mechanisms of triplex-mediated gene regulation including chromatin rearrangement, recruitment of epigenetic modifiers, and inhibition of translation initiation (1,3–8). Additionally, intramolecular RNA triplexes are requisite for the function of the spliceosome (9), telomerase RNA (10,11), –1 programmed ribosomal frameshifting elements (12) and the Campylobacter jejuni trans-activating CRISPR RNA (13). These intramolecular triplex interactions are short (typically <6 base triples) and often require additional intramolecular structural interactions, cations, or protein binding partners to support their functional roles. Even relatively long DNA or DNA–RNA triple helices exhibit limited stability due to charge repulsion stemming from the close proximity of the three polyanionic strands. Necessarily, studies designing triplex forming oligonucleotides for therapeutic intervention have explored the role of (poly)cations and chemical modifications to promote triplex stability and function (14–19).
Recently, a new class of long intramolecular RNA triplexes was identified at the 3′ termini of several viral and human long noncoding RNAs (lncRNA) (20–22). A conserved core, called the element for nuclear expression (ENE) (23–25), contains two U-rich sequences which form a parallel pyrimidine-purine-pyrimidine triplex upon association with a 3′ terminal A-rich sequence. Sequestration of the lncRNA 3′ end within this triplex protects the entire transcript from 3′-5′ exonucleolytic degradation in a deadenylation-independent manner (20–22). The ENE triplex is a transferable stability element, conferring significant stabilization to mRNA transcripts when encoded in the 3′ UTR (20,22,23). The 3′-terminal 94 nucleotides of the human lncRNA MALAT1 adopt this protective ENE triplex, leading to the high cellular abundance and persistent oncogenic activity of the 7-kb lncRNA in several cancer types. Specific triplex-disrupting mutations lead to MALAT1 degradation and loss of accumulation (20,22) and reduced levels of the lncRNA significantly inhibit tumor formation, migration, and metastasis (26,27). Therefore, this triplex is an important target for therapeutic discovery and selective triplex inhibition may slow proliferation and metastasis in several cancer types (26,28–30).
Interestingly, the MALAT1 triple helix is not located at the 3′ end of the primary MALAT1 transcript. Instead, the terminal sequence of the nascent transcript adopts a short tRNA-like domain of unknown function (31). Cleavage of this domain by RNase P liberates a mature 3′ end comprising the A-rich sequence required for triplex formation. In the absence of this maturation step, the ENE and 3′ A-rich tail sequences are insufficient to promote transcript abundance in vivo (20). Therefore, rather than targeting the triplex structure directly, preclusion of triplex formation represents an alternative therapeutic approach to functional inhibition of MALAT1. However, thorough studies evaluating the formation, stability, and dynamics of the MALAT1 ENE triple helix, herein denoted M1TH, have not been reported. Here, we present a thorough biochemical and biophysical investigation of the structural and environmental factors regulating the protective function of M1TH. Using FRET, differential scanning fluorimetry (DSF) and FRET (DS-FRET), and UV thermal melts we generated empirical conformational and stability landscapes probing the effects of structural deletions and the ionic environment on triplex formation. Our results show that the entire triplex region is metastable and highly dynamic in near-physiological ionic conditions. Large conformational dynamics in wild-type and mutated M1TH disrupt the triplex and lead to 3′-5′ exonucleolytic degradation in vitro by RNase R. Overall, our results indicate that finely tuned triplex dynamics, and not simply triplex stability, are critical for the protective function of M1TH. Furthermore, peripheral structural elements on M1TH serve as important regulators of triplex dynamics. We conclude that biological triplexes may generally depend on structural elements outside the base triple interactions to support functional stability and activity.
MATERIALS AND METHODS
Preparation of unlabeled RNAs
Short RNA oligos M1A, M1Tail and M1Bs, were purchased from IDT. Deprotected synthetic RNAs were resuspended in water and stored at −20°C. All other RNAs were transcribed from PCR products using T7 RNA polymerase. PCR primers were designed and purchased from IDT for amplification of a plasmid containing the MALAT1 ENE triple helix sequence. All reverse primers contained two 2′-OMe nucleotides at the 5′ end in order to limit non-templated nucleotide addition by T7 RNAP at the 3′ end of the transcript (32). The RNAs were purified using preparative scale denaturing PAGE. RNA bands of interest were excised and electroeluted overnight (Elutrap, GE). The eluate was concentrated by ethanol precipitation and the pellet was resuspended in water and stored at −20°C.
Triplex purification using size exclusion chromatography
All RNAs used in UV, FRET, DS-FRET, and DSF experiments were purified on ENrich SEC 70 (Bio-Rad) or Superdex 75 (GE Healthcare) size-exclusion chromatography columns. Unimolecular RNAs were prepared in refolding buffer containing 20 mM HEPES–KOH, pH 7.4 (22°C), and 1 mM MgCl2. RNA was heated at 95°C for 2 min, snap-cooled on ice for 5 min, and allowed to equilibrate at room temperature for at least 1 h. The refolded RNAs were purified on a sizing column that was equilibrated in the refolding buffer. To monitor RNA elution, absorbance was recorded at 260 nm. For the bimolecular RNAs, a 1.5 molar excess of the shorter RNA (M1Tail or M1A) was added to the longer RNA (M1ENE or M1B, respectively), and then annealed in the refolding buffer as above. The RNAs were purified similar to the unimolecular RNA above while monitoring the absorbance signal at 260, 550 (Cy3) and 650 nm (Cy5) to ensure complex formation. In each case, the two-piece RNA triplex complex was well separated from the excess of short RNA. For the tri-molecular M1A-Bs-T, a 1.5-fold excess of the M1Tail RNA and 1.1-fold excess of M1Bs was added to M1A in refolding buffer and annealed as above. The three-stranded complex was well separated from the excess M1Bs and M1Tail on the sizing column.
UV thermal melts
RNAs were annealed and then purified using an ENrich SEC 70 (Bio-Rad) size exclusion chromatography column in 20 mM HEPES–KOH, pH 7.4 (22°C), 1 mM MgCl2. Using this SEC purified RNA, a 500–700 μl sample was prepared in the refolding buffer with a final RNA concentration of 5–10 μg ml−1, and the desired magnesium (0.1–1 mM) and total monovalent concentrations (2.6–742.6 mM). NaCl and KCl were equimolar except for the 2.6 mM KCl introduced from the HEPES–KOH buffer. All UV melt experiments were performed in a stoppered 1-cm quartz cuvette. Absorbance was monitored at 260 nm using an Agilent 8453 UV-Vis spectrophotometer with an Agilent 89090A temperature controller across a temperature range from 20°C to 85°C with 0.1°C increment/cycle. Because the total experimental time was ∼3 h, we calculate an average ramp rate of ∼0.42°C/min. Raw melting profiles and derivatives were plotted using Origin2015 software (OriginLab). The derivative curves were smoothed using Fast Fourier Transform (FTT) 70-point smoothing. The Tm,1 and Tm,2 were determined from the smoothed derivative plot using a peak finding algorithm within the software.
Preparation of fluorescently labeled RNAs
Fluorescently labeled RNAs were purchased from the Keck Foundation Biotechnology Resource Facility at Yale University. The ENE motif was synthesized with a 3′ terminal Cy3. M1Tail was synthesized with a 5′ terminal Cy5. M1A was synthesized with a 3′ terminal Cy3. M1B was synthesized containing a 3′ terminal Cy5. Synthetic RNA oligonucleotides were resuspended in water. The ENE-Cy3 and M1B-Cy5 RNAs were purified by denaturing PAGE and electroeluted overnight. The eluate RNA was ethanol precipitated, resuspended in water, and stored at −20°C until use. M1Tail and M1A RNAs were stored at −20°C until use without further purification.
Sample preparation for multiplexed FRET experiments
Five-fold concentrated stocks of eight different magnesium concentrations (0–5 mM) in 20 mM HEPES–KOH, pH 7.4 were prepared. Five-fold concentrated stocks of eight different monovalent concentrations (0–1000 mM) containing equimolar NaCl and KCl in 20 mM HEPES–KOH, pH 7.4 were prepared. 24 μl of ∼200 nM SEC purified RNA containing <0.1 mM MgCl2 and 20 mM HEPES–KOH, pH 7.4 were pipetted into an 8 × 8 grid on a black 384-well plate (Corning). 8 μl of MgCl2 stock were added to appropriate rows using a multichannel pipette. 8 μl of monovalent stock were added to appropriate columns using a multichannel pipette. Each individual final reaction contained ∼100 nM RNA, a single MgCl2 concentration between 0.05 and 1 mM MgCl2, and a single total monovalent concentration between 2.6 and 202.6 mM (including 2.6 mM KOH from the HEPES–KOH buffer). The microplate was sealed with an adhesive seal, covered with aluminum foil, centrifuged at 1000 × g for 2 min at room temperature, and incubated for 1–2 h. The adhesive seal was removed prior to recording Cy3 and Cy5 fluorescence.
FRET measurements at room temperature
Room temperature fluorescence was recorded using a Synergy H1MF (BioTek) multimode microplate reader using one excitation filter for Cy3 (540 ± 25 nm) and two emission filters for Cy3 (590 ± 20 nm) and Cy5 (680 ± 30 nm). An apparent EFRET was calculated using EFRET = ICy5/(ICy5 + ICy3), where ICy5 and ICy3 represent the fluorescence intensities of Cy5 and Cy3, respectively. Origin2015 software (OriginLab) was used to create 3D plots of the EFRET values together with a 3D surface. Experiments were performed in triplicate.
