Abstract
Poly (ADP-ribose) polymerase 1 (PARP1) has emerged as an attractive target for cancer therapy due to its key role in DNA repair processes. Inhibition of PARP1 in BRCA-mutated cancers has been observed to be clinically beneficial. Recent genome-mapping experiments have identified a non-canonical G-quadruplex-forming sequence containing bulges within the PARP1 promoter. Structural features, like bulges, provide opportunities for selective chemical targeting of the non-canonical G-quadruplex structure within the PARP1 promoter, which could serve as an alternative therapeutic approach for the regulation of PARP1 expression. Here we report the G-quadruplex structure formed by a 23-nucleotide G-rich sequence in the PARP1 promoter. Our study revealed a three-layered intramolecular (3+1) hybrid G-quadruplex scaffold, in which three strands are oriented in one direction and the fourth in the opposite direction. This structure exhibits unique structural features such as an adenine bulge and a G·G·T base triple capping structure formed between the central edgewise loop, propeller loop and 5′ flanking terminal. Given the highly important role of PARP1 in DNA repair and cancer intervention, this structure presents an attractive opportunity to explore the therapeutic potential of PARP1 inhibition via G-quadruplex DNA targeting.
INTRODUCTION
Poly (ADP-ribose) polymerase-1 (PARP1) is a nuclear enzyme known for its key role in the DNA repair processes (1). PARP1 binds to DNA breaks and recruits DNA repair proteins by catalyzing the formation of poly (ADP-ribose) scaffolds (1,2). Additionally, PARP1 has also been implicated in other cellular processes such as transcriptional regulation, chromatin remodelling, cell signaling and cell death (3–5). Synthetic lethality induced by PARP1 inhibition has emerged as a promising method for targeting tumor cells with defective homologous recombination pathways (6). Inhibition of PARP1 activity in homologous recombination deficient cancer cells causes accumulation of unrepaired single-strand DNA breaks which are potentially converted to double-stranded DNA breaks, causing cell death selectively in those cancer cells (7).
Consequently, several PARP1 inhibitors were developed showing promising results, with olaparib being the first one to be approved clinically for treating BRCA1/2 mutated cancers (8,9). However, the use of these drugs would be limited in light of increasing evidence showing acquired resistance to PARP1 inhibitors, mediated by multiple molecular mechanisms including changes in the expression of PARP1 itself (10). Alternative strategies are required to selectively regulate PARP1 expression that can be used in combination with current PARP1 inhibitors or alone.
A recent G-quadruplex-specific sequencing study (G4-seq) has identified non-canonical G-quadruplex-forming sequences in the promoter region of the PARP1 gene (11), suggesting a potential for interfering with PARP1 regulation via G-quadruplex targeting. A link between ligand-mediated stabilization of G-quadruplexes in gene promoters and transcriptional regulation has been proposed for several oncogenes (12–14). Moreover, PARP1 has also been linked with transcriptional regulation of c-KIT, c-MYC and KRAS oncogenes by interacting with G-quadruplex structures in the promoter (15–17). Chemical inhibition of PARP1 synergized with the G-quadruplex ligand RHPS4 in HT29 xenografts could further inhibit tumor cell growth by preventing the repair of uncapped telomere ends (18). Transcriptional repression of PARP1 could, therefore, help in the prevention of diseases that occur due to overexpression of PARP1 and subsequently influence the expression of other oncogenes. To investigate the opportunity of G-quadruplex mediated PARP1 regulation, we set to confirm that the G-rich sequence element in the PARP1 promoter indeed forms a stable G-quadruplex and if so, determine its folded structure. In particular, we aimed to elucidate unique structural features of the PARP1 G-quadruplex that could, in principle, be exploited for selective chemical targeting of this promoter, given the therapeutic relevance of PARP1.
G-quadruplexes are generally four-stranded structures formed by G-rich nucleic acids comprising a stack of multiple guanine(G)-tetrads. A G-tetrad is a square planar arrangement of four guanines stabilized by Hoogsteen hydrogen bonds with a metal ion coordination in the center (19,20). A wide variety of G-quadruplex structures have been elucidated, differing in terms of strand orientations, glycosylic conformations, groove widths, connecting loops, molecularity and the number of tetrads (21–23). A widely used regular sequence motif for the prediction of potential G-quadruplex-forming sequences (pG4) is G3+N1–7G3+N1–7G3+N1–7G3+, where tracts of three or more continuous guanines will form G-quadruplex columns and linker sequences will form the connecting loops (24,25). As many as 376 000 G-quadruplex-forming sequences have been reported to exist in the human genome using this algorithm (24,25). However, it is now evident that non-canonical G-quadruplex-forming sequence motifs exist that include bulges (26–28), G-triads (29), long connecting loops (30,31), structured loops (32–35), left-handed (36) and GAGA/GCGC-tetrad containing quadruplexes (37). G4-seq has recently identified 716 310 G-quadruplex-forming sequences in the human genome, among which 451 646 sequences were non-canonical including those containing long loops and bulges (11) not predicted by previous bioinformatics searches using regular pG4 motifs (24,25).
