Abstract
Previous works have reported significant effects of macromolecular crowding on the structure and behavior of biomolecules. The crowded intracellular environment, in contrast to in vitro buffer solutions, likely imparts similar effects on biomolecules. The enzyme serving as the gatekeeper for the genome, RNA polymerase (RNAP), is among the most regulated enzymes. Although it was previously demonstrated that macromolecular crowding affects association of RNAP to DNA, not much is known about how crowding acts on late initiation and promoter clearance steps, which are considered to be the rate-determining steps for many promoters. Here, we demonstrate that macromolecular crowding enhances the rate of late initiation and promoter clearance using in vitro quenching-based single-molecule kinetics assays. Moreover, the enhancement’s dependence on crowder size notably deviates from predictions by the scaled-particle theory, commonly used for description of crowding effects. Our findings shed new light on how enzymatic reactions could be affected by crowded conditions in the cellular milieu.
INTRODUCTION
The cellular environment is distinct from that of dilute buffer. Cells contain many macromolecules in a small volume, resulting in a highly condensed environment (1–3). For example, in Escherichia coli, macromolecules take up to ∼40% of the overall volume of the cytoplasm (3). Such a dense environment is expected to alter biological reactions, as compared to the same reactions in buffer (4–7), because of greatly increased viscosity that slows macromolecular motions and kinetics (8,9). In addition, as macromolecules occupy a large fraction of the cellular space, the available volume for other molecules decreases because each macromolecule excludes other molecules from its vicinity. The associated excluded volume (6,10,11) of a macromolecule is a function of its shape and size as well as those of other nearby molecules (10). This volume exclusion noticeably affects the thermodynamics and/or kinetics of reacting molecules, especially when the reaction causes a change in the volume of reactants. It has previously been shown that excluded volume of macromolecules in the crowded environment considerably enhances the kinetics and thermodynamics of the association reactions, and also affects the size of intrinsically disordered proteins, the stability of native protein structures, and the folding behavior of proteins and RNAs (12–14).
Transcription is a highly regulated and crucial first step in gene expression, and therefore has been extensively studied (15–19). The great complexity of in vivo transcription directed many researchers to take a reductionist approach, focusing on in vitro-reconstituted transcription systems in buffer (16,18,19). However, this approach sometimes gives rise to results that differ from in vivo assays, both quantitatively and qualitatively, in part due to environmental factors such as the crowding effect (20,21).
Recently, several works reported that transcription reactions could be affected by macromolecular crowding (20,22–24). For example, studies utilizing cell-free protein expression systems have shown that transcription by T7 RNA polymerase (T7 RNAP) was markedly enhanced under crowding conditions (20,22). This enhancement was thought to be mainly attributable to an increase in affinity between promoter DNA and T7 RNAP as a result of crowding, with the assumption that the association is the rate -determining step of the entire transcription reaction (20,22). However, the effects of crowding on late initiation and promoter clearance are still poorly understood; most studies on transcription under crowding conditions have focused on multiple cycles of transcription, specifically on binding events of DNA and RNAP to form the RNAP–Promoter open complex (RPO). Moreover, recent studies reported that these steps could be rate-limiting for some promoters (e.g. with unique recognition and initial transcribing regions (25,26)). A more detailed understanding of how crowding affects these initiation steps is therefore required.
In this work, we utilized an in vitro single-round quenched-kinetics transcription assay using single-molecule detection (26,27). Our assay is based on the hybridization of a doubly end-labeled DNA oligo to the RNA transcript produced by the transcription reaction. Using this assay, we were able to monitor transcription reactions in various controlled environments that mimic the crowded cellular environment (Figure 1). We studied the effect of the size and concentration of various macromolecular crowding agents on stable open complexes (RPICT=2, see details in ‘Materials and Methods’ section #2), and validated these studies using an RNA-binding-dye assay. We demonstrate that single-round transcription kinetics—starting from the point after RNAP binds the promoter DNA specifically to form an open transcription bubble—are noticeably affected by crowders. However, the dependence of the kinetics on crowder size is inconsistent with the prediction from the hard-sphere-model-based scaled-particle theory (SPT), the prevalent framework for elucidating the effect of macromolecular crowding (10,14,28).
Figure 1.
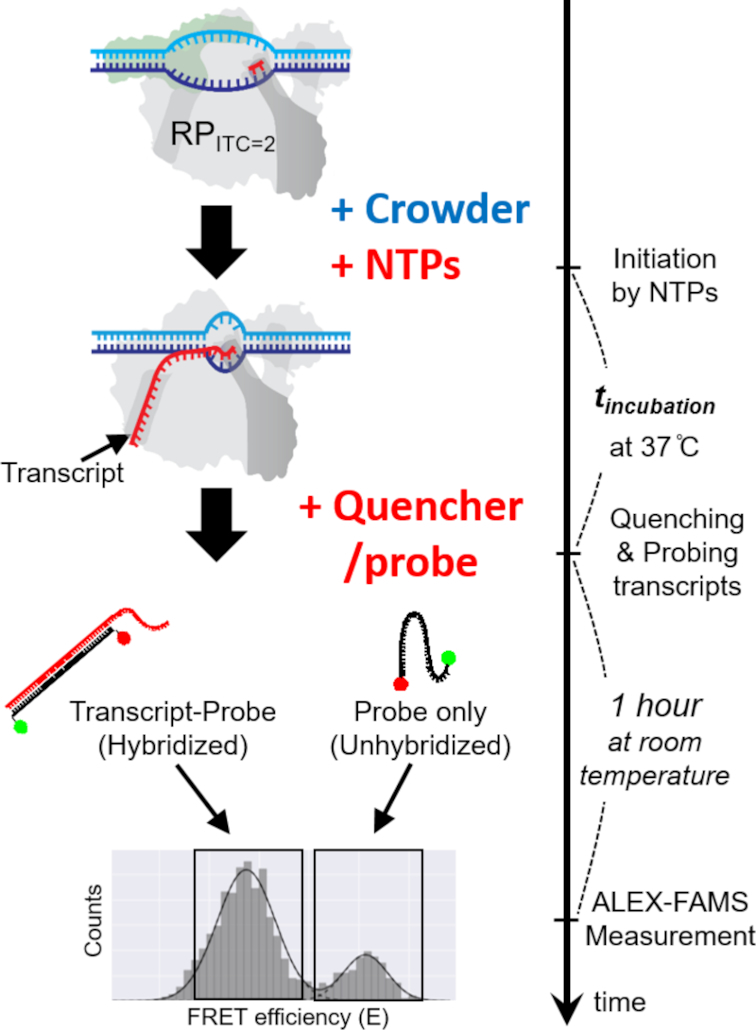
A schematic of the in vitro single-round quenched-kinetics transcription assay. The transcription from stable open complexes (RPITC=2, prepared in buffer) starts with addition of NTPs under crowding conditions and proceeds until quenched by a reaction quencher. RNA transcripts produced by single-round transcription reactions during the incubation time (tincubation) are hybridized with the ssDNA probe (black line with two dots) that has sequence complementary to part of the transcript (Supplementary Figure S1). The hybridized fraction can be detected by ALEX-FAMS because the stretched structure of hybridized probe shows lower FRET efficiency compared to that of an unhybridized probe (26,27,36).