Differential scanning FRET
Following measurement of room temperature fluorescence, 25 μl of each sample were transferred into a clear qPCR plate (SP-1384, Thermo Scientific), sealed with an optically clear adhesive seal (Thermo Scientific), covered with aluminum foil, and centrifuged at 1000 × g for 2 min. Thermal melts were performed using a QuantStudio 7 Flex (Applied Biosystems) from 20 to 85°C with a 0.015°C/s ramp rate. One excitation filter (x2: 520 ± 10 nm) was used to excite Cy3 and two emission filters were used to record fluorescence of Cy3 (m3: 586 ± 10 nm) and Cy5 (m5: 682 ± 14 nm). For each experiment, raw melting profiles of Cy3 and Cy5 were plotted and analyzed using Origin2015. The FRET curve (EFRET = ICy5/(ICy5 + ICy3)), the first derivative of the EFRET, and smoothed derivative (Fast Fourier Transform, 70-point smoothing) were also plotted. The Tm,1 was determined from the smoothed derivative curve using a peak finding algorithm within the software. Finally, a 3D thermostability plot was generated using the individual Tm,1 values from each of the 64 experiments. Experiments were performed in triplicate.
Differential scanning fluorimetry (DSF)
Differential scanning fluorimetry (DSF) experiments were performed using RiboGreen dye and unlabeled full-length M1TH. Following SEC purification, the RNA was mixed with RiboGreen at concentrations of 900 and 500 nM, respectively, in 20 mM HEPES, pH 7.4. 24 μl of this sample were pipetted into 64 wells in an 8 × 8 format in a black 384-well plate (Corning) using a multichannel pipette. Five-fold concentrated MgCl2 and monovalent concentrations were prepared as described for the multiplexed FRET experiment above. 8 μl of MgCl2 stock were pipetted in successive columns. 8 μl of monovalent salt stocks were pipetted in successive rows. The final experiments contained 0.54 μM RNA, 0.3 μM RiboGreen, a single MgCl2 concentration between 0.1 and 1.1 mM, and a single total monovalent concentration between 2.6 and 392.6 mM (including 2.6 mM KOH from the HEPES–KOH buffer). The microplates were sealed with an adhesive seal, covered with aluminum foil, centrifuged at 1000 × g for 2 min, and incubated for 1–2 h prior to experimentation.
25 μl of each sample were transferred into a clear qPCR plate (SP-1384, Thermo Scientific), sealed with an optically clear adhesive seal (Thermo Scientific), and centrifuged at 1000 x g for 2 min. Thermal melts were performed on a QuantStudio 7 Flex (Applied Biosystems) using one excitation filter (x1: 470 ± 15 nm) and one emission filter (m1: 520 ± 15 nm). RiboGreen fluorescence was recorded from 20 to 95°C with a ramp rate of 0.015°C/s. Raw fluorescence signal and its first derivative were plotted using Origin2015 software (OriginLab). A 70-point Fast Fourier Transform (FTT) smoothing function was applied on the first derivative to aid in peak identification. The Tm,1 and Tm,2 peaks were identified using a peak finding algorithm in Origin2015. For the Tm,1 values a 3D thermostability plot was generated across the 64 ionic conditions. Experiments were performed in triplicate.
RNase R degradation experiments
For single time-point RNase R degradation reactions, 0.5 μg RNA was incubated in buffer containing 20 mM HEPES–KOH, pH 7.4, and final MgCl2 and monovalent concentrations as indicated in a total volume of 10 μl for 1 h at room temperature. To initiate the reaction, 10 μl of either 0.1 U/μl RNase R (Lucigen) (for 1 U enzyme in the final reaction) or 0.5 U/μl RNase R (for 5 U enzyme in the final reaction) in 20 mM HEPES–KOH, pH 7.4, and final MgCl2 and monovalent concentrations as indicated (Conditions I-IV) was added to the RNA. Reactions were incubated for 5 h at 37°C. Reactions were stopped with the addition of 20 μl RNA loading dye (5 mM EDTA, 95% formamide) and either immediately analyzed using denaturing PAGE or frozen at −20°C until analysis. For time-course reactions, 3 μg of RNA were incubated in 20 mM HEPES–KOH, pH 7.4, and final MgCl2 and monovalent concentrations as indicated (Conditions I-IV) in a total volume of 120 μl for 1 h at room temperature. Either 0.3 μl of RNase R (for 1 U/0.5 μg RNA total in the final reaction) or 1.5 μl of RNase R (for 5 U/0.5 μg RNA total in the final reaction) were added directly as a stock to initiate the reaction. Reactions were incubated at 37°C. 20 μl aliquots were removed immediately after enzyme addition for time zero, at 30 min, 1, 2 and 5 h and stopped with the addition of 20 μl RNA loading dye. Reactions were analyzed by denaturing polyacrylamide gel electrophoresis stained with ethidium bromide.
RESULTS
The basal linker and apical stem significantly impact triplex stability
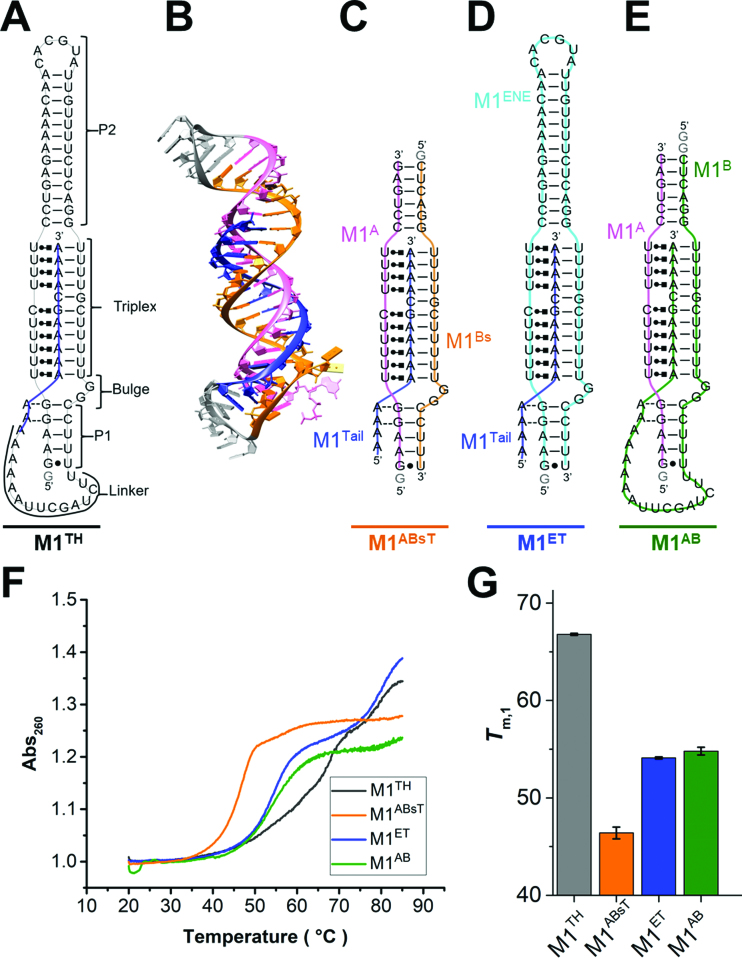
A recent crystal structure of the M1TH core (33) revealed the unique architecture that specifically supports this exceptionally long triplex comprising ten base triples and an intervening G-C base pair (Figure 1). Methyltransferase-like protein 16 (METTL16) binds M1TH both in vitro and in vivo, as demonstrated by gel shift analysis and cross-linking, respectively, in a manner dependent on the ENE triplex sequence and structure (34). Therefore, the ENE triplex adopts a similar well-formed structure in these environments. The structure of the full-length 94-nt wild-type M1TH (Figure 1A) adopts an extended stack of 29 base pairs and triples. This rod-like structure includes two helices (P1 and P2) of the ENE (Figure 1, M1ENE) abutting either end of the triplex. The triplex is formed between two conserved U-rich regions and the A-rich sequence of the 3′ tail (Figure 1B). Specifically, the tail (Figure 1, M1Tail) forms Watson–Crick interactions with the 3′-most U-rich region while the 5′-most U-rich region forms base triple interactions with the Hoogsteen edges of nucleobases in the 3′ tail. Formation of a C+•G-C triple interaction is expected to be sensitive to changes in the protonation state of C11. Indeed, Brown et al. performed UV melts in pH 5 relative to pH 7 buffer and demonstrated that the triplex melting temperature is stabilized by ∼5°C at the lower pH (20) (see also Supplementary Information, Supplementary Figure S1).
Figure 1.