In this study, G-quadruplex structure formation in the PARP1 promoter DNA sequence TGGGGGCCGAGGCGGGGCTTGGG (termed as TP3) located 125 nucleotide (nt) upstream of the transcription start site (TSS) is examined by NMR spectroscopy. TP3 was chosen among other sequences identified in the PARP1 gene due the fact that it displays a well-resolved NMR spectrum which is indicative of a well-defined structure, further facilitating structural determination (Supplementary Figure S1). Our study shows the formation of a (3+1) hybrid, intramolecular, three-layered G-quadruplex topology with unique structural features.
MATERIALS AND METHODS
Sample preparation
Unlabeled and site-specific labelled DNA oligonucleotides were chemically synthesized on an ABI 394 DNA synthesizer using products from Glen Research and Cambridge Isotope Laboratories and then purified following the protocol from Glen Research. The samples (concentration, 0.2−2 mM) were dialyzed successively against water, 25 mM KCl, and water again. DNA oligonucleotides were frozen, lyophilized and dissolved in buffer containing 70 mM KCl and 20 mM KPi (pH 7) buffer. The DNA concentration was expressed in strand molarity using the nearest neighbor approximation for the 260 nm molar extinction coefficient of the unfolded species.
CD/UV melting experiments
The stability of G-quadruplexes was measured via CD/UV melting experiments conducted on a JASCO V-650 spectrophotometer or a JASCO-815 spectropolarimeter. Experiments were performed with 1-cm path length quartz cuvettes. The absorbance at 295 nm was recorded as a function of temperature ranging from 15 to 95°C. Heating and cooling were done at a rate of 0.2°C/min. The DNA concentration ranged from 4 to 6 μM and experiments were carried out in buffer containing 70 mM KCl, 20 mM KPi (pH 7), as well as other buffer conditions in which the concentration of KCl and KPi were varied and KCl was substituted with LiCl (data not shown). To calculate the DNA folded fraction, all UV melting curves are manually fitted for two-state folding where two baselines are drawn at low and high temperatures corresponding to completely folded and unfolded states. The difference in the Tm values from folding and unfolding was <2°C.
Circular dichroism
Circular dichroism (CD) spectra were recorded on a JASCO-815 spectropolarimeter using 1-cm or 0.01-cm path length quartz cuvettes in a reaction volume of 480 or 30 μl, respectively, at 25°C. Scans from 220 to 320 nm were performed with a rate of 200 nm/min, 1-nm pitch, and 2-nm bandwidth. DNA samples with concentrations of approximately 6 μM or 250 μM, dissolved in a buffer containing 70 mM KCl, 20 mM KPi (pH 7), were used for experiments. For each measurement, an average of three scans was taken, the spectral contribution of the buffer was subtracted and data were zero-corrected at 320 nm.
Gel electrophoresis
The molecular size of the oligonucleotides was visualized by non-denaturing gel electrophoresis. Oligonucleotides were prepared in the mentioned buffer conditions. The samples were loaded on a 20% polyacrylamide gel, supplemented with 50 mM KCl and 40% sucrose was added just before loading. The gel was viewed by UV shadowing.
NMR spectroscopy
NMR experiments were performed on Bruker Avance spectrometers operating at 600/700/800 MHz at 25°C, unless otherwise specified. The DNA concentration for NMR experiments was typically 0.2−2.0 mM in 70 mM KCl, 20 mM KPi (pH 7) at 25°C, unless otherwise specified. Note different baseline levels of the presented TP3 spectra, which are sensitive to the sample concentration and time after dilution (see below). Assignment of the imino protons of guanine residues was obtained by 15N-filtered experiments using 4% site-specific labelled samples. Assignment of aromatic protons of guanine residues was obtained using both 2H (100%) and 15N (4%) site-specific labeling. In the 2H (100%) site-specific labeling approach, a simple 1D NMR spectrum was recorded and compared with the reference spectrum to identify the missing peak. In the 15N (4%) site-specific labeling approach, a 1D 15N–1H HMQC experiment was performed to filter out proton signals not coupled with 15N. The two approaches are complementary: the site-specific deuterium (2H) labeling approach requires less sample and NMR recording time, but may suffer from spectral overlap. Assignment of thymine residues was further facilitated by site-specific T-to-U substitutions. Spectral assignments were made by carrying out through-space (NOESY) and through-bond correlation experiments (TOCSY). Spectra analyses were performed using the SpinWorks (http://home.cc.umanitoba.ca/∼wolowiec/spinworks/) and SPARKY (https://www.cgl.ucsf.edu/home/sparky/) programs.
