Abstract
ASCE ATPases include ring-translocases such as cellular helicases and viral DNA packaging motors (terminases). These motors have conserved Walker A and B motifs that bind Mg2+-ATP and a catalytic carboxylate that activates water for hydrolysis. Here we demonstrate that Glu179 serves as the catalytic carboxylate in bacteriophage λ terminase and probe its mechanistic role. All changes of Glu179 are lethal: non-conservative changes abrogate ATP hydrolysis and DNA translocation, while the conservative E179D change attenuates ATP hydrolysis and alters single molecule translocation dynamics, consistent with a slowed chemical hydrolysis step. Molecular dynamics simulations of several homologous terminases suggest a novel mechanism, supported by experiments, wherein the conserved Walker A arginine ‘toggles’ between interacting with a glutamate residue in the ‘lid’ subdomain and the catalytic glutamate upon ATP binding; this switch helps mediate a transition from an ‘open’ state to a ‘closed’ state that tightly binds nucleotide and DNA, and also positions the catalytic glutamate next to the γ-phosphate to align the hydrolysis transition state. Concomitant reorientation of the lid subdomain may mediate mechanochemical coupling of ATP hydrolysis and DNA translocation. Given the strong conservation of these structural elements in terminase enzymes, this mechanism may be universal for viral packaging motors.
INTRODUCTION
Molecular motor proteins that convert the energy of nucleotide hydrolysis into mechanical work are ubiquitous in biology. These include motors involved in cell motility, cell division, electromotive force, protein degradation and cargo transport (1–5). The motors are fueled by ATP hydrolysis, and many of them including multi-subunit ring translocases and helicases are members of the ‘Additional Strand, Conserved E’ (ASCE) superfamily of P-loop ATPases (4,6–9). The ATPase domains contain conserved Walker A (WA) and Walker B (WB) motifs that participate in binding and positioning the γ-phosphate of ATP for hydrolysis (10–19). An additional feature of ASCE enzymes is that they have a conserved glutamate immediately downstream of the classical WB motif that is proposed to position and activate a water molecule for nucleophilic attack on the γ-phosphate in the hydrolysis transition state (4,20,21). This model is based on structural data obtained for various ASCE ATPases showing that the glutamate is positioned close to the γ-phosphate, and in many cases is supported by biochemical data showing that changes of this residue or nearby residues adversely affect the rate of ATP hydrolysis and/or motor function (20,22–25). Ostensibly, the catalytic role of the glutamate supported by the data presented here is conserved across ASCE ATPases that couple ATP hydrolysis to motor movement.
Terminase enzymes are molecular motors that package viral genomes into a pre-assembled procapsid shell, fueled by ATP hydrolysis (26–30). The DNA substrate is typically an end-to-end concatemer of viral chromosomes. A site-specific restriction endonuclease-like activity of terminase initiates packaging by cleaving the duplex at the packaging initiation sequence (pac or cos) in the concatemer. The terminase-DNA complex next assembles on the portal vertex of the procapsid and translocation of downstream DNA (the genome) into the shell ensues. Packaging is terminated by a second DNA cleavage event such that a single (mature) genome is packaged to a density approaching that of crystalline DNA. Thus, terminases perform two essential functions: (i) excision of an individual genome from the concatemer using an endonuclease activity (genome maturation), and (ii) packaging of the duplex into the shell using an ATP-fueled DNA translocation activity (genome packaging) (26,27,29–31). Terminases have been classified as ASCE ATPases and genome packaging shares functional similarities to the cellular ASCE motors described above. Indeed, the fundamental principles of energy transduction are likely conserved in all of the translocating biological motors, both viral and cellular (1,4,7,11,17,21,24,32).
Terminases are generally heterooligomers of a small genome-recognition subunit (TerS) and a large catalytic subunit (TerL) that contains the maturation and packaging activities of the enzyme (see Figure 1) (26,33). Herein we use the nomenclature TerLx to denote TerL subunits from the indicated phage ‘x’. For instance, TerLλ is the phage λ TerL subunit, TerLT4 is the phage T4 TerL subunit, etc. The TerL subunits are composed of a C-terminal endonuclease domain and an N-terminal domain that contains the packaging ATPase center (14). The catalytic activities of these two structural domains can be separated genetically and biochemically, which allows detailed mechanistic dissection of motor function (11,24,25,31,34–41). Early sequence alignment studies revealed that terminase ATPase domains contain sequences identified as the signature WA and WB motifs, and a putative catalytic carboxylate residue conserved in the ASCE superfamily (see Table 1) (11–13). Accompanying genetic and biochemical analysis led to identification of the WA and WB motifs in TerLT4 (11,24). Importantly, the biochemical studies showed that changing the glutamate adjacent to the classical WB motif abrogated in vitro DNA packaging, but did not alter endonuclease activity or abrogate binding of the ATP analog azido-ATP (24). Subsequent X-ray crystallography studies of TerLT4 supported the assignment of this residue as the catalytic glutamate due to its proximity to the ATP phosphates (14,15). Structural studies have further provided evidence for catalytic glutamates in TerL proteins from several other phages including Sf6, P74-26, D6E and for the packaging ATPase from ϕ29 (16–19). Genetic and/or biochemical studies support these structural assignments, showing that changing the identified residues abrogates ATP hydrolysis by the mutant proteins (17–19). Recently, it was also shown that changing hydrophobic residues near the TerLT4 glutamate residue slowed hydrolysis and translocation, consistent with perturbation of the ability of the glutamate to activate a lytic water molecule (25). However, the exact mechanism of action of the glutamate and the potential roles of other nearby residues that may interact with it are not fully known.
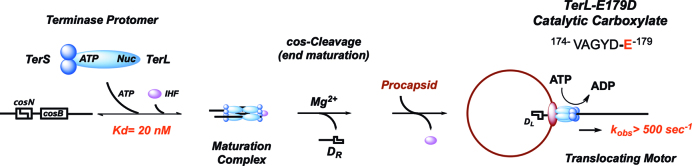
Figure 1.
The Lambda Genome Packaging Pathway. One TerL subunit (cyan) and two TerS subunits (blue) tightly associate in the terminase protomer. TerL maturation (Nuc) and packaging (ATP) domains are indicated. Pathway details are provided in the text. Putative Walker B motif (VAGYD) and the conserved glutamate (E179) are indicated.
Table 1.
Conserved Walker A (WA), Walker B (WB) and Lid Subdomain Sequences in Terminase TerL Subunits
| Phage | WA Motif | WB Motif | Lid |
|---|---|---|---|
| Lambda | KSA-R79-VGYS | VAGYD-E79 | 327W-D342-T364,a |
| T4 | LS-R162-QLGKT | MIYID-E256 | 314I-E351-E357 |
| P74-26 | LG-R39-QSGKS | FVILD-E150 | 221S-E247-V251 |
| Sf6 | GG-R24-GSGKS | ICWVE-E119 | 173N-E187-A210 |
Our laboratories have spent significant effort in elucidating the genetics, biochemistry and biophysics of phage λ terminase, a prototypical viral packaging enzyme (9,26,33,38–44). We recently confirmed the identities of WA motif residues in the TerLλ subunit and elucidated their mechanistic roles in ATP binding, catalysis of hydrolysis and mechanochemical coupling through analysis of a large collection of mutants (41). These studies provide experimental evidence that the λ motor protein undergoes a conformational change upon docking of ATP that is required for both tight binding of the nucleotide and tight DNA gripping (referred to as the tight binding transition, see Supplementary Figure S1); similar models have been proposed for the ϕ29, T4 and P74-26 packaging motors based on single-molecule or biochemical data (17,31,45,46). Here, we extend these studies and report our integrated genetic, biochemical, biophysical and computational studies to identify and characterize the role of the conserved glutamate 179 in TerLλ. Our results confirm that TerLλ-E179 is essential for virus development, ATP hydrolysis and genome packaging activities of terminase, commensurate with published studies in other viral systems. In addition, our studies provide several new mechanistic insights into how this residue is involved in catalyzing ATP hydrolysis and in the mechanochemical cycle. We also present molecular dynamics studies that provide evidence, supported by functional data on mutants, for a novel mechanism wherein the ASCE glutamate and a WA arginine conserved in the terminase enzymes act in concert to facilitate the ATP tight-binding transition. This ‘open’ to ‘closed’ transition positions the glutamate next to the γ-phosphate of ATP where it can activate a water molecule to catalyze ATP hydrolysis. After hydrolysis and product release, the arginine toggles to interact with a different glutamate in the lid subdomain, which has been implicated in mechanochemical coupling (17,41). Given the strong conservation of these residues amongst the terminase enzymes, these results may have broad biological implications for motor translocation by the viral genome packaging motors, both prokaryotic and eukaryotic.