Peripheral elements regulate thermostability of the MALAT1 ENE triple helix. (A) Secondary structure of the full-length MALAT1 ENE triple helix (M1TH) including Watson–Crick interactions (—), Hoogsteen interactions ( ), A-minor interactions (−−), and a wobble base pair (•). A 5′ GTP (grey) is added to the sequence to facilitate transcription. The A-rich tail is colored blue and structural regions are indicated in brackets. (B) The crystal structure of the MALAT1 triple helix core (PDB: 4PLX) (33). The three strands are colored orange, pink, and blue. The two short loops (gray) were engineered to facilitate crystallization. (C) Secondary structure of trimolecular M1ABsT. M1Bs is colored orange, M1A is colored pink, and M1Tail is colored blue, as in B. (D) Secondary structure of bimolecular M1ET, which eliminates the basal linker. The ENE motif is colored cyan and the M1Tail is colored as in C. (E) Secondary structure of bimolecular M1AB, wherein the basal linker is included and the apical P2 helix is truncated. M1A is colored as in C and M1B is colored green. In C–E, a 5′ GTP (gray) is added to the sequence to facilitate transcription. (F) Normalized UV melt profiles for M1TH (gray), M1ABsT (orange), M1ET (blue), M1AB (green) from 20 to 85°C in 20 mM HEPES•KOH, pH 7.4, 1 mM MgCl2, 25 mM NaCl and 25 mM KCl. Triplex formation for all constructs is explicitly demonstrated in Supplementary Figure S2. Triplex and duplex melting occur in the same transition for the bimolecular M1AB. All experiments were performed in duplicate. A single representative melting profile is plotted. (G) Triplex melting temperatures (Tm,1) for the unimolecular, bimolecular, and trimolecular constructs, colored as in F. Large changes in Tm,1 for the bi- and tri-molecular RNA relative to the full-length RNA demonstrate the contributions of peripheral elements to triplex stability.
), A-minor interactions (−−), and a wobble base pair (•). A 5′ GTP (grey) is added to the sequence to facilitate transcription. The A-rich tail is colored blue and structural regions are indicated in brackets. (B) The crystal structure of the MALAT1 triple helix core (PDB: 4PLX) (33). The three strands are colored orange, pink, and blue. The two short loops (gray) were engineered to facilitate crystallization. (C) Secondary structure of trimolecular M1ABsT. M1Bs is colored orange, M1A is colored pink, and M1Tail is colored blue, as in B. (D) Secondary structure of bimolecular M1ET, which eliminates the basal linker. The ENE motif is colored cyan and the M1Tail is colored as in C. (E) Secondary structure of bimolecular M1AB, wherein the basal linker is included and the apical P2 helix is truncated. M1A is colored as in C and M1B is colored green. In C–E, a 5′ GTP (gray) is added to the sequence to facilitate transcription. (F) Normalized UV melt profiles for M1TH (gray), M1ABsT (orange), M1ET (blue), M1AB (green) from 20 to 85°C in 20 mM HEPES•KOH, pH 7.4, 1 mM MgCl2, 25 mM NaCl and 25 mM KCl. Triplex formation for all constructs is explicitly demonstrated in Supplementary Figure S2. Triplex and duplex melting occur in the same transition for the bimolecular M1AB. All experiments were performed in duplicate. A single representative melting profile is plotted. (G) Triplex melting temperatures (Tm,1) for the unimolecular, bimolecular, and trimolecular constructs, colored as in F. Large changes in Tm,1 for the bi- and tri-molecular RNA relative to the full-length RNA demonstrate the contributions of peripheral elements to triplex stability.
Only ∼1/3 of the 94-nt ENE triplex protective element is involved in formation of the parallel pyrimidine–purine–pyrimidine base triples. Mutational analyses supported by the crystal structure of the triple helix core have revealed the molecular basis for functional stability within the triplex region (33,35). The remaining 61 nucleotides of M1TH are involved in peripheral structural elements, including the 5-bp P1 helix, the 13-bp P2 apical helix, a 17-nucleotide basal linker, and a 2-nt bulge (Figure 1A). These elements are semi-conserved in several related triplexes (20), but their contributions to M1TH formation and stability in vitro have been minimally examined. To evaluate the role of these regions in stabilizing the triplex we generated several unimolecular, bimolecular, and trimolecular RNA constructs comprising various mutations and truncations (Figure 1, and Supplementary Information). Triplex formation was confirmed for each of our bi- and tri-molecular RNAs using native gel shift analysis (Supplementary Figure S2A–D), consistent with previously published gel shift results (20,35). Additionally, each RNA construct was purified by size-exclusion chromatography to remove aggregates and isolate only the appropriate triplex-containing fraction for experimentation (Supplementary Figure S2E).
We employed thermal UV melts to assess the contributions of various structural elements to triplex stability. Triplex melting disrupts stacking interactions and leads to increased absorbance (260 nm) resulting from the hyperchromicity of single-stranded RNA relative to duplex or triplex RNA. We first evaluated the UV melting profile for the unimolecular M1TH, which exhibits two transitions (Figure 1F, black), corresponding to sequential melting events (Supplementary Figure S3A). Our thermal melting experiments performed in HEPES buffer, which has a small but not negligible temperature dependent pKa, slightly overestimate the Tm due to small changes in protonation of the C+•G-C triple interaction (see Supplementary Information; Supplementary Tables S1, S2; and Supplementary Figure S1). The transition at very high temperature, hereafter identified as Tm,2 (Tm,2 = 78.3 ± 0.05°C, Table 1), is consistent with secondary structure melting under our reaction conditions (1 mM MgCl2, 25 mM NaCl, 25 mM KCl). The earlier melting transition, identified as Tm,1, occurs at 66.6 ± 0.2°C (Figure 1G, gray) and includes triplex melting (20). However, the amplitude of this transition is ∼2.5 times greater than the duplex transition (Figure 1F). Melting of the short P1 helix and the basal linker may occur together with the triplex.
Table 1.
Melting temperatures determined by UV melts. Melting temperatures for the two transitions in 1 mM MgCl2, 25 mM KCl, 25 mM NaCl, 20 mM HEPES–KOH, pH 7.4, are reported for the various RNA constructs used in this study. The pH of HEPES buffer lowers slightly as the temperature increases, leading to small inaccuracies in the absolute melting temperature (see Supplementary Information).
| RNA | T m,1 (°C) | T m,2 (°C) |
|---|---|---|
| M1TH | 66.7 ± 0.1 | 78.2 ± 0.2 |
| M1Δbulge1 | 65.4 ± 0.4 | 80.1 ± 0.0 |
| M1Δbulge2 | 64.3 ± 0.5 | 79.8 ± 0.2 |
| M1BsT | 35.4 ± 0.4 | N/A |
| M1ABsT | 46.4 ± 0.6 | 58.5 ± 0.6 |
| M1ET | 54.1 ± 0.1 | 80.3 ± 0.1 |
| M1ENE | 47.0 ± 1.1 | 79.3 ± 0.4 |
| M1AB | 54.8 ± 0.6 | N/A◊ |
◊M1AB melting profile exhibits a single transition in which both triplex and duplex regions melt concomitantly.
To assess the role of bulged nucleotides (G59, G60), we deleted one (Δbulge1) or both (Δbulge2) of these nucleotides and monitored changes in thermal stability. The single and double mutation RNAs only reduced the triplex melting transition by 1.4 and 2.3°C, respectively (Table 1). Therefore, the bulge does not significantly contribute to triplex stability in vitro. Its conservation in several triplexes may indicate its role as a structural anchor point at the base of the triplex, which may be important for the recruitment of binding partners within the cellular milieu.
We next performed UV melt analysis using a bottom-up approach to assess the contribution of various structural elements to triplex stability. First, we examine melting events in a truncated triplex, denoted M1ABsT (Figure 1C), comprising the A-rich tail (M1Tail), the third strand (M1A), and the duplex strand (M1Bs, where Bs indicates ‘B short’). Within this triplex core, M1Bs and M1Tail form Watson–Crick interactions (Figure 1C, orange and blue) while M1A forms interactions with the Hoogsteen edge of M1Tail and Watson–Crick interactions with the M1Bs (Figure 1C, pink). Hybridization and UV thermal melt analysis of the two short RNA strands, M1Bs and the M1Tail (Figure 1C, orange and blue strands), reveal only a weak interaction within this duplex (Table 1, Tm = 35.3 ± 0.4°C, Supplementary Figure S3B) compared to the secondary structure transition in full-length M1TH (Tm,2 = 78.2 ± 0.2°C). Addition of M1A results in triplex formation (Supplementary Figure S2C–E) together with the short duplexes abutting either side (Figure 1C). The M1ABsT melting profile reveals a two-transition process akin to M1TH (Figure 1F, Table 1). The large melting transition (Tm,1 = 46.4 ± 0.6°C, Figure 1G, orange) indicates that the triplex is more stable than the duplex by >10°C. A small second transition (Tm,2 = 58.5 ± 0.6°C, Table 1) may involve disruption of one (or both) of the short duplexes; the truncated P2 helix likely melts in this transition owing to its longer length and more G-C rich base pairs relative to P1.
To evaluate the contributions of apical stem loop, we next performed thermal melts using a bimolecular ENE-tail construct (Figure 1D, M1ET), which includes the entire P2 helix. A previous study used this construct to characterize the role of specific base triple interactions on overall triplex stability (35). In comparison to M1ABsT, both melting transitions are stabilized in M1ET (Figure 1F, Table 1). Not surprisingly, inclusion of the full P2 helix and its loop increases the secondary structure melting by nearly 22°C. More interestingly, this peripheral duplex stabilizes the triplex transition by 8°C, highlighting an important role for structural regions distal to the triplex. The long, stable P2 helix may aid pre-formation of the ENE (Figure 1D, cyan) prior to triplex folding. To assess the ENE structure in the absence of the A-rich tail, we evaluated the melting profile of the ENE alone and identify two transitions (Table 1, Supplementary Figure S3B). The transition at high temperature is consistent with P2 helix melting, as in M1TH and M1ET (Table 1). A transition at low temperature (Tm,1 = 47.0 ± 0.1°C) suggests some residual structure within P1 and the U-rich internal loop is present at physiological temperature (Table 1, Supplementary Figure S3B). The broad profile of this transition over ∼35°C (30–65°C) is consistent with multiple structural interactions which do not melt cooperatively (Supplementary Figure S3B, inset). In contrast, the presence of the A-rich tail in M1ET, and hence the triplex, results in a sharp transition, consistent with a cooperative unfolding event (Supplementary Figure S3A).
