Abstract
Targeted DNA double-strand breaks (DSBs) with CRISPR–Cas9 have revolutionized genetic modification by enabling efficient genome editing in a broad range of eukaryotic systems. Accurate gene editing is possible with near-perfect efficiency in haploid or (predominantly) homozygous genomes. However, genomes exhibiting polyploidy and/or high degrees of heterozygosity are less amenable to genetic modification. Here, we report an up to 99-fold lower gene editing efficiency when editing individual heterozygous loci in the yeast genome. Moreover, Cas9-mediated introduction of a DSB resulted in large scale loss of heterozygosity affecting DNA regions up to 360 kb and up to 1700 heterozygous nucleotides, due to replacement of sequences on the targeted chromosome by corresponding sequences from its non-targeted homolog. The observed patterns of loss of heterozygosity were consistent with homology directed repair. The extent and frequency of loss of heterozygosity represent a novel mutagenic side-effect of Cas9-mediated genome editing, which would have to be taken into account in eukaryotic gene editing. In addition to contributing to the limited genetic amenability of heterozygous yeasts, Cas9-mediated loss of heterozygosity could be particularly deleterious for human gene therapy, as loss of heterozygous functional copies of anti-proliferative and pro-apoptotic genes is a known path to cancer.
INTRODUCTION
CRISPR–Cas9-assisted genome editing requires the simultaneous presence of the Cas9 endonuclease and a guide-RNA (gRNA) that confers target-sequence specificity (1). A gRNA consists of a structural domain and a variable sequence homologous to the targeted sequence (1–4). A Cas9–gRNA complex introduces a DSB when the gRNA binds to its reverse complement sequence on the 5′ side of a PAM sequence (NGG). Imperfect gRNA complementarity and/or absence of a PAM sequence strongly reduce editing efficiencies (5). CRISPR–Cas9 enables specific editing of any sequence proximal to a PAM sequence, with minimal off-targeting effects (5). The introduction of a DSB facilitates genome editing by increasing the rate of repair by homologous recombination (6). When a repair fragment consisting of a DNA oligomer with homology to regions on both sides of the introduced DSB is added, it is integrated at the targeted locus by homologous recombination, resulting in replacement of the original sequence and repair of the DSB (2–4). In Saccharomyces cerevisiae, double stranded DNA oligomers with 60 bp of homology are sufficient to obtain accurate gene-editing in almost 100% of transformed cells (3). By inserting sequences between the homologous regions of the repair oligonucleotide, heterozygous sequences of up to 35 kb could be inserted at targeted loci (7). While such gene editing approaches have been very efficient in haploid and homozygous diploid yeasts, the accurate introduction of short DNA fragments can be tedious in heterozygous yeast. In homozygous diploid and polyploid eukaryotes, CRISPR–Cas9 introduces DSBs in all alleles of a targeted sequence (8). In heterozygous genomes, gRNAs can be designed for allele-specific targeting if heterozygous loci have different PAM motifs and/or different 5′ sequences close to a PAM motif (8,9), enabling allele-specific gene editing using Cas9. In such cases, a DSB is introduced in only one of the homologous chromosomes while the other homolog remains intact. However, the presence of intact homologous chromosomes facilitates repair of DSBs by homology-directed repair (HDR) using mechanisms such as homologous recombination (HR), or break-induced repair (BIR) (10–12). In particular, HDR of DSBs can induce chromosome recombinations and even loss of heterozygosity (LOH) in diploid genomes (9,13–16). Therefore, the presence of an intact homologous chromosome could compete with an intended gene-editing event, resulting in reduced editing efficiency and possibly in extensive genetic changes due to LOH. So far, no systematic analysis has been performed of the efficiency of Cas9-mediated gene editing at heterozygous loci. To investigate if Cas9 gene editing works differently in heterozygous diploid yeast, we tested if allele-specific targeting of heterozygous loci using Cas9 enables accurate gene editing in an interspecies Saccharomyces hybrid, and investigated the resulting transformants. In addition, we systematically investigated the efficiency of Cas9-mediated genome editing when targeting various homozygous and heterozygous loci in diploid laboratory S. cerevisiae strains while monitoring genetic changes.
MATERIALS AND METHODS
Strains, plasmids, primers and statistical analysis
S. cerevisiae strains used in this study are derived from the laboratory strains CEN.PK113-7D and S288C (17,18). Yeast strains, plasmids and oligonucleotide primers used in this study are provided in Tables S3–S5. Statistical significance was determined using two-tailed unpaired Student's t-tests in GraphPad Prism 4 (GraphPad, La Jolla, CA, USA).
Media and growth conditions
Plasmids were propagated overnight in Escherichia coli XL1-Blue cells in 10 ml LB medium containing 10 g/l peptone, 5 g/l Bacto Yeast extract, 5 g/l NaCl and 100 mg/l ampicillin at 37°C. Unless indicated otherwise, yeast strains were grown at 30°C and 200 RPM in 100 ml flat-bottom flasks containing 50 ml YPD medium, containing 10 g/l Bacto yeast extract, 20 g/l Bacto peptone and 20 g/l glucose. Alternatively, strains were grown in synthetic medium (SM) containing 3.0 g/l KH2PO4, 5.0 g/l (NH4)2SO4,0.5 g/l MgSO47H2O, 1 ml/l trace elements, 1 ml/l vitamin solution and 20 g/l glucose (19). For uracil auxotrophic strains, SM-derived media were supplemented with 150 mg/l uracil (20). Solid media were supplemented with 20 g/l agar. Selection for the amdSYM marker was performed on SM-AC: SM medium with 0.6 g/l acetamide and 6.6 g/l K2SO4 as nitrogen and sulfur sources instead of (NH4)2SO4 (21). The amdSYM marker was lost by growth on YPD and counter-selected on SM-FAC: SM supplemented with 2.3 g/l fluoroacetamide (21). Yeast strains and E. coli containing plasmids were stocked in 1 ml aliquots after addition of 30% (v/v) glycerol to the cultures and stored at −80°C.
Flow cytometric analysis
Overnight aerobic cultures in 100 ml flat-bottom flasks on 20 mL YPD medium were vortexed thoroughly to disrupt cell aggregates and used for flow cytometry on a BD FACSAria™ II SORP Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA) equipped with 355, 445, 488, 561 and 640 nm lasers and a 70 μm nozzle, and operated with filtered FACSFlow™ (BD Biosciences). Cytometer performance was evaluated prior to each experiment by running a CST cycle with CS&T Beads (BD Biosciences). Drop delay for sorting was determined by running an Auto Drop Delay cycle with Accudrop Beads (BD Biosciences). Cell morphology was analysed by plotting forward scatter (FSC) against side scatter (SSC). The fluorophore mRuby2 was excited by the 561 nm laser and emission was detected through a 582 nm bandpass filter with a bandwidth of 15 nm. The fluorophore mTurquoise2 was excited by the 445 nm laser and emission was detected through a 525 nm bandpass filter with a bandwidth of 50 nm. The fluorophore Venus was excited by the 488 nm laser and emission was detected through a 545 nm bandpass filter with a bandwidth of 30 nm. For each sample, 100,000 events were analysed and the same gating strategy was applied to all samples of the same strain. First, ‘doublet’ events were discarded on a FSC-A/FSC-H plot, resulting in at least 75′000 single cells for each sample. Of the remaining single cells, cells with and cells without fluorescence from Venus were selected in a FSC-A/Venus plot. For both these groups, cells positive for mRuby2 and mTurquoise2, cells positive for only mRuby2, cells positive for only mTurquoise2 and cells negative for mRuby2 and mTurquoise2 were gated. The same gating was used for all samples of each strain. Sorting regions (‘gates’) were set on these plots to determine the types of cells to be sorted. Gated single cells were sorted in 96-well microtiter plates containing YPD using a ‘single cell’ sorting mask, corresponding to a yield mask of 0, a purity mask of 32 and a phase mask of 16. FACS data was analysed using FlowJo® software (version 3.05230, FlowJo, LLC, Ashland, OR, USA). Separate gating strategies were made for IMX1555, IMX1557 and IMX1585 to account for possible differences in cell size, shape and morphology.