MATERIALS AND METHODS
Preparation of the template DNA and probe for in vitro single-round quenched kinetics transcription assays
The DNA template used for the transcription reaction was designed to produce a transcript containing a sequence complementary to the probe sequence (Supplementary Figure S1). Thus, an RNA produced by the transcription reaction would be efficiently hybridized with the single-stranded DNA (ssDNA) probe. The transcription detection probe is a doubly-labeled ssDNA with a donor–acceptor FRET pair—a donor 5-carboxytetramethylrhodamine (TMR) at its 5′ end and an acceptor (Alexa Fluor 647) at its 3′ end. In order for efficient hybridization with transcripts, we designed the ssDNA probe with 20 consecutive deoxythymidines (20 dT) to be unstructured (i.e. with no stable secondary structure) in solution (26,27). Since the persistence length of unstructured ssDNA is short (∼1.5 nm in 2 M NaCl, ∼3 nm in 25 mM NaCl) (29), the two dyes are close to each other, yielding a single high FRET population with a peak FRET efficiency of E ∼ 0.8. However, when hybridized to the run-off mRNA transcripts, the probe-mRNA assumes a rigid double-stranded DNA (dsDNA)–RNA conformation (persistence length for dsDNA ∼50 nm (30), for dsRNA ∼64 nm (31)) and the distance between the two dyes is increased, leading to the appearance of a second FRET sub-population with a peak FRET efficiency of ∼0.3.
dsDNA template, lacCONS-GTG-20dA (Supplementary Figure S1), was prepared by hybridizing ssDNAs (Integrated DNA Technologies, Coralville, IA, USA). The strands were hybridized in hybridization buffer (40 mM Tris–HCl, pH 8, 150 mM Magnesium Chloride (MgCl2)) with a thermocycler (with temperature increased to 95°C and then slowly decreased to 21°C).
Preparation of a stable RNAP–Promoter open complex (RPITC=2)
The transcription open complex (RNAP–Promoter open complex, RPO) can be further stabilized once it forms RPITC=2 (i.e. initial transcribing complex (ITC) at +2 register in reference to the transcription start site) by addition of a dinucleotide. For the preparation of RPITC=2 with the lacCONS promoter, the dinucleotide ApA was added to RPO according to the initial transcribing sequence (ITS in Supplementary Figure S1). Although RPITC=2 is technically an ITC rather than the open complex, in this study, RPITC=2 is referred to as the stable open complex and used as a starting point of the assays because (i) our assays need a complex that is stable for a long time, (ii) there is not a recognizable difference in structure between RPITC=2 and RPO (32–34).
RNAP open complex (RPO) was prepared by incubating 3 μl E. coli RNAP holoenzyme (NEB, Ipswich, MA, USA, M0551S; 1.6 μM), 10 μl 2× transcription buffer (80 mM HEPES KOH, 100 mM KCl, 20 mM MgCl2, 2 mM dithiotreitol, 2 mM 2-mercaptoethylamine-HCl, 200 μg/ml bovine serum albumin, pH 7), 1 μl of 1 μM lacCONS-GTG-20A promoter and 6 μl of water at 37°C for 30 min. To remove nonspecifically-bound RNAP–DNA complexes and free RNAPs, 1 μl of 100 mg/ml Heparin-Sepharose CL-6B beads (GE Healthcare, Little Chalfont, Buckinghamshire, UK) was added to the RPO solution together with 10 μl of pre-warmed 1× transcription buffer followed by 1 min incubation at 37°C and centrifugation for 45 s at 6000 rpm to pellet Heparin-Sepharose. A total of 20 μl of the supernatant containing RPO was transferred to a new tube, and 1.5 μl of 10 mM dinucleotide primer Adenylyl(3′-5′) adenosine (ApA, Ribomed, Carlsbad, CA, USA) was added to the RPo solution, which was then incubated at 37°C for 20 min to form RPITC=2, stabilizing the open complex. The RPITC=2 solution was used as a stock for all transcription reactions. A total of 2 μl of RNase inhibitor (NEB, Ipswich, MA, USA, M0314S) was added to the solution to protect transcribed RNA molecules from possible degradation.
In vitro single-round quenched kinetics transcription assays
Prepared stock solution of RPITC=2 was diluted in transcription buffer containing a given concentration of the crowders. Transcription reactions were initiated by 100 μM of high-purity nucleoside triphosphates (NTPs), (GE Healthcare, Little Chalfont, Buckinghamshire, UK) at 37°C. The transcription reaction was stopped at a given time t by addition of 1.5× volumes of quencher/probe solution containing 1.25 M Guanidium Chloride (GdmCl) and 250 pM ssDNA probe (final concentration of GdmCl and ssDNA probe were 500 mM and 100 pM, respectively). Then, the quenched-probed solution was incubated for an additional 1 h at room temperature in order to establish complete hybridization between transcribed RNA molecules and ssDNA probes. The numbers of hybridized probes (to transcripts) and non-hybridized probes are accurately determined using alternating-laser excitation (ALEX)-based fluorescence-aided molecule sorting (ALEX-FAMS) (35,36).
Experiments were performed with a low concentration of FRET probe (∼100 pM) (within the optimal concentration range for ALEX-FAMS). This is a much lower concentration than the RNAP–Promoter binding affinity (Kd = 10∼100 nM at low temperature (10°C and below) and physiological ionic strength (37)).
Supplementary Figure S10 shows that starting with free 1 nM RNAP holoenzyme and free 1 nM lacCONS-GTG-20dA, there is no detectable amount of transcripts being produced during 15 min incubation time, which is the same incubation time as used for single-point quenched kinetics assays. For transcription reactions starting from pre-formed RPO, a second round of transcription can only occur after (i) finishing the first round of transcription (then RNAP core will be dissociated from σ70 and template DNA), (ii) RNAP core associates with σ70 to form a holoenzyme, (iii) the holoenzyme binds with DNA to form the open complex. (iv) RNAP escapes from the promoter. Therefore, this will take more time than single-round transcription reaction starting from free RNAP holoenzyme and free template DNA. More importantly, the concentration of RPITC=2 used for quenched-kinetics assays is lower than 1 nM (based on template DNA concentration). In this regard, we insure that our assays are based on a single-round run-off transcription reaction (re-association of RNAP to a template dsDNA is unlikely after it dissociates from the template). This, in turn, allows us to quantify single-round transcription efficiencies at different time points after initiation of the reaction by simply counting the number of transcripts. We could therefore study how crowders affect the efficiency of transcription for varying concentrations and the effect of sizes and types of different crowders.