Because a 12.5°C reduction in triplex stability is attributed to the removal of the basal linker in M1ET (Figure 1G, M1ET versus M1TH), we generated a new bimolecular construct, M1AB, containing the linker (Figure 1E). In this construct the wild-type linker is added to the M1ABsT core, joining M1Bs and M1T in cis to form M1B. Both M1A and M1B are required for triplex formation as well as formation of duplexes P1 and P2, which immediately stack on either end of the triplex. Gel shift and size-exclusion chromatography analyses demonstrate proper triplex formation upon association of these two RNAs (Supplementary Figure S2B and E). This RNA melts in a single transition (Tm = 54.8 ± 0.6) suggesting that the small secondary structure transition observed in M1ABsT (Figure 1F, orange) is subsumed into the stabilized triplex melting transition (Figure 1F, Table 1). Addition of the linker stabilizes the triplex by slightly more than 8°C (Figure 1G, green). Loop closure between M1Bs and M1T is likely not sufficient to account for the important stability contribution of this peripheral linker; long unstructured loops have been shown to significantly destabilize structure formation (36). Alternatively, specific structural interactions, yet undetermined, in the wild-type basal linker may significantly promote triplex stability (N.J.B. unpublished results).
Removal of either the apical stem or basal linker leads to similar losses in thermal stability (Table 1). Analysis at lower magnesium concentrations (<0.2 mM) reveals that linker removal is slightly more destabilizing than truncation of the P2 helix (Supplementary Figure S3C). Overall, our bottom-up analysis of the MALAT1 ENE triplex stability underscores the important contributions of peripheral elements to the stability of the triplex core. Our results provide a functional rationale for the reported semi-conservation of sequence and length in these peripheral elements from human to zebrafish (22).
Highly variable triplex stabilities result from changes in ionic conditions
We reasoned that the increased charge density of the three-stranded ENE triplex may destabilize this region, leading to a metastable structure in physiological conditions. Because triplex stability is important for its protective function (35), we next examined the range of stability (Tm,1) attained in various cation concentrations. Previous studies on DNA triplexes revealed multiphasic thermal stability responses to cation concentration and valency (14–16). We performed UV melt analysis of M1TH, M1ET and M1AB in solution mixtures containing various monovalent (2.6–742.6 mM) and magnesium (0.05–1 mM) concentrations (Supplementary Figure S4). We employ equimolar monovalent salt concentrations (KCl and NaCl) to generally assess the role of monovalent cations on triplex formation and stability. The Tm,1 values increase monotonically with increased MgCl2 concentrations at all but the highest monovalent concentrations (742.6 mM). In contrast, triplex stability first decreases and then increases with increasing monovalent concentrations. However, because UV thermal melts are a low-throughput experimental approach, only minimal analysis is afforded by the limited number of solution conditions examined.
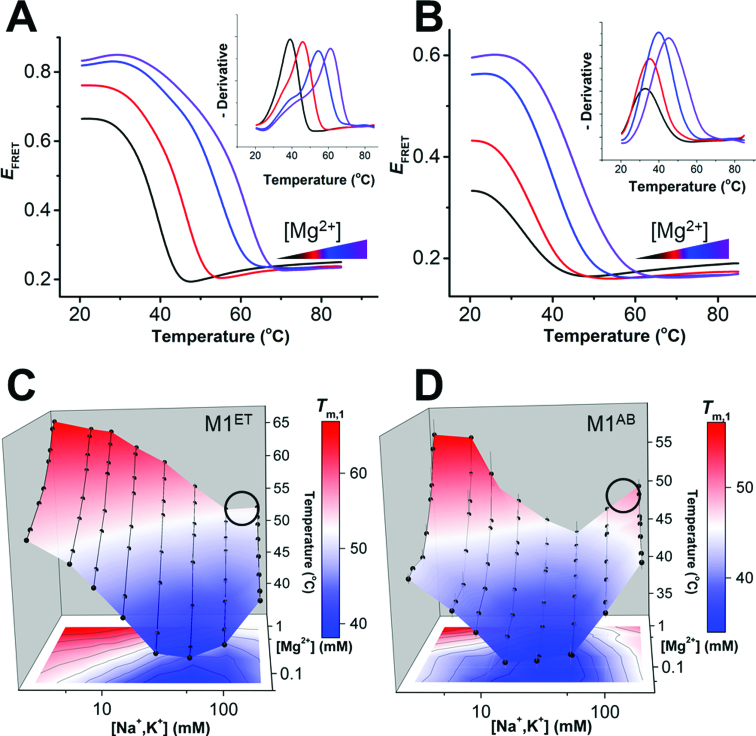
To overcome the throughput limitations of UV melt experiments, we turned to thermal melting experiments monitored using differential scanning fluorescence resonance energy transfer (DS-FRET). We recently developed DS-FRET to monitor RNA thermal denaturation for up to 384 experiments simultaneously on a traditional quantitative PCR (qPCR) instrument (37). We designed site-specific fluorophore pairs (Cy3 and Cy5) for both M1ET and M1AB (Supplementary Figure S5). Using a qPCR machine, we excite the Cy3 donor and record fluorescence signals of the two dyes from 20°C to 85°C (Supplementary Figures S5). An apparent FRET efficiency (EFRET) is calculated using ICy5/(ICy5 + ICy3), where ICy5 and ICy3 represent the fluorescence intensities of Cy5 and Cy3, respectively. Thermal unfolding increases the distance between the donor and acceptor dyes leading to a lower EFRET signal across the melting transition (Figure 2A and B). The dye pair positions report only on triplex melting transitions, denoted as Tm,1 to maintain consistency with our UV melt analysis. No change in EFRET signal is observed for the duplex melting transition at higher temperatures, precluding Tm,2 determination with this method. We determine the Tm,1 for each DS-FRET experiment from the peak in the negative derivative of the EFRET curve (Figure 2A and B, insets), and accomplish rapid evaluation of triplex stability across a range of solution conditions (Figure 2A and B).
Figure 2.
Triplex stabilities vary across a matrix of monovalent and divalent concentrations. Melting profiles monitored by DS-FRET for M1ET (A) and M1AB (B) under four different magnesium concentrations (∼0.05, 0.2, 0.5 and 1 mM, with increasing concentrations colored from black to purple) in 25 mM total monovalent salt (equimolar NaCl and KCl). See Supplemental Figure S5 for samples of raw Cy3 and Cy5 melting profiles. Inset: Peaks in a plot of the negative derivative of the raw melting profile are used to determine Tm,1 for each experiment. (C, D) Tertiary thermal stability (Tm,1) landscapes of M1ET (C) and M1AB (D) monitored using DS-FRET across multiplexed ionic conditions (2.6, 5.7, 8.8, 15.1, 27.6, 52.6, 102.6 and 202.6 mM total monovalent salt (equimolar KCl and NaCl); and ∼0.05, 0.1, 0.2, 0.3, 0.5, 0.6, 0.8 and 1 mM MgCl2). n = 3 experiments; S.D. A black circle on each stability landscape indicates approximate physiological ionic conditions (∼150 mM monovalent, 1 mM MgCl2).
To evaluate the role of the ionic environment on the MALAT1 ENE triplex stability, we generate an empirical thermal stability landscape over a matrix of ionic conditions containing MgCl2 (0.1–1 mM) and monovalent salts (2.6–202.6 mM total, equimolar amounts of NaCl and KCl) (Figure 2C and D). Across 64 multiplexed ionic solution conditions we record highly variable thermal stabilities for both M1AB and M1ET. Differences in landscape shape or absolute Tm values between the two constructs can be attributed to the distinct fluorophore pair locations, which report on local changes near the top (M1AB) or bottom (M1ET) of the triplex. Each landscape has a chevron-like shape in response to increasing monovalent concentrations at low MgCl2 concentrations. The minimum Tm,1 occurs between 20 and 50 mM monovalent salt for both RNAs. The generally similar shapes of these two Tm,1 landscapes indicate that DS-FRET directly reports on structural changes within the triplex region, rather than an artefactual interaction between the dye and RNA. Our results provide the first thorough characterization of multimodal triplex stabilities and are consistent with the lower-throughput studies of DNA triplexes performed decades ago (15,38). Owing to the distinct locations of the FRET pairs in these two constructs, direct comparison of stabilities is not straightforward. Nonetheless, the stability of either construct spans a temperature range >22°C for the conditions examined; changes in RNA stability are expected commensurate with changes in cation concentrations. Of important note for the MALAT1 triplex, significant stability increases are recorded in response to significant monovalent cation concentration decreases. The complex interplay of ionic conditions and triplex stability leads to maximal triplex stability under conditions nearly devoid of monovalent cations (1 mM MgCl2, 2.6 mM monovalent cations). In near-physiological conditions, (Figure 2C and D; black circles), neither M1ET nor M1AB are maximally stable. For closer comparison of Tm values within a landscape, we calculate a ‘relative stability’ of the triplex, defined as [(P – Min) / (Max – Min)] × 100, where P is the stability of the triplex at one condition, and Max and Min are the maximal and minimal triplex stabilities, respectively, representing the maximum stability range (Max – Min) recorded on the landscape (Table 2). Under near-physiological conditions the relative stabilities of M1ET and M1AB are only 52% and 62%, respectively, suggesting that either (1) a carefully tuned metastable triplex is sufficient to achieve functional protection, or (2) the removal of a peripheral element in either bimolecular construct produces an artefactual metastability.
