Abstract
Fungal tRNA ligase (Trl1) is an essential enzyme that repairs RNA breaks with 2′,3′-cyclic-PO4 and 5′-OH ends inflicted during tRNA splicing and non-canonical mRNA splicing in the fungal unfolded protein response. Trl1 is composed of C-terminal cyclic phosphodiesterase (CPD) and central GTP-dependent polynucleotide kinase (KIN) domains that heal the broken ends to generate the 3′-OH,2′-PO4 and 5′-PO4 termini required for sealing by an N-terminal ATP-dependent ligase domain (LIG). Here we report crystal structures of the Trl1-LIG domain from Chaetomium thermophilum at two discrete steps along the reaction pathway: the covalent LIG-(lysyl-Nζ)–AMP•Mn2+ intermediate and a LIG•ATP•(Mn2+)2 Michaelis complex. The structures highlight a two-metal mechanism whereby a penta-hydrated metal complex stabilizes the transition state of the ATP α phosphate and a second metal bridges the β and γ phosphates to help orient the pyrophosphate leaving group. A LIG-bound sulfate anion is a plausible mimetic of the essential RNA terminal 2′-PO4. Trl1-LIG has a distinctive C-terminal domain that instates fungal Trl1 as the founder of an Rnl6 clade of ATP-dependent RNA ligase. We discuss how the Trl1-LIG structure rationalizes the large body of in vivo structure–function data for Saccharomyces cerevisiae Trl1.
INTRODUCTION
Fungal tRNA ligase Trl1 is an essential agent of the repair of programmed tRNA and mRNA breaks with 2′,3′-cyclic phosphate and 5′-OH ends that are generated during tRNA splicing and non-canonical mRNA splicing in the fungal unfolded protein response (1,2). Trl1 executes three sequential repair reactions catalyzed by its three component enzymatic domains: (i) the 2′,3′-cyclic phosphate (>p) end is hydrolyzed to a 3′-OH,2′-PO4 by a C-terminal cyclic phosphodiesterase (CPD) module that belongs to the 2H phosphoesterase superfamily; (ii) the 5′-OH end is phosphorylated by a central GTP-dependent polynucleotide kinase module of the P-loop phosphotransferase superfamily; and (iii) the 3′-OH,2′-PO4 and 5′-PO4 ends are sealed by an N-terminal ATP-dependent RNA ligase module, of the covalent lysine nucleotidyltransferase superfamily, to form a 2′-PO4, 3′-5′ phosphodiester splice junction (3–9).
Fungal Trl1 is considered a promising target for anti-fungal drug discovery because: (i) its domain structure and biochemical mechanism are unique compared to the RtcB-type tRNA repair systems elaborated by metazoa, archaea, and many bacteria (10–18) and (ii) there are no homologs of the Trl1 ligase domain in mammalian proteomes. To fortify the case for Trl1 as a drug target, our laboratory has characterized Trl1 from the budding yeasts Saccharomyces cerevisiae and Kluyveromyces lactis and from several fungi that cause human disease: Aspergillus fumigatus, Candida albicans and Coccidioides immitis (5–9,19). The fungal tRNA ligases are biochemically very similar.
The lack of crystal structures of fungal Trl1 at key steps along the reaction pathway is a major knowledge gap in tRNA metabolism and an impediment to inhibitor design and discovery. We aim to close this gap by attaining structures of Trl1′s component domains. To that end, we recently reported the 2.2 Å crystal structure of the autonomous polynucleotide kinase (KIN) domain of C. albicans Trl1 in complex with GDP and magnesium (19). The Candida KIN structure revealed a distinctive ‘G-loop’ motif, conserved in other fungal tRNA ligases, that accounts for the shared GTP phosphate donor specificity of fungal Trl1 enzymes (5,8,9,19,20).
Our goal in the present study was to determine the structure of the RNA ligase (LIG) domain of a fungal Trl1. Sealing of RNA ends by Trl1-LIG requires ATP and proceeds via three divalent cation-dependent adenylate transfer steps. First, LIG reacts with ATP to form a covalent LIG-(lysyl-Nζ)–AMP intermediate and displace pyrophosphate. Second, LIG transfers AMP to the 5′-PO4 RNA terminus (that was generated by the Trl1 kinase) to form an RNA-adenylate intermediate (A5′pp5′RNA). Third, LIG directs the attack of an RNA 3′-OH on AppRNA to form the splice junction and displace AMP. A defining feature of Trl1-LIG and its plant homolog is the requirement for a 2′-PO4 to synthesize a 3′-5′ phosphodiester bond (3,21).
Here we present the crystal structure of a catalytically active LIG domain of Chaetomium thermophilum Trl1, as the covalent LIG-(lysyl-Nζ)–AMP intermediate in complex with a catalytic metal ion at the AMP phosphate and as a Michaelis complex with ATP and two bound metal ions. The structures illuminate the mechanism of Trl1 adenylylation, pinpoint the conserved and distinctive structural elements of Trl1 vis-à-vis other RNA ligases, suggest how the 2′-PO4 might be recognized, and rationalize a large body of mutational data that has been generated for the S. cerevisiae Trl1-LIG (5,6).
MATERIALS AND METHODS
Recombinant Chaetomium Trl1-FL and Trl1-LIG proteins
A synthetic codon-optimized open reading frame (ORF) encoding the full-length 846-amino acid Trl1 protein (Trl1-FL) from C. thermophilum DSM1495 (UniProtKB G0S6G2) was purchased from Genscript and inserted into the NdeI and XhoI sites of T7 RNA-polymerase-based expression plasmid pET28a to generate plasmid pET28-Trl1-FL. The N-terminal RNA ligase domain (LIG; aa 1–407) was PCR-amplified from the full-length ORF using primers designed to introduce a BamHI site at the translation start codon and a HindIII site immediately 3′ of the stop codon. The Trl1-LIG PCR product was digested with BamHI and HindIII and then inserted into BamHI/HindIII-cut pET28a to generate plasmid pET28-Trl1-LIG. The codon for the motif I lysine nucleophile (Lys148) was mutated in pET28-Trl1-LIG to introduce methionine in lieu of Lys148, thereby generating plasmid pET28-Trl1-LIG-K148M. The pET28-Trl1-FL, pET28-Trl1-LIG and pET28-Trl1-LIG-K148M plasmids encode the respective Trl1 polypeptides fused to an N-terminal His6 tag. The plasmid inserts were sequenced to confirm that no unwanted coding changes were introduced during PCR amplification and cloning.
pET28-Trl1-FL, pET28-Trl1-LIG, and pET28-Trl1-LIG-K148M were transformed into Escherichia coli BL21(DE3). Cultures (1000 ml) derived from single kanamycin-resistant transformants were grown at 37°C in LB medium containing 50 μg/ml kanamycin until the A600 reached 0.5 to 0.6, at which time the cultures were adjusted to 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) and then incubated for 16 h at 18°C with continuous shaking. Cells were harvested by centrifugation, and the pellets were resuspended in 25 ml lysis buffer (50 mM Tris–HCl, pH 8.0, 1 M NaCl, 20 mM imidazole, 10% glycerol) and stored at −80°C pending purification. All subsequent purification procedures were performed at 4°C. The thawed cell suspensions in lysis buffer were adjusted to 1 mg/ml lysozyme and supplemented with one protease inhibitor tablet (Roche) and incubated for 30 min. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation for 45 min at 14000 rpm. The soluble extract was mixed for 1 h with 5 ml of Ni-NTA agarose (Qiagen) that had been equilibrated in lysis buffer. The resin was collected by centrifugation and resuspended in 50 ml of buffer A (50 mM Tris–HCl, pH 8.0, 300 mM NaCl) containing 20 mM imidazole, then re-collected by centrifugation and suspended in 30 ml of 50 mM Tris–HCl, pH 8.0, 3 M KCl. The resin was recovered again and resuspended in 50 ml of buffer A containing 50 mM imidazole. The resin was then poured into a gravity flow column and the bound material was serially step-eluted with 150, 250 and 500 mM imidazole in buffer A. The elution profiles were monitored by SDS-PAGE. The peak Trl1-containing fractions were pooled, concentrated by centrifugal ultrafiltration, and then subjected to gel-filtration through a 120-ml Superdex 200 column that was equilibrated with 20 mM HEPES–NaOH, pH 7.0, 200 mM NaCl, 1 mM DTT, 5% glycerol. Peak fractions were pooled and concentrated by centrifugal ultrafiltration and stored at –80°C. Protein concentrations were determined by using the BioRad dye reagent with bovine serum albumin as the standard.