Plasmid assembly
Plasmid pUD574 was de novo synthesised at GeneArt (Thermo Fisher Scientific, Waltham, MA, USA) containing the sequence 5′ GGTCTCGCAAAATTACACTGATGAGTCCGTGAGGACGAAACGAGTAAGCTCGTCTGTAATATCTTAATGCTAAAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTGGCCGGCATGGTCCCAGCCTCCTCGCTGGCGCCGGCTGGGCAACATGCTTCGGCATGGCGAATGGGACACAGCGAGACC 3′.
Plasmids pUD429 was constructed in a 10 μl golden gate assembly using T4 ligase (Thermo Fisher Scientific) and BsaI (New England BioLabs, Ipswich, MA, USA) from 10 ng of parts pYTK002, pYTK047, pYTK067, pYTK079, pYTK081 and pYTK083 of the yeast toolkit as described previously (22). Similarly, pUD430 was constructed from pYTK003, pYTK047, pYTK068, pYTK079, pYTK081 and pYTK083, and pUDP431 from pYTK004, pYTK047, pYTK072, pYTK079, pYTK081 and pYTK083. Plasmid pUDE480 expressing mRuby2 was constructed from GFP dropout plasmid pUD429 with pYTK011, pYTK034 and pYTK054 using golden gate assembly as described previously (22). Similarly, pUDE481 expressing mTurquoise2 was constructed from pUD430, pYTK009, pYTK032 and pYTK053, and pUDE482 expressing Venus from pUD431, pYTK013, pYTK033 and pYTK055.
Plasmids pUDR323, pUDR324, pUDR325, pUDR358, pUDR359, pUDR360, pUDR361 and pUDR362, expressing gRNAs targeting SIT1, FAU1, spcas9, UTR2, FIR1, AIM9, YCK3 and intergenic region 550K respectively, were constructed using NEBuilder® HiFi DNA Assembly Master Mix by assembling the 2 μm fragment amplified from pROS11 with primers 12230, 12235, 9457, 12805, 12806, 12807, 12808, 12809 respectively, and the plasmid backbone amplified from pROS11 with primer 6005 as described previously (3,23).
Plasmid pUDP045, expressing gRNAMAL11 and cas9, was constructed by Golden Gate cloning by digesting pUDP004 and pUD574 using BsaI and ligating with T4 ligase (24). Correct assembly was verified by restriction analysis using PdmI.
Strain construction
Yeast strains were transformed according to the high-efficiency protocol by Gietz et al. (25). IMX1544 was constructed by transforming IMX581 with 1 μg pUDR323 and 1 μg of a repair fragment amplified from pUD481 using primers 12233 and 12234 containing an expression cassette for mTurquoise2 and 60 bp homology arms with the FAU1 locus. IMX1555 was constructed by transforming IMX1544 with 1 μg pUDR324 and 1 μg of repair fragment amplified from pUD480 using primers 12228 and 12229 containing an expression cassette for mRuby2 and 60 bp homology arms with the SIT1 locus. Transformants were selected on SM-AC plates, three single colony isolates were grown overnight on YPD an streaked on SM-FAC plates. Genomic DNA of a single colony was extracted, insertion of mTurquoise2 in FAU1 was confirmed by PCR using primers 12236 and 12237, and insertion of mRuby2 in SIT1 was confirmed by PCR using primers 12231 and 12232 followed by digestion with PvuII and XhoI digestion. IMX1557 was constructed by adding 10 μl of stationary phase culture of IMX1555 and of IMK439 in 1 ml of SM medium, incubating overnight at 30°C and plating on SM plates with 10 mg/l clonNAT and 100 mg/l G418. IMX1585 was constructed by adding 10 μl of stationary phase culture of IMX1555 and of S288C in 1 ml of SM medium, incubating overnight at 30°C and plating on SM plates with 10 mg/l clonNAT without added uracil. All constructed strains were grown overnight in YPD and fluorescence corresponding to mRuby2 and mTurquoise2 was verified by flow cytometry.
Cas9 mediated targeting in S. cerevisiae x eubayanus hybrid IMS0408
IMX1421, IMX1422, IMX1423 and IMX1424 were constructed by transforming IMS0408 with 1 μg pUDP045 and 1 μg of a 120 bp repair fragment constructed by annealing primers 10813 and 10814 as described previously (8). Transformants were selected on SM-AC plates, genomic DNA of 10 single colonies was extracted, but no band could be obtained when amplifying the MAL11 locus using primer sets 1084/1470 and 1657/1148. The exact same procedure was performed without the addition of the 120 bp repair fragment. Four randomly selected colonies transformed with repair fragment were re-streaked three times on YPD agar, the plasmid was counter-selected for by plating on SM-FAC and the isolates were stocked as IMX1421, IMX1422, IMX1423 and IMX1424.
Cas9 mediated introduction of DSBs in S. cerevisiae strains
DSBs were introduced by transforming yeast strains using 1 μg of purified gRNA expression plasmid and 1 μg of gel-purified double stranded repair fragment. The expression of gRNAs was done with plasmids pMEL11 to target CAN1, pUDR325 to target cas9, pUDR358 to target UTR2, pUDR359 to target FIR1, pUDR360 to target AIM9, pUDR361 to target YCK3 and pUDR362 to target 550K according to Mans et al. (3,23). Repair fragments containing Venus expression cassettes were PCR amplified from plasmid pUDE482 with primers with an overlap of ∼20 bp with the nucleotides flanking the targeted open reading frame and purified on a 1% agarose gel (Supplementary Table S5). Upon transformation, the cells were transferred to 100 mL flat-bottom flasks containing 20 ml SM-AC medium and grown until stationary phase at 30°C and 200 RPM to select cells transformed with the gRNA expression plasmid. After about 72 h, 0.2 ml of these cultures was transferred to fresh SM-AC and grown under the same conditions to stationary phase to dilute any remaining untransformed cells. After about 48 h, 0.2 ml of these cultures was transferred to 100 ml flat-bottom flasks containing 20 ml YPD medium and grown for ∼12 h under the same conditions to obtain optimal fluorescence signals.