Table 2.
Relative triplex thermostability and folded population
| T m,1 (°C) | E FRET | ||||
|---|---|---|---|---|---|
| aM1ET | aM1AB | bM1TH | M1ET | M1AB | |
| Maximum signal | 66.6 | 57.1 | 78.5 | 0.81 | 0.55 |
| Minimum signal | 38.4 | 35.2 | 59.2 | 0.57 | 0.23 |
| Signal in near-physiological ionic conditions | 53.2 | 48.8 | 63.8 | 0.75 | 0.46 |
| Relativec stability (Tm,1) or folded population (EFRET) | 52% | 62% | 24% | 75% | 72% |
a T m,1 calculated using DS-FRET assay.
b T m,1 calculated using DSF assay.
cRelative thermostability or folded population was calculated using [(P – Min)/(Max – Min)] × 100, where Max, Min and P indicate maximum signal, minimum signal, and the signal in near-physiological ionic conditions (∼150 mM monovalent, 1 mM MgCl2), respectively. P was calculated from the average of the signal obtained in 102.6 mM and 202.6 mM monovalent concentrations.
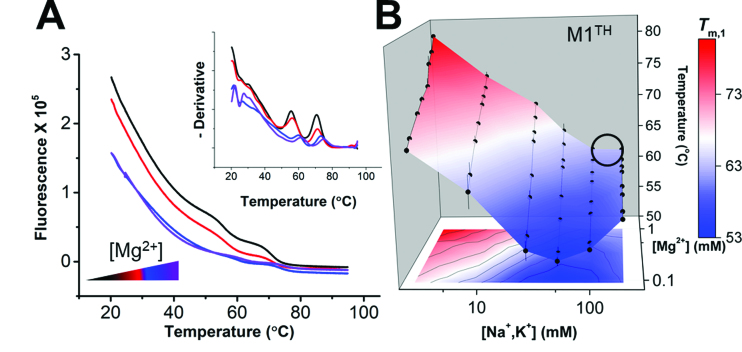
To assess these possibilities, we aimed to generate a stability landscape for the full-length M1TH. Preparation of a dual-labeled 94-nt RNA at the scale required for our thorough FRET landscape analysis is limited by yield, time, and cost. Therefore, we turned to an orthogonal fluorescence-based thermal shift assay. Differential scanning fluorimetry (DSF) is a label-free approach to monitor RNA stability at much higher throughput than traditional UV thermal melts (39). RiboGreen (ThermoFisher) fluorescence increases significantly upon binding to single-stranded RNA (ssRNA), but not double-stranded RNA. A monotonic decrease in RiboGreen fluorescence over the temperature range results from dye dissociation from the RNA (39) or changes in dye fluorescence quantum yield with increased temperature. RNA melting transitions, which liberate ssRNA, result in increased fluorescence (or, at least, not monotonically decreased) over the temperature range of the transition (Figure 3A). Similar to UV melts, and contrasting with DS-FRET melts, DSF monitors global changes in RNA structure. Tm,1 values are determined from a negative derivative plot of the raw fluorescence (Figure 3A, inset), and are identical to Tm values determined from traditional UV melts (Supplementary Table S3). Across the same 64 multiplexed solution conditions, the DSF Tm,1 stability landscape (Figure 3B) reproduces the chevron-shaped responses observed by DS-FRET (Figure 2C and D). The stability of the full-length M1TH is very high overall; the lowest Tm,1 is 53°C. Again, in near-physiological conditions (Figure 3B, black circle), M1TH is only metastable though its Tm,1 (62.7°C) is still much higher than physiological temperature. Surprisingly, this full-length, highly structured MALAT1 ENE triplex has a very low ‘relative stability’ near physiological conditions (24%) (Table 2). A limited UV melt analysis under similar conditions yielded similar results (Supplementary Figure S4, Supplementary Table S4). Overall, the DSF landscape reveals Tm,1 values much higher than physiological temperatures, suggesting that the triplex should be stably formed in vivo to protect the entire ∼7-kb lncRNA from degradation. However, a recent report concluded that in vitro stability is necessary but not sufficient to explain functional protection by the MALAT1 ENE triple helix (35).
Figure 3.
Stability landscape of M1TH determined by differential scanning fluorimetry. (A) Melting profiles of M1TH monitored by RiboGreen fluorescence using DSF under four different magnesium concentrations (0.1, 0.2, 0.3 and 0.6 mM MgCl2, with magnesium concentrations increased from black to purple) in 51 mM total monovalent salt (equimolar NaCl and KCl). Inset: The negative derivative of the raw melting profile is used to determine the melting temperature transitions. Two melting transitions are recorded by DSF, with Tm values consistent with UV melt experiments (Table S2). (B) Tertiary thermal stability landscape of M1TH monitored by DSF under multiplexed ionic conditions (2.6, 5.7, 8.8, 15.1, 27.6, 52.6, 102.6 and 202.6 mM total monovalent salt; and 0.1, 0.2, 0.3, 0.5, 0.6, 0.8 and 1 mM MgCl2). n = 3 experiments; S.D. Thermostability of the unimolecular M1TH is considerably higher than physiological temperatures. A circle on the stability landscape indicates approximate physiological ionic conditions (∼150 mM monovalent, 1 mM MgCl2).
Dynamic conformations of the MALAT1 ENE triple helix at room temperature
Functional regulation of RNA-controlled biological processes is often coupled to inherent RNA structural dynamics and conformational changes (40–47). At least two structural descriptions are consistent with the observed M1TH triplex stability changes across the landscape of ionic conditions. First, the RNA structure itself may be identical in each of the solution conditions but with differential stability depending on the ionic environment. Second, the RNA may adopt distinct conformations with correspondingly distinct Tm,1 values. For example, thermal melts performed in low magnesium concentrations with buffers of slightly different pH resulted in triplex stabilities less sensitive to the protonation state of the cytosine in the C+•G-C triple (Figure 1A), suggesting deformation of at least that base triple (Supplementary Figure S1B). Even so, neither mechanism can be directly addressed by thermal melt analyses alone. To determine the structural origin of the variations in triplex stability, we evaluated FRET at room temperature. If the RNA forms a similar triplex but with differential stabilities, an identical EFRET signal should be observed in all experimental conditions at room temperature. In contrast, distinct EFRET signals would indicate the presence of alternative or dynamically interchanging RNA conformations.
We monitored Cy3 and Cy5 fluorescence at room temperature using a microplate fluorimeter to generate EFRET conformational landscapes for M1ET and M1AB (Figure 4). Wide-ranging changes in EFRET are observed across each landscape. The absolute range of EFRET differs between M1ET (Figure 4A) and M1AB (Figure 4B) due to the distinct fluorophore positions in each construct (Supplementary Figure S5). Nonetheless, the chevron-shaped EFRET landscapes confirm that the ionic environment dictates significant structural changes in the RNA. Our ensemble FRET measurements are consistent with dynamic structural interconversions between at least two different conformations, a largely folded triplex and an unfolded triplex, which are populated to varying degrees throughout the EFRET landscape.
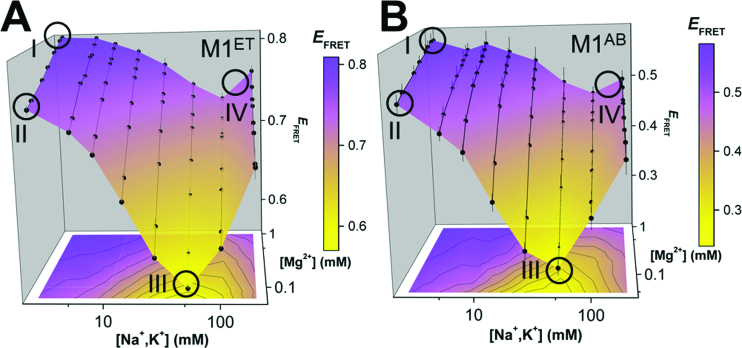
Figure 4.
Triplex conformational variability. Conformational landscapes of (A) M1ET and (B) M1AB monitored by FRET (EFRET = ICy5 / (ICy3 + ICy5) at room temperature in different ionic conditions (2.6, 5.7, 8.8, 15.1, 27.6, 52.6, 102.6 and 202.6 mM total monovalent salt (equimolar NaCl and KCl); and ∼0.05, 0.1, 0.2, 0.3, 0.5, 0.6, 0.8 and 1 mM MgCl2). n = 3 experiments; S.D. EFRET at Conditions I, II, III and IV are circled (Condition I: 2.6 mM total monovalent salt, 1 mM MgCl2; Condition II: 2.6 mM total monovalent salt, 0.1 mM MgCl2; Condition III: 52.6 mM total monovalent salt, 0.1 mM MgCl2; Condition IV: ∼150 mM total monovalent salt, 1 mM MgCl2). Condition IV represents near-physiological ionic conditions. Maximal EFRET is observed in high MgCl2 concentration and low monovalent concentration (Condition I, top left of each panel). Minimal EFRET is recorded in low MgCl2 concentration and medium monovalent concentration (Condition III, bottom middle of each panel).