Although the same amounts of reagents were used to generate the RNAP–Promoter open bubble, the efficiency of formation could vary for many reasons. In this context, the concentration of the RNAP–DNA complex was calibrated in order for transcription efficiency at equilibrium (t = ∞) not to exceed the dynamic range of the transcription assay [The concentration of transcripts cannot be more than that of the probe (∼100 pM) at equilibrium.].
For all in vitro quenched kinetics transcription assays, data were acquired for 15∼20 minutes using the ALEX-FAMS setup, as described in Kapanidis et al. (36) with two single-photon Avalanche photodiodes (SPADs, Perkin Elmer Inc., Waltham, MA, USA) and 532 and 638 nm CW lasers (Coherent Inc., Santa Clara, CA, USA) operating at powers of 170 and 80 μW, respectively. All FRET data analyses in this work have been done using a Python-based open-source burst analysis toolkit for confocal single-molecule FRET, FRETBursts (38) and a Python package for non-linear least-squares minimization and curve-fitting, lmfit (zenodo.org/record/11813).
mRNA detection by RNA binding dye assay
The template DNA for RNA binding dye assays contains lacCONS promoter (as used for in vitro single-round quenched kinetics transcription assays) and a short elongation region with no known pausing sequence (Sequence of the non-template strand: 5′-AGGCTTGACACTTTATGCTTCGGCTCGTATAATGTGTGGAATTGTGAGAGCGGAAAAAAAAAAAAAAAAAAAAA-3). For 50-μl reactions, prepared stock solutions of stable RPITC=2 were diluted to the final concentration of 1 nM (based on the concentration of promoter DNA) in transcription buffer containing different types and concentrations of crowders. Transcription reactions were initiated by 500 nM of NTPs and incubated at 37°C for 15 min. DNase I was added with DNase buffer (NEB, Ipswich, MA, USA, M0303S) to each sample, resulting in a total volume of 100 μl, to digest the DNA template and stop the transcription reaction at 37°C for 1 h. Pre-prepared 100 μl of Quant-iT™ OliGreen® solution (Thermo Fisher Scientific Inc, Canoga Park, CA, USA) was then added to each sample to measure the amount of mRNA produced by transcription reactions. Fluorescence signals were obtained with a Tecan Infinte M1000 Plate Reader (excitation wavelength 480 nm, emission wavelength 520 nm).
Microviscosity measurements using fluorescence correlation spectroscopy (FCS)
We performed fluorescence correlation spectroscopy (FCS) measurements to measure microviscosity for RNAP–Promoter complexes under given crowding conditions. lacCONS-GTG-20dA promoter labeled with Atto550 at −5 register on the non-template strand and Atto647N at −8 register (in reference to the transcription start site) on the template strand (26,39) was used for preparation of dual-labeled stable RNAP–Promoter complexes (RPITC=2) as described in #2 Above. The dual-labeled RPITC=2 solution was analyzed by ALEX-FAMS before the FCS measurements to ensure that the final solution contains mostly (≥ 90%) RPITC=2 (and not free promoter DNA, see Supplementary Figure S6).
All FCS measurements were performed at 25°C using a home-built confocal microscope based on an Olympus IX71 with CW laser at 532 nm (Melles Griot Inc., Carlsbad, CA, USA). The emitted photon stream from the labeled complexes was split by a dichroic mirror (635LP, Chroma, VT, USA) into two different color channels (one for the fluorescence from Atto550, and the other for the fluorescence from Atto647N) equipped with SPADs (Perkin Elmer Inc., Waltham, MA, USA). Detected fluorescence signals were sent to an ALV-6000 MultiCorr digital real-time correlator and then cross-correlated to eliminate the unwanted signal from singly-labeled species and detector after-pulsing. Each sample was measured three times.
RESULTS
In vitro transcription quenched-kinetics assays
Our in vitro transcription quenched-kinetics assay is a hybridization-based assay that quantifies the number of transcripts at each time point, defined by the time a reaction quencher is added (26). Once the transcription reaction is stopped by the quencher, the added ssDNA FRET probes hybridize to the transcribed RNAs (during an incubation period). Since the distance between the donor (D) and acceptor (A) dyes on the ssDNA probe increases upon hybridization, a new sub-population of detected molecules with reduced FRET efficiency (E) appears in an ALEX-FAMS 2D histogram—see Figure 1, allowing quantification of the number of transcripts (26,27).
The efficiency of transcription during a given incubation period could be extracted by normalizing the number of single-molecule events for the hybridized sub-population (low E) to the sum of the non-hybridized probe sub-population (high E) and hybridized sub-population (low E). Since the concentrations of probe and RPITC=2 are identical in all measurements, a greater fraction of hybridized probes indicates a higher number of transcripts, that is, higher efficiency of transcription for a given time point (26,27) (Figure 1).
Crowders affect single-round transcriptional kinetics starting from the open complex
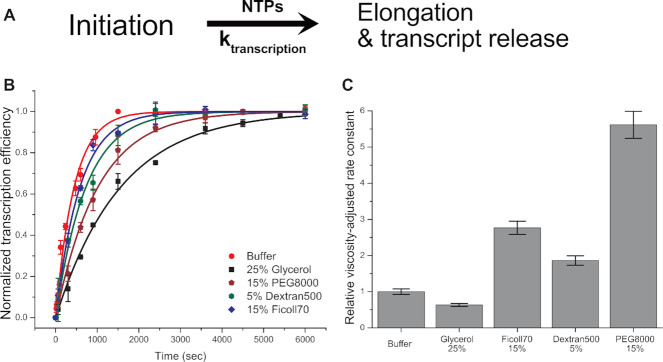
Single-round transcription kinetics (see ‘Materials and Methods’ section #3 for details) starting from stable open complex (RPITC=2), in the presence of various crowding conditions, were measured using the in vitro transcription quenched-kinetics assay (26) to test whether crowders act through modulation of transcription kinetics or through modulation of the activity of RNAP in transcription (i.e. modulating the number of active RNAP–DNA complexes) after RPITC=2 formation. Transcription kinetics were tested with 25% Glycerol, 15% PEG 8000, 15% Ficoll 70 and 5% Dextran 500 (w/v). Transcription kinetics in buffer (i.e. in the absence of viscogen or macromolecular crowders) were also measured as a reference.
Figure 2B shows that the same steady-state levels (t = ∞) were reached for reactions in buffer as in the presence of tested crowders, regardless of their size and type. This shows that crowders do not affect the activity of RNAP. Surprisingly, however, transcription kinetics in polymer solutions (containing PEG, Dextran and Ficoll) are faster than the kinetics in 25% glycerol, even though the viscosities of the polymer solutions are much higher than that of 25% glycerol. Previous studies have demonstrated that the effective viscosity in a crowded medium (referred to as microviscosity) could differ from the bulk viscosity of the medium (40–42). We therefore performed FCS experiments to estimate the actual viscosities (microviscosities) experienced by RPITC=2 for the various crowding environments. These FCS measurements demonstrated that microviscosities for the large crowders Ficoll 70 and Dextran 500 are much smaller than their macroviscosities, while the Dextran10 and PEG 8000 microviscosities are comparable to their macroviscosities (Supplementary Figure S7).