DNA extraction and whole genome analysis
IMX1557, IMX1585, IMX1596-IMX1635, IMS0408 and IMX1421-IMX1424 were incubated in 500 ml flat-bottom flasks containing 100 ml liquid YPD medium at 30°C on an orbital shaker set at 200 RPM until the strains reached stationary phase with an OD660 between 12 and 20. Genomic DNA was isolated using the Qiagen 100/G kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions and quantified using a Qubit® Fluorometer 2.0 (Thermo Fisher Scientific). Between 11.5 and 54.6 μg genomic DNA was sequenced by Novogene Bioinformatics Technology Co., Ltd (Yuen Long, Hong Kong) on a HiSeq 2500 (Illumina, San Diego, CA, USA) with 150 bp paired-end reads using TruSeq PCR-free library preparation (Illumina). For IMX1557, IMX1585 and IMX1596-IMX1635, reads were mapped onto the S. cerevisiae CEN.PK113-7D genome (17) using the Burrows–Wheeler Alignment tool (BWA) and further processed using SAMtools and Pilon for variant calling (26–28). Homozygous SNPs from IMX1585 were subtracted from the list of homozygous SNPs of each strain and a list of homozygous SNPs on chromosome V was compiled per strain. Based on the list of heterozygous SNPs in IMX1585, all homozygous SNPs corresponded to the nucleotide from S288C while the nucleotide from IMX1557 was lost, and regions were identified in which all contiguous heterozygous SNPs lost heterozygosity for each strain. LOH was confirmed by visualising the generated .bam files in the Integrative Genomics Viewer (IGV) software (29). Regions mapped as having lost heterozygosity correspond to regions between the first and last nucleotide which lost heterozygosity. For IMS0408 and IMX1421-IMX1424, reads where aligned to a reference genome obtained by combining the reference genome of CEN.PK113-7D (17) and the reference genome of S. eubayanus strain CBS12357 (30) as they are closely related to the haploid parents of IMS0408. Regions affected by LOH were defined as regions in which reads did not align to the S. cerevisiae reference chromosome VII while reads aligned to the corresponding region of the S. eubayanus reference chromosome VII with approximately double the normal coverage.
RESULTS
Targeting of a heterozygous gene in a S. cerevisiae x S. eubayanus hybrid
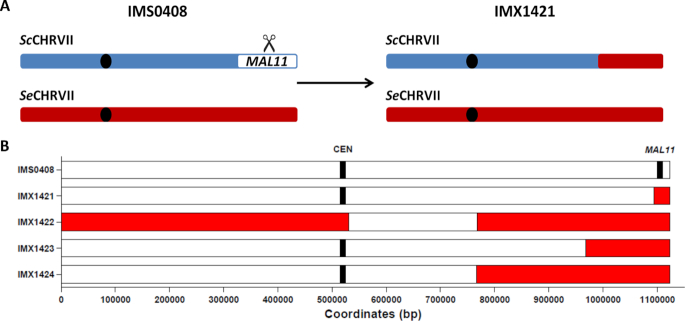
To investigate Cas9 gene editing in a genetic context with extensive heterozygosity, we targeted a heterozygous locus in an interspecies S. cerevisiae × eubayanus hybrid. The hybrid IMS0408 was constructed previously by mating a haploid S. cerevisiae laboratory strain and a haploid spore from the S. eubayanus type strain CBS 12357, resulting in an allodiploid strain with ∼85% nucleotide identity between corresponding chromosomes of the two subgenomes (31). The MAL11 gene encodes a membrane transporter located on chromosome VII in S. cerevisiae, which is absent in the S. eubayanus CBS 12357 genome. Therefore, the S. cerevisiae chromosome VII could be specifically targeted using Cas9 and a gRNA targeting MAL11. IMS0408 was transformed with plasmid pUDP045, expressing Cas9 and a gRNA targeting MAL11, with and without a repair fragment with 60-bp of homology to sequences adjacent to the 5′ and 3′ ends of the coding region of MAL11. Normally, selection for the presence of the Cas9/gRNA expression plasmid is sufficient to obtain accurate gene editing in almost 100% of transformed cells without the need of a selection marker incorporated in the repair fragment in Saccharomyces yeast (3,8). In common laboratory strains, replacement of a sequence with a repair DNA can be detected by diagnostic PCR. However, in the hybrid strain IMS0408, multiple attempts failed to yield the expected fragments after transformation with the gRNA targeting MAL11 and a repair fragment. Therefore, the genomes of four random transformants, named IMX1421 to IMX1424, were sequenced using 150 bp paired-end Illumina reads and aligned to a haploid S. cerevisiae × S. eubayanus reference genome. While reads of strain IMS0408 aligned unambiguously to the MAL11 locus on chromosome VII of the S. cerevisiae sub-genome, MAL11 DNA was absent in transformants IMX1421-IMX1424. Absence of MAL11 was associated with loss of large regions of chromosome VII, ranging from 29 to 356 kbp (Figure 1). For IMX1422-IMX1424, the corresponding regions on the S. eubayanus chromosome VII devoid of MAL11 ortholog showed double sequence coverage, indicating that targeting of MAL11 using Cas9 resulted in replacement of varying regions of the targeted S. cerevisiae chromosome by regions from the corresponding S. eubayanus chromosome (Supplementary Figure S1). The recombination in IMX1423 occurred between the S. cerevisiae HSV2 gene and its S. eubayanus HSV2 ortholog. The recombination events in IMX1422 and IMX1424 both occurred between the S. cerevisiae YGR125W gene and its S. eubayanus YGR125W ortholog. The exact coordinates of the recombination within YGR125W were separated by more than 1000 nucleotides. For IMX1421, the loss of S. cerevisiae chromosome VII started at the IMA2 gene. However, the presence of other IMA genes with high identity to IMA2 prevented unique read alignment. Therefore, read pairing information did not reveal with which sequence the right arm of chromosome VII was replaced. In addition, the subtelomeric position of other IMA genes in the genome prevented identification of this duplicated sequence by sequencing coverage analysis, as sequencing depth is highly irregular in subtelomeric regions due to the abundance of repetitive elements (17). While the MAL11 locus was targeted in all four strains, the recombination events leading to LOH occurred at four unique loci. The distance of these loci to the targeted site varied between 7 and 334 kbp, possibly reflecting different degrees of DNA resection at the DSB site. The sequence similarities of the S. cerevisiae and S. eubayanus orthologs of HSV2 and YGR125W were 80% and and 82%, which is lower than the average 85% identity between the two subgenomes. This observation indicates that recombination events did not only occur in regions with particularly high homology. It should be noted that in IMX1422, LOH did not only affect the right arm of S. cerevisiae chromosome VII, but also the first 530 kbp of the left arm of S. cerevisiae chromosome VII (Figure 1). Since no segmental aneuploidies were observed on non-targeted chromosomes in IMX1421-IMX1424 (Supplementary Figure S1), the observed LOH is likely due to the targeting of MAL11. These results indicated that genome editing using Cas9 caused LOH rather than the intended gene editing when targeting a locus present on just one of two homologous chromosomes in a heterozygous yeast.
Figure 1.