To investigate triplex conformational variability further, we examine the EFRET changes at four distinct conditions on the landscape (Figure 4: (I) 2.6 mM monovalent, 1 mM MgCl2; (II) 2.6 mM monovalent, 0.1 mM MgCl2; (III) 52.6 mM monovalent, 0.1 mM MgCl2; (IV) ∼150 mM monovalent, 1 mM MgCl2 (near-physiological conditions). For M1ET, the highest EFRET signal (0.81, Condition I) represents the most folded triplex, placing the 5′-Cy5-labeled A-rich tail in close proximity to the 3′-Cy3-labeled ENE P1 helix (Supplementary Figure S5). EFRET decreases (∼0.7, Condition II) commensurate with reductions in MgCl2 concentration indicate either the unwinding of the A-rich tail from the 5′ end of the triplex or the unfolding of the P1 helix (Figure 4A). The minimum EFRET (∼0.57, Condition III) represents a distinct, largely unfolded triplex, wherein the lower portion of the triplex together with P1 are unfolded. In near-physiological conditions (Condition IV), the RNA is overall more folded (EFRET ∼0.76) but does not achieve a maximally folded population. Similar EFRET signal between Condition II and Condition IV may indicate that these distinct ionic conditions support a similar fraction of folded triplex or that the RNA structure is in fact distinct in these two conditions (Supplementary Figure S5).
M1AB exhibits similar relative EFRET changes under each of the four conditions (Figure 4B). However, the interpretation of the conformational changes are vastly different. The fluorophore pair in M1AB is positioned at the 3′ ends of M1A (Cy3) and M1B (Cy5). In this construct, FRET reports on structural changes near the apex of the triplex. For example, the minimum EFRET (∼0.23, Condition III) indicates unwinding of 3′ end of the A-rich tail. Therefore, owing to the distinct dye positions in M1ET and M1AB, the EFRET changes suggest unfolding at both the apex and nadir of the triplex in Condition III. However, because Cy5 fluorescence signal, which depends on proximity to Cy3 for energy transfer, is recorded under all conditions for both constructs, our results demonstrate that the two RNA strands do not completely unwind or dissociate. Similar to the stability landscape analyses, the conformational landscapes indicate that only ∼75% of the population contains a maximally formed triplex near physiological conditions (Table 2). Therefore, the MALAT1 triplex is thermodynamically stable but samples both the folded and partially unfolded RNA conformations in this condition. These conformational changes may affect the protective function of this triplex and suggest that MALAT1 abundance in vivo may be regulated in part by RNA structural dynamics near the 3′ end.
Full-length M1TH is susceptible to 3′-5′ degradation
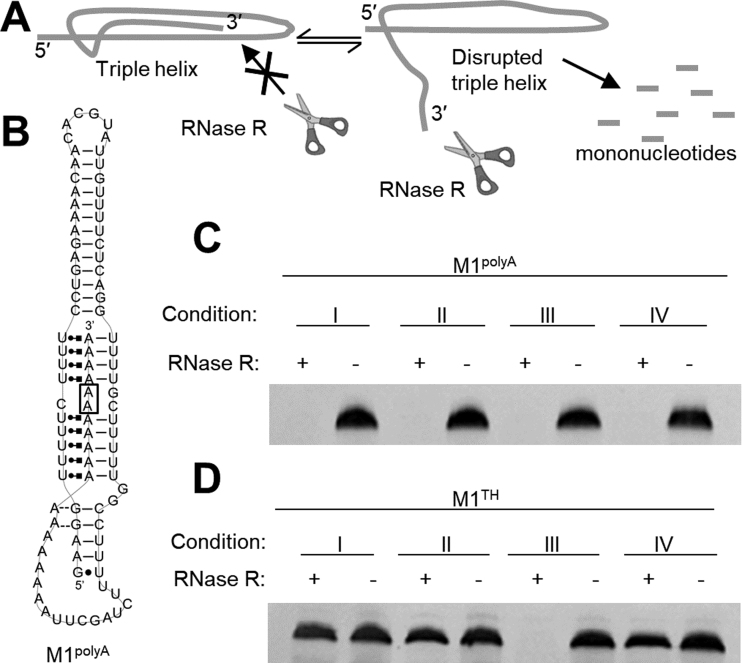
To assess the functional relevance of triplex conformational sampling and dynamics we developed an RNase degradation assay using RNase R, a processive and highly active 3′-5′ exonuclease (Figure 5A). We first evaluated RNase activity in various ionic conditions to ensure robust activity. Using denaturing gel electrophoresis we monitored degradation of an unstructured 94-nt ssRNA in Conditions I-IV at 37°C. The complete degradation of the control ssRNA in each condition confirms high activity and processivity of RNase R across our matrix of ionic conditions (Supplementary Figure S6A).
Figure 5.
3′-5′ exonucleolytic degradation of the MALAT1 ENE triplex. (A) Schematic of exonucleolytic triplex degradation by RNase R. Sequestration of the 3′ end precludes degradation by 3′-5′ exonuclease RNase R while a highly dynamic, disrupted 3′ end leads to degradation of the entire RNA. (B) Secondary structure of M1polyA, which includes two mutations, G88A and C89A (boxed) in middle of the triple helix. In (C) and (D) 0.5 μg RNA was incubated in 20 mM HEPES, pH 7.4 in ionic Conditions I-IV with 5 units of RNase R for 5 hours prior to analysis using 6% denaturing PAGE stained with ethidium bromide. (Condition I: 2.6 mM total monovalent salt, 1 mM MgCl2; Condition II: 2.6 mM total monovalent salt, 0.1 mM MgCl2; Condition III: 52.6 mM total monovalent salt, 0.1 mM MgCl2; Condition IV: 152.6 mM total monovalent salt, 1 mM MgCl2). (C) A representative denaturing PAGE image of the M1polyA mutant reveals that the RNA is completely degraded under all four conditions tested. (D) A representative denaturing PAGE image demonstrates that M1TH is not appreciably degraded in Conditions I, II, and IV but is completely degraded in Condition III (for full gel image, see Supplementary Figure S7). Additional replicates of experiments in C and D containing 1 U RNase R are shown in Supplementary Figure S6F and G.
To evaluate the functional protection against 3′-5′ degradation afforded by triplexes, we tested the wild-type sequence (M1TH) and two mutants, which disrupt or eliminate the triplex interaction. A truncated sequence (M1trunc) lacking the A-rich tail (Supplementary Figure S6B), is incapable of forming the triplex and is degraded by RNase R in all conditions tested (Supplementary Figure S6C). A double mutant (M1polyA) disrupts the C+•G-C triplet and C-G base pair in the middle of the triple helix (Figure 5B), which leads to complete degradation of a triplex-containing reporter construct in vivo (20). UV melt analysis reveals that the M1polyA triplex is significantly destabilized relative to M1TH (Tm,1 = 49.2°C, near Condition IV; Supplementary Figure S6D), but should remain stably formed at physiological temperature. Nonetheless, the double mutant is completely degraded by RNase R in all four conditions (Figure 5C, Supplementary Figure S6F), consistent with loss of accumulation in vivo (20). Hence, although the M1polyA triplex is thermodynamically stable, the mutation must sufficiently increase RNA dynamics near the 3′ end to promote degradation.
In contrast, the full-length M1TH affords complete protection from RNase R-mediated degradation in Conditions I, II and IV (Figure 5D). In Condition III, however, the RNA is completely degraded (Figure 5D). Even with reduced amounts of RNase R, M1TH is nearly completely degraded in Condition III over a 5-hour time course (Supplementary Figure S6E and G). Strikingly, the Tm,1 recorded by UV melt and DSF in Condition III is nearly 55°C. Thus, thermodynamic stability alone is insufficient to describe the mechanism of protection by M1TH. Dynamic conformational fluctuations and loss of structure near the 3′ terminus of the triplex are responsible for the increased susceptibility to exonucleolytic degradation recorded under this condition. In Condition IV, though the triplex is not maximally formed (Table 2 and Figure 4A and B) the level of structural dynamics is sufficiently tuned to confer protection against degradation.