Figure 2.
Transcription kinetics at different crowding conditions measured by the in vitro single-round quenched-kinetics transcription assay. (A) Simplified transcription model used for extraction of kinetic constants from the results of in vitro quenched-kinetics assays. (B) Transcription kinetics in the presence of 25% glycerol (black square), 15% PEG 8000 (wine pentagon), 5% Dextran500 (olive hexagon) and 15% Ficoll70 (blue diamond) compared to transcription kinetics in buffer (red circle). All transcription efficiencies were normalized to the reaction at 25 min without a crowder. (C) The relative viscosity-adjusted rate constants (compared to the rate constant without a crowder). In (B), error bars represent the standard deviation of triplicate repeats. Error bars in (C) were calculated based on microviscosities for crowders and standard errors obtained from the fitting procedure.
To better understand the effects of crowders of various sizes on transcription kinetics, a unidirectional first-order kinetics model (Figure 2A, See Supplementary Data for the details) was used to fit and extract kinetic rate constants. To isolate the crowder’s excluded-volume contribution to transcription kinetics, we adjusted extracted rates by the viscosity using Kramers kinetic theory (which inversely relates rate constants to viscosity) (43). Kramers theory provides a theoretical framework for describing how the viscosity of the reaction medium affects the kinetics of chemical reaction. Several reports demonstrated that the effect of viscosity on some protein folding kinetics follow Kramers theory (44–46). We assume that the kinetics of transcription is similarly modulated by viscosity. The theory allows the effects of viscosity to be decoupled from those due to volume exclusion.
Surprisingly, the viscosity-adjusted rate constants exhibit a recognizable acceleration by a factor of ∼2 (Dextran 500 and Ficoll 70) or ∼6 (PEG 8000) in transcription kinetics for large size crowders (Figure 2C). On the other hand, the viscosity-adjusted kinetics was only marginally affected by 25% glycerol (as compared to reaction in buffer) as expected for glycerol, known as a viscogen that enhances only the viscosity of the medium.
Only large crowders affect the transcription kinetics in other ways than viscosity and therefore act as macromolecular crowders
Since all transcription kinetics measurements exhibited first-order (exponential) kinetics (Figure 2B) and the same steady-state levels for all crowding conditions (regardless of crowder type), it was possible to study the effect of crowder sizes and concentration by simpler (and higher-throughput) single-time-point transcription assays (transcription reaction quenched at a single constant-time point).
Transcription reactions were initiated by incubation with NTPs for a constant time of 15 min for various crowder sizes and concentrations. A 15-min single time point was chosen since it exhibited the highest sensitivity for changes (as function of crowding conditions) in transcription efficiencies (Figure 2B). Since the reaction conditions were supposed to be for a single round of transcription starting from an RNAP bound to a promoter DNA, the assay examines how crowding conditions affect initial transcription and promoter clearance (i.e. excluding the RNAP–promoter binding step).
Our results show that regardless of crowder size, the number of transcribed RNAs at a given time (as compared to an identical reaction with no crowder) decreases with increase in the crowder concentration (Figure 3B and C). However, this reduction is not due to decreased RNAP activity. Rather it is due to the crowder modulating the kinetics of the transcription reaction (note that all kinetic curves reach the same steady-state transcript number, Figure 2B). This result is expected because according to Kramers theory kinetics depend reciprocally on viscosity (43).
Figure 3.
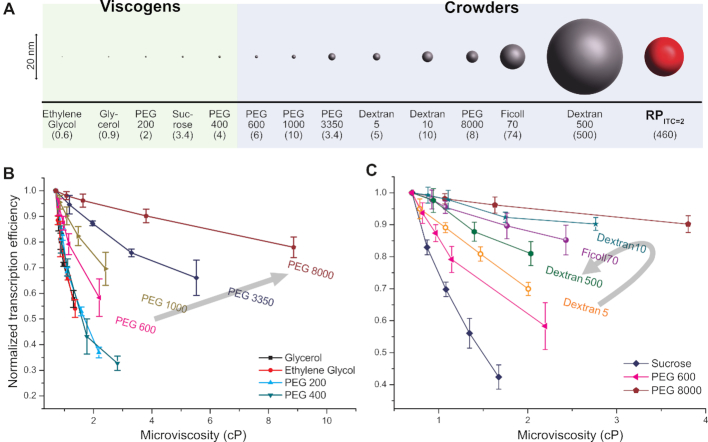
Normalized transcription efficiencies (at a single time point, tincubation = 900 s) for different crowding conditions, as a function of microviscosity. (A) Sizes of crowders used in this study. The sizes of crowders are listed in Supplementary Table S1 and RPITC=2 size (RH, 8.1 nm) was estimated by FCS measurements (see Supplementary Data for details). The average molecular weights (in kDa) appear in the parentheses. (B andC) Transcription efficiencies measured by single-time-point-transcription quenched-kinetics assays for various sizes of PEGs (B) and for Dextran and Ficoll (C). Transcription efficiency values are normalized to the reaction without crowders (buffer only). Error bars represent standard deviations of triplicate runs. Lines are guides to the eye. The black arrows in (B) and (C) indicate the direction of increase in the crowder size.
If the effect of crowders was solely to increase viscosity, the same transcription efficiency should have been observed for all crowder sizes at the same viscosity. Indeed we found that transcription efficiencies at a constant viscosity are similar for small crowders (i.e. no size dependence for Ethylene glycol, PEG 200, Sucrose and PEG 400, similar to glycerol, see bottom four curves in Figure 3B and the a bottom curve in Figure 3C). For large crowders, however, reductions in transcription efficiency (at a constant viscosity) were size dependent (Figure 3B and C). For large PEGs (PEG 600∼PEG 8000), the reduction in transcription efficiency was noticeably smaller (as compared to that of small crowders) and exhibited an inverse relation with respect to size (top four curves in Figure 3B).
To corroborate these results, we performed additional transcription assays with RNA binding dye. This assay showed similar trends distinguishing between small (e.g. Glycerol and PEG 400) and large (e.g. PEG 3500 and 8000) crowders (Supplementary Figure S5).
Large polysaccharide crowders, Dextrans and Ficoll, displayed transcription efficiencies lower than PEG 8000 but higher than PEG 600 at a given microviscosity. For a given microviscosity, the effect of crowder size on the reduction in transcription efficiency is non-monotonic, unlike the observations for large PEGs (Figure 3B and C). Dextran 10, which is larger than Dextran 5 but smaller than Ficoll 70 or Dextran 500, shows the least reduction in transcription efficiency (Figure 3C).