MATERIALS AND METHODS
Experimental Methods
The genetic, biochemical and single molecule studies were performed as previously published and are outlined in Supporting Materials.
Simulation methodology
We performed all-atom molecular dynamics (MD) simulations to develop a better understanding of the dynamic reorganization of the ATP binding pocket and provide a framework for interpreting our experimental results, as follows. Because there is no crystal structure available for TerLλ, we used the crystal structures of three other large terminases: TerLT4, TerLSf6, and TerLP74-26 (14–17), which have homologous WA and WB motifs, and catalytic glutamate residue assignments supported by experimental findings. We simulated both the apo and ATP-bound states of the three terminases as well as specific mutants chosen to mimic those studied experimentally with phage λ terminase. We note that while terminases are known to function as oligomeric complexes, there is no definitive atomic-resolution oligomeric structure available for any TerL. Therefore, we simulated all structures as single subunits, based on the available crystal structures. As such the simulations, like the monomeric crystal structures from which they were derived, do not shed light on the potential role of trans-acting residues, such as trans-acting ‘arginine fingers’ essential for ATP hydrolysis in the P74-26, ϕ29 and D6E motor proteins (17–19,47) and in many oligomeric ASCE ATPases (4,32).
All MD simulations were performed in AMBER16 (48) using the AMBER ff14SB force field (49) with a 2 fs time-step integrator; details for each simulation are presented in Supporting Material. All bond lengths connecting hydrogens to other atoms were held rigid by using the SHAKE algorithm (50). A 10 Å cutoff was used for calculating non-bonded interactions and the particle mesh Ewald method (51) was used for computing long-range electrostatic interactions. ATP parameters were taken from the Amber Parameter Database (http://research.bmh.manchester.ac.uk/bryce/amber).
The proteins were centered in a truncated octahedral periodic box of TIP3P (52) water with a minimum padding of 14 Å. We neutralized all systems with appropriate amounts of Na+ or Cl− counterions, and then added additional ions to reach the physiological concentration of 150 mM NaCl. The systems were energy-minimized using 300 steps of steepest descent and conjugate gradient algorithms. The systems were heated slowly from 100 K to 310 K in a canonical ensemble (NVT) over 100 ps restraining the backbone of the proteins. This was followed by 15 ns of equilibration in the isothermal-isobaric (NPT) ensemble held at 1 bar using the Monte Carlo barostat (1.0 ps relaxation time), and at 310 K using the Langevin thermostat (2.0 ps−1 collision frequency) with no backbone restraints. Finally, 100-ns production runs were carried out in the NPT statistical ensemble at 310 K and 1 bar using the same coupling scheme as described above. Each TerL system was simulated in triplicate (three independent simulation runs, departing from each other at the heating stage) to ensure that the computed results were reliable and that all simulated structures were stable (Supplementary Figure S2).
RESULTS
Genetic analysis
We first used a genetic approach to investigate effects of changing the proposed catalytic glutamate, E179, in TerLλ. We mutagenized codon 179 to create a collection of mutants and the activity of each enzyme was quantified using an in vivo genetic complementation assay previously described (41). In brief, a plasmid complementation vector is used to supply TerLλ (WT or mutant) to a prophage whose production of TerLλ is blocked by amber mutations in the viral genome. If the supplied TerLλ is functional, phages are produced and plaques are counted by plating on an amber suppressing host. The phage yield (phages/cell) reflects the level of enzyme activity supplied by the mutant TerLλ and is normalized to the WT enzyme. The complementation assay is highly sensitive and can detect phage yields for mutants down to 107-fold lower than WT.
Eleven TerLλ mutants were tested in which E179 was changed to D, A, C, G, I, L, N, P, Q, R and V; this comprises a wide range of both chemically conservative and non-conservative changes. Strikingly, none support detectable phage production (Supplementary Table S2), including the ‘sterically conservative’ E179Q change that retains steric bulk and hydrogen bonding potential, but not the negative carboxylate functional group. Even the chemically conservative change E179D, which preserves negative charge and similar side-chain bulk, results in no measurable activity. Thus, even a minor perturbation of this conserved residue drastically affects virus yield, presumably due to loss of ATPase activity and motor function. These findings are consistent with the predicted crucial role of TerLλ-E179.
While the genetic studies clearly demonstrate that E179 is essential for virus viability, they do not define the mechanism for the defect. In addition to the predicted defects in ATP hydrolysis and translocation, impairments could be caused by defects in protein folding, motor assembly, impaired endonuclease activity and/or procapsid binding interactions (see Figure 1). Even if ATP hydrolysis and translocation were primarily affected, they could be affected for many possible reasons, including impaired ATP binding, catalysis of hydrolysis or mechanochemical coupling. Finally, a mutant terminase could be totally defective in the complementation assay, yet still have weak ATPase and DNA translocation activity that could shed light on the motor mechanism (25,38,39,53). Thus, to define the mechanistic basis for the in vivo virus assembly defects, we carried out additional biochemical and biophysical studies.
Biochemical analysis
Structural characterization
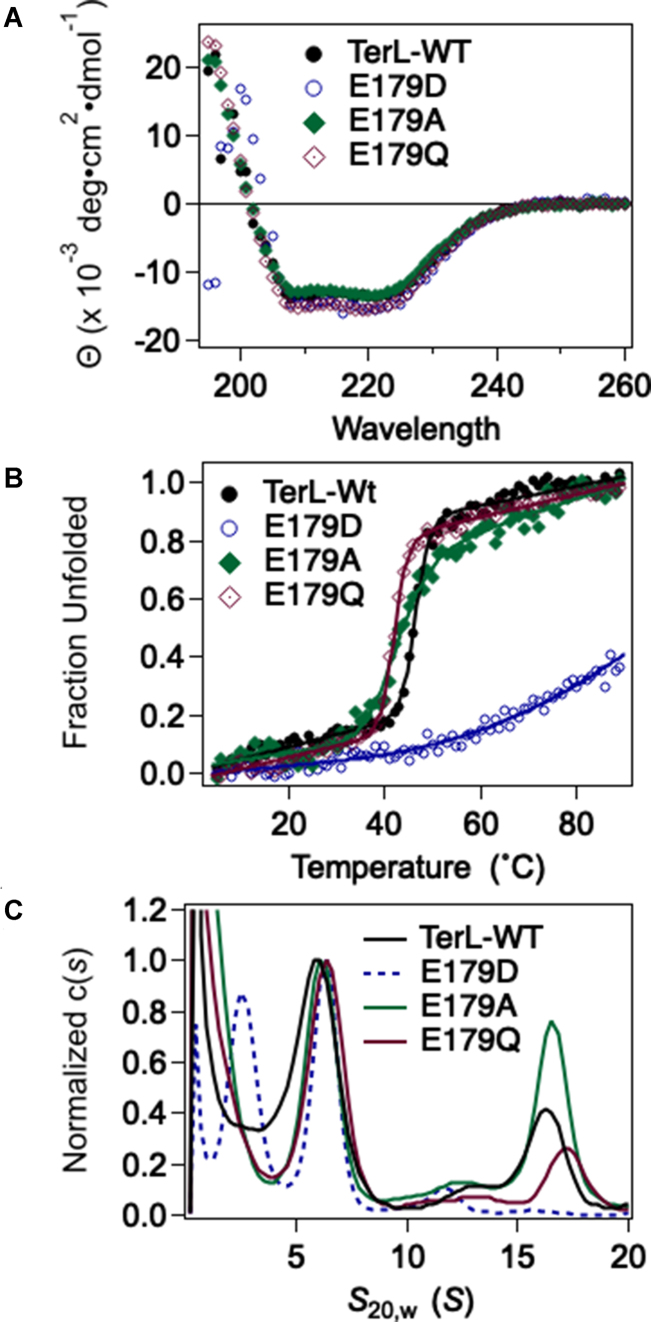
We chose three mutant enzymes for biochemical interrogation: the conservative E179D, the sterically conservative E179Q and the non-conservative E179A. The proteins purified in a similar fashion as the WT enzyme (not shown) indicating that protein structure is not grossly affected. Protein secondary structure was characterized by CD spectroscopy, which showed that all three mutants possess α-helical structures similar to the WT protein (Figure 2A). The E179A and E179Q terminase mutants exhibit highly cooperative unfolding transitions with thermal stabilities similar to WT, indicative of compact and well-folded structures (Figure 2B, Supplementary Table S3). In contrast, a broad, non-cooperative denaturation curve is observed with E179D suggesting that this change results in a loosely-folded structure and/or a heterogeneous population of structures.