Preparation of 3′ 32P-labeled RNA2′p substrate
HORNA3′p oligonucleotides labeled with 32P at the penultimate phosphate were prepared by T4 Rnl1-mediated addition of [5′-32P]pCp to a 9-mer synthetic oligoribonucleotide. The 10-mer HORNA3′p oligonucleotide was treated with E. coli RNA 3′-terminal phosphate cyclase (RtcA) and ATP to generate the 2′,3′-cyclic phosphate derivative, HORNA>p, which was then converted to HORNA2′p by reaction with the 2′,3′-cyclic phosphodiesterase component of plant tRNA ligase AtRNL. The labeled RNA was gel-purified, eluted from an excised gel slice, and recovered by ethanol precipitation. The 32P-labeling and end-modification reactions were performed as described previously (12,21,22).
RNA ligase assay
Reaction mixtures (10 μl) containing 50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 100 μM ATP, 100 μM GTP, 0.5 pmol 32P-labeled 10-mer HORNA2′p substrate, and enzyme as specified were incubated at 37°C for 30 min. The reactions were quenched by adding 10 μl of 90% formamide, 50 mM EDTA. The products were analyzed by electrophoresis through a 40-cm 20% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris-borate, 2.5 mM EDTA). The radiolabeled RNAs were visualized by autoradiography.
Crystallization of Trl1-LIG and structure determination
Trl1-LIG crystals were grown at 22°C by sitting drop vapor diffusion. A 1 μl solution of Trl1-LIG (30 mg/ml) was mixed with an equal volume of precipitant solution containing 0.1 M Tris–HCl, pH 8.5, 0.2 M trimethylamine-N-oxide, 20% (w/v) PEG-MME-2000. Crystals appeared after 1 day. The crystals were transferred briefly to precipitant solution containing 25% glycerol prior to flash-freezing in liquid nitrogen. Selected crystals were soaked for 1 h in precipitant solution containing 25% glycerol and 1.5 mM thimerosal prior to freezing. Other crystals were soaked in precipitant solution containing 25% glycerol and 50 mM MnCl2 prior to freezing. X-ray diffraction data were collected from single crystals at the Advanced Photon Source beamline 24ID-C. Indexing and merging of the diffraction data were performed in HKL2000 (23). The phases were obtained in SHELX (24) by single isomorphous replacement with anomalous scattering (SIRAS) using data from single crystals of wild-type Trl1-LIG and a thimerosal-soaked Hg derivative, both in space group C2. Four heavy atom sites were identified. The phases were applied to solve the Trl1-LIG structure, using diffraction data at 2.5 Å resolution from a crystal of wild-type Trl1-LIG that had been soaked in manganese. Interactive model building was performed using O (25). Refinement was accomplished with Phenix (26). Data collection and refinement statistics are summarized in Table 1. The refined model (Rwork/Rfree = 0.212/0.256) comprised two LIG protomers in the asymmetric unit, each with AMP covalently attached to Lys148.
Table 1.
Crystallographic data and refinement statistics
| Native LIG | Hg derivative | LIG Mn2+ soak | LIG-K148M•ATP•Mn2+ | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | APS 24-ID-E | APS 24-ID-C | APS 24-ID-C | APS 24-ID-E |
| Space group | C 2 | C 2 | C 2 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 155.2, 114.2, 67.8 | 155.5, 112.5, 68.3 | 155.2, 114.1, 68.2 | 49.3, 56.7, 172.7 |
| α, β, γ (°) | 90, 100.8, 90 | 90, 99.9, 90 | 90, 100.4, 90 | 90, 109.1, 90 |
| Resolution (Å) | 50–2.4 (2.45–2.4) | 50–3.0 (3.05–3.0) | 50–2.5 (2.55–2.5) | 50–2.5 (2.55–2.5) |
| Wavelength (Å) | 0.9791 | 0.9778 | 0.9791 | 0.9791 |
| R pim | 0.056 (0.309) | 0.076 (0.571) | 0.052 (0.436) | 0.044 (0.436) |
| CC(1/2) | 0.993 (0.735) | 0.992 (0.560) | 0.996 (0.713) | 0.991 (0.827) |
| <I>/<σI> | 16.3 (1.9) | 12.9 (1.5) | 22.2 (2.0) | 21.4 (2.5) |
| Completeness (%) | 97.3 (92.5) | 99.1 (99.4) | 97.8 (98.3) | 99.9 (99.3) |
| Redundancy | 3.9 (3.1) | 4.3 (4.1) | 3.4 (3.4) | 7.6 (5.4) |
| Unique reflections | 43858 | 22933 | 39376 | 17102 |
| Phasing | ||||
| Heavy atom sites | 4 | |||
| Figure of merit | 0.577 | |||
| Refinement | ||||
| R work/Rfree | 0.212/0.256 | 0.178/0.247 | ||
| B-factors (Å2) average/Wilson | 61.7/47.2 | 51.0/42.1 | ||
| RMS deviations | ||||
| bond lengths (Å) | 0.01 | 0.007 | ||
| bond angles (°) | 1.24 | 1.01 | ||
| Ramachandran plot | ||||
| % favored | 95.3 | 95.2 | ||
| % allowed | 4.7 | 4.5 | ||
| outliers | 0 | 0.3 | ||
| Model contents | ||||
| Protomers/ASU | 2 | 1 | ||
| Protein residues | 788 | 383 | ||
| Ions | 2 Mn | 2 Mn, 1 sulfate | ||
| Water | 148 | 139 | ||
| PDB ID | 6N0V | 6N0T |
Values in parentheses refer to the highest resolution shell.
R free set consists of 10% of data chosen randomly against which structures were not refined.
Crystals of Trl1-LIG with ATP and metal cofactor were grown at 22°C by hanging drop vapor diffusion. A solution of Trl1-LIG-K148M (10 mg/ml) was adjusted to 1 mM ATP and 10 mM MnCl2 and incubated for 60 min on ice before aliquots (3 μl) were mixed with 3 μl of precipitant solution containing 0.1 M HEPES, pH 7.6, 2.5 M ammonium sulfate. Crystals grew overnight. Single crystals were transferred briefly to precipitant solution containing 1 mM ATP, 50 mM MnCl2, and 25% glycerol prior to flash-freezing in liquid nitrogen. Diffraction data at 2.5 Å resolution was collected from a single Trl1-LIG-K148M crystal in space group P212121. The structure was solved by molecular replacement using the structure of wild-type Trl1-LIG as the search model. Data collection and refinement statistics are compiled in Table 1. The refined model (Rwork/Rfree = 0.178/0.247) contained a single LIG-K148M protomer in the asymmetric unit, with ATP and two metal ions in the active site.
RESULTS AND DISCUSSION
Chaetomium thermophilum tRNA ligase
Full-length 846-aa Chaetomium Trl1 is composed of N-terminal ligase (LIG), central kinase (KIN), and C-terminal CPD domains (Figure 1A). We predicted a distal margin of the N-terminal LIG domain at aa 407 (Figure 1A), based on sequence alignments to other fungal Trl1s (6), previous genetic and biochemical characterization of active Trl1-LIG domains from other fungi (5,9), and the structurally defined proximal margin of the Candida Trl1 kinase domain (19). We produced recombinant Trl1-FL and Trl1-LIG in E. coli as His6 fusions and purified them from soluble extracts by sequential Ni-affinity chromatography and Superdex-200 gel filtration steps. Trl1-FL and Trl1-LIG eluted as monomers during gel filtration. SDS-PAGE indicated that whereas Trl1-LIG was highly pure with respect to the recombinant polypeptide (Figure 1B); the Trl1-FL preparation consisted of Trl1-FL as the major species, plus several smaller polypeptides as minority species (Figure 1B). The minor species co-eluted with Trl1-FL during gel filtration. Such impurities (perhaps arising via scission of Trl1 by E. coli proteases) have been noted for recombinant tRNA ligase enzymes from several fungal and plant sources (9,27).