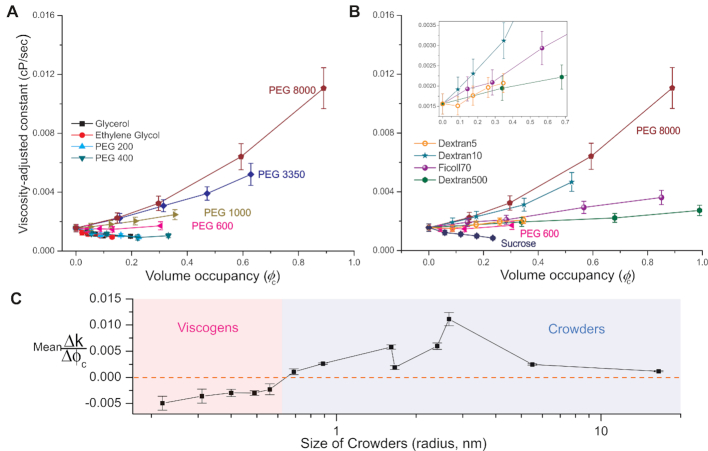
The crowding effect stems from volume exclusion. We therefore plotted in Figure 4 the viscosity-adjusted rate constants (k) as a function of crowder volume occupancy (φc). To estimate the rate constants while decoupling viscosity effects from the data acquired at a single time point (900 s), we derived an equation (Supplementary Equation S6) based on Kramers theory and a first-order kinetics model (Figure 2A). Supplementary Equation S4 was used to calculate volume occupancies.
Figure 4.
Viscosity-adjusted rate constants as a function of crowder’s volume occupancy. Viscosity-adjusted rate constants were calculated according to Supplementary Equation S6 (using results reported in Figure 3). (A and B) Small crowders (< PEG 600) do not affect transcription kinetics (excluding effect of viscosity). (A) Large PEGs (PEG 600∼8000) show strong dependence on size and volume occupancy. (B) Although large polysaccharide crowders (Dextran 5, 10, 500, Ficoll70) show dependence on size and volume occupancy, the trend of size dependence is non-monotonic, which is different from large PEGs. Zoomed-in curves for Dextran 5 (orange hollow circle), Dextran 10 (teal star), Ficoll 70 (violet sphere) and Dextran500 (green hexagon) are shown in the inset. Errors are propagated according to Supplementary Equation S6. (C) Linear fits to all curves in (A) and (B) against crowders were performed to show qualitatively the trend of transcription kinetics as a function of crowder size. Lines are guides to the eye.
After decoupling the viscosity effect, we found that small crowders affected transcription rate constants (as a function of volume occupancy) only marginally, i.e. they do not act as macromolecular crowders, but rather as viscogens (they only increase the viscosity of the medium). In contrast, large crowders (large PEGs, Dextrans and Ficoll 70) strongly affected transcription rate constants. The rate constants depended on size as well as on volume occupancy of the crowders, therefore identifying them as macromolecular crowders (Figure 4).
Although both types (PEGs and polysaccharides) of large crowders showed an overall acceleration of transcription that is proportional to volume occupancy, the dependence of their size on the acceleration of transcription is different. We found that for large PEGs the enhancement of transcription kinetics (due to the crowders) monotonically increases with increasing PEG size while large polysaccharides exhibit a biphasic trend for the transcription enhancement with respect to the crowder size, at a given volume occupancy (Figure 4A and B).
It is noteworthy to mention that (i) similar to the other small crowders used (size smaller than PEG 600), sucrose, a small (disaccharide) crowder, had no effect on transcription kinetics other than viscosity, (ii) at a given volume occupancy, the trend of the size dependence for large polysaccharide crowders is the same as the one for large PEGs up to Dextran 10 whose size is comparable to that of PEG8000. We therefore conclude that (as expected) the observed crowding effect is a function of the crowder’s size and volume occupancy rather than a function of crowder’s type.
Some macromolecular crowding conditions facilitate a phase separation
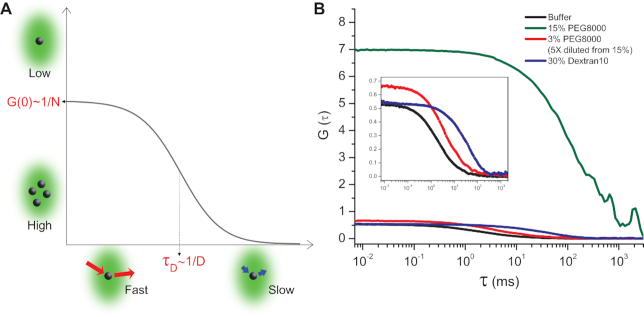
Another interesting crowding-related phenomenon was observed for high concentrations of PEG8000 whereby RNAP–Promoter complexes reversibly aggregated (Figure 5). FCS measurements for RNAP–Promoter complexes, with added PEG8000 at concentrations of 7.5% (Supplementary Figure S7A) and above, exhibited: (i) Exceptionally long residence times, τD. Despite the fact that the bulk viscosities of 15% PEG8000 and 30% Dextran10 are comparable, the residence time for 15% PEG8000 was much longer (compare blue and green curves in Figure 5B); (ii) Large correlation amplitude at zero time delay, G(0), the correlation amplitude for 15% PEG8000, is ∼10 times higher than for all other tested conditions (compare blue, black and green curves in Figure 5B). Once the RPITC=2 solution containing 15% PEG8000 was diluted to 3% PEG8000, these features disappeared (compare green and red curves in Figure 5B). The detailed mechanism for this aggregation is of great interest since previous studies have demonstrated that macromolecular crowding could cause a phase separation that controls enzymatic activities (20,41,47). Surprisingly, this aggregation did not affect transcription rates and yields. We plan to explore this regime further in a future study.
Figure 5.
FCS measurements show that RNAP–Promoter complexes could form reversible aggregates in the presence of PEG8000. (A) A schematic of a fluorescence correlation curve for a single species in a solution. The amplitude of the correlation curve extrapolated to zero time delay (i.e. G(0)) is inversely proportional to the mean number of fluorescent molecules in the detection volume. The characteristic resident time in the detection volume τD is inversely proportional to the diffusion coefficient of the molecules. (B) FCS curves for RPITC=2 in buffer (black), 15% PEG8000 (olive), 3% PEG8000 (red) obtained from 5× dilution of 15% PEG8000 whose correlation curve is shown in olive, 30% Dextran10 (blue). Zoomed-in curves for buffer (black), 3% PEG8000 (red) and 30% Dextran10 (blue) are shown in the inset.
DISCUSSION
We have used an in vitro single-round quenched-kinetics transcription assay (26,27) to investigate the kinetics of E. coli RNAP transcription reactions starting from the stable open complex RPITC=2, in the presence of various crowders. Our measurements focused on the combined kinetics of abortive initiation, promoter clearance and elongation. They excluded the kinetics of promoter binding and bubble opening since crowders were added only after bubble formation was established and since very low concentrations of RNAP–DNA complexes were used (preventing re-association of RNAP to DNA within the measured kinetic times, see ‘Materials and Methods’ section #3 for details). We note that the effect of crowding on the RNAP–DNA association reaction is well documented (20,22).