Figure 2.
Structural characterization of the mutant terminase enzymes. (A) Far-UV CD spectra of wild type and mutant terminase enzymes indicate that they all possess similar secondary structures. (B) Thermal denaturation of WT and mutant terminases. Solid lines are the best fits of the data to obtain Tm presented in Supplementary Table S3. (C) Sedimentation Velocity Analytical Ultracentrifugation analysis. The WT enzyme assembles into a protomer species (6S) that is in slow equilibrium with a ring-like tetramer of protomers (16S). The E179A and E179Q enzymes show native-like assembly behavior but the E179D mutant displays significant defects. The novel 3.3 species is likely the isolated TerL subunit which indicates dissociation of the mutant terminase protomer. The isolated TerS subunit is not observed in the sedimentation experiment because it avidly aggregates (73) and it has pelleted from solution during the AUC run.
We next investigated whether the introduced changes affect quaternary interactions required for protomer and/or motor complex assembly using analytical ultracentrifugation. The analysis for WT terminase reveals two species, 6S and 16S, which represent the stable protomer (TerL•TerS2) and a tetramer of protomers, respectively (Figure 2C) (40,41,44). Neither the E179A nor E179Q changes significantly affect the solution distribution of protomer or tetramer species; this indicates that neither tertiary nor quaternary interactions of the enzymes have been affected by the changes. In contrast, the conservative E179D change has significant effects. First, the 16S species is not detected, indicating that the change strongly impairs protomer assembly into a functional tetrameric motor complex. Second, a novel 3.3S species is observed, which is not observed with the WT enzyme. This likely represents the TerLλ-E179D subunit free in solution, suggesting that the protomer is unstable and has dissociated into the composite subunits. In sum, the data indicate that while the E179A and E179Q changes little affect protein folding or motor assembly, the highly conservative E179D protein is severely compromised, most likely due to a folding defect. This likely accounts for much of the impairment in viral assembly activity observed genetically.
cos-Cleavage endonuclease activity
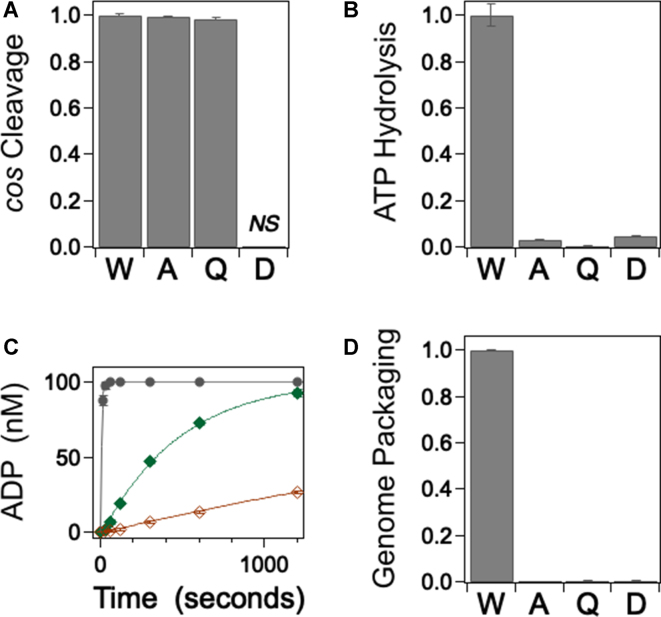
While the E179A and E179Q changes do not impart any major structural defects (Figure 2), they could feasibly introduce modest structural effects that result in a global loss of function and also affect the endonuclease activity centered in the C-terminal domain. To address this question, we examined the site-specific cos-cleavage endonuclease activity of the mutant enzymes, which should remain intact if the introduced change specifically affects only ATP hydrolysis (35). The cos-cleavage reaction entails site-specific nicking of both duplex strands within the cos sequence of λ DNA and separation of the annealed duplex with a so-called ‘helicase’ activity (54). Figure 3A shows that neither the E179A nor the E179Q change affects nuclease activity to any detectable extent; this indicates that the C-terminal endonuclease domain is folded and functional. In contrast, the conservative E179D mutant enzyme possesses a vigorous non-specific nuclease activity (data not shown) consistent with the observed folding defects. Notwithstanding this promiscuity, faint specific product bands are also observed with this mutant suggesting that a fraction of the protein retains site-specific cos-cleavage activity (not shown). This is discussed further below.
Figure 3.
Biochemical Characterization of the Mutant Terminase Enzymes. (A) cos-cleavage endonuclease activity was quantified as described in Methods. The TerLλ-E179D enzyme possesses a strong, non-specific nuclease activity that precludes accurate quantitation of specific cleavage at the cos site (not shown). Each bar represents the normalized average activity of at least three independent experiments with standard deviations indicated with error bars. (B) Steady state ATPase activity quantified as described in Methods. Each bar represents the normalized average activity of at least three independent experiments with standard deviation indicated. (C) Single turnover ATP hydrolysis quantified as described in Methods: black •, wild type terminase; green ♦, E179A mutant terminase; red ◊, E179Q mutant terminase. Each data point represents the average of at least three independent experiments with standard deviations indicated (in some cases the error bars are obscured by the data point). The solid lines represent the best fit of the data which affords the rate constants presented in Table 2. (D) DNA packaging activity was quantified as described in Methods. Each bar represents the normalized average activity of at least three independent experiments with standard deviations indicated with error bars.
ATPase activity
The residue changes were introduced into the putative catalytic carboxylate of TerLλ and were designed to specifically affect ATPase activity. As anticipated, steady-state ATP hydrolysis by the E179A and E179Q enzymes is strongly impaired (Figure 3B, Table 2). To further probe the reaction, we employed a single-turnover ATP hydrolysis assay in which the enzyme concentration is in excess of the ATP substrate. Under these conditions, single catalytic turnover is observed, which captures ATP binding, the tight-binding transition and ATP hydrolysis events, but not subsequent conformational change or product release steps. The data presented in Figure 3C and Table 2 demonstrate that the E179A and E179Q mutant enzymes possess severe defects in single-turnover hydrolysis that parallel their observed effects on steady-state ATPase activity; this indicates that ATP binding and/or hydrolysis steps are strongly affected by the changes. Importantly, there is no observable lag in the single-turnover kinetic time course as we observed with some WA mutants (41). In those cases, we attributed the lag to reflect a significantly slowed rate for the tight-binding transition (>75-fold). The absence of a lag with the E179A and E179Q enzymes suggests that the ATP tight-binding step is not significantly affected by the changes. Thus, the ensemble of data indicates that the primary defect in these mutants is neither protein folding, motor assembly nor DNA maturation events needed to initiate packaging. Rather, the defect resides in the chemical step, which indicates that E179 is directly involved in ATP hydrolysis. Finally, we note that the TerLλ-E179D mutant retains a weak steady state ATPase activity (Figure 3B) in addition to the weak endonuclease activity described above. Thus, despite the fact that the conservative E179D change results in a severe folding defect, a fraction of enzyme does nevertheless fold and assemble into a complex that is capable of maturing DNA and hydrolyzing ATP. This is discussed further below.
Table 2.