DISCUSSION
Mechanism of triplex formation
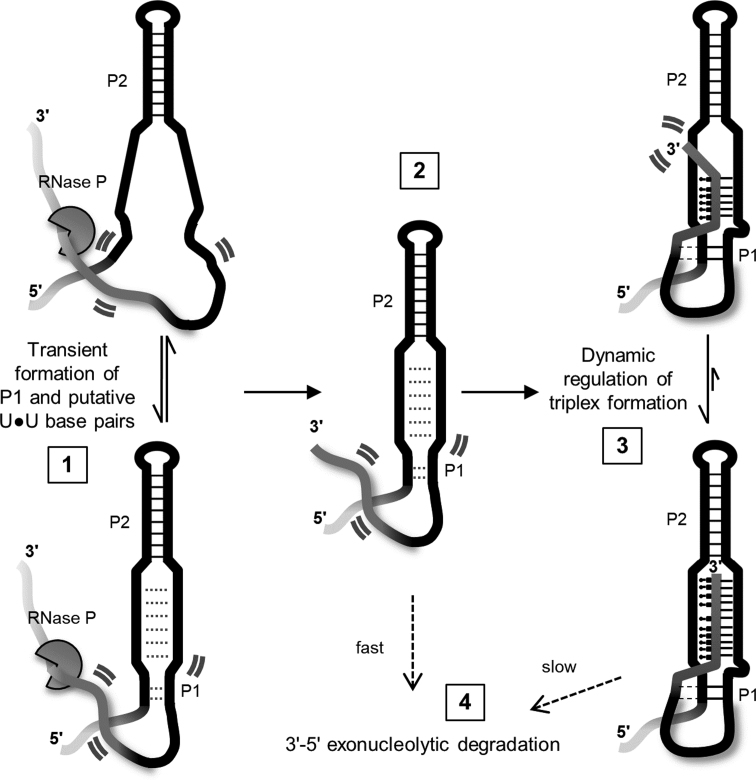
Despite lacking a canonical polyA tail, the human lncRNA MALAT1 achieves high levels of abundance and persistent oncogenic activity in several cancer types (26,28–30). Following maturation by RNase P, a triple helix formed between the ENE motif and mature A-rich 3′ tail is functionally responsible for protecting the lncRNA against 3′-5′ exonucleolytic degradation. Proper 3′ end maturation and subsequent triplex formation is essential to maintain high cellular abundance (20). Therefore, inhibition of triplex formation represents a potentially attractive mode of therapeutic intervention. We propose a mechanism for MALAT1 ENE triplex formation consistent with our conformational and stability data (Figure 6). (i) Prior to RNase P cleavage of the nascent 3′ terminus, the P2 apical helix of the ENE motif is stably formed. At physiological temperatures, the U-rich regions may form transient interactions including U•U base pairs (Supplementary Figure S3B; N.J.B. unpublished results, manuscript in preparation) and the relatively weak P1 helix is only partially formed (Supplementary Figure S3B). The triplex is not yet formed due to sequestration of the 3′ end within the RNase P active site. (ii) Liberation of the 3′ end following RNase P cleavage initiates a structural rearrangement of the A-rich tail. (iii) Triplex formation leads to a dynamic, rod-like duplex-triplex-duplex stacked structure. Triplex-mediated protection from degradation is dynamically controlled (35). (iv) Degradation is slow when the triplex is sufficiently formed and fast when the 3′ end is not sequestered within the triplex.
Figure 6.
Maturation, folding, and degradation of the 3′ terminus of lncRNA MALAT1. The schematic depicts the ENE triplex region; faded lines indicate 5′ and 3′ ends not to scale. (1) The P2 helix is stably formed near a 3′ terminal tRNA-like domain in the nascent transcript (not shown). Transient, weak formation of putative U•U base pairs and a short P1 helix in the ENE motif occurs prior to triplex formation (dotted lines). Highly dynamic regions of the RNA structure are denoted by two curved lines. (2) Cleavage by RNase P produces the mature 3′ end containing an A-rich tail (dark gray). (3) In a concerted step, the A-rich tail forms triplex interactions with the U-rich regions and A-minor interactions (dashed lines) with the P1 helix. The continuous duplex-triplex-duplex stacking interactions (P1–triplex–P2) significantly restrict 3′ end dynamics. (4) 3′-5′ exonucleolytic degradation is slow when the triplex is well-formed. However, structural disruption or increased triplex dynamics lead to rapid degradation of the RNA.
Our data are consistent with the concerted formation of the triplex with both U-rich strands, rather than a sequential association with each U-rich strand. Specifically, the simple AU-rich duplex formed between the A-rich tail and one of the U-rich regions (M1BsT; Figure 1C, orange and blue strands) is not stable at physiological temperatures, underscoring the need for additional structure formation concomitant with this interaction to promote a stable interaction. Indeed, the three-stranded triplex (M1ABsT) significantly stabilizes this region and supports co-incident formation of a three-stranded structure (Table 1). A concerted mechanism would require close proximity of the two U-rich regions in the ENE motif prior to release of the A-rich tail upon cleavage by RNase P. This could be achieved by formation of putative, semi-stable U•U base pairs as suggested by the melting profile of the ENE motif (Supplementary Figure S3B) and ongoing structural simulations in our lab (unpublished results, manuscript in preparation). Herein lies a novel approach for therapeutic intervention targeting M1TH: preclusion of triple helix formation by inhibitors that bind U•U pairs and prevent association of the A-rich tail. Indeed, successful small molecule inhibitors have been semi-rationally developed against disease-related RNAs harboring putative U•U pairs (48–51).
Maximal triplex formation and stability are not required for its protective function
Stable formation of the MALAT1 ENE triple helix is achieved across a broad range of multiplexed monovalent and MgCl2 solution conditions (Figures 2 and 3). Previous work examining DNA triplexes concluded that magnesium stabilizes and potassium destabilizes triplex formation above millimolar concentrations (16). Facilitated by higher throughput techniques, our empirical fluorescence and FRET thermal stability landscapes reveal a more nuanced cation competition affecting RNA triplex stability. The triplex melting (Tm,1) landscape exhibits a chevron-like dependence in response to increasing monovalent salts concentrations at all Mg2+ concentrations. Magnesium displaces monovalent cations and confers increased triplex stability (Figures 2 and 3). Thus, similar to several other structured RNAs, monovalent-divalent competition (52,53) impacts the structural interactions within the triplex. Within the range of ionic conditions tested the MALAT1 ENE triplex is minimally stable in Condition III (52.6 mM monovalent, 0.1 mM MgCl2). Interestingly, in near-physiological ionic conditions (Condition IV: ∼150 mM monovalent, 1 mM MgCl2), the MALAT1 ENE triplex is not maximally stable. Maximal triplex stability is only achieved in 1 mM MgCl2 and minimal monovalent salt (2.6 mM). Using our empirical landscapes, we define a ‘relative stability’ range and determine that only 24% of the maximal M1TH stability is achieved in near-physiological ionic conditions (Table 2). The observed metastability of M1TH in near-physiological conditions is consistent with previous studies of DNA triplexes (15,16). Even so, all in vitro measured stabilities are much higher than physiological temperatures, indicating that the triplex is thermostable in vivo, regardless of the specific ionic environment. Differences in stability between our unimolecular and bimolecular constructs suggest a role for peripheral elements in regulating the triplex stability and function. Indeed, Brown et al. reported a ∼15% decrease in RNA levels in vivo upon truncation of the P2 hairpin, forming a unimolecular version of our M1AB construct (20).
Because conformational dynamics regulate many RNA functions throughout biology, we reasoned that structural fluctuations may likewise regulate M1TH-mediated protection against exonucleolytic degradation. Triplex interactions monitored by FRET at room temperature reveal structural changes at both ends of this long triplex region (Figure 4). Under near physiological ionic conditions we determine that the triplex population is approximately 75% well-formed at room temperature (Table 2). Taken together with our thermal stability analyses, the MALAT1 ENE triplex is neither maximally formed nor maximally stable in near-physiological ionic conditions. Furthermore, at physiological temperatures, RNA dynamics are expected to increase and lead to an overall lower percent of well-formed triplex. Even so, the full-length M1TH sufficiently protects against 3′-5′ exonucleolytic degradation by RNase R at 37°C (Figure 5). This protective function is effectively inhibited by either significantly higher structural dynamics (M1polyA, Figure 5) or disruption of the triplex induced by changes in the ionic environment (M1TH, Figure 5).
Despite its relatively simple structure, we show here that the MALAT1 ENE triplex exhibits complex conformational variations which are exquisitely controlled to achieve protection against degradation. Structural elements outside the triple helix itself significantly tune M1TH stability and conformational dynamics in near-physiological conditions. Our stability and conformational landscape analyses across a vast array of ionic environments affords quantitative descriptions of these properties and their mechanistic roles in protecting M1TH from exonucleolytic degradation by RNase R. In fact, the overall rod-like duplex–triplex–duplex stacked structure (Figure 1) may control triplex function as much as the specific base triple interactions. A triplex alone is not likely sufficient to achieve protection. Indeed, mutations eliminating even just the Watson–Crick base pairs immediately above or below the triplex lead to >70% degradation of a reporter mRNA (20). Our results may provide a functional explanation for the semi-conserved helical elements directly above and below the homologous ENE triplexes from MALAT1, PAN and MENβ (21,22). More recent bioinformatic searches have identified >200 other ENE motifs in transposable elements and viral genomes with similar patterns of conservation outside the triplex region (25). Northern blot analyses of a β-globin reporter mRNA confirmed the protective role of these ENE triplexes. Several tandem helix-ENE-helix-ENE-helix structures were identified. In light of our results, we suggest that this tandem arrangement may further enhance protection through formation of a large continuously stacked structure incorporating polyA sequences and peripheral helices.
Within the cellular context, additional processes such as protein binding or chemical modifications may be important for regulating the dynamic properties and protective function of the MALAT1 ENE triplex. Though no modifications have been reported within the triplex region of M1TH, a small N6-methyladenosine modification of unknown function was recently reported on A8290 (involved in the terminal base pair at the apex of P2) (54). Methyltransferase-like protein 16 (METTL16), a confirmed methyltransferase (55,56), was recently shown to bind specifically to M1TH, though its contribution to the mechanism of triplex-mediated protection was not evaluated (34). While it is intriguing to consider potential chemical modification by METTL16 within the triplex, it remains unknown whether MALAT1 is a substrate for METTL16. Nonetheless, our results would be consistent with a METTL16 binding event serving to regulate triplex dynamics and further obscure the 3′ end from exonucleolytic degradation.