We found that the activity of RNAP (starting from RPITC=2) is negligibly affected by macromolecular crowding (since kinetic curves for all crowders reached the same steady-state transcription level as in buffer) whereas transcription reaction kinetics are strongly affected by crowders.
Since crowders also act as viscogens, the first effect of crowding is to increase viscosity. This, in turn, slows the reaction kinetics as predicted by Kramers theory (43). We found that the microviscosities for large crowders such as Ficoll 70 and Dextran 500 are much lower than their bulk viscosities. In agreement with FCS microviscosity measurements, we found faster apparent kinetics for Ficoll 70 or Dextran 500 than for PEG 8000 (Figure 2 and Supplementary Figure S7).
The second, and the more interesting observation, is the non-monotonic dependence of reaction kinetics on crowder size. When viscosity effects are decoupled, regardless of the type, small crowders exhibit a negligible effect on transcription kinetics and thus act solely as viscogens. However large crowders exhibit an acceleration in transcription kinetics as their volume fractions increase and act both as viscogens and as macromolecular crowders. Evaluating the effect of different size crowders on the enhancement in transcription kinetics at a given volume occupancy revealed that the trend is biphasic. This indicates an optimal relative size of the crowder to enhance reaction kinetics. Interestingly, comparing different crowders showed accelerated kinetics of transcription with increasing size that peaked when using PEG8000. Crowders larger than PEG8000, Dextran 500 and Ficoll 70, exhibit an opposite trend: the acceleration for Dextran 500 (500 kDa) is smaller than the acceleration for Ficoll 70 (70 kDa) (Figure 4C).
According to the transcription kinetic model (19,48,49) (shown in Supplementary Figure S9), promoter escape is the rate-limiting step of transcription (excluding promoter binding and bubble-opening steps). We hypothesize that the acceleration of the reaction kinetics by crowders is caused by an induced conformational change to a more compact conformation (or stabilization of a more compact conformation) in the RNAP–Promoter initial transcribing complexes, resulting in a reduction in the activation barrier for promoter escape.
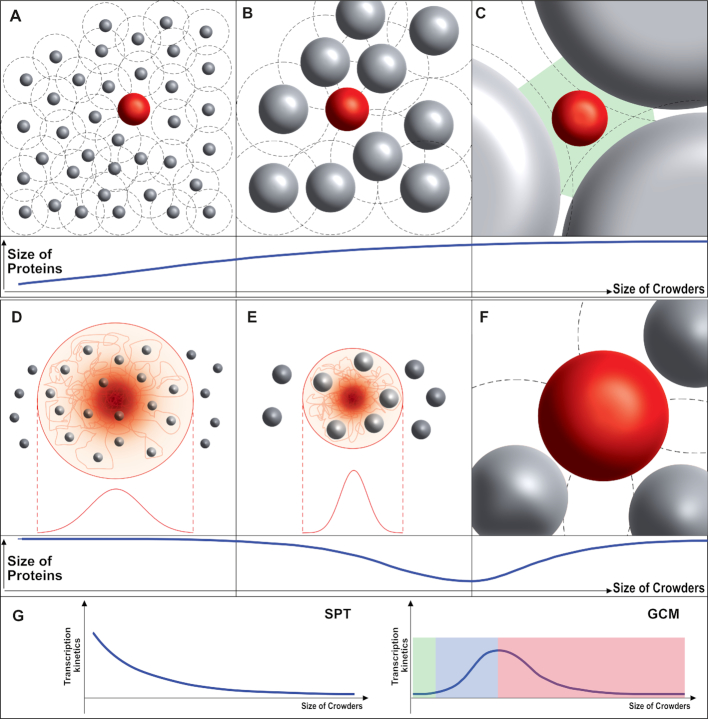
SPT based on the hard-sphere model has been commonly and successfully used to explain the effect of macromolecular crowding in terms of volume exclusion (Figure 6A–C) (10,28). This theory estimates thermodynamic properties of a reaction based on the energy cost for creating a cavity whose size is the same as the one of a newly introduced molecule. According to SPT the energy gained by compacting a newly introduced molecule is reduced as crowder size increases because larger crowders have larger interstitial cavities. These larger cavities could accommodate expanded molecules more easily. SPT therefore predicts that larger crowders would exclude less volume at a fixed volume occupancy as compared to smaller crowders (Figure 6A–C).
Figure 6.
The effect of crowding according to the SPT (top) and GCM (bottom). (A–C, top) The SPT-based hard-sphere model predicts that an unfolded protein chain (red sphere) in the presence of macromolecular crowding becomes less compact as the size of crowder (gray spheres) increases. (A) When the size of a crowder is small compared to the size of the protein, the excluded volume (the volume not accessible to the protein chain, black-dashed circle) is much larger than the volume fraction of the crowder. This leads to the compaction of the protein. (B) As the size of the crowder increases, the excluded volume approaches the volume of the crowder, resulting in less compaction (compared to a smaller crowder at the same volume fraction). (C) When the size of the crowder is much larger than the protein, interstitial cavities (green area) can accommodate the unfolded protein chain without compaction so that it will have the same size (or same protein conformation) as in buffer. (D–F, bottom) GCM predicts a non-monotonic trend for the size of a protein chain under crowding. (D) A crowder smaller than a given (lower-threshold) size would seldom affect the size of the protein (since it permeates the chain, red line). (E) As the crowder size increases (above the lower threshold), less volume would be available for the protein chain (red line) since the chain cannot occupy the same volume as the rigid crowders (gray sphere) in the protein cloud. This induces a compaction of the protein. (F) As the size of the crowder increases further, the GCM eventually turns into the SPT (i.e. the crowder is no longer in the cloud of protein residues). (G) With the hypothesis that folded/compacted structure enhances the transcription kinetics, SPT predicts monotonic decrease in enhancement of transcription kinetics and eventually no effect as the size of crowder increases while GCM predicts (i) marginal effect when crowder’s size is smaller than the lower threshold (highlighted in green), (ii) increase in the enhancement with increasing size when the size is in between lower and upper threshold (highlighted in blue), (iii) decrease in the enhancement and eventually no effect as seen in SPT model when the size is larger than upper threshold (highlighted in red).
With our RNAP conformational change hypothesis, SPT would predict a decrease in enhancement of transcription rates for larger crowders, since larger crowders will not compact RNAP–Promoter complexes by much as compared to smaller crowders, at a constant volume occupancy. This is indeed the case for Dextran 500 and Ficoll 70 (Figure 4B and C). We observed, however, an opposite trend for crowders smaller than Dextran 500 and Ficoll 70, which cannot be explained by SPT. This disagreement could be due to the hard-sphere description for biomolecules and crowders, which fails when (i) the shape of the crowder or of the biomolecule deviates significantly from a sphere (10) or when (ii) the crowder penetrates the biomolecule (i.e. the biomolecule can no longer be approximated by a hard sphere) (14,28).