Kinetic analysis of wild type and mutant terminase enzymes. Steady state ATP hydrolysis was performed as described in Materials and Methods. 100% relative steady state activity corresponds to kobs = 2.7 ± 0.1 μM/min. Single turnover ATP hydrolysis was performed as described in Materials and Methods and the data presented in Figure 3C were fit to a single exponential time course to afford kobs; the best fit of each data set is indicated as a solid line in the Figure. 100% relative activity corresponds to kobs= (140 ± 4) x 10−3 s−1
| Enzyme | Steady state ATP hydrolysis (relative) | Single turnover ATP hydrolysis (relative) |
|---|---|---|
| Wild Type | 2.7 ± 0.1 μM/min (100%) | (140 ± 4) x 10−3 s−1 (100%) |
| E179A | 0.09 ± 0.01 μM/min (3.3%) | (1.9 ± 0.1) x 10−3 s−1 (1.4%) |
| E179Qa | 0.02 ± 0.06 μM/min (0.7%) | (0.25 ± 0.02) x 10−3 s−1 (0.2%) |
| E179D | 0.13 ± 0.2 μM/min (4.8%) | - |
aWe note that the ATPase activity observed with the most severely defective mutant enzyme (E179Q) represents an upper limit of activity and which could feasibly be associated with a contaminating cellular ATPase.
Genome packaging activity
The DNA packaging activity of terminase is powered by ATP hydrolysis and defects in ATPase activity are expected to concomitantly impact DNA translocation by the motor. To confirm this hypothesis, we next examined the genome packaging activity of the mutant enzymes. In this assay, the full-length λ genome (48.5 kb) is translocated into the procapsid shell and is rendered resistant to DNase digestion (55). The data presented in Figure 3D confirms that packaging activity is strongly impaired by all of the E179 residue changes examined, commensurate with their effect on ATP hydrolysis.
Single-molecule translocation measurements
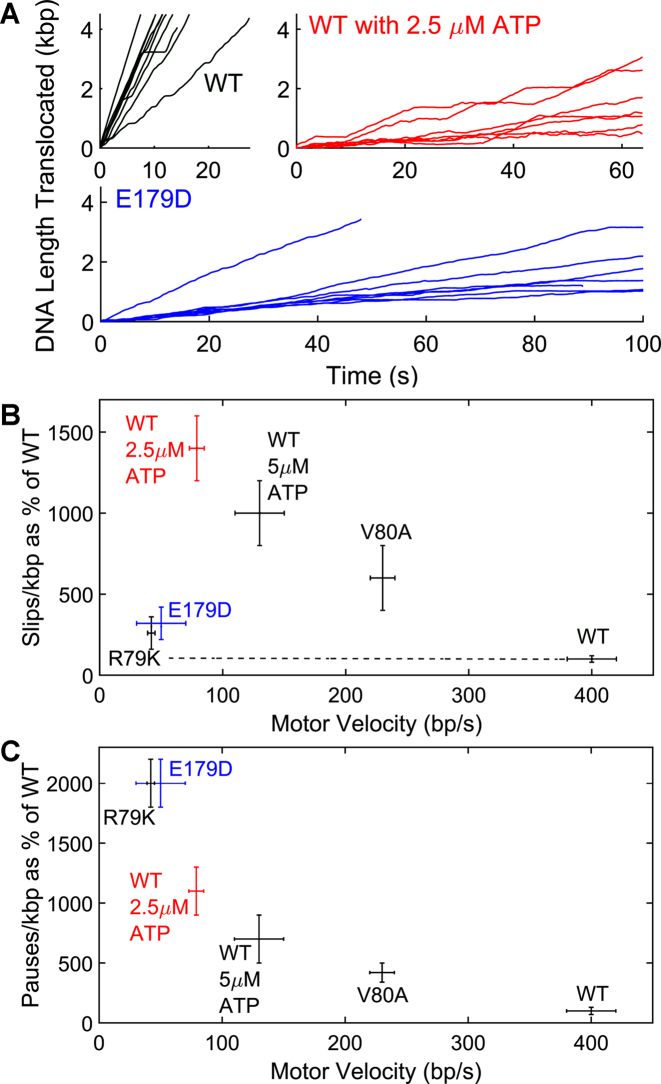
The biochemical studies demonstrate that while all of the E179 mutant enzymes examined possess weak, but detectable ATPase activity, there is no detectable genome packaging. The ensemble biochemical assay, however, probes full-genome-length packaging and we considered that the mutant motors might possess partial DNA translocation activity not detected by this stringent assay. Therefore, we used a single-molecule optical tweezers assay to directly probe DNA translocation activity in single procapsid-motor complexes (1,28,56–60). To give the mutant motors the best chance to exhibit activity, these measurements were made under the ‘least taxing’ conditions, with saturating ATP (500 μM), a low applied load force (5 pN) and low capsid filling (0–20% of the genome length) to minimize resistive forces (57,61–64). Consistent with the ensemble biochemical studies, we detected no translocation with the E179A and E179Q proteins despite several hundreds of trials (Table 3). Interestingly, however, DNA translocation events were detected with the conservative TerLλ-E179D mutant enzyme. The single molecule studies revealed that alterations in the motor function caused by the E179D change are clearly evident (Figure 4A). There is an overall ∼13-fold reduction in packaging rate which detailed analysis of the data shows is caused by an ∼8-fold reduction in average motor velocity (translocation rate not including pauses and slips), a ∼3-fold increase in average slipping frequency (Figure 4B), a ∼20-fold increase in average pausing frequency (Figure 4C) and a ∼70% increase in average pause duration (Table 3). Thus, while the genetic data clearly show that this change is lethal and biochemical data demonstrate that the mutant protein has structural defects, the single molecule data clearly show that a fraction of enzyme can assemble active complexes that are capable of translocating DNA, albeit with altered dynamics. The single molecule data directly implicate residue TerLλ-E179 as playing a direct role in governing the translocation kinetics. Further, the observed translocation phenotype, exhibiting a large decrease in motor velocity but without a proportionally large increase in slipping, provides evidence that the residue change primarily affects the ATP hydrolysis step, not the ATP binding step (41). To our knowledge, this is the first example of viral packaging motor mutant for which changing the conserved glutamate in an otherwise wild-type motor has been shown to result in complexes that retain DNA translocation activity but exhibit alterations in the translocation kinetics consistent with a slowed hydrolysis step. Analogous mutants could potentially be identified in the other viral motors and may be useful tools for biophysical studies.
Table 3.
Metrics of single DNA molecule packaging dynamics with wild type terminase (WT) and mutants determined by analysis of the optical tweezers measurements. Results for Walker A mutants V80A and R79K obtained previously (41) are shown for comparison. Except where indicated all measurements were recorded with 500 μM ATP. Quantities are reported as averages over all events and reported uncertainties are standard errors in the means
| Pauses | |||||
|---|---|---|---|---|---|
| Enzyme | Events | Motor velocity (bp/s) | Slip frequency (slips/kb) | Frequency (pauses/kb) | Duration (s) |
| WT | 53 | 400 ± 20 | 0.5 ± 0.1 | 1.2 ± 0.4 | 2 ± 0.1 |
| WT 5 μM ATP | 9 | 130 ± 20 | 5 ± 1 | 8 ± 2 | 1.6 ± 0.1 |
| WT 2.5 μM ATP | 13 | 79 ± 6 | 7 ± 1 | 13 ± 2 | 1.8 ± 0.1 |
| E179A | 0 | - | - | - | - |
| E179Q | 0 | - | - | - | - |
| E179D | 25 | 50 ± 20 | 1.6 ± 0.5 | 24 ± 2 | 3.3 ± 0.2 |
| R79K | 5 | 42 ± 3 | 1.3 ± 0.5 | 24 ± 2 | 3.4 ± 0.3 |
| V80A | 51 | 230 ± 10 | 3 ± 1 | 5 ± 1 | 4.4 ± 0.3 |
Figure 4.
Single DNA Molecule Translocation Measurements. (A) Examples of single DNA molecule translocation measurements for wild type (WT) terminase with saturating ATP (500 μM) (top left), wildtype terminase with 2.5 μM ATP (top right) and E179D mutant terminase with 500 μM ATP (bottom). (B) Slipping frequency vs. motor velocity for WT and mutant terminases. (C) Pausing frequency vs. motor velocity for WT and mutant terminases. The results for WA V80A and R79K mutants published previously (41) are shown for comparison. Unless otherwise indicated, measurements were with saturating ATP (500 μM).