Figure 1.
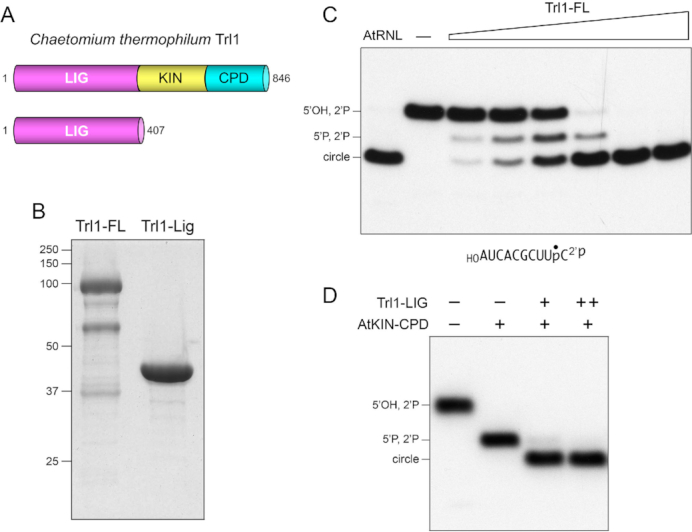
Chaetomium Trl1 ligase activity. (A) Cartoon depiction of the domain organization of full-length (FL) 846-aa C. thermophilum Trl1 consisting of N-terminal ligase (LIG), central kinase (KIN), and C-terminal cyclic phosphodiesterase (CPD) domains. An N-terminal 407-aa segment is shown here to comprise an autonomous LIG module. (B) Aliquots (10 μg) of the Superdex fractions of recombinant Trl1-FL and Trl1-LIG were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (C) RNA ligase reaction mixtures (10 μl) containing 50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 100 μM GTP, 100 μM ATP, 0.5 pmol (50 nM) 32P-labeled 10-mer HORNA2′p (shown at the bottom with the 32P-label indicated by •), and either no enzyme (lane –), 1.25 μg plant tRNA ligase (AtRNL), or 0.625, 1.25, 2.5, 5, 10 or 25 ng Trl1-FL (from left to right in the titration series) were incubated at 37°C for 30 min. The labeled RNAs were resolved by urea-PAGE and visualized by autoradiography. The positions of the 5′-OH, 2′-PO4 RNA substrate, 5′-PO4, 2′-PO4 kinase reaction product, and the 10-mer circle product of intramolecular ligation are indicated on the left. (D) Reactions mixtures (10 μl) containing 50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 2 mM DTT, 10 mM MgCl2, 100 μM GTP, 100 μM ATP, 0.5 pmol (50 nM) 32P-labeled 10-mer HORNA2′p, 250 ng AtKIN-CPD (where indicated by +), and 5 or 10 pg Trl1-LIG (where indicted by + and ++, respectively) were incubated at 37°C for 30 min. The labeled RNAs were resolved by urea-PAGE and visualized by autoradiography.
To assay the activity of Trl1-FL, we used a 10-mer RNA with 5′-OH and 2′-PO4 ends and a single radiolabel between the 3′-terminal and penultimate nucleosides (Figure 1C). The HORNA2′p substrate (50 nM) was reacted with increasing amounts of Trl1-FL, or with plant tRNA ligase AtRNL as a positive control, in the presence of 10 mM Mg2+, 0.1 mM ATP and 0.1 mM GTP. The products were analyzed by urea-PAGE. AtRNL completely converted the labeled HORNA>p substrate into a circular RNA product that migrated ahead of the substrate strand (Figure 1C). Trl1-FL effected a concentration-dependent conversion of HORNA2′p to circular product that was complete at saturating enzyme. At limiting enzyme concentration, Trl1-FL generated a mixture of species, consisting of the circular product and a 5′-phosphorylated pRNA2′p splicing intermediate formed by the Trl1 kinase domain that migrates between the HORNA2′p substrate and the product circle (Figure 1C).
Having verified the kinase and ligase activity of Trl1-FL, we proceeded to test the ligase activity of Trl1-LIG, in a reaction with the HORNA2′p substrate in which the requisite 5′ kinase activity was provided by the C-terminal end healing domain of plant tRNA ligase (AtKIN-CPD) (27). As expected, KIN-CPD per se converted the HORNA2′p substrate to a more rapidly migrating pRNA2′p species (Figure 1D). Inclusion of Trl1-LIG effected conversion of pRNA2′p to a ligated circle product (Figure 1D). Thus we affirmed that Trl1-LIG is an autonomous RNA ligase domain.
Crystal structures of Trl1-LIG
Crystals of wild-type Trl1-LIG were grown by sitting drop vapor diffusion after mixing a sample of the protein solution (30 mg/ml) with an equal volume of precipitant solution containing 0.1 M Tris–HCl, pH 8.5, 0.2 M trimethylamine-N-oxide, 20% (w/v) PEG-MME-2000. The crystals, cryo-protected as such or during a 1 h soak in 50 mM MnCl2, belonged to space group C2. SIRAS phasing was accomplished using diffraction data from a native crystal and a crystal that had been soaked in 1.5 mM thimerosal (Table 1). The refined model at 2.5 Å resolution of native manganese-soaked native Trl1-LIG contained two LIG protomers in the asymmetric unit, each of which had AMP linked covalently to Lys148. The A and B protomers in the asymmetric unit had virtually identical folds (rmsd = 0.6 Å at 395 Cα positions). Our discussion below of the LIG–AMP structure will focus on the B protomer of the LIG–AMP•Mn2+ crystal.
To capture Trl1-LIG as a Michaelis complex with ATP and metal cofactors, we mutated the Lys148 nucleophile to methionine (to prevent covalent adenylylation) and preincubated LIG-K148M (10 mg/ml) with 1 mM ATP and 10 mM MnCl2 prior to mixture with an equal volume of precipitant solution containing 0.1 M HEPES, pH 7.6, 2.5 M ammonium sulfate. Crystals grown by hanging drop vapor diffusion diffracted to 2.5 Å resolution and belonged to space group P212121 with one LIG protomer in the asymmetric unit. The refined model (Table 1), which is shown in stereo view in Figure 2A, contained one ATP and two manganese ions in the active site. The ‘Mn1’ ion engages the ATP α phosphate; the ‘Mn2’ ion bridges the β and γ phosphates. A sulfate anion is bound to the enzyme adjacent to the ATP•(Mn2+)2 complex (Figure 2A).
Figure 2.