Generalizations of hard-sphere models have been proposed to account for such cases (10,14,28). The Gaussian cloud model (GCM), proposed by Minton (28), takes into account the fact that a crowder can permeate a biomolecule. In this model, an unfolded protein is treated as a time-averaged spherically symmetric cloud of residues whose density distribution is described by a Gaussian function centered on the protein’s center of mass (Figure 6D and E), with the rigid (hard-sphere) crowder allowed to penetrate the cloud of protein residues (Figure 6D and E). If the crowder size is small, the density distribution of protein residues would be only slightly affected (Figure 6D). As the size of the crowder increases it becomes harder for the unfolded chain to avoid crowders and it will therefore compact itself (Figure 6E). As the size of the crowder increases further, however, it becomes too big to penetrate the protein cloud (28) and the GCM converges to SPT (Figure 6F). Note that this non-monotonic effect (on protein size) of increasing crowder size is not seen in the SPT progression depicted in Figure 6A–C.
Applied to the transcription reaction, the GCM would predict that small crowders will have little effect on transcription kinetics. As the crowder size increases (above a lower threshold), the transcription kinetics would accelerate (as long as the stabilized conformation allows faster kinetics). This acceleration, however, will start to decrease and the kinetics eventually stop accelerating with the increase in size once the crowder’s size transitions to the SPT regime (Figure 6G). This predicted non-monotonic dependence is consistent with our observations (Figure 4C). Because GCM deviates from SPT only when the size of the crowder is small, it is worth noting that PEG 8000 is much smaller than the RNAP–DNA complex (Figure 3A).
The polymeric nature of a protein suggests that it could be partially or fully infiltrated by a crowder, unlike an interaction with a hard sphere. Although a previous report demonstrated that the T7 DNA polymerase complex became more compacted under macromolecular crowding conditions (47), it is still possible that the size of the entire RNAP–Promoter complex will be only nominally affected by crowders since the majority of the complex is well-structured. However, the complex does contain a few partially unstructured domains, or hinged structured domains that are crucial for the enzyme's functionality. For example, the trigger loop is partially disordered and its folding propensity determines the rate of polymerization throughout all transcription stages (after promoter open-complex formation) (50,51). σR3.2 forms an unstructured σ finger loop and its displacement is known to be a common rate-determining step for promoter escape (25,26,52). Also, recent studies (25,26) have suggested that RNAP could be paused in late initiation stages and this pause could be controlled by σR3.2. We therefore argue that macromolecular crowders could compact partially disordered/hinged domains of RNAP that in turn affect transcription kinetic rates. Future studies should examine whether the entire RNAP–DNA complex is affected by crowders, or identify which of the RNAP domains is most affected.
Other than viscosity and volume exclusion, crowders may contribute to physico-chemical properties of biomolecules in various ways. One interesting way is that crowding could affect the characteristics of biomolecules through changing dielectric and thermodynamic properties of solvent. Recently, several studies reported that crowders such as Dextran, Ficoll and PEG notably alter the acidity, alkalinity and polarity of water relative to those in pure water, depending on the polymer type and concentration (53). This aspect of crowding should not be ruled out because change in those properties of water could drastically alter the status of electrostatic interaction and hydrogen bonding of enzymes. We will investigate this possibility in a future study.
At last, we emphasize that since the cellular milieu contains various sizes of crowders such as proteins, glycans and oligonucleotides, our observation of non-monotonic dependence of crowders’ size on the transcription reaction provides valuable insight into elucidating how the crowded cell environment plays a role in the transcription process (specifically in regulatory pathways) in the cell.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Xavier Michalet and Dr. Antonino Ingargiola for fruitful scientific discussions and consultations. The authors also thank Maya Lerner for help in preparation of the illustrations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [GM069709 to S.W.]; NSF [MCB-1244175 to S.W., CHE-1051507 to W.M.G., C.M.K. and MCB-1716925 to W.M.G.]. Funding for open access charge: NSF [MCB-1842951 to S.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fulton A.B. How crowded is the cytoplasm. Cell. 1982; 30:345–347. [DOI] [PubMed] [Google Scholar]
- 2. Zimmerman S.B., Trach S.O.. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991; 222:599–620. [DOI] [PubMed] [Google Scholar]
- 3. Ellis R.J., Minton A.P.. Cell biology: join the crowd. Nature. 2003; 425:27–28. [DOI] [PubMed] [Google Scholar]
- 4. Tabaka M., Kalwarczyk T., Szymanski J., Hou S., Holyst R.. The effect of macromolecular crowding on mobility of biomolecules, association kinetics, and gene expression in living cells. Front. Phys. 2014; 2:54. [Google Scholar]
- 5. Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001; 26:597–604. [DOI] [PubMed] [Google Scholar]
- 6. Kuznetsova I.M., Turoverov K.K., Uversky V.N.. What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 2014; 15:23090–23140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Minton A.P. Macromolecular crowding. Curr. Biol. 2006; 16:R269–R271. [DOI] [PubMed] [Google Scholar]
- 8. Hagen S.J. Solvent viscosity and friction in protein folding dynamics. Curr. Protein Pept. Sci. 2010; 11:385–395. [DOI] [PubMed] [Google Scholar]
- 9. de Sancho D., Sirur A., Best R.B.. Molecular origins of internal friction effects on protein-folding rates. Nat. Commun. 2014; 5:4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou H.-X., Rivas G., Minton A.P.. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008; 37:375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minton A.P. How can biochemical reactions within cells differ from those in test tubes. J. Cell Sci. 2006; 119:2863–2869. [DOI] [PubMed] [Google Scholar]
- 12. Dupuis N.F., Holmstrom E.D., Nesbitt D.J.. Molecular-crowding effects on single-molecule RNA folding/unfolding thermodynamics and kinetics. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung M.S., Klimov D., Thirumalai D.. Molecular crowding enhances native state stability and refolding rates of globular proteins. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:4753–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soranno A., Koenig I., Borgia M.B., Hofmann H., Zosel F., Nettels D., Schuler B.. Single-molecule spectroscopy reveals polymer effects of disordered proteins in crowded environments. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murakami K.S., Darst S.A.. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 2003; 13:31–39. [DOI] [PubMed] [Google Scholar]
- 16. Busby S., Kolb A., Buc H.. Buc H, Strick T. Where it all Begins: An Overview of Promoter Recognition and Open Complex Formation. RNA Polymerases as Molecular Motors. 2009; Cambridge: RSC Publishing; 13–37. [Google Scholar]