Loss of heterozygosity observed by whole genome sequencing upon Cas9-targeting of MAL11 on the Saccharomyces cerevisiae derived chromosome VII in the S. cerevisiae × S. eubayanus hybrid IMS0408. IMS0408 was transformed with a 120 bp repair fragment with 60 bp flanks corresponding to the sequence upstream and downstream of the MAL11 ORF, and with plasmid pUDP045 expressing Cas9 and a gRNA targeting the S. cerevisiae specific gene MAL11 gene. Upon plating on selective medium, four randomly picked colonies were selected and sequenced using 150 bp pair-end reads and mapped against a reference genome composed of chromosome level assemblies of S. cerevisiae and of S. eubayanus. The centromere and targeted gene MAL11 are shown at their exact coordinates, but their size is not at scale. Loss of heterozygosity is shown in red and was defined as regions in which reads did not align to the S. cerevisiae reference chromosome VII while reads aligned to the corresponding region on the S. eubayanus reference chromosome VII with approximately double the normal coverage.
Targeting of heterozygous loci in a mostly homozygous diploid S. cerevisiae strain
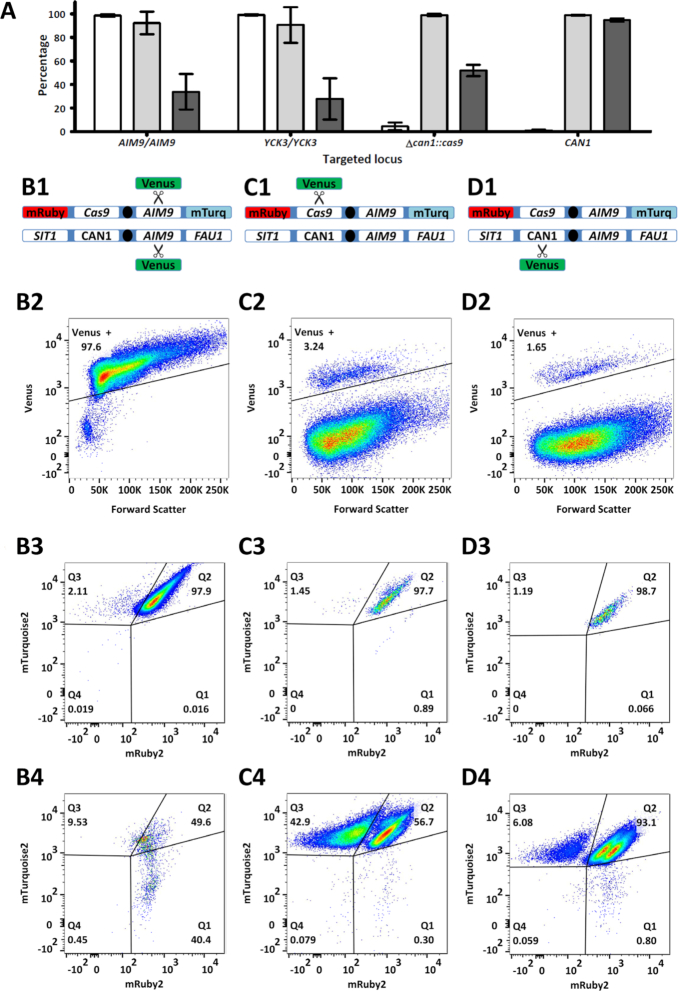
To investigate if the observed lack of efficient gene editing was specific to this highly heterozygous S. cerevisiae × eubayanus hybrid, we systematically investigated the impact of target-sequence heterozygosity on the efficiency of gene editing in S. cerevisiae strains. To this end, DSBs were introduced at homozygous and heterozygous loci on chromosome V of several strains that carried a Cas9 expression cassette integrated at the CAN1 locus. Plasmid-based gRNA expression was performed as described previously (3). Use of a repair fragment expressing the fluorescent protein Venus enabled analysis of editing efficiency by flow cytometry (22). To verify functional Cas9 and gRNA expression, the Δcan1::Spcas9 locus was first targeted in the haploid S. cerevisiae strain IMX1555, resulting in integration of the repair fragment in 98.3 ± 1.3% of cells (Supplementary Table S1). Subsequently, the homozygous alleles of AIM9 and YCK3 were targeted in the congenic diploid S. cerevisiae strain IMX1557, resulting in integration of the repair fragment in 98.6 ± 0.8% and 99.2 ± 0.4% of cells, respectively (Figure 2A). In contrast, when individually editing each allele of the heterozygous CAN1/Δcan1::cas9 locus in the diploid strain IMX1557, the repair fragment was integrated in only 4.4 ± 2.5% of cells when targeting the Δcan1::cas9 allele, and 0.9 ± 0.6% of the cells when targeting the CAN1 allele (Figure 2A). These results indicated that gene editing efficiencies were up to 99-fold lower for heterozygous target loci than for homozygous target loci (P < 10−4). Since IMX1557 was homozygous in most of its genome, except the targeted locus, the introduction of a DSB in only one of two homologous chromosomes rather than genome heterozygosity itself, impeded accurate and efficient gene editing using Cas9.
Figure 2.
Cas9-mediated gene editing of homozygous and heterozygous loci on chromosome V of S. cerevisiae. (A) Average fluorescence of cell populations in which the homozygous AIM9 and YCK3 alleles and the heterozygous cas9 and CAN1 alleles were targeted in the diploid strain IMX1557. The percentage of cells expressing Venus (white), the percentages of cells expressing both mTurquoise2 and mRuby2 in the Venus positive (light gray) and in Venus negative cells (dark gray) are shown. For each target, averages and standard deviation for biological triplicates are shown. When targeting the hemizygous CAN1 allele, LOH manifests itself in a lower percentage of cells expressing Venus, but it does not affect mTurquoise2 and mRuby2 fluorescence, as these fluorophores are located on the non-targeted chromosome copy. (B–D) Fluorescence profiles obtained when targeting AIM9, cas9 and CAN1 in IMX1557. (row 1) Schematic representation of both copies of chromosome V in IMX1557, with the alleles at the SIT1, CAN1, AIM9 and FAU1 loci and scissors indicating Cas9 targeting. While one chromosome copy has the wildtype alleles for all loci, the other copy has mRuby2 integrated in SIT1, cas9 integrated in CAN1 and mTurquoise2 integrated in FAU1. (rows 2, 3 and 4) Flow cytometry profiles of targeted cells. Each gene was targeted in three biological replicates and flow cytometric data for a representative replicate is shown. After transformation, 100 000 cells were analysed by flow cytometry and single cells were selected based on a FSC-A/FSC-H plot to avoid multicellular aggregates. For each replicate, at least 75 000 single cells remained and the fluorescence corresponding to Venus was used to determine gene-editing efficiency (row 2). For each gene, the fluorescence corresponding to mRuby2 and mTurquoise2 is plotted for the cells with expression of Venus (row 3) and for cells without expression of Venus (row 4). Fluorescence results for all samples are provided in Supplementary Table S1.