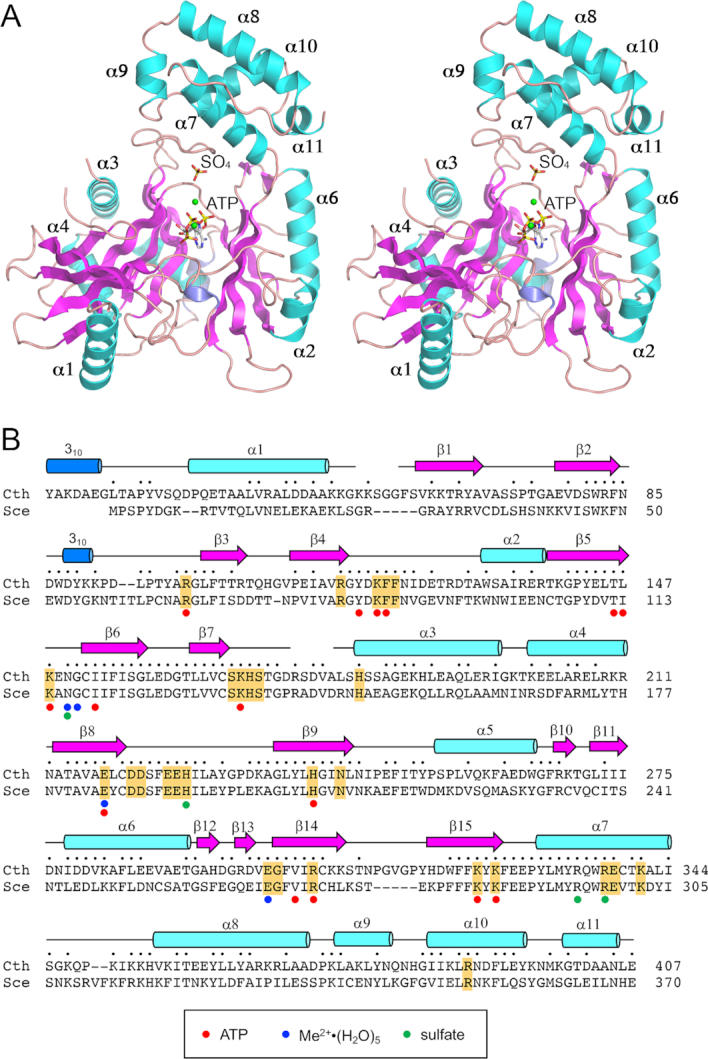
Structure of Trl1-LIG. (A) Stereo view of the LIG tertiary structure, depicted as a ribbon model with magenta β strands, cyan α helices (numbered sequentially), and blue 310 helices. The ATP in the active site and a nearby sulfate anion are rendered as stick models. Mn2+ ions are depicted as green spheres. (B) Secondary structure elements (colored as in panel A) are displayed above the C. thermophilum (Cth) Trl1-LIG primary structure, which is aligned to the primary structure of the LIG domain of S. cerevisiae (Sce) Trl1. Positions of amino acid side chain identity or similarity are indicated by dots above the Cth sequence. Gaps in the alignment are indicated by dashes. Amino acids in Trl1-LIG that make atomic contacts to ATP are denoted by red dots below the alignment. Amino acids that coordinate the hydrated metal complex are indicated by blue dots below the alignment. Amino acids that coordinate the sulfate anion are indicated by green dots below the alignment. The amino acids in S. cerevisiae Trl1-LIG that were identified by alanine scanning as essential for LIG activity in vivo (6) are highlighted in gold shading.
The modeled LIG protein in the Michaelis complex consists of three continuous polypeptide segments from Ser14 to Gly55, Phe61 to Asp174, and Leu180 to Leu406, organized as 15 β strands, 11 α helices, and two 310 helices. The secondary structure elements are displayed above the primary structure in Figure 2B. Alignment of the amino acid sequence of Chaetomium Trl1-LIG to that of the N-terminal LIG domain of S. cerevisiae Trl1 highlights 215 positions of side chain identity/similarity (Figure 2B). Chaetomium LIG has an N-terminal leader segment (aa 1–28) that is not present in Trl1 enzymes from S. cerevisiae (Figure 2B) and other model fungi (e.g. Candida, Aspergillus, etc.). This leader peptide in the structure adopts an extended conformation, punctuated by a 310 helix, that drapes across the back surface of the enzyme in the view in Figure 2A. The LIG tertiary structure consists of two domains: (i) an adenylyltransferase module (from aa 39 to 327, embracing α1 to β15) homologous to the N-terminal adenylyltransferase domain of T4 RNA ligase 1 (Rnl1) and (ii) a C-terminal module (aa 328–406, composed of helices α7 to α11) that is unique to Trl1-LIG.
The LIG adenylyltransferase domain is itself composed of two sub-domains. The segment from aa Gly117 to Glu328 is homologous to the core nucleotidyltransferase (NTase) domain found in ATP-dependent RNA ligases, ATP-dependent DNA ligases, NAD+-dependent DNA ligases, and GTP-dependent RNA capping enzymes that together comprise the covalent lysine nucleotidyltransferase superfamily (28). The shared NTase domain includes six peptide motifs (I, Ia, III, IIIa, IV and V), constituents of which engage the NTP substrate and the catalytic metal ion. Motif I (KxNG in Trl1-LIG) contains the lysine that becomes covalently attached to the NMP. The NTase sub-domain of Trl1-LIG consists of two central β-sheets: a 5-strand sheet (with topology β7↑•β6↓•β8↑•β9↓•β10↓) and a 4-strand sheet (with topology β11↓•β5↑•β14↓•β15↑) that together form the adenylate-binding pocket (Figure 2A). The segment from Asp37 to Arg116 is homologous to the N-terminal sub-domain of T4 Rnl1 and Methanobacterium thermoautotrophicum RNA ligase (MthRnl) (29–31). This sub-domain of Trl1-LIG comprises a four-strand antiparallel β-sheet (topology β1↑•β2↓•β3↑•β4↓) against which the α1 helix packs (Figure 2A).
Trl1-LIG exemplifies a distinct clade of RNA ligase
At present, there are five structurally characterized RNA ligase families, exemplified by: T4 RNA ligase 1 (Rnl1 family) (29,30); T4 RNA ligase 2 (Rnl2 family) (32); MthRn1 and Pyrococcus abyssi RNA ligase (Rnl3 family) (31,33); Clostridium thermocellum RNA ligase CthPnkpC-Hen1N (Rnl4 family) (34,35); and Naegleria gruberi RNA ligase NgrRnl (Rnl5 family) (36). These families are defined by the distinctive structural domains that are appended to their NTase cores. A DALI search (37) with the Trl1-LIG structure identified T4 Rnl1 (pdb 5TT6; in complex with ATP and two Mg2+ ions) as the closest homolog (Z score 21.6; 3.7 Å rmsd at 252 Cα positions with 19% amino acid identity) followed by MthRnl (pdb 5DLO) as the second best hit (Z score 17.2; 4.0 Å rmsd at 257 Cα positions with 15% amino acid identity). The structural similarity between Trl1-LIG, T4 Rnl1 and MthRnl spans both sub-domains of their respective adenylyltransferase domains, but does not embrace the C-domain of Trl1-LIG. A superposition in Figure 3A of the adenylyltransferase domains of Trl1-LIG (in blue) and T4 Rnl1 (in beige) highlights their structural similarity and the virtually identical positions of the AMP moiety of ATP in their respective active sites. The metal ions engaged to the ATP α phosphate also occupy identical positions. By contrast, the positions of the ATP γ phosphate and the second metal that bridges the β and γ phosphates differ by 2.3 Å (γ-PO4) and 3.3 Å (M2) respectively. This comparison suggests a subtle difference in these two RNA ligases in the trajectory of the pyrophosphate leaving group during the ligase adenylylation step.
Figure 3.
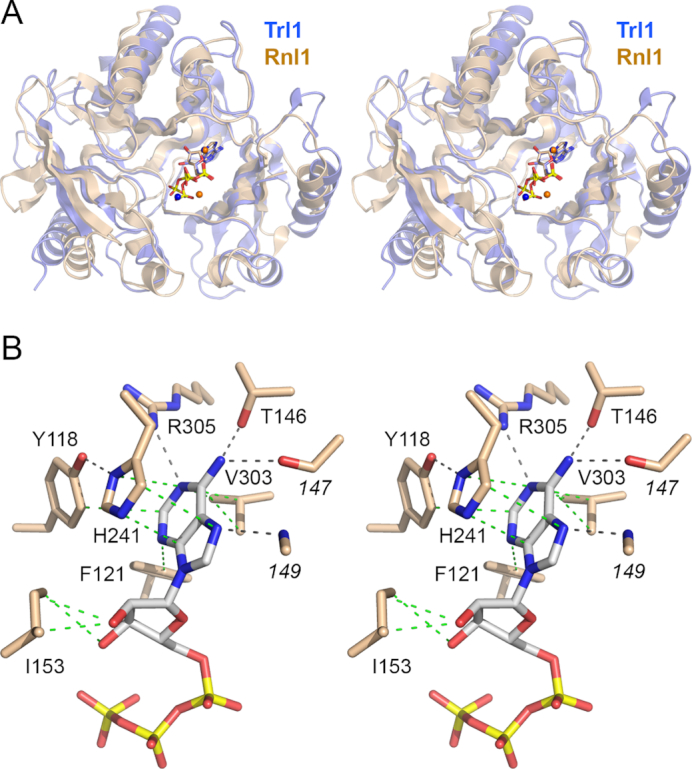
Homology to T4 Rnl1 and basis for adenine nucleotide specificity. (A) Stereo view of the superimposed structures of the adenylyltransferase domains of Trl1-LIG (blue) and T4 Rnl1 (beige) in their respective complexes with ATP (stick models) and divalent cations (shown as blue spheres for Trl1-LIG and beige spheres for Rnl1). (B) Stereo view of the adenylate binding pocket highlighting contacts to the adenine nucleobase and ribose sugar. Amino acids and ATP are shown as stick models with beige and gray carbons, respectively. Atomic contacts are indicated by black dashed lines (hydrogen bonds) or green dashed lines (van der Waals contacts).