Molecular dynamics simulations
The biochemical and single-molecule studies are consistent with each other and support that TerLλ-E179 is directly involved in ATP hydrolysis by the enzyme. To provide additional insight into the molecular mechanism of ATP hydrolysis by terminase enzymes, we carried out all-atom MD simulations. Unfortunately, there is no structure available for TerLλ; however, the predicted WA and WB motifs and the catalytic carboxylate residue in its core ATPase domain are strongly homologous with other TerL subunits whose structures are known (Table 1). Therefore, we used crystal structures of TerLT4 (14,15), TerLP74-26 (17) and TerLSf6 (16) to model the binding pocket geometry around the glutamate, its dynamic rearrangement in solution to accommodate ATP binding and the effects of introduced changes. The ASCE ATPase domains of T4 and P74-26 terminases have the canonical conserved aspartate in their WB motifs that is immediately followed by the identified catalytic glutamate (ΦΦΦΦD-E, where Φ represents a hydrophobic residue); this is also observed in TerLλ (Table 1). In contrast, TerLSf6 has a ‘deviant EE’ sequence (ΦΦΦΦE-E). Thus, we reasoned that simulations of TerLT4 and TerLP74-26 would be most useful in providing a structural/dynamic framework for interpreting the experimental results obtained herein with λ, and that TerLSf6 simulations might further provide a general mechanistic insight into catalysis by the ASCE ATPase motors.
Active site conformation in apo state
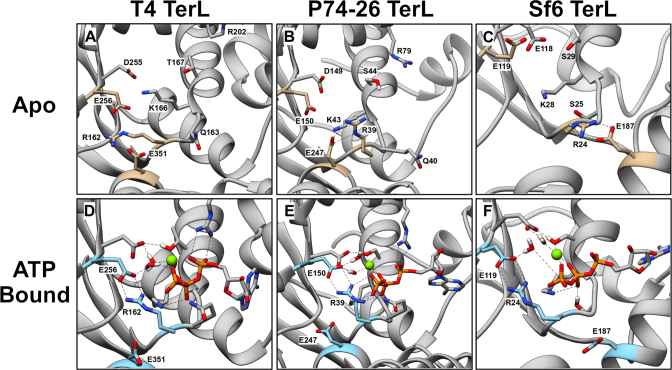
MD simulation of the apo structures shows that in all three TerL systems the identified catalytic glutamate points out of the ‘open’ ATP binding pocket (Figures 5A–C). It is also notable that in all three systems a conserved WA arginine also points out of the active site and typically interacts with a second glutamate residue located in what has been termed the ‘lid’ subdomain (Table 1) by analogy with other ASCE ATPases (17). Lid subdomains have been found to change conformation upon ATP hydrolysis and also to interact with adjacent subunits in multi-subunit complexes (21,65–67). Structural studies of the P74-26 and Sf6 terminases provide evidence that their lid subdomains undergo a rotation upon nucleotide binding which has been proposed to be critical for mechanochemical coupling and translocation (16,17). This is discussed further below.
Figure 5.
Conformations of the ATP-Binding Pocket in WT TerL obtained from MD Simulations. Representative conformations of the apo binding pocket of TerLT4 (A), TerLP74-26 (B) and TerLSf6 (C). Walker motif residues and lid subdomain glutamate (see Table 1) are labeled and depicted as sticks. The WA arginine, catalytic glutamate and lid subdomain glutamate are highlighted in tan. Representative conformations of the ATP-bound binding pocket of TerLT4 (D), TerLP74-26 (E) and TerLSf6 (F). ATP is depicted as sticks, and Mg2+ is a green sphere. The WA arginine, catalytic glutamate and lid subdomain glutamate are labeled and are highlighted in cyan. In all three terminases, the conserved WA arginine (R162, R39, and R24 in TerLT4, TerLP74-26, and TerLSf6, respectively) interacts with the catalytic glutamate (E256, E150, E119 in TerLT4, TerLP74-26, and TerLSf6) near the γ-phosphate of ATP.
Conformational changes associated with ATP binding
Consistent with structural studies, MD simulation of the ATP-bound complex indicates that in all three TerLs the conserved WA and WB residues mediate interactions expected of conserved ASCE motifs (4,15,21): the WA critical lysine interacts with the β- and γ-phosphates of ATP, while WA (T/S) and WB (D/E) residues chelate Mg2+ (Figures 5D–F). Importantly, the proposed catalytic glutamate residues, which were pointing out of the active site in the apo conformation, move into the active site and position a water molecule next to the γ-phosphate of ATP (compare Figure 5A–C, D–F); this ‘closed’ arrangement would allow the glutamate to polarize a water molecule for nucleophilic attack of the γ-phosphate, as required for a bona fide catalytic glutamate residue (4,20,24,25).
We note that the nucleotide-bound crystal structures of TerLSf6 found that the catalytic glutamate identified based on sequence homology (E119) points out of the binding pocket, away from the nucleotide (Supplementary Figure S3) (16). On the basis of this structure, the authors proposed that the ‘deviant’ WB glutamate (E118) might serve as the catalytic carboxylate rather than the conserved E119 (16); however, our simulations predict that when the protein is allowed to undergo thermal motion in solution, E119 changes conformation such that it does coordinate with the γ-phosphate of the bound ATP. Thus, our simulations suggest that WB E118 performs the conventionally postulated role of chelating the Mg2+-ATP and that E119 is actually the catalytic glutamate.
An ‘arginine toggle’
In addition to the anticipated interactions described above, the simulations predict a novel interaction induced by ATP binding that has not been reported in any of the static TerL structures that were crystallized with nucleotides or nucleotide analogs (the T4 structure being of an inactive mutant and the P74-26 structure with a non-hydrolyzable ATP analog) (14–17). All three simulations predict that when these inactive structures are reverted to their WT, ATP-bound forms and allowed to undergo thermal motion in solution, the conserved WA arginine breaks its interaction with the lid subdomain glutamate and moves into the binding pocket to interact with the catalytic glutamate positioned adjacent to the γ-phosphate of ATP (compare Figure 5A–C, D–F; Supplementary Figures S4 and S5). The simulations further reveal significant movement of the lid subdomain in the transition from the open apo to the closed ATP-bound state. In particular, principal component analysis of the MD trajectories of TerLT4 and TerLSf6 starting from equilibrated apo structures with docked ATP show that the lid subdomain exhibits a concerted motion towards the ATP-binding site (Figure 6, Supplementary Figure S6). In the case of TerLP74-26, the simulations were started from a closed nucleotide-bound crystal structure whose lid has already undergone this conformational transition (17) and thus the transition was not sampled (see Supplementary Figure S7). Given the likely role of lid subdomain motion in mechanochemical coupling (17,41) and because we observe the dynamic repositioning of this subdomain synchronously with the rearrangement of the binding pocket upon ATP binding, the MD simulations implicate residues involved in mechanochemical coupling. Specifically, the conserved WA arginine acts as a ‘toggle’ that breaks its interaction with the lid subdomain glutamate and rotates to interact solely with the catalytic glutamate. We note that this arginine toggle mechanism should not be confused with the “arginine finger" residue also commonly found in oligomeric ATPase enzymes (4,17). The latter residue conventionally acts in trans, directly interacting with the γ-phosphate of an ATP molecule bound at an adjacent subunit and acts to stabilize the hydrolysis transition state.
Figure 6.
Principal Component Analysis of TerLT4 Reveals Lid Subdomain Motion Upon ATP Binding. (A) Principal component analysis (PCA) of TerLT4 apo simulation shows little concerted motion of the lid subdomain (also called the NII-subdomain in TerLT4 literature), circled by black dashed circle. Residues are colored based on the magnitude of root-mean-square fluctuations during the simulation, with red representing least mobile residues and blue representing most mobile residues. Arrows represent the projection of the individual residue motions onto the first principal component. (B) Principal component analysis of the first 10 ns of TerLT4 ATP-bound simulation shows concerted rotation of the lid subdomain towards the ATPase active site consistent with prior experimental studies of Sf6 and P74-26 terminases. (C) Principal component analysis of the last 10 ns of TerLT4 ATP-bound simulation shows little lid subdomain motion, indicating that the lid subdomain is stable in the closed conformation once the rotation is complete.