To further investigate if Cas9 gene editing resulted in LOH, as observed in the hybrid IMS0408, the presence of both chromosome arms of the targeted chromosome homolog was monitored by flow cytometry. IMX1557 expressed the fluorophores mRuby2 and mTurquoise2 from the SIT1 and FAU1 loci of the chromosome V copy harboring the Δcan1::cas9 allele, but not from the non-modified homologous chromosome (Figure 2, B1–D1). Loss of the left and right arms of the copy of chromosome V harboring Δcan1::cas9 could therefore be monitored by measuring fluorescence corresponding to respectively mRuby2 and mTurquoise2 (22). For all targeted loci, when the expression of Venus indicated correct gene-editing, over 97.7% of cells expressed both mTurquoise2 and mRuby2 (Figure 2, panels A and B3–D3). However, when targeting the Δcan1::cas9 allele on the chromosome harboring mRuby2, 42.9% of cells which did not integrate Venus had lost mRuby2 fluorescence, while mTurquoise2 was still expressed (Figure 2, quadrant Q3 in panel C4). These results indicated that targeting of the heterozygous Δcan1::cas9 allele resulted in LOH of the targeted chromosome arm harboring mRuby2, but did not affect the opposite chromosome arm. In addition, when targeting the CAN1 allele on the chromosome without mRuby2, an additional population expressing both mRuby2 and mTurquoise2 emerged among the cells which did not integrate Venus (Figure 2, quadrant Q2 in panel D4). Within quadrant Q2, the two adjacent populations had the same average mTurquoise2 fluorescence, but their average mRuby2 fluorescence differed by a factor of 2. The difference in fluorescence suggested a duplication of mRuby2, consistent with replacement of the targeted non-fluorescent chromosome by an additional copy of the chromosome harboring mRuby2. Loss of mRuby2 fluorescence upon transformation with a gRNA targeting Δcan1::cas9 and doubling of mRuby2 fluorescence when targeting CAN1 were also observed in the absence of a co-transformed repair fragment (Supplementary Table S1). These results indicated that introduction of a DSB at a heterozygous locus caused LOH through replacement of a targeted chromosome segment by duplication of the corresponding segment from its homologous chromosome, as was observed when targeting MAL11 in the S. cerevisiae x eubayanus hybrid IMS0408.
Elucidation of genetic changes caused by Cas9-targeting using whole genome sequencing
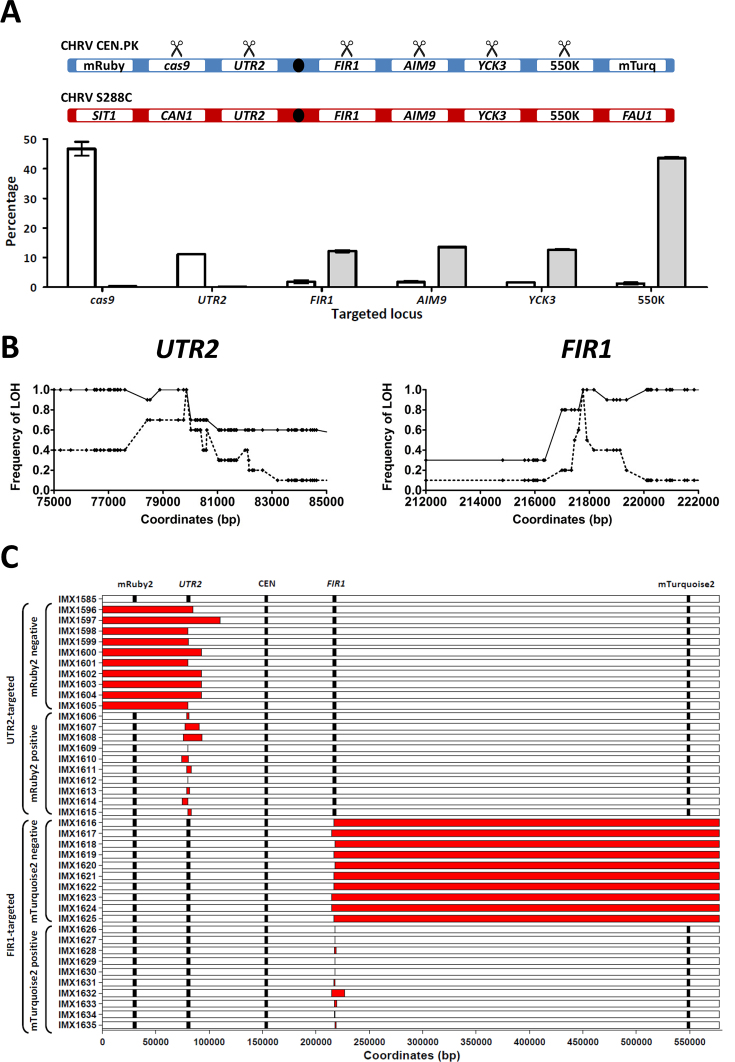
Chromosome-arm LOH has previously been reported upon introduction of a DSB in one of two homologous chromosomes, but was considered rare and has not been described as disruptive to gene-editing approaches (9,13,32). To investigate the extent and nature of the LOH caused by Cas9-editing of heterozygous loci, a strain with an average of four heterozygous SNPs or INDELs per kbp was generated by mating IMX1555 (CEN.PK genetic background, expressing Cas9, mRuby2 and mTurquoise2 from chromosome V) with S288C (Supplementary Table S6). LOH could be monitored at the chromosome arm level by flow cytometry and at the nucleotide level by whole-genome sequencing. By using PAM sequences absent in S288C, we specifically targeted the CEN.PK-derived chromosome V, which carried expression cassettes for mRuby2 and mTurquoise2 on its left and right arms, respectively, at the CAN1, UTR2, FIR1, AIM9 and YCK3 loci and at intergenic coordinate 549603, referred to as 550K. Upon targeting of the CAN1 and UTR2 loci, mRuby2 fluorescence was lost in 46.7 ± 2.4 and 11.2 ± 0.2% of cells, respectively, while mTurquoise2 fluorescence was unaffected in at least 99.6 ± 0.2% of the cells (Figure 3A). Targeting of the FIR1, AIM9, YCK3 or 550K loci caused loss of mTurquoise2 fluorescence in 12.2 ± 0.4, 13.6 ± 0.1, 12.7 ± 0.2 and 43.6 ± 0.3% of cells, respectively, while mRuby2 fluorescence was conserved in at least 98.1 ± 0.5% of cells (Figure 3A). As the centromere is located between UTR2 and FIR1, these results confirm that, for all investigated loci, a large fraction of cells lost the targeted chromosome arm. Fluorescence-activated cell sorting (FACS) was subsequently used to isolate 10 single cells each from the following populations: UTR2-targeted cells with mRuby2 fluorescence (IMX1606-IMX1615), UTR2-targeted cells without mRuby2 fluorescence (IMX1596-IMX1605), FIR1-targeted cells with mTurquoise2 fluorescence (IMX1626-IMX1635), and FIR1-targeted cells without mTurquoise2 fluorescence (IMX1616-IMX1625). Whole-genome sequencing and alignment of reads to the CEN.PK113-7D genome sequence (17) revealed LOH of the targeted locus in all 40 isolates (Figure 3B). In cell lines that did not lose a fluorophore, LOH was local, affecting regions ranging from 3 to 17 495 nucleotides for UTR2-targeted cells and regions ranging from 1 to 11 900 nucleotides for FIR1-targeted cells, corresponding to up to 79 heterozygous nucleotides (Figure 3C and Supplementary Table S2). In isolates that did lose a fluorophore, LOH affected the chromosome arm harboring the targeted locus, affecting 79 859 to 110 289 nucleotides for UTR2-targeted cells and 359 841 to 362 790 nucleotides for FIR1-targeted cells, corresponding to up to 1697 heterozygous nucleotides (Figure 3C and Supplementary Table S2). Absence of newly introduced SNPs at targeted loci indicated that repair of DSBs did not involve non-homologous end joining (33).