DALI also recovered hits to exemplars of the other RNA ligase families, as follows: NgrRnl (pdb 5COU; Z score 12.0; 3.8 Å rmsd at 196 Cα positions with 14% amino acid identity); T4 Rnl2 (pdb 2HVS; Z score 11.9; 3.3 Å rmsd at 187 Cα positions with 11% amino acid identity); and CthPnkpC-Hen1N (pdb 4E6N; Z score 8.4; 5.1 Å rmsd at 206 Cα positions with 12% amino acid identity). In these cases, the similarity to Trl1-LIG embraces the core NTase sub-domain, but not the N sub-domain or C-domain of Trl1-LIG.
A separate DALI search with just the C-domain of Trl1-LIG (from aa 326 to 406) recovered no hits with a Z score greater than 4.9. Many of the low-scoring hits were to transcription factors with α-helical folds; there were no C-domain hits identified that had any apparent connections to nucleic acid enzymes. Thus, we suggest that Trl1-LIG be considered the founding member of an Rnl6 clade of ATP-dependent RNA ligases by virtue of its unique C-domain.
Trl1-LIG active site with bound ATP
The adenosine nucleoside of ATP is in the syn conformation (Figure 3B). The adenine ring is sandwiched between the aromatic ring of His241 (nucleotidyltransferase motif IIIa) and the hydrophobic side chain of Val303 (motif IV; EGFVIR). Adenine specificity is conferred by hydrogen bonds to adenine-N6 from Thr146-Oγ (motif I; TLKENG) and the Leu147 main-chain carbonyl and to adenine-N1 from Arg305 (motif IV; EGFVIR). There is an additional hydrogen bond to adenine-N7 from the Glu149 main-chain amide (Figure 3B). Phe121-Cζ makes van der Waals contacts to the adenine-N3 atom. Tyr118 makes a van der Waals contact from Cϵ to adenine-C2 and also makes a hydrogen bond from its OH to His241-Nδ (Figure 3B). Ile153 makes van der Waals contacts to the ATP ribose O2′ and O3′ atoms (Figure 3B). The ATP ribose 2′-OH makes a hydrogen bond to Glu218 (motif III) (Figure 4A).
Figure 4.
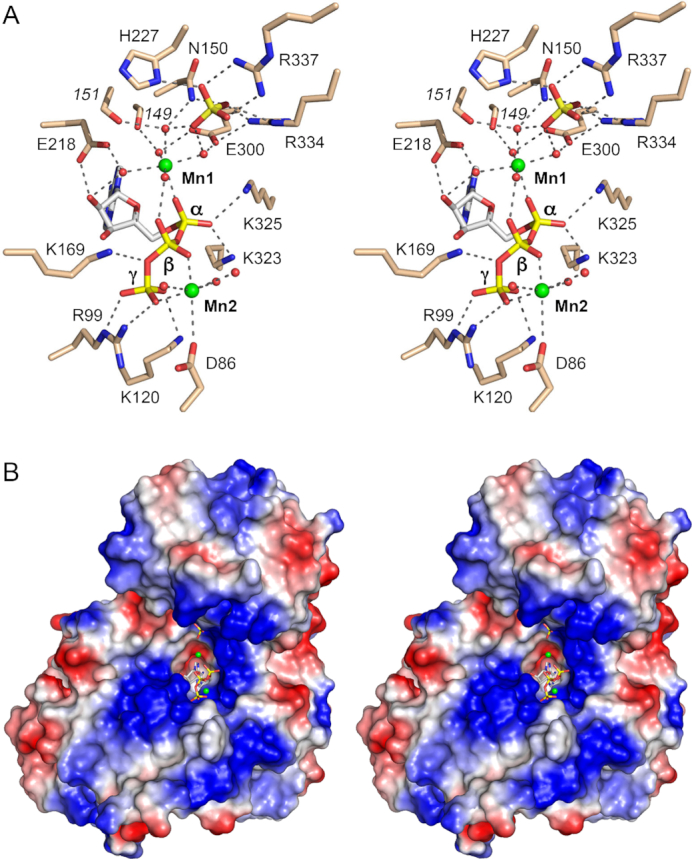
LIG interactions with ATP•Mn2+ and surface electrostatics. (A) Stereo view of the active site highlighting contacts to the ATP phosphates, two associated manganese ions (green spheres), and a nearby sulfate anion (stick model). Amino acids and ATP are shown as stick models with beige and gray carbons, respectively. Waters are depicted as red spheres. Atomic contacts are indicated by black dashed lines. (B) Stereo view of a surface electrostatic model of Trl1-LIG (generated in Pymol). ATP in the active site and a nearby sulfate anion are depicted as a stick models. Manganese ions are green spheres.
The ATP α phosphate is engaged by a ‘catalytic’ manganese (Mn1) (Figure 4A). Waters occupy five of the ligand sites in the octahedral Mn2+ complex. Trl1-LIG binds the Mn2+(H2O)5 complex via water-mediated contacts to Asn150 (motif I), Glu218 (motif III), Glu300 (motif IV), and the Glu149 and Gly151 main-chain carbonyls. One of the metal-bound waters makes a bridging contact to the ATP β phosphate. The sixth Mn2+ ligand site is occupied by one of the non-bridging α phosphate oxygens (Figure 4A). The other non-bridging α phosphate oxygen is contacted by Lys323 and Lys325 (motif V; FFKYK). The structure suggests that Lys323, Lys325, and the catalytic metal stabilize a pentavalent transition state of the ATP α phosphate during the lysine adenylylation reaction.
There are no direct enzymic contacts to the ATP β phosphate. Direct contacts to the ATP γ phosphate are made by Arg99, Lys120, and Lys169 (motif Ia; SKHS) (Figure 4A). The ‘non-catalytic’ manganese (Mn2) is coordinated to ATP β and γ phosphate oxygens, the Asp86 carboxylate, and three waters (Figure 1B). The Mn1 and Mn2 metal ions are 8.3 Å apart.
Surface electrostatics
A surface electrostatic model of Trl1-LIG prepared in Pymol (Figure 4B) highlights a broad swath of positive potential flanking the deep (hydrophobic/negative) pocket into which the adenosine moiety of ATP fits. The positive surface tracking leftward and downward from the ATP is contributed by the N subdomain, especially residues Lys54, Lys56, Lys57, Lys64, Arg83, Lys91, Arg99 and Lys120. The positive surface extending rightward and upward from the AMP is contributed by the C domain, via Arg334, Arg337, Lys341, Lys347, Lys350, Lys352 and Lys353. These surfaces provide, in principle, a favorable interface with the negatively charged phosphodiester backbone of the 5′-PO4-terminated and 3′-OH/2′-PO4-terminated RNA strands that are sealed by Trl1-LIG.
Trl1-LIG covalent lysyl–AMP intermediate
The modeled LIG–AMP structure comprises a continuous polypeptide from Ala13 to Glu407. The LIG–AMP and LIG•ATP structures align with a rmsd of 1.4 Å at 383 Cα positions. Superposition of the LIG–AMP and LIG•ATP structures with respect to their adenylyltransferase domains reveals a rigid-body movement of the C-domain away from the adenylyltransferase domain in the transition from the LIG•ATP state to the LIG–AMP state (Figure 5A). This transition entails a 6.2 Å shift of the position of Gly346 in the C-domain.