- 17. Rojo F. Repression of transcription initiation in bacteria. J. Bacteriol. 1999; 181:2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Losick R. In vitro transcription. Annu. Rev. Biochem. 1972; 41:409–446. [DOI] [PubMed] [Google Scholar]
- 19. Dangkulwanich M., Ishibashi T., Bintu L., Bustamante C., Choy J.L.. Molecular mechanisms of transcription through single-molecule experiments. Chem. Rev. 2014; 114:3203–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sokolova E., Spruijt E., Hansen M.M.K., Dubuc E., Groen J., Chokkalingam V., Piruska A., Heus H. A., Huck W.T.S.. Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11692–11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen M.M.K., Meijer L.H.H., Spruijt E., Maas R.J.M., Rosquelles M.V., Groen J., Heus H.A., Huck W.T.S.. Macromolecular crowding creates heterogeneous environments of gene expression in picolitre droplets. Nat. Nanotechnol. 2015; 11:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ge X., Luo D., Xu J.. Cell-free protein expression under macromolecular crowding conditions. PLoS One. 2011; 6:e28707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsuda H., Putzel G.G., Backman V., Szleifer I.. Macromolecular crowding as a regulator of gene transcription. Biophys. J. 2014; 106:1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Demidenko A.A., Lee J., Powers T.R., Nibert M.L.. Effects of viscogens on RNA transcription inside reovirus particles. J. Biol. Chem. 2011; 286:29521–29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duchi D., Bauer D.L.V., Fernandez L., Evans G., Robb N., Hwang L.C., Gryte K., Tomescu A., Zawadzki P., Morichaud Z. et al. . RNA polymerase pausing during initial transcription. Mol. Cell. 2016; 63:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lerner E., Chung S., Allen B.L., Wang S., Lee J., Lu S.W., Grimaud L.W., Ingargiola A., Michalet X., Alhadid Y. et al. . Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6562–E6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S., Streets A.M., Lin R.R., Quake S.R., Weiss S., Majumdar D.S.. High-throughput single-molecule optofluidic analysis. Nat. Methods. 2011; 8:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minton A.P. Models for excluded volume interaction between an unfolded protein and rigid macromolecular cosolutes: macromolecular crowding and protein stability revisited. Biophys. J. 2005; 88:971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murphy M.C., Rasnik I., Cheng W., Lohman T.M., Ha T.. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004; 86:2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brinkers S., Dietrich H.R.C., de Groote F.H., Young I.T., Rieger B.. The persistence length of double stranded DNA determined using dark field tethered particle motion. J. Chem. Phys. 2009; 130:215105. [DOI] [PubMed] [Google Scholar]
- 31. Abels J.A., Moreno-Herrero F., van der Heijden T., Dekker C., Dekker N.H.. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys. J. 2005; 88:2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zuo Y., Steitz T.A.. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell. 2015; 58:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kapanidis A.N., Margeat E., Ho S.O., Kortkhonjia E., Weiss S., Ebright R.H.. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006; 314:1144–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basu R.S., Warner B. a, Molodtsov V., Pupov D., Esyunina D., Fernandez-Tornero C., Kulbachinskiy A., Murakami K.S.. Structural basis of transcription initiation by bacterial RNA Polymerase holoenzyme. J. Biol. Chem. 2014; 289:24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kapanidis A.N., Laurence T.A., Nam K.L., Margeat E., Kong X., Weiss S.. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005; 38:523–533. [DOI] [PubMed] [Google Scholar]
- 36. Kapanidis A.N., Lee N.K., Laurence T.A., Doose S., Margeat E., Weiss S.. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:8936–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pieter L., Haseth D.E., Zupancic M.L.. MINIREVIEW RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1998; 180:3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ingargiola A., Lerner E., Chung S., Weiss S., Michalet X.. FRETBursts: an open source toolkit for analysis of freely-diffusing single-molecule FRET. PLoS One. 2016; 11:e0160716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lerner E., Ingargiola A., Lee J.J., Borukhov S., Michalet X., Weiss S.. Different types of pausing modes during transcription initiation. Transcription. 2017; 8:242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rashid R., Chee S.M.L., Raghunath M., Wohland T.. Macromolecular crowding gives rise to microviscosity, anomalous diffusion and accelerated actin polymerization. Phys. Biol. 2015; 12:034001. [DOI] [PubMed] [Google Scholar]
- 41. Hou S., Trochimczyk P., Sun L., Wisniewska A., Kalwarczyk T., Zhang X., Wielgus-Kutrowska B., Bzowska A., Holyst R.. How can macromolecular crowding inhibit biological reactions? The enhanced formation of DNA nanoparticles. Sci. Rep. 2016; 6:22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kalwarczyk T., Ziebacz N., Bielejewska A., Zaboklicka E., Koynov K., Szymański J., Wilk A., Patkowski A., Gapiński J., Butt H.J. et al. . Comparative analysis of viscosity of complex liquids and cytoplasm of mammalian cells at the nanoscale. Nano Lett. 2011; 11:2157–2163. [DOI] [PubMed] [Google Scholar]
- 43. Kramers H.A. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940; 7:284–304. [Google Scholar]
- 44. Plaxco K.W., Baker D.. Limited internal friction in the rate-limiting step of a two-state protein folding reaction. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:13591–13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jas G.S., Eaton W.A., Hofrichter J.. Effect of Viscosity on the Kinetics of α-Helix and β-Hairpin Formation. J. Phys. Chem. B. 2001; 105:261–272. [Google Scholar]
- 46. Pabit S.A., Roder H., Hagen S.J.. Internal friction controls the speed of protein folding from a compact configuration. Biochemistry. 2004; 43:12532–12538. [DOI] [PubMed] [Google Scholar]
- 47. Akabayov B., Akabayov S.R., Lee S.-J., Wagner G., Richardson C.C.. Impact of macromolecular crowding on DNA replication. Nat. Commun. 2013; 4:1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bai L., Shundrovsky A., Wang M.D.. Buc H, Strick T. Kinetic modeling of transcription elongation. RNA Polymerases as Molecular Motors. 2009; Cambridge: RSC Publishing; 263–280. [Google Scholar]
- 49. Geszvain K., Landick R. 2005. Higgins N. The structure of bacterial RNA polymerase. The bacterial chromosome. 2005; Washington, D.C.: ASM Press; 283–296. [Google Scholar]
- 50. Mejia Y.X., Nudler E., Bustamante C.. Trigger loop folding determines transcription rate of Escherichia coli's RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E.. A ratchet mechanism of transcription elongation and its control. Cell. 2005; 120:183–193. [DOI] [PubMed] [Google Scholar]
- 52. Samanta S., Martin C.T.. Insights into the mechanism of initial transcription in Escherichia coli RNA polymerase. J. Biol. Chem. 2013; 288:31993–32003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kuznetsova I.M., Zaslavsky B.Y., Breydo L., Turoverov K.K., Uversky V.N.. Beyond the excluded volume effects: mechanistic complexity of the crowded milieu. Molecules. 2015; 20:1377–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.