Prevalence of the arginine toggle
The MD studies suggest that an arginine toggle mechanism may couple ATP hydrolysis to motor movement in the three terminase enzymes examined. Rao et al. noted that the WA motifs in TerL subunits contained a conserved arginine residue at position 3 or 4 (12,13,34), which suggests that the arginine toggle mechanism may be a general feature of terminase enzymes. To further explore this possibility, we used TerL sequences of well-studied bacteriophages in BLASTP searches against members of the Tailed Bacteriophage taxid. These searches revealed that arginine residues are nearly universally conserved at positions 3 or 4 of the WA motif (Supplementary Table S4); 138/138 of the T4-like phages and 76/76 of the P74-26-like phages conserve this arginine residue in the WA motif. This strict sequence conservation is similarly observed in the eukaryotic terminase enzymes, where all of the herpes virus packaging motors possess a WA arginine. Although there are exceptions in the P22- and λ-like phages, a strictly conserved glutamine appears in these motors and could perform an analogous role. In contrast, alignments of selected classes of ATPases associated with diverse cellular activities (AAA+ family of ASCE ATPases) reveal that few of the enzymes contained a conserved WA arginine (Supplementary Table S4). This is discussed further below.
Conformational changes associated with the catalytic E→D mutant
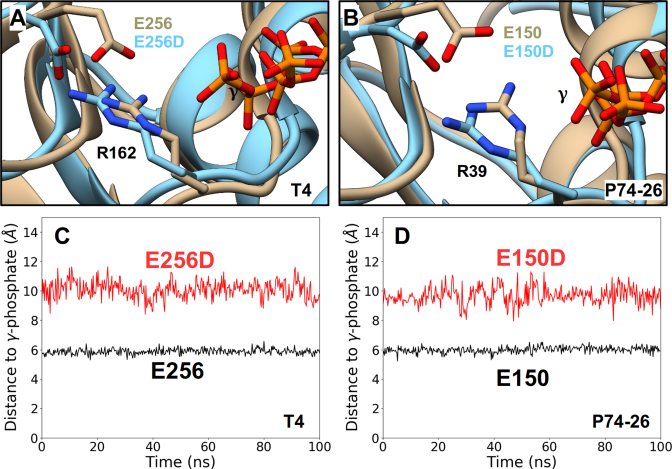
Finally, we modeled and simulated ATP-bound TerLT4-E256D and TerLP74-26-E150D mutant enzymes wherein the catalytic glutamate has been changed to aspartate; these studies were designed to provide insight into the effects of the analogous TerLλ-E179D mutant studied experimentally herein. In both cases, we find that the mutant Asp carboxylates retain their interactions with the WA arginines as is observed in the WT ATP-bound enzymes (Figure 7A, B); however, in both cases the carboxylate functional groups are positioned ∼2–5 Å further away from the γ-phosphate (Figure 7C, D and Supplementary Figure S8). This is caused, in part, by the 1.5 Å shorter length of the Asp vs. Glu side chain but is also due to significant reorientation of the binding-pocket to accommodate the mutated residue. This affects the ability of other WA residues, such as the critical lysine, to properly bind the ATP and further increases the distance between the catalytic carboxyl group and the γ-phosphate. This likely perturbs positioning and activation of the water molecule, thereby hindering the chemical step of hydrolysis. We suggest that a similar perturbation occurs in the E179D change of TerLλ and explains the slower rate of ATP hydrolysis and coupled packaging observed experimentally.
Figure 7.
Effects of the Conservative ASCE E→D Mutation Obtained from MD Simulations. Representative conformations of the ATP-binding pocket of ATP-bound WT TerLs (tan) superimposed onto the binding pocket conformations of the corresponding catalytic E→D mutant TerLs (cyan). Such analysis carried out for TerLT4 (A) and TerLP74-26 (B) structures show significant displacement of the labeled carboxylate functional group and miscoordination of the labeled γ-phosphate of ATP in both mutant structures. Separation distances, obtained from an equilibrium MD trajectory, between the carboxylate functional group and the γ-phosphate for WT (black) and catalytic E→D mutants (red) of TerLT4 (C) and TerLP74-26 (D). In both cases, the E→D change results in a significant increase this distance and the hindered ATP hydrolysis observed experimentally in the E→D mutants is attributed to this increased distance.
DISCUSSION
While a number of studies have shown that change of the identified catalytic carboxylate in ASCE ATPase motors affects their function (17,18,24,68,69), a detailed mechanistic dissection of the defect has not been performed in most cases. In the present study, we have identified E179 as the catalytic glutamate in the λ terminase ASCE ATPase domain. The genetic data clearly show that all mutations of TerLλ-E179, including the conservative E179D, are profoundly lethal in vivo. The lethal phenotype could reflect multiple defects including protein misfolding, disrupted motor assembly, attenuated ATP hydrolysis, a reduced DNA translocation rate and/or an inability to package the entire genome. The biochemical and biophysical data show that TerLλ-E179D retains weak ATPase and DNA translocation activity but is nevertheless lethal even using our highly sensitive genetic assay. The differences between the in vivo genetic studies and the in vitro biochemical and optical tweezer studies can be reconciled as follows: The single-molecule assay follows the early stages of DNA packaging, ensemble genome packaging assesses the packaging of full-length genomes into the capsid and the genetic studies measure the final product, an infectious virus particle. The combined genetic, biochemical and single-molecule data indicate that while TerLλ-E179D can initiate packaging, the slowed motor velocity and increased pausing and slipping prevents packaging of the full-length genome, which is required to assemble an infectious virus. Thus, the integrated studies presented here provide insight into the mechanistic defects responsible for the observed biological outcome.
Mechanistic insight derived from TerLλ mutant analysis
Our previous findings suggest an ATP tight binding transition model where the λ terminase motor undergoes a conformational change upon binding of ATP that is required for both tight binding of the nucleotide and tight DNA gripping during duplex translocation (Supplementary Figure S1) (41). This is based, in part, on the observation that decreasing [ATP] slows the WT terminase motor and also causes an increase in slipping (41). Some WA mutants, such as TerLλ-V80A follow a similar trend of slowed motor velocity and increased slipping even at saturating concentrations of ATP and have been implicated as having a slowed ATP tight binding transition. In contrast, the TerLλ-E179D catalytic carboxylate mutant studied here slows the motor velocity but does not cause a proportionally large increase in slipping (Figure 4B). Thus, the slowed translocation of E179D is not attributable to a slowed ATP tight binding transition. Rather, the phenotype is virtually identical to that of the TerLλ-R79K WA mutant studied previously, which was implicated as having a slowed hydrolysis step (41). The virtually identical effects of the E179D and R79K residue changes, both chemically conservative changes that slightly shorten the side-chain lengths, also support the prediction of the molecular dynamics simulations that an interaction between these two residues is critical to position the catalytic glutamate adjacent to the γ-phosphate of the ATP.
Mechanochemical coupling in terminase enzymes
Structural studies in the TerLP74-26 system reveal interactions between WA and lid subdomain residues that are remodeled upon binding of the ATP mimic ADP•BeF4 (17). This is accompanied by a ∼13° rotation between the ATPase and lid subdomains which has been proposed to be central to communication of ATP hydrolysis in one subunit to the next, and in DNA translocation (17). Similar observations have been made in TerLSf6; comparison of the ATP-γ-S and ADP-bound forms reveals a change in the interaction of the conserved WA Arg and lid residue E187, with concomitant 0.5 Å movement of the lid subdomain relative to the ATPase domain (16). As with the P74-26 model, the small lid subdomain movement is proposed to be amplified through additional domain movements to produce the ∼8 Å movement needed to translocate 2.5 bp/ATP hydrolysis.