Structure calculations
NOE distance restraints
Inter-proton distances for TP3-T6 were obtained from NOESY experiments performed in H2O and D2O at various mixing times (100, 250 and 300 ms). For non-exchangeable protons, the peaks were classified as strong, medium and weak corresponding to the distance restraints of (2.7 ± 0.8), (3.8 ± 0.9) and (5.5 ± 1.7) Å respectively. Distances from exchangeable protons were classified as strong, medium and weak corresponding to the distance restraints of (3.6 ± 0.9), (4.8 ± 1.2) and (5.5 ± 1.7) Å respectively.
Dihedral restraints
Dihedral angle restraints were imposed to the dihedral angle formed by O4′–C1′–N9–C4 of guanine residues. Anti guanine residues were restricted to an angle of (240 ± 70)° or (240 ± 40)° for the outer tetrad and inner tetrad guanines respectively. Syn guanine residues were restricted to an angle of (60 ± 70)° or (60 ± 40)° for the outer tetrad and inner tetrad guanines respectively.
Hydrogen bond restraints
Hoogsteen hydrogen bonds between guanines were restrained using H21–N7, N2–N7, H1–O6 and N1–O6 distances, which were set to (2.0 ± 0.2), (2.9 ± 0.3), (2.0 ± 0.2) and (2.9 ± 0.3) Å respectively. The hydrogen bond restraints from the two interacting guanines G14 and G2 were (2.0 ± 0.2) for G14(H21)–G2(O6) and G14(H1)–G2(N7), as well as (2.9 ± 0.3) Å for G14(N2)-G2(O6) and G14(N1)-G2(N7). The hydrogen bond from G2(H1) was assigned ambiguously towards either T20(O2), T20(O4) or T20(O4′) with restraints of (2.0 ± 0.2) Å.
Planarity restraints
Planarity restraints were used for the G3–G21–G15–G12, G4–G11–G16–G22 and G5–G9–G17–G23 tetrads and G14–G2 base pair.
Repulsion restraints
Repulsion restraints between T(20)–H7 to G(14)–H1 and G(2)–H1 of (>3.5 Å) were imposed following the non-existence of the corresponding cross-peaks in the H2O NOESY spectra (both in 10°C and 25°C).
Distance geometry simulated annealing
An initial extended conformation of TP3-T6 was generated using the XPLOR-NIH program (38). The system was then subjected to distance geometry simulated annealing by incorporating distance, dihedral, hydrogen bond, planarity and repulsion restraints. One hundred structures were generated and subjected to further refinement.
Distance-restrained molecular dynamics refinement
The 100 structures obtained from the simulated annealing step were refined with a distance-restrained molecular dynamics protocol incorporating all distance restraints. The system was heated from 300 to 1000 K in 14 ps and allowed to equilibrate for 6 ps, during which force constants for the distance restraints were kept at 2 kcal.mol−1.Å−2. The force constants for non-exchangeable proton and exchangeable proton restraints were then increased to 16 and 8 kcal.mol−1.Å−2 respectively in 20 ps before another equilibration at 1000 K for 50 ps. Next, the system was cooled down to 300 K in 42 ps, after which an equilibration was performed for 18 ps. Coordinates of the molecule were saved every 0.5 ps during the last 10.0 ps and averaged. The average structure obtained was then subjected to minimization until the gradient of energy was less than 0.1 kcal.mol−1. Dihedral (50 kcal.mol−1.rad−2) and planarity (1 kcal.mol−1.Å−2 for tetrads) restraints were maintained throughout the course of refinement. Ten-lowest energy structures were generated.
Data deposition
The coordinates for the ten lowest-energy structures of the TP3-T6 G-quadruplex have been deposited in the Protein Data Bank (PDB ID: 6AC7).