Taken together with previous studies of DNA triplexes, we conclude that metastability and dynamic structural fluctuations are general properties of nucleic acid triple helices. Within biological systems the most stable triplexes will include additional structural elements surrounding the triplex to facilitate an extended rod-like structure akin to M1TH. In lieu of such an extended structural network, triplexes may require small molecule or protein binding partners to overcome inherently weak and flexible interactions (34,41). In this way, triplexes may generally function as specific structural beacons guiding the assembly of larger functional complexes involving other RNAs, proteins, or small molecules to regulate biological processes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stuart LeGrice, Michael Yonkunas, and Michael Bruist for helpful discussions. We thank Sevde Yilmaz and Klaudio Mehmeti for assistance with RNA purification and UV melt experimentation, and Hoa Nguyen for assistance with RNA purification and DSF experiments. We thank the reviewers for insightful comments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of the Sciences (to N.J.B.); National Institutes of Health [K22-HL121113A to N.J.B.]; Saudi Arabian Cultural Mission and Jazan University to A.A.A. Funding for open access charge: National Institutes of Health [K22-HL121113A].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bacolla A., Wang G., Vasquez K.M.. New perspectives on DNA and RNA triplexes as effectors of biological activity. PLos Genet. 2015; 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Conrad N.K. The emerging role of triple helices in RNA biology. WIREs RNA. 2014; 5:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilbert S.D., Rambo R.P., Van Tyne D., Batey R.T.. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat. Struct. Mol. Biol. 2008; 15:177–182. [DOI] [PubMed] [Google Scholar]
- 4. Huang L., Wang J., Wilson T.J., Lilley D.M.J.. Structure of the guanidine III riboswitch. Cell Chem. Biol. 2017; 24:1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Y., Syed J., Sugiyama H.. RNA–DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 2016; 23:1325–1333. [DOI] [PubMed] [Google Scholar]
- 6. Smith K.D., Shanahan C.A., Moore E.L., Simon A.C., Strobel S.A.. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7757–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eichhorn C.D., Kang M., Feigon J.. Structure and function of preQ1 riboswitches. Biochim. Biophys. Acta. 2014; 1839:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liberman J.A., Suddala K.C., Aytenfisu A., Chan D., Belashov I.A., Salim M., Mathews D.H., Spitale R.C., Walter N.G., Wedekind J.E.. Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3485–E3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fica S.M., Mefford M.A., Piccirilli J.A., Staley J.P.. Evidence for a group II intron–like catalytic triplex in the spliceosome. Nat. Struct. Mol. Biol. 2014; 21:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cash D.D., Cohen-Zontag O., Kim N.-K., Shefer K., Brown Y., Ulyanov N.B., Tzfati Y., Feigon J.. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:10970–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shefer K., Brown Y., Gorkovoy V., Nussbaum T., Ulyanov N.B., Tzfati Y.. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 2007; 27:2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen G., Chang K.-Y., Chou M.-Y., Bustamante C., Tinoco I.. Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12706–12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamada M., Watanabe Y., Gootenberg J.S., Hirano H., Ran F.A., Nakane T., Ishitani R., Zhang F., Nishimasu H., Nureki O.. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol. Cell. 2017; 65:1109–1121. [DOI] [PubMed] [Google Scholar]
- 14. Plum E.G. Thermodynamics of oligonucleotide triple helices. Biopolymers. 1997; 44:241–256. [Google Scholar]
- 15. Plum G.E., Breslauer K.J.. Thermodynamics of an intramolecular DNA triple helix: a calorimetric and spectroscopic study of the pH and salt dependence of thermally induced structural transitions. J. Mol. Biol. 1995; 248:679–695. [DOI] [PubMed] [Google Scholar]
- 16. Singleton S.F., Dervan P.B.. Equilibrium association constants for oligonucleotide-directed triple helix formation at single DNA sites: linkage to cation valence and concentration. Biochemistry. 1993; 32:13171–13179. [DOI] [PubMed] [Google Scholar]
- 17. Floris R., Scaggiante B., Manzini G., Quadrifoglio F., Xodo L.E.. Effect of cations on purine.purine.pyrimidine triple helix formation in mixed-valence salt solutions. Eur. J. Biochem. 1999; 260:801–809. [DOI] [PubMed] [Google Scholar]
- 18. Li H., Broughton-Head V.J., Peng G., Powers V.E., Ovens M.J., Fox K.R., Brown T.. Triplex staples: DNA double-strand cross-linking at internal and terminal sites using psoralen-containing triplex-forming oligonucleotides. Bioconjug. Chem. 2006; 17:1561–1567. [DOI] [PubMed] [Google Scholar]
- 19. Rusling D.A., Peng G., Srinivasan N., Fox K.R., Brown T.. DNA triplex formation with 5-dimethylaminopropargyl deoxyuridine. Nucleic Acids Res. 2009; 37:1288–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown J.A., Valenstein M.L., Yario T.A., Tycowski K.T., Steitz J.A.. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:19202–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tycowski K.T., Shu M.-D., Borah S., Shi M., Steitz J.A.. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012; 2:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.-D., Joshua-Tor L., Sharp P.A.. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012; 26:2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conrad N.K., Steitz J.A.. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005; 24:1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Conrad N.K., Mili S., Marshall E.L., Shu M.D., Steitz J.A.. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell. 2006; 24:943–953. [DOI] [PubMed] [Google Scholar]
- 25. Tycowski K.T., Shu M.D., Steitz J.A.. Myriad triple-helix-forming structures in the transposable element RNAs of plants and fungi. Cell Rep. 2016; 15:1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. et al. . The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amodio N., Stamato M.A., Juli G., Morelli E., Fulciniti M., Manzoni M., Taiana E., Agnelli L., Cantafio M.E.G., Romeo E. et al. . Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018; 32:1948–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arun G., Diermeier S., Akerman M., Chang K.-C.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L. et al. . Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016; 30:34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 30. Chakravarty D., Sboner A., Nair S.S., Giannopoulou E., Li R., Hennig S., Mosquera J.M., Pauwels J., Park K., Kossai M. et al. . The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014; 5:5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilusz J.E., Freier S.M., Spector D.L.. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008; 135:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kao C., Zheng M., Rüdisser S.. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999; 5:1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown J.A., Bulkley D., Wang J., Valenstein M.L., Yario T.A., Steitz T.A., Steitz J.A.. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014; 21:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A.. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:14013–14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A.. Hoogsteen-position pyrimidines promote the stability and function of the MALAT1 RNA triple helix. RNA. 2016; 22:743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuznetsov S.V., Ren C., Woodson S.A., Ansari A.. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008; 36:1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baird N.J., Inglese J., Ferré-D’Amaré A.R.. Rapid RNA-ligand interaction analysis through high-information content conformational and stability landscapes. Nat. Comm. 2015; 6:8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Darby R.A., Sollogoub M., McKeen C., Brown L., Risitano A., Brown N., Barton C., Brown T., Fox K.R.. High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res. 2002; 30:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silvers R., Keller H., Schwalbe H., Hengesbach M.. Differential scanning fluorimetry for monitoring RNA stability. ChemBioChem. 2015; 16:1109–1114. [DOI] [PubMed] [Google Scholar]
- 40. Wood S., Ferré-D’Amaré A.R., Rueda D.. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem. Biol. 2012; 7:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haller A., Rieder U., Aigner M., Blanchard S.C., Micura R.. Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 2011; 7:393–400. [DOI] [PubMed] [Google Scholar]
- 42. Panja S., Hua B., Zegarra D., Ha T., Woodson S.A.. Metals induce transient folding and activation of the twister ribozyme. Nat. Chem. Biol. 2017; 13:1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ali M., Lipfert J., Seifert S., Herschlag D., Doniach S.. The ligand-free state of the TPP riboswitch: a partially folded RNA structure. J. Mol. Biol. 2010; 396:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baird N.J., Ferré-D’Amaré A.R.. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010; 16:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baird N.J., Ferré-D’Amaré A.R.. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA. 2013; 19:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baird N.J., Kulshina N., Ferré-D’Amaré A.R.. Riboswitch function: flipping the switch or tuning the dimmer. RNA Biol. 2010; 7:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kulshina N., Baird N.J., Ferré-D’Amaré A.R.. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009; 16:1212–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen L., Lee J., Wong C.-H.H., Zimmerman S.C.. Small molecules that target the toxic RNA in myotonic dystrophy type 2. ChemMedChem. 2014; 9:2455–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nguyen L., Luu L.M., Peng S., Serrano J.F., Chan H.Y., Zimmerman S.C.. Rationally designed small molecules that target both the DNA and RNA causing myotonic dystrophy type 1. J. Am. Chem. Soc. 2015; 137:14180–14189. [DOI] [PubMed] [Google Scholar]
- 50. Rzuczek S.G., Colgan L.A., Nakai Y., Cameron M.D., Furling D., Yasuda R., Disney M.D.. Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat. Chem. Biol. 2017; 13:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Childs-Disney J.L., Hoskins J., Rzuczek S.G., Thornton C.A., Disney M.D.. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem. Biol. 2012; 7:856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lambert D., Leipply D., Shiman R., Draper D.E.. The influence of monovalent cation size on the stability of RNA tertiary structures. J. Mol. Biol. 2009; 390:791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shiman R., Draper D.E.. Stabilization of RNA tertiary structure by monovalent cations. J. Mol. Biol. 2000; 302:79–91. [DOI] [PubMed] [Google Scholar]
- 54. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017; 169:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Hobartner C., Sloan K.E., Bohnsack M.T.. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017; 18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.