Figure 5.
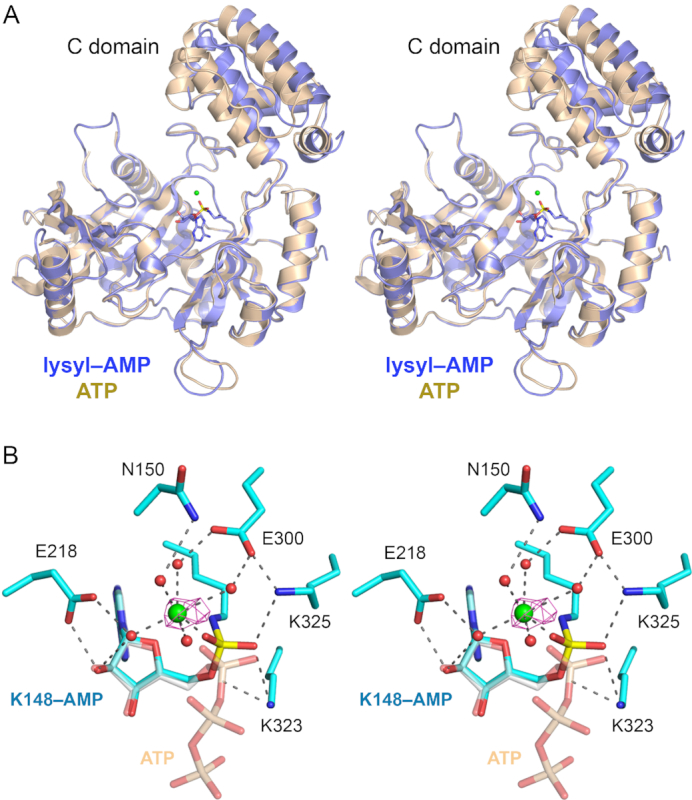
Trl1-LIG–AMP covalent intermediate. (A) Stereo view of the tertiary structures of the Tr1LIG–AMP intermediate (in blue) and the LIG–ATP Michaelis complex (in beige) superimposed with respect to their N-terminal adenylyltransferase domains. The image highlights a rigid body movement of the C-terminal domain in the transitions from LIG•ATP to LIG–AMP. The lysyl-adenylate adduct in the LIG–AMP structure is shown as a stick model. The catalytic Mn2+ ion is rendered as a green sphere. (B) Stereo view of the active site of LIG–AMP. Amino acids and lysyl–AMP are shown as stick models with cyan carbons; the AMP phosphorus atom is colored yellow. A catalytic Mn2+ ion and associated waters are depicted as green and red spheres, respectively. The anomalous difference peak overlying the manganese ion, contoured at 4σ, is depicted in magenta mesh. Atomic contacts are indicated by dashed lines. The superimposed ATP from the Michaelis complex structure is shown as a semi-transparent stick model with gray carbons and beige phosphorus atoms (to highlight the stereochemical inversion of the α phosphorus center after lysine adenylylation).
Figure 5B shows a stereo view of the active site of the lysyl–AMP intermediate highlighting atomic interactions relevant to catalysis. For comparison, we included in the figure the superimposed ATP molecule from the Michaelis complex. The LIG–AMP structure has a single manganese ion adjacent to the AMP phosphate, as affirmed by the anomalous difference density overlying the metal atom (Figure 5B). The octahedral Mn2+ complex that engages the AMP phosphate in the lysyl–AMP intermediate is virtually identical to the catalytic Mn1 complex that engages the ATP α phosphate in the Michaelis complex. The Lys148-Nζ of lysyl-AMP is situated 3.0 Å from the superimposed ATP α phosphorus, in a favorable, nearly apical orientation to the pyrophosphate leaving group (Nζ–Pα–O3α angle = 150°). Consistent with a single-step in-line mechanism, we see that the α phosphate undergoes stereochemical inversion during the transition from ATP Michaelis complex to lysyl–AMP intermediate (Figure 5B). The lysyl–AMP phosphate oxygens are engaged by the Lys323 and Lys325 side chains.
Sulfate anion as potential 2′-PO4 mimetic
The Michaelis complex, which was crystallized in precipitant solution containing ammonium sulfate, contains a single well-ordered sulfate anion situated 5.3 Å away from the ATP α phosphate (Figure 4A). Insofar as sulfate is a steric and electrostatic analog of phosphate, we infer that this sulfate mimics the position and interactions of a phosphate in the RNA substrate that Trl1-LIG seals. The sulfate is engaged to the ligase and to the catalytic Mn1 complex by a network of atomic contacts to all four of the sulfate oxygens. To wit, three of the sulfate oxygens collectively receive bidentate hydrogen bonds from Arg337 NH1 and NH2, bifurcated hydrogen bonds from Arg334 NH2 and Nϵ, a hydrogen bond from His227-Nδ, and a hydrogen bond from Asn150-Nδ (Figure 4A). The fourth sulfate oxygen occupies the secondary sphere of the Mn1 complex, making bridging contacts to two of the Mn1-bound waters (Figure 4B). The intimacy of the sulfate and the active site suggest that the sulfate is a mimetic of a terminal phosphate of the RNA ligation substrate. Because the amino acids in CthTrl1-LIG that contact the sulfate are conserved in SceTrl1 (Figure 2B) and in Trl1 enzymes from other fungi (6), and because these amino acids are unique to the Trl1-LIG clade (i.e. they are not present in other families of RNA ligases), we hypothesize that they and their sulfate/phosphate interactions underpin the distinctive biochemical property of fungal tRNA ligases vis-à-vis other end-joining enzymes: their requirement for a 3′-OH,2′-PO4 terminus for RNA sealing. Thus, we suggest that the sulfate in the crystal structure is a likely mimetic of the terminal 2′-PO4 of the RNA substrate. This idea is consistent with mutational studies of S. cerevisiae Trl1-LIG showing that alanine substitutions at the corresponding sulfate-binding amino acids His193 and Arg298 (equivalent to CthTrl1 residues His227 and Arg337) are lethal in vivo (6). We disfavor the alternative view that the sulfate mimics the RNA 5′-PO4. Rather, we suspect, based on the structure of T4 Rnl2 in complex with its AppRNA reaction intermediate (32) in which the 5′-PO4 is engaged by the signature basic side chain of motif Ia (Arg55 in Rnl2), that the RNA 5′-PO4 in the Trl1-LIG reaction is also recognized by the corresponding motif Ia residue, which is Lys169 in CthTrl1. This is sensible, given that Lys169 contacts the bridging β-γ phosphate oxygen of ATP in the Michaelis complex (Figure 4A) and that the RNA 5′-PO4 (the nucleophile in step 2 of the ligase reaction leading to the AppRNA intermediate) is thought to occupy the same position as the PPi leaving group in the step 1 reaction.
Chaetomium Trl1-LIG structure rationalizes mutational data for Saccharomyces Trl1
An extensive alanine scanning mutational analysis of SceTrl1-LIG, targeting amino acids conserved among fungal Trl1 proteins, identified 28 residues as essential for Trl1-LIG activity in vivo (6). These amino acids, and their conserved counterparts in CthTrl1-LIG, are listed in Table 2. We can now account for the important contributions of these conserved elements based on their atomic contacts revealed by the CthTrl1-LIG crystal structure. The essential amino acids fall into two categories: (i) those in the active site that contact either ATP, the metal cofactors, or the sulfate anion (a putative mimetic of the RNA 2′-PO4) and are therefore deemed to be important for ligase activity (designated as ‘A’ for activity in Table 2) and (ii) those that contact other amino acids in the ligase protein and thereby help stabilize the Trl1-LIG tertiary structure or properly position components of the active site (this class is designated as ‘S’ for structural in Table 2).
Table 2.