Figure 3.
Loss of heterozygosity caused by Cas9-mediated gene editing at heterozygous loci in the heterozygous S. cerevisiae diploid IMX1585. Mating of the haploid S. cerevisiae strains IMX1555 (CEN.PK-derived) and S288C yielded the heterozygous diploid strain IMX1585 (on average four heterozygous nucleotides per kbp on chromosome V). The CEN.PK-derived chromosome harbors the fluorophores mRuby2 and mTurquoise2, enabling detection of the loss of each arm of the CEN.PK-derived chromosome V by flow cytometry. DSBs were introduced specifically in the CEN.PK–derived chromosome and loss of heterozygosity was monitored at the population level using flow cytometry and in single cell isolates by whole genome sequencing. (A) Population-level loss of heterozygosity after targeting cas9, UTR2, FIR1, AIM9, YCK3 and 550K in IMX1585. The targeted loci on the CEN.PK-derived and S288C-derived chromosome V of IMX1585 are represented schematically. The SIT1, CAN1, UTR2, FIR2, AIM9, YCK3, 550K and FAU1 loci and the centromeres are indicated. The CEN.PK-derived chromosome harbors mRuby2 at the SIT1 locus and mTurquoise2 at the FAU1 locus. Scissors indicate CEN.PK-derived alleles which were specifically targeted using Cas9. In the graph, the percentage of cells having lost mRuby2 fluorescence (white) and mTurquoise2 (gray) is shown for each targeted locus. Averages and standard deviations were calculated from biological triplicates. (B) Loss of heterozygosity at the nucleotide level in single isolates obtained by targeting UTR2 and FIR1 in IMX1585. For both targeted loci, the frequency at which LOH was observed for each heterozygous nucleotide in a 10 kbp region around the targeted site was determined by whole genome sequencing. For UTR2, the 79,857th nucleotide was targeted and frequencies are indicated for 10 isolates with intact fluorescence (dashed line, isolates IMX1606-IMX1615) and 10 isolates having lost fluorescence corresponding to mRuby2 (continuous line, IMX1596-IMX1605). For FIR1, the 217,767th nucleotide was targeted and frequencies are indicated for 10 isolates with intact fluorescence (dashed line, isolates IMX1626-IMX1635) and 10 isolates having lost fluorescence corresponding to mTurquoise2 (continuous line, IMX1616-IMX1625). (C) Loss of heterozygosity at the chromosome scale in single isolates obtained by targeting UTR2 and FIR1 in IMX1585. Whole genome sequencing data was used to identify regions of chromosome V affected by loss of heterozygosity in isolates after targeting of UTR2 (IMX1596-IMX1615) and of FIR1 (IMX1616-IMX1635). For each strain, the fluorophores mRuby2 and mTurquoise2, the targeted genes UTR2 and FIR1 and the centromere are shown at their exact coordinates, but their size is not at scale. Loss of heterozygosity was defined as regions in which nucleotides which were heterozygous in IMX1585 were no longer heterozygous in the isolate (in red). In isolates which lost the fluorophore mRuby2 after targeting of UTR2 or which lost the fluorophore mTurquoise2 after targeting of FIR1, entire chromosome arms were affected by LOH. Exact coordinates are provided in Supplementary Table S2.
Identification of repair patterns corresponding to homology-directed repair
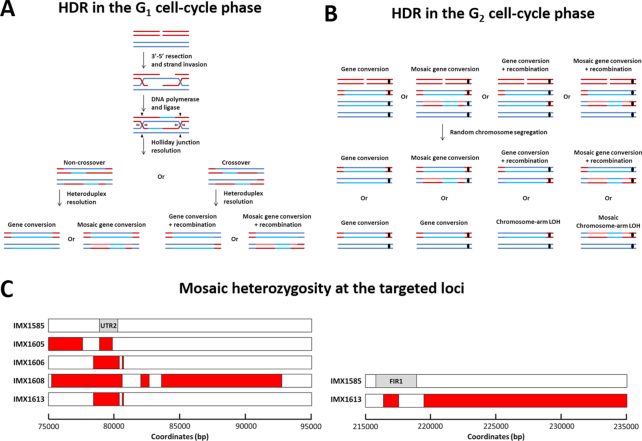
We conclude that introduction of a DSB at a heterozygous locus results in low gene-editing efficiencies due to a competing repair mechanism that causes local or chromosome-arm LOH. In eukaryotes, repair using homologous chromosomes typically relies on BIR or HR (34), which occur by distinct mechanisms and yield different results (10–12). In the case of BIR, the entire targeted chromosome arm is lost and an additional copy of its homolog is generated from the 5′ strand by replication, using the homolog as a polymerase template. Depending on the degree of strand resection prior to BIR, this mechanism results in complete loss of heterozygosity for varying portions of the targeted chromosome arm, including the locus in which a DSB was introduced (35). In the case of HR, the DSB is repaired by strand invasion, strand elongation, ligation, Holiday junction resolution and heteroduplex resolution (Figure 4A). The Holiday junction can be resolved by crossover (CO), resulting in gene conversion with a chromosomal recombination, or by non-crossover (NCO), resulting in gene conversion only (Figure 4A). In addition the resolution of heteroduplex DNA can result in mosaic LOH patterns due to a combination of gene conversion and some restoration (Figure 4A). Such mosaic patterns can also result from template switching during repair synthesis. Of the strains sequenced in this study, IMX1606-IMX1615 and IMX1626-IMX1635 lost heterozygosity only in the region surrounding the targeted DSB, indicating HR had occurred (Figure 3). In these strains, mosaic patterns resulting from heteroduplex resolution were observed in strains IMX1606, IMX1608 and IMX1613 (Figure 4C and Supplementary Table S2). Since strains IMX1596-IMX1605 and strains IMX1616-1625 lost heterozygosity of entire chromosome arms (Figure 3), repair could have occurred by BIR. However, mosaic patterns corresponding to heteroduplex resolution were observed in strains IMX1605 and IMX1619 (Figure 4C and Supplementary Table S2). While BIR does not cause mosaic LOH, chromosome-arm LOH is not a commonly-recognized result of HR (Figure 4A) (10–12). Therefore, we propose a repair mechanism that involves HR of at least one of the targeted chromatids at the G2 stage of the cell cycle (Figure 4B). The proposed mechanism would result in daughter cells with either local LOH or chromosome-arm LOH, with and without mosaic heterozygosity at the targeted locus. The proposed mechanism is consistent with all phenotypes and genotypes encountered in this study as well as in previous studies involving hemizygous introduction of DSBs (9,13–16,32,36). While HR at the G2 stage of the cell cycle could explain all observed genotypes, HR at the G1 stage and BIR could also contribute.
Figure 4.