Based on this published data and the experimental and simulation data presented herein, a general model can be proposed. The conserved WA arginines found in ATP-binding pocket of terminase enzymes perform two essential functions: (i) they play an important role in ATP hydrolysis by appropriately positioning the catalytic glutamate in the transition state, and (ii) they communicate the nucleotide bound state of the ATPase domain to the lid subdomain to mediate mechanochemical coupling. In addition, the open to closed transition is associated with a tight binding enzyme-substrate transition that has been proposed for several terminase enzymes and that may be required for processive DNA packaging by the enzymes. We note that a pre-catalytic tight binding transition is common in biochemistry and similar models have been proposed for DNA polymerase enzymes in which the transition modulates the fidelity of nucleotide incorporation (70,71).
Arginine toggle
The wide conservation of the WA arginine among viral packaging motors suggests that these enzymes have developed use of an arginine toggle to couple ATP binding and hydrolysis to motor movement. In contrast, a number of ASCE ATPases employ a ‘sensor II motif arginine’ that is located within the lid subdomain and moves into the ATP binding pocket upon nucleotide-binding (65); this binding switch directly confers motion to the lid subdomain and performs an analogous role as the arginine toggle reported here. Of note, terminase enzymes lack a conserved sensor II motif arginine, which led Kelch and co-workers to propose that the WA arginine of TerLP74-26 terminase performs an analogous role (17); however, the specific mechanism and interactions with the lid subdomain and catalytic glutamate residues could not be identified in the structural data. Our MD data suggest that this residue, Arg39 in TerLP74-26, is indeed conceptually analogous to the sensor II arginine found in ASCE ATPase family, but is mechanistically distinct and conserved in the viral packaging motors.
CONCLUSIONS
The present study represents a detailed dissection of the role of TerLλ-E179 in the packaging mechanism of λ terminase. Our experimental results confirm that this residue plays an essential role in the function of the enzyme and the ensemble of data supports the conclusion that E179 is a catalytic residue involved in the hydrolysis of ATP. Assignment of this residue as the catalytic glutamate firmly places λ terminase within the ASCE family of ATPases and among homologous terminase enzymes such as those of T4 and P74-26. In addition, our computational results predict a general mechanism in the terminase enzymes, supported by our experimental findings, whereby the catalytic glutamate acts in concert with the conserved WA arginine in a transition from an open to closed conformation that binds ATP tightly. This tight-binding conformation (i) positions ATP, the catalytic carboxylate and a water molecule for activation and in-line attack on the γ-phosphate in the transition state and (ii) mediates mechanochemical coupling when, after hydrolysis and product release, the arginine toggles to interact with a Glu residue in the lid subdomain. Although little overall sequence identity is found for TerL proteins, strong conservation of the ASCE ATPase motifs (12,13,15–18) and other structural features, such as the arginine toggle residue and the lid subdomain (4,21,31), suggest that the mechanical output of these viral motors is based on a unitary fundamental mechanism.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Brian Kelch, whose thoughtful comments significantly strengthened the paper. We dedicate this paper to the memory of our friend and colleague, Dr Shelley Grimes, whose unbridled enthusiasm and ground-breaking research in the field always brought out the best in us.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [5R01GM088186 to C.E.C., M.F., D.S., R01GM118817 to D.S., G.A.]; A.V. was supported by the NSF-REU program at University of Iowa [DBI-7290775]; computational resources were provided by the NSF XSEDE Program [ACI-1053575] and the Duke Computer Cluster. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu S., Chistol G., Bustamante C.. Mechanical operation and intersubunit coordination of ring-shaped molecular motors: insights from single-molecule studies. Biophys. J. 2014; 106:1844–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartman M.A., Spudich J.A.. The myosin superfamily at a glance. J. Cell Sci. 2012; 125:1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vale R.D. The molecular motor toolbox for intracellular transport. Cell. 2003; 112:467–480. [DOI] [PubMed] [Google Scholar]
- 4. Lyubimov A.Y., Strycharska M., Berger J.M.. The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol. 2011; 21:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allemand J.-F., Maier B., Smith D.E.. Molecular motors for DNA translocation in prokaryotes. Curr. Opin. Biotechnol. 2012; 23:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iyer L.M., Makarova K.S., Koonin E.V., Aravind L.. Comparative genomics of the FtsK -HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004; 32:5260–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burroughs A.M., Iyer L.M., Aravind L.. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007; 3:48–65. [DOI] [PubMed] [Google Scholar]
- 8. Iyer L.M., Leipe D.D., Koonin E.V., Aravind L.. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004; 146:11–31. [DOI] [PubMed] [Google Scholar]
- 9. Rao V.B., Feiss M.. The bacteriophage DNA packaging motor. Annu. Rev. Genetics. 2008; 42:647–681. [DOI] [PubMed] [Google Scholar]
- 10. Walker J.E., Saraste M., Runswick M.J., Gay N.J.. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982; 1:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitchell M.S., Rao V.B.. Functional analysis of the bacteriophage T4 DNA-packaging ATPase motor. J. Biol. Chem. 2006; 281:518–527. [DOI] [PubMed] [Google Scholar]
- 12. Mitchell M.S., Rao V.B.. Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology. 2004; 321:217–221. [DOI] [PubMed] [Google Scholar]
- 13. Mitchell M.S., Matsuzaki S., Imai S., Rao V.B.. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002; 30:4009–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun S., Kondabagil K., Draper B., Alam T.I., Bowman V.D., Zhang Z., Hegde S., Fokine A., Rossmann M.G., Rao V.B.. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008; 135:1251–1262. [DOI] [PubMed] [Google Scholar]
- 15. Sun S., Kondabagil K., Gentz P.M., Rossmann M.G., Rao V.B.. The structure of the ATPase that powers DNA packaging into bacteriophage t4 procapsids. Mol. Cell. 2007; 25:943–949. [DOI] [PubMed] [Google Scholar]
- 16. Zhao H., Christensen T.E., Kamau Y.N., Tang L.. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8075–8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hilbert B.J., Hayes J.A., Stone N.P., Duffy C.M., Sankaran B., Kelch B.A.. Structure and mechanism of the ATPase that powers viral genome packaging. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3792–E3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mao H., Saha M., Reyes-Aldrete E., Sherman M.B., Woodson M., Atz R., Grimes S., Jardine P.J., Morais M.C.. Structural and molecular basis for coordination in a Viral DNA packaging motor. Cell Rep. 2016; 14:2017–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu R.-G., Jenkins H.T., Antson A.A., Greive S.J.. Structure of the large terminase from a hyperthermophilic virus reveals a unique mechanism for oligomerization and ATP hydrolysis. Nucleic Acids Res. 2017; 45:13029–13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Story R.M., Steitz T.A.. Structure of the recA protein–ADP complex. Nature. 1992; 355:374. [DOI] [PubMed] [Google Scholar]
- 21. Erzberger J.P., Berger J.M.. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006; 35:93–114. [DOI] [PubMed] [Google Scholar]
- 22. Hörtnagel K., Voloshin O.N., Kinal H.H., Ma N., Schaffer-Judge C., Camerini-Otero R.D.. Saturation mutagenesis of the E. coli RecA loop L2 homologous DNA pairing region reveals residues essential for recombination and recombinational repair1. J. Mol. Biol. 1999; 286:1097–1106. [DOI] [PubMed] [Google Scholar]
- 23. Park M.-Y., Omote H., Maeda M., Futai M.. Conserved Glu-181 and Arg-182 residues of Escherichia coli H+-ATPase (ATP synthase) β subunit are essential for catalysis: properties of 33 mutants between βGlu-161 and βLys-201 residues. J. Biochem. 1994; 116:1139–1145. [DOI] [PubMed] [Google Scholar]
- 24. Goetzinger K.R., Rao V.B.. Defining the ATPase center of bacteriophage T4 DNA packaging machine: requirement for a catalytic glutamate residue in the large terminase protein gp17. J. Mol. Biol. 2003; 331:139–154. [DOI] [PubMed] [Google Scholar]