Rationale for structure-activity relations in budding yeast Trl1-LIG
| Essential SceTrl1 aa | CthTrl1 equivalent | Contacts | Rationale (A or S) |
|---|---|---|---|
| Arg66 | Arg99 | ATP γ-PO4 | A |
| Arg82 | Arg116 | Asp119-Oδ, Gly117-O, Ile247-O, Pro248-O | S |
| Lys86 | Lys120 | ATP γ-PO4 | A |
| Phe87 | Phe121 | ATP adenine (VDW) | A |
| Phe88 | Phe122 | Lys120, Glu126, Thr127 (VDW) | S |
| Lys114 | Lys148 | AMP phosphate (covalent) | A |
| Ser134 | Ser168 | Ser171-O | S |
| Lys135 | Lys169 | ATP γ-PO4 | A |
| His136 | His170 | Arg83, Lys169 (VDW) | S |
| Ser137 | Ser171 | Tyr68-OH, Arg175-NH1 | S |
| His148 | His182 | Glu225-Oϵ | S |
| Glu184 | Glu218 | ATP ribose-O2′, His241-Nϵ, Metal complex | A |
| Asp187 | Asp221 | His189-Nϵ | S |
| Asp188 | Asp222 | Tyr231-N | S |
| Glu191 | Glu225 | His182-Nϵ | S |
| Glu192 | Glu226 | Gly384-N, Ile385-N, Ile386-N | S |
| His193 | His227 | SO4 (RNA) | A |
| His207 | His241 | ATP adenine, Tyr118-OH, Glu218-Oϵ | A |
| Asn210 | Asn244 | R116-O, Thr252-Oγ | S |
| Glu266 | Glu300 | Metal complex, Arg334 | A |
| Gly267 | Gly301 | Phe326, Thr324 (VDW), Met148-N | S |
| Arg271 | Arg305 | ATP adenine-N1, H241-O | A |
| Lys284 | Lys323 | ATP α-phosphate | A |
| Lys286 | Lys325 | ATP α-phosphate | A |
| Arg298 | Arg337 | SO4 (RNA) | A |
| Glu288 | Glu338 | Arg337 | S |
| Lys302 | Lys341 | N382-Oδ | S |
| Arg352 | Arg389 | Ile228-O, Tyr333 (π-cation) | S |
The constellation of ‘activity’ residues includes constituents of five of the covalent nucleotidyltransferase motifs in the adenylyltransferase domain that had been predicted based on sequence alignments (6). These are (using CthTrl1 numbering) Lys148 in motif I; Lys135 in motif Ia; Glu218 in motif III; Glu300 and Arg305 in motif IV; and Lys323 and Lys325 in motif V. Up to now, it was not possible to identify on the basis of sequence alignments the Trl1 counterpart of motif IIIa (6), which in other covalent nucleotidyltransferases is a β strand that contributes an essential aromatic residue that makes a π-stack on the purine nucleobase of the NTP substrate (28). The Trl1-LIG structure now clarifies that motif IIIa resides in strand β9 and that His241 is the relevant amino acid that stacks on the adenine of ATP (Figure 3B), with the corresponding histidine in SceTrl1-LIG being essential for function in vivo (6).
Three additional imputed activity residues are contributed by the Rnl1-like N-terminal sub-domain. These are (in CthTrl1 numbering) Arg99 and Lys120 that contact the ATP γ phosphate (Figure 4A) and Phe121 that contacts the adenine base (Figure 3B). Indeed, the equivalent residues in T4 Rnl1 (Arg54, Lys75 and Phe76) make analogous contacts to the ATP γ phosphate and adenine in the crystal structure of the T4 Rnl1 Michaelis complex (30). Mutational analyses of T4 Rnl1 establish that Arg54 and Lys75 are essential for its ligase activity in vitro (38).
Among the putative ‘structural’ residues that were found by alanine scanning to be essential for SceTrl1-LIG activity in vivo, His148 (equivalent to His182 in CthTrl1) is noteworthy in light of its importance in implicating tRNA ligase as a catalyst of unconventional mRNA splicing in the yeast unfolded protein response (UPR) (2). Sidrauski et al. conducted a genetic screen for yeast mutants that failed to mount a transcriptional response to tunicamycin treatment and thereby identified a single H148Y missense change in Trl1 as responsible for the UPR defect, which was caused by a failure to splice HAC1 mRNA after Ire1 had excised the cryptic intron (2). Because TRL1-H148Y cells grew at normal rates and did not accumulate unligated tRNA halves during a pulse-label with 32Pi, it was suggested that the H148Y mutation selectively impairs HAC1 mRNA splicing. The CthTrl1-LIG structure reveals that the corresponding His182 residue is located in helix α3 and is remote from the active site (i.e. 10–11 Å away from ATP, Mn1, and the sulfate). His182 is deemed structural because it is conserved among fungal ligases and it donates a hydrogen bond to the Glu225 carboxylate (the equivalent of Glu191 in SceTrl1), which is also conserved among fungal ligases and is located in a loop connecting strands β8 and β9 (Figure 1B). In SceTrl1-LIG, alanine substitutions for His148 or Glu191 are lethal in vivo (6), highlighting the importance of the His-Glu interaction. We infer that a tyrosine in lieu of the histidine can sustain this interaction with Glu, so that H148Y yeast cells are viable. However, because it is not obvious from the structure how alteration of the His or the His-Glu pair might act as a discriminator between tRNA and HAC1 substrates, we are inclined to regard H148Y as a hypomorph.
CONCLUSIONS
The present study extends our knowledge of fungal tRNA splicing enzymology by providing the structure of a catalytically active ligase domain of Trl1. We show that Trl1-LIG exemplifies a new RNA ligase clade—which we designate Rnl6—by virtue of its structurally distinctive C-terminal module appended to an N-terminal adenylyltransferase domain shared by the Rnl1 and Rnl4 ligase clades. We propose that the Rnl1, Rnl4 and Rnl6 ligases evolved from a common ancestor, possibly a stand-alone adenylyltransferases that has the Rnl1-like N-terminal sub-module, via independent acquisitions of distinct C-terminal domain fusions. In the case of T4 Rnl1, the C domain is responsible for the high specificity of Rnl1 for joining broken 3′-OH and 5′-PO4 ends in the tRNA anticodon loop (39). tRNA repair is the physiological function of T4 Rnl1 during phage infection (40). By analogy, we envision that the C-domain of Trl1-LIG, which is essential for Trl1-LIG activity in vivo in yeast (5), confers specificity for repairing RNA breaks with 3′-OH,2′-PO4 and 5′-PO4 ends. As discussed above, we posit that C-domain amino acids Arg334 and Arg337 contribute to an enzymic binding site for the 2′-PO4.
A trifunctional RNA ligase, composed of LIG, KIN, and CPD domains, is the agent of tRNA splicing in plants (41) and kinetoplastid protozoa (42). Although the kinetoplastid Trl1 has not been characterized biochemically, its LIG domain shares the conserved features of the fungal Trl1-LIG (6), including counterparts of the Rnl1-like N subdomain, the NTase motifs, and an RxxR motif flanking motif V of the NTase domain that is a candidate counterpart of 334RxxR337 in CthTrl1-LIG that binds the sulfate anion we invoke as a 2′-PO4 mimetic. Therefore we speculate that the kinetoplastid LIG domain belongs to the Rnl6 clade.
The tRNA ligase from the plant Arabidopsis thaliana (AtRNL) has been well characterized. AtRNL complements S. cerevisiae trl1Δ (7) and shares the requirement of fungal Trl1-LIG for a 2′-PO4 terminus (21,41), but it does not display the specificity of the fungal Trl1-KIN domain for GTP as the phosphate donor (8,21). There is substantial amino acid sequence divergence between the plant and fungal tRNA ligases (41). Mutational analysis places the AtRNL-LIG domain in the covalent lysine nucleotidyltransferase superfamily (7), but we are unable to judge whether the plant LIG might be in the same ligase clade as fungal Trl1-LIG.