Proposed mechanism for Cas9-mediated loss of heterozygosity in a diploid genome based on homologous recombination (HR) between homologous chromosomes. (A) Possible outcomes of HR in cells with one chromosome complement during the G1 stage of the cell cycle. (B) Possible outcomes of HR in cells with two chromosome complements during the G2 stage of the cell cycle. The targeted chromosome (red), its homolog (blue) and the centromere are indicated (black, where relevant). Newly synthesized DNA is shown in a lighter shade. Heteroduplex resolution occurs prior to chromatid segregation and the strand with the targeted NGG PAM sequence is always discarded due to Cas9 activity. For HR during the G2 stage of the cell cycle, HR occurs between one chromatid of the targeted and one chromatid of the non-targeted chromosome, as with HR during the G1 stage. The chromatids subsequently segregate according to their centromere pairing, with one red and one blue centromere in each daughter cell. For simplicity, only repair of one targeted chromatid is shown in the figure. Repair of both targeted sister chromatids results in the same genome alterations as shown here. As indicated in the figure, HR during the G2 stage of the cell cycle could yield local as well as chromosome-arm LOH by mitotic crossover, both with and without mosaic structures. (C) Mosaic loss of heterozygosity at the targeted loci in single isolates obtained by targeting UTR2 and FIR1 in IMX1585. Strains with mosaic loss of heterozygosity are at the UTR2 locus (left) and at the FIR1 locus (right) are indicated. The location of the ORFs is indicated in IMX1585 (gray). For each strain, heterozygous sequence is indicated in red. Exact coordinates of heterozygous nucleotides are indicated in Supplementary Table S2.
DISCUSSION
The efficiency of gene editing using Cas9 can decrease by almost two orders of magnitude when targeting only one of two homologous chromosomes due to a competing repair mechanism causing either local or chromosome-arm scale LOH. In previous work, Cas9-mediated gene editing was reported to cause large deletions at the targeted loci (37), which sometimes resulted in loss of heterozygosity. Here, the observed LOH consisted not only of loss of genetic material from the targeted chromosome, but also of replacement of the affected sequence by an additional copy of sequence homologous to the targeted site. While such LOH upon introduction of a hemizygous DSB has been observed in the yeasts S. cerevisiae and Candida albicans (9,13), this study demonstrates that repair by LOH is not only possible, but occurs at rates which impede gene editing approaches based on integration of repair fragments. Gene-editing was similarly inhibited in an S. cerevisiae diploid with 99% homozygosity and in an interspecies S. cerevisiae × eubayanus hybrid with 85% homozygosity. In addition, the recombination events occurred at loci with homologies as low as 80%. The lack of necessity for high identity suggests that Cas9-mediated gene editing may also cause LOH by translocations resulting from recombination events between paralogous genes. However, such translocations were not observed in this study, and Cas9-mediated gene-editing has been applied successfully to delete various paralogs without resulting in translocations (38,39).
Cas9-mediated LOH is likely to contribute to a lesser genome accessibility of heterozygous yeasts relative to laboratory strains, which tend to be haploid or homozygous. Therefore, these results are likely to affect the genome editing of hybrids, industrial yeasts and natural isolates due to their frequent heterozygosity (40), and should be used to update guidelines for designing gene editing strategies. We strongly recommend to design gRNAs targeting homozygous nucleotides stretches when targeting heterozygous genomes. When allele-specific gene editing is required, we recommend the use of repair fragments with integration markers such as the Venus fluorophore in this study, since accurate gene editing is not impossible, simply inefficient. When the use of a marker is not permissible, extensive screening of transformants for correct gene editing may be required.
While the HDR machinery is well conserved in eukaryotes (11,12), further research is required to determine if LOH occurs at similar rates in eukaryotes other than S. cerevisiae, and if it impedes gene editing. While DSB-mediated LOH was observed in S. cerevisiae, C. albicans, Drosophilia melanogaster and Mus musculus (9,13,32,36), relative contributions of HR, BIR and NHEJ to DSB repair vary across species. However, since integration of a repair fragment and repair by LOH both involve HR (41,42), targeting heterozygous loci likely causes low gene-editing efficiencies and LOH in other eukaryotes as well, regardless of the efficiency of NHEJ.
Targeting of heterozygous loci is common in gene editing, for example during allele propagation of gene drives and disease allele correction in human gene therapy (41,42). Although gene drives are based on LOH by HR (42), the extent of LOH beyond the targeted locus has not been systematically studied but could, by analogy with the present study, potentially affect entire chromosome arms. Allele-specific gene editing generally aims at repair by HR using a co-transformed repair fragment instead of a homologous chromosome. Reports of LOH after targeting a heterozygous allele in human embryos despite availability of an adequate repair fragment, are consistent with Cas9-induced LOH extending beyond the targeted locus, as described here (41). While, in the human-embryo study, repair by LOH was perceived as a success, the reported role of LOH in cancer development (43) indicates that large-scale LOH can have important phenotypic repercussions. Therefore we recommend avoiding allele-specific gene editing when possible until further research determines if it is a risk in other eukaryotes. Based on the proposed HR mechanism for CRISPR/Cas9-mediated LOH (Figure 4B), the risk of LOH can be mitigated by designing gRNAs that cut all alleles of heterozygous loci, even if only a single allele needs to be edited. Eventually, CRISPR–Cas9 editing could become safer by favouring DSB-independent gene-editing methods such as guided nickases and base-editing strategies for preventing or reducing the incidence of LOH (44–47).
DATA AVAILABILITY
The sequencing data were deposited at NCBI (https://www.ncbi.nlm.nih.gov/) under the Bioproject PRJNA471787.