- 25. Lin S., Alam T.I., Kottadiel V.I., VanGessel C.J., Tang W.-C., Chemla Y.R., Rao V.B.. Altering the speed of a DNA packaging motor from bacteriophage T4. Nucleic Acids Res. 2017; 45:11437–11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rao V.B., Feiss M.. Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol. 2015; 2:351–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Black L.W. Old, new, and widely true: The bacteriophage T4 DNA packaging mechanism. Virology. 2015; 479:650–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chemla Y.R., Smith D.E.. Rao V, Rossmann MG. Single-Molecule Studies of Viral DNA Packaging. Viral Molecular Machines. Advances in Experimental Medicine and Biology. 2012; 726:Boston: Springer; 549–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Casjens S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011; 9:647–657. [DOI] [PubMed] [Google Scholar]
- 30. Catalano C.E. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. 2005; NY: Kluwer Academic/Plenum Press; 1–4. [Google Scholar]
- 31. Hilbert B.J., Hayes J.A., Stone N.P., Xu R.-G., Kelch B.A.. The large terminase DNA packaging motor grips DNA with its ATPase domain for cleavage by the flexible nuclease domain. Nucleic Acids Res. 2017; 45:3591–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tafoya S., Bustamante C.. Molecular switch-like regulation in motor proteins. Phil. Trans. R. Soc. B. 2018; 373:20170181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feiss M., Rao V.B.. Rao V, Rossmann MG. The Bacteriophage DNAPackaging Machine. Viral Molecular Machines. Advances in Experimental Medicine and Biology. 2012; 726:Boston: Springer; 489–509. [DOI] [PubMed] [Google Scholar]
- 34. Rao V.B., Mitchell M.S.. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J. Mol. Biol. 2001; 314:401–411. [DOI] [PubMed] [Google Scholar]
- 35. Duffy C., Feiss M.. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J. Mol. Biol. 2002; 316:547–561. [DOI] [PubMed] [Google Scholar]
- 36. Kondabagil K.R., Zhang Z., Rao V.B.. The DNA translocating ATPase of bacteriophage T4 packaging motor. J. Mol. Biol. 2006; 363:486–499. [DOI] [PubMed] [Google Scholar]
- 37. Alam T.I., Draper B., Kondabagil K., Rentas F.J., Ghosh-Kumar M., Sun S., Rossmann M.G., Rao V.B.. The headful packaging nuclease of bacteriophage T4. Mol. Microbiol. 2008; 69:1180–1190. [DOI] [PubMed] [Google Scholar]
- 38. Tsay J.M., Sippy J., DelToro D., Andrews B.T., Draper B., Rao V., Catalano C.E., Feiss M., Smith D.E.. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 2010; 285:24282–24289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsay J.M., Sippy J., Feiss M., Smith D.E.. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:14355–14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Andrews B.T., Catalano C.E.. Strong subunit coordination drives a powerful viral DNA packaging motor. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5909–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. delToro D., Ortiz D., Ordyan M., Sippy J., Oh C.S., Keller N., Feiss M., Catalano C.E., Smith D.E.. Walker-A motif acts to coordinate ATP hydrolysis with motor output in viral DNA packaging. J. Mol. Biol. 2016; 428:2709–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feiss M., Catalano C.. Catalano C. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. 2005; NY: Klewer Academic/Plenum Press; 5–39. [Google Scholar]
- 43. Sippy J., Patel P., Vahanian N., Sippy R., Feiss M.. Genetics of critical contacts and clashes in the DNA packaging specificities of bacteriophages λ and 21. Virology. 2015; 476:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang T.-C., Ortiz D., Yang Q., De Angelis R.W., Sanyal S.J., Catalano C.E.. Physical and functional characterization of a viral genome maturation complex. Biophys. J. 2017; 112:1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chemla Y.R., Aathavan K., Michaelis J., Grimes S., Jardine P.J., Anderson D.L., Bustamante C.. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005; 122:683–692. [DOI] [PubMed] [Google Scholar]
- 46. Kottadiel V.I., Rao V.B., Chemla Y.R.. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:20000–20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao Z., De-Donatis G.M., Schwartz C., Fang H., Li J., Guo P.. Arginine finger regulates sequential action of asymmetrical hexameric ATPase in dsDNA translocation motor. Mol.Cell. Biol., MCB. 2016; 36:2514–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Case D., Betz R., Cerutti D., Cheatham T., Darden T., Duke R.. AMBER16. 2016; San Francisco. [Google Scholar]
- 49. Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C.. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015; 11:3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ryckaert J.-P., Ciccotti G., Berendsen H.J.. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977; 23:327–341. [Google Scholar]
- 51. Darden T., York D., Pedersen L.. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993; 98:10089–10092. [Google Scholar]
- 52. Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L.. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983; 79:926–935. [Google Scholar]
- 53. Migliori A.D., Keller N., Alam T.I., Mahalingam M., Rao V.B., Arya G., Smith D.E.. Evidence for an electrostatic mechanism of force generation by the bacteriophage T4 DNA packaging motor. Nat. Commun. 2014; 5:4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang J.R., Andrews B.T., Catalano C.E.. Energy-independent helicase activity of a viral genome packaging motor. Biochemistry. 2012; 51:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang Q., Maluf N.K., Catalano C.E.. Packaging of a Unit-Length viral Genome: The role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage lambda. J. Mol. Biol. 2008; 383:1037–1048. [DOI] [PubMed] [Google Scholar]
- 56. Smith D.E., Tans S.J., Smith S.B., Grimes S., Anderson D.L., Bustamante C.. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001; 413:748–752. [DOI] [PubMed] [Google Scholar]
- 57. Fuller D.N., Raymer D.M., Rickgauer J.P., Robertson R.M., Catalano C.E., Anderson D.L., Grimes S., Smith D.E.. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007; 373:1113–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fuller D.N., Raymer D.M., Kottadiel V.I., Rao V.B., Smith D.E.. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:16868–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith D.E. Single-molecule studies of viral DNA packaging. Curr. Opin. Virol. 2011; 1:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Keller N., delToro D., Smith D.E.. Lavelle C. Single-Molecule Measurements of Motor-Driven Viral DNA Packaging in Bacteriophages Phi29, Lambda, and T4 with Optical Tweezers. Molecular Motors. Methodsin Molecular Biology. 2018; 1805:NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- 61. Liu S., Chistol G., Hetherington C.L., Tafoya S., Aathavan K., Schnitzbauer J., Grimes S., Jardine P.J., Bustamante C.. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014; 157:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Berndsen Z.T., Keller N., Smith D.E.. Continuous allosteric regulation of a viral packaging motor by a sensor that detects the density and conformation of packaged DNA. Biophys. J. 2015; 108:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Keller N., Berndsen Z.T., Jardine P.J., Smith D.E.. Experimental comparison of forces resisting viral DNA packaging and driving DNA ejection. Phys. Rev. E. 2017; 95:052408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Keller N., Grimes S., Jardine P.J., Smith D.E.. Repulsive DNA-DNA interactions accelerate viral DNA packaging in phage phi29. Phys. Rev. Lett. 2014; 112:248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Erzberger J.P., Mott M.L., Berger J.M.. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006; 13:676–683. [DOI] [PubMed] [Google Scholar]
- 66. Glynn S.E., Martin A., Nager A.R., Baker T.A., Sauer R.T.. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009; 139:744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kelch B.A., Makino D.L., O’Donnell M., Kuriyan J.. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011; 334:1675–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Orelle C., Dalmas O., Gros P., Di Pietro A., Jault J.-M.. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 2003; 278:47002–47008. [DOI] [PubMed] [Google Scholar]
- 69. Männikkö R., Flanagan S.E., Sim X., Segal D., Hussain K., Ellard S., Hattersley A.T., Ashcroft F.M.. Mutations of the same conserved glutamate residue in NBD2 of the sulfonylurea receptor 1 subunit of the KATP channel can result in either hyperinsulinism or neonatal diabetes. Diabetes. 2011; 60:1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johnson K.A. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta (BBA)-Proteins Proteomics. 2010; 1804:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schlick T., Arora K., Beard W.A., Wilson S.H.. Perspective: pre-chemistry conformational changes in DNA polymerase mechanisms. Theor. Chem. Acc. 2012; 131:1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Andrews B.T., Catalano C.E.. The enzymology of a viral genome packaging motor is influenced by the assembly state of the motor subunits. Biochemistry. 2012; 51:9342–9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang Q., Berton N., Manning M.C., Catalano C.E.. Domain structure of gpNu1, a phage lambda DNA packaging protein. Biochemistry. 1999; 38:14238–14247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.