Our structures of the Trl1-LIG Michaelis complex and LIG–AMP intermediate fortify the findings from recent Rnl1 and Rnl5 structures (30,36), which together engender the conclusion that all ATP-dependent RNA ligases employ a two-metal mechanism of lysine adenylylation, in which a catalytic metal stabilizes the transition state of the ATP α phosphate (aided by basic amino acids that contact the α phosphate) and then remains in place after step 1 catalysis. A second non-catalytic metal bridges the β and γ phosphates and (together with basic amino acids that engage the γ phosphate) ensures a conformation of the ATP triphosphate moiety conducive to expulsion of the PPi•Me2+ leaving group.
The catalytic metal in Trl1-LIG is a penta-hydrated divalent cation in which the sixth position in the metal coordination complex is filled by an ATP or AMP α phosphate oxygen. All of the contacts between the catalytic metal complex and Trl1-LIG are water-bridged and involve amino acids in motifs I, III, and IV. A penta-hydrated catalytic metal complex at the AMP phosphate coordinated by amino acids in motifs I, III, and IV is a mechanistic feature shared with other members of the covalent lysine nucleotidyltransferase superfamily, i.e., NgrRnl (36), T4 Rnl1 (30), MthRnl (31), CthPnkpC-Hen1N (35) and E. coli DNA ligase (30).
Of course, a fuller understanding of the mechanism of fungal tRNA ligase hinges on obtaining structural snapshots of the LIG enzyme at the several discrete stages of the reaction pathway subsequent to lysine adenylylation. This is technically challenging at present, insofar as it will require the large-scale synthesis of defined RNA substrates with 5′-PO4 and 3′-OH,2′-PO4 termini, or chemical modifications thereof that will arrest the ligation reactions at desired points.
DATA AVAILABILITY
Structural coordinates have been deposited in Protein Data Bank under accession codes 6N0V and 6N0T.
FUNDING
National Institutes of Health [R35-GM126945 to S.S.]; Geoffrey Beene Cancer Research Center (to S.S.); The MSKCC structural biology core laboratory is supported by National Cancer Institute [P30-CA008748]; X-ray diffraction data were collected at synchrotron facilities supported by grants and contracts from the National Institutes of Health [P41GM103403, HEI-S10RR029205]; Department of Energy [DE-AC02-06CH11357]. Funding for open access charge: NIH [R35-GM126945].
Conflict of interest statement. None declared.
REFERENCES
- 1. Abelson J., Trotta C.R., Li H.. tRNA splicing. J. Biol. Chem. 1998; 273:12685–12688. [DOI] [PubMed] [Google Scholar]
- 2. Sidrauski C., Cox J.S., Walter P.. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996; 87:405–413. [DOI] [PubMed] [Google Scholar]
- 3. Greer C.L., Peebles C.L., Gegenheimer P., Abelson J.. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983; 32:537–546. [DOI] [PubMed] [Google Scholar]
- 4. Apostol B.L., Westaway S.K., Abelson J., Greer C.L.. Deletion analysis of a multifunctional yeast tRNA ligase polypeptide: identification of essential and dispensable functional domains. J. Biol. Chem. 1991; 266:7445–7455. [PubMed] [Google Scholar]
- 5. Sawaya R., Schwer B., Shuman S.. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003; 278:43298–43398. [DOI] [PubMed] [Google Scholar]
- 6. Wang L.K., Shuman S.. Structure-function analysis of yeast tRNA ligase. RNA. 2005; 11:966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang L.K., Schwer B., Englert M., Beier H., Shuman S.. Structure-function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res. 2006; 34:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Remus B.S., Shuman S.. Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases. RNA. 2014; 20:462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Remus B.S., Schwer B., Shuman S.. Characterization of the tRNA ligases of pathogenic fungi Aspergillus fumigatus and Coccidioides immitis. RNA. 2016; 22:1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka N., Shuman S.. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011; 286:7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka N., Chakravarty A.K., Maughan B., Shuman S.. A novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011; 286:43134–43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanaka N., Meineke B., Shuman S.. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011; 286:30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chakravarty A.K., Subbotin R., Chait B.T., Shuman S.. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6072–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakravarty A.K., Shuman S.. The sequential 2′,3′ cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res. 2012; 40:8558–8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., Eom S.H., Koningsberg W.H., Söll D., Wang J.. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl Acad. Sci. U.S.A. 2012; 109:15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desai K.K., Bingman C.A., Phillips G.N., Raines R.T.. Structure of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013; 52:2518–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Lürhmann R., Söll D. et al. . HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011; 331:760–764. [DOI] [PubMed] [Google Scholar]
- 18. Kosmaczewski S.G., Edwards T.J., Han S.M., Eckwahl M.J., Meyer B.I., Peach S., Hesselberth J.R., Wolin S.L., Hammarlund M.. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 2014; 15:1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Remus B.S., Goldgur Y., Shuman S.. Structural basis for the GTP specificity of the RNA kinase domain of fungal tRNA ligase. Nucleic Acids Res. 2017; 45:12945–12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Westaway S.K., Belford H.G., Apostol B.L., Abelson J., Greer C.L.. Novel activity of a yeast ligase deletion polypeptide: evidence for GTP-dependent tRNA splicing. J. Biol. Chem. 1993; 268:2435–2443. [PubMed] [Google Scholar]
- 21. Remus B.S., Shuman S.. A kinetic framework for tRNA ligase and enforcement of a 2′-phosphate requirement for ligation highlights the design logic of an RNA repair machine. RNA. 2013; 19:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das U., Shuman S.. Mechanism of RNA 2,3-cyclic phosphate end healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res. 2013; 41:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 24. Sheldrick G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst. 2010; D66:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M.. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991; A47:110–119. [DOI] [PubMed] [Google Scholar]
- 26. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nandakumar J., Schwer B., Schaffrath R., Shuman S.. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol. Cell. 2008; 31:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shuman S., Lima C.D.. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004; 14:757–764. [DOI] [PubMed] [Google Scholar]
- 29. El Omari K., Ren J., Bird L.E., Bona M.K., Klarmann G., LeGrice S.F., Stammers D.K.. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006; 281:1573–1579. [DOI] [PubMed] [Google Scholar]
- 30. Unciuleac M.C., Goldgur Y., Shuman S.. Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD+-dependent polynucleotide ligases. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2592–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gu H., Yoshinari S., Ghosh R., Ignatochkina A.V., Gollnick P.D., Murakami K.S., Ho C.K.. Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis. Nucleic Acids Res. 2016; 44:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nandakumar J., Shuman S., Lima C.D.. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006; 127:71–84. [DOI] [PubMed] [Google Scholar]
- 33. Brooks M.A., Meslet-Cladiére L., Graille M., Kuhn J., Blondeau K., Myllykallio H., van Tilbeurgh H.. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008; 17:1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith P., Wang L.K., Nair P.A., Shuman S.. The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang P., Chan C.M., Christensen D., Zhang C., Selvadurai K., Huang R.H.. Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:13248–13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Unciuleac M.C., Goldgur Y., Shuman S.. Structure and two-metal mechanism of a eukaryal nick-sealing RNA ligase. Proc. Natl. Acad. Sci. USA. 2015; 112:13868–13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holm L., Kaariainen S., Rosenstrom P., Schenkel A.. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008; 24:1780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L.K., Ho C.K., Pei Y., Shuman S.. Mutational analysis of bacteriophage T4 RNA ligase 1: different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J. Biol. Chem. 2003; 278:29454–29262. [DOI] [PubMed] [Google Scholar]
- 39. Wang L.K., Nandakumar J., Schwer B., Shuman S.. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA. 2007; 13:1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amitsur M., Levitz R., Kaufman G.. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987; 6:2499–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Englert M., Beier H.. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005; 33:388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lopes R.R., Silveira G.O., Eitler R., Vidal R.S., Kessler A., Hinger S., Paris Z., Alfonzo J.D., Polycarpo C.. The essential function of the Trypanosoma brucei Trl1 homolog in procyclic cells is maturation of the intron-containing tRNATyr. RNA. 2016; 22:1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Structural coordinates have been deposited in Protein Data Bank under accession codes 6N0V and 6N0T.