Supplementary Material
ACKNOWLEDGEMENTS
ARGdV conceived the study and designed the experiments. ARGdV and LGFC performed plasmid and strain construction. ARGdV, LGFC, PdlTC and JtH performed the experimental work. ARGdV and MvdB performed bioinformatics analysis. ARGdV, JTP and JMGD supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
We thank Liset Jansen for drawing our attention to the difficulty to edit a heterozygous gene, Robert Mans for his expertise with gene editing in Saccharomyces cerevisiae, Melanie Wijsman for constructing and Pascale Daran-Lapujade for sharing plasmids pUDE480, pUDE481 and pUDE482, Sai T. Reddy for his insights in the potential impact for human gene therapy and Nick Brouwers, Alex Salazar, Xavier D.V. Hakkaart, Ioannis Papapetridis, Niels G.A. Kuijpers, Jan-Maarten Geertman and Thomas Abeel for their critical input.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
BE-Basic R&D Program (grant numbers TKIBE01003 and TKIBE01001, http://www.be-basic.org/), which was granted a TKI-subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). Funding for open access charge: BE-Basic.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sander J.D., Joung J.K.. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014; 32:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mans R., van Rossum H.M., Wijsman M., Backx A., Kuijpers N.G., van den Broek M., Daran-Lapujade P., Pronk J.T., van Maris A.J.A., Daran J.-M.G.. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015; 15:fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DiCarlo J.E., Norville J.E., Mali P., Rios X., Aach J., Church G.M.. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013; 41:4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klein M., Eslami-Mossallam B., Arroyo D.G., Depken M.. Hybridization kinetics explains CRISPR-Cas Off-Targeting rules. Cell Rep. 2018; 22:1413–1423. [DOI] [PubMed] [Google Scholar]
- 6. Jasin M., Rothstein R.. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013; 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuijpers N.G.A., Solis-Escalante D., Luttik M.A., Bisschops M.M.M., Boonekamp F.J., van den Broek M., Pronk J.T., Daran J.-M., Daran-Lapujade P.. Pathway swapping: Toward modular engineering of essential cellular processes. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:15060–15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorter de Vries A.R., de Groot P.A., van den Broek M., Daran J.-M.G.. CRISPR–Cas9 mediated gene deletions in lager yeast Saccharomyces pastorianus. Microb. Cell. Fact. 2017; 16:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sadhu M.J., Bloom J.S., Day L., Kruglyak L.. CRISPR-directed mitotic recombination enables genetic mapping without crosses. Science. 2016; 352:1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li X., Heyer W.-D.. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008; 18:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haber J.E. Partners and pathways: repairing a double-strand break. Trends Genet. 2000; 16:259–264. [DOI] [PubMed] [Google Scholar]
- 12. Moynahan M.E., Chiu J.W., Koller B.H., Jasin M.. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999; 4:511–518. [DOI] [PubMed] [Google Scholar]
- 13. Feri A., Loll-Krippleber R., Commere P.-H., Maufrais C., Sertour N., Schwartz K., Sherlock G., Bougnoux M.-E., d’Enfert C., Legrand M.. Analysis of repair mechanisms following an induced double-strand break uncovers recessive deleterious alleles in the Candida albicans diploid genome. MBio. 2016; 7:e01109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Charles J.S., Hazkani-Covo E., Yin Y., Andersen S.L., Dietrich F.S., Greenwell P.W., Malc E., Mieczkowski P., Petes T.D.. High-resolution Genome-wide Analysis of Irradiated (UV and gamma rays) diploid yeast cells reveals a high frequency of genomic Loss of Heterozygosity (LOH) events. Genetics. 2012; 190:1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Argueso J.L., Westmoreland J., Mieczkowski P.A., Gawel M., Petes T.D., Resnick M.A.. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hum Y.F., Jinks-Robertson S.. Mitotic gene conversion tracts associated with repair of a defined double-strand break in Saccharomyces cerevisiae. Genetics. 2017; 207:115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salazar A.N., Gorter de Vries A.R., van den Broek M., Wijsman M., de la Torre Cortés P., Brickwedde A., Brouwers N., Daran J.-M.G., Abeel T.. Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN. PK113-7D. FEMS Yeast Res. 2017; 17:fox074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goffeau A., Barrell B.G., Bussey H., Davis R., Dujon B., Feldmann H., Galibert F., Hoheisel J., Jacq C., Johnston M.. Life with 6000 genes. Science. 1996; 274:546–567. [DOI] [PubMed] [Google Scholar]
- 19. Verduyn C., Postma E., Scheffers W.A., van Dijken J.P.. Physiology of Saccharomyces Cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 1990; 136:395–403. [DOI] [PubMed] [Google Scholar]
- 20. Pronk J.T. Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 2002; 68:2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Solis-Escalante D., Kuijpers N.G., Nadine B., Bolat I., Bosman L., Pronk J.T., Daran J.-M.G., Daran-Lapujade P.. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 2013; 13:126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E.. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 2015; 4:975–986. [DOI] [PubMed] [Google Scholar]
- 23. Mans R., Wijsman M., Daran-Lapujade P., Daran J.-M.G.. A protocol for introduction of multiple genetic modifications in Saccharomyces cerevisiae using CRISPR/Cas9. FEMS Yeast Res. 2018; 18:foy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engler C., Kandzia R., Marillonnet S.. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008; 3:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gietz R.D., Woods R.A.. Methods Enzymol. 2002; 350:Elsevier; 87–96. [DOI] [PubMed] [Google Scholar]
- 26. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K.. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brickwedde A., Brouwers N., van den Broek M., Gallego Murillo J.S., Fraiture J.L., Pronk J.T., Daran J.-M.G.. Structural, physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357T. Front. Microbiol. 2018; 9:1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hebly M., Brickwedde A., Bolat I., Driessen M.R.M., de Hulster E.A.F., van den Broek M., Pronk J.T., Geertman J.-M., Daran J.-M.G., Daran-Lapujade P.. S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 2015; 15:fov005. [DOI] [PubMed] [Google Scholar]
- 32. Heinze S.D., Kohlbrenner T., Ippolito D., Meccariello A., Burger A., Mosimann C., Saccone G., Bopp D.. CRISPR–Cas9 targeted disruption of the yellow ortholog in the housefly identifies the brown body locus. Sci. Rep. 2017; 7:4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R.. Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543. [DOI] [PubMed] [Google Scholar]
- 34. Jasin M., Haber J.E.. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair. 2016; 44:6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Llorente B., Smith C.E., Symington L.S.. Break-induced replication: what is it and what is it for. Cell cycle. 2008; 7:859–864. [DOI] [PubMed] [Google Scholar]
- 36. Henson V., Palmer L., Banks S., Nadeau J.H., Carlson G.A.. Loss of heterozygosity and mitotic linkage maps in the mouse. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:6486–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wijsman M., Świat M.A., Marques W.L., Hettinga J.K., van den Broek M., de la Torre Cortés P., Mans R., Pronk J.T., Daran J.-M.G., Daran-Lapujade P.. A toolkit for rapid CRISPR-SpCas9 assisted construction of hexose-transport-deficient Saccharomyces cerevisiae strains. FEMS Yeast Res. 2018; 19:foy107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marques W.L., Mans R., Marella E.R., Cordeiro R.L., van den Broek M., Daran J.-M.G., Pronk J.T., Gombert A.K., van Maris A.J.A.. Elimination of sucrose transport and hydrolysis in Saccharomyces cerevisiae: a platform strain for engineering sucrose metabolism. FEMS Yeast Res. 2017; 17:fox006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gorter de Vries A.R., Pronk J.T., Daran J.-M.G.. Industrial relevance of chromosomal copy number variation in Saccharomyces yeasts. Appl. Environ. Microbiol. 2017; 83:e03206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma H., Marti-Gutierrez N., Park S.-W., Wu J., Lee Y., Suzuki K., Koski A., Ji D., Hayama T., Ahmed R.. Correction of a pathogenic gene mutation in human embryos. Nature. 2017; 548:413–419. [DOI] [PubMed] [Google Scholar]
- 42. Champer J., Buchman A., Akbari O.S.. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 2016; 17:146. [DOI] [PubMed] [Google Scholar]
- 43. Naylor S.L., Johnson B.E., Minna J.D., Sakaguchi A.Y.. Loss of heterozygosity of chromosome 3p markers in small-cell lung cancer. Nature. 1987; 329:451. [DOI] [PubMed] [Google Scholar]
- 44. Kim K., Ryu S.-M., Kim S.-T., Baek G., Kim D., Lim K., Chung E., Kim S., Kim J.-S.. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 2017; 35:435. [DOI] [PubMed] [Google Scholar]
- 45. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X.. Efficient genome modification by CRISPR–Cas9 nickase with minimal off-target effects. Nat. Methods. 2014; 11:399. [DOI] [PubMed] [Google Scholar]
- 47. Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y.. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data were deposited at NCBI (https://www.ncbi.nlm.nih.gov/) under the Bioproject PRJNA471787.