RESULTS AND DISCUSSION
TP3 forms a (3+1) intramolecular G-quadruplex structure
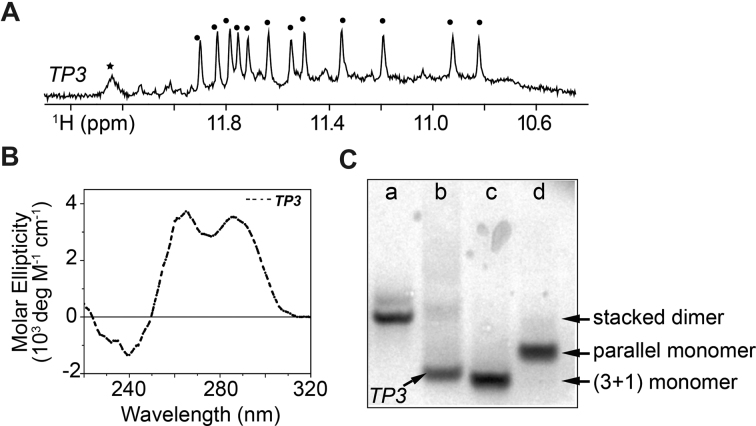
TP3 shows twelve major sharp imino proton peaks from 10.8 to 12.0 ppm (Figure 1), indicative of the formation of a major G-quadruplex fold containing three G-tetrads. An additional major peak was observed at ∼12.2 ppm (see further discussion below).
Figure 1.
(A) Imino proton spectrum of TP3. (B) CD spectrum of TP3. (C) Native PAGE mobility shift analysis of TP3 (lane-b) with other references; (lane-a) T95 (d(GGGTGGGTGGGTGGGT), stacked dimer G-quadruplex), (lane-c) HT (d(TTGGGTTAGGGTTAGGGTTAGGGA), three-layer, monomer (3+1) hybrid G-quadruplex) and (lane-d) T95-2T (d(TTGGGTGGGTG GGTGGGT), three-layer, monomer, propeller-type parallel G-quadruplex).
Unambiguous assignments of guanine imino (H1) and aromatic (H8) protons for the major species of TP3 (Supplementary Figures S2 and S3) were carried out via site-specific 4% 15N-labeling and site-specific 2H substitution approaches; some representative imino proton spectral assignments are shown in Figure 2 (39). Imino proton assignments revealed the twelve guanines that are involved in the formation of the G-tetrad core (underlined, Figure 2A). The CD spectrum of the TP3 G-quadruplex displays two positive peaks at 265 and 290 nm along with a trough at 240 nm, as previously reported for (3+1) hybrid G-quadruplexes (40–42). Native PAGE analysis compares the migration of the TP3 G-quadruplex structure with respect to other reference G-quadruplex structures. The migration of TP3 is comparable to that of the human telomeric (HT) sequence forming a three-layered, monomeric (3+1) hybrid G-quadruplex (43) and faster than other reference samples T95-2T (three-layered, monomeric and propeller-type parallel G-quadruplex) and T95 (stacked dimer parallel G-quadruplex) (44). Altogether, these data suggest that TP3 forms a three-layered monomeric G-quadruplex.
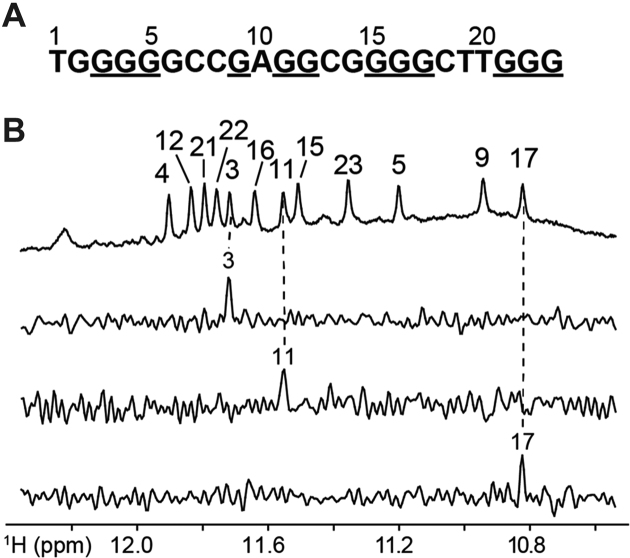
Figure 2.
(A) TP3 DNA sequence with the guanines forming G-tetrads underlined. (B) Imino proton resonance assignment of TP3 indicated on the reference spectrum (top). Below are some examples of the spectral assignment of individual guanine imino protons using samples with 4% 15N-labeled guanines at indicated positions.
However, due to poor long-term stability of the G-quadruplex structure in TP3 at room temperature (Supplementary Figure S4), we could not execute long two-dimensional NMR experiments to determine the structure of the TP3 G-quadruplex. Temporal instability of the G-quadruplex fold in the TP3 sequence could probably arise due to the formation of higher-order aggregate structures over time, as indicated by the appearance of a background hump in the imino proton NMR spectra, enhanced 265-nm peak in the CD profile and low-mobility smear bands in native PAGE (Figure 1 and Supplementary Figure S4). Despite its poor temporal stability, the TP3 monomer displays a relatively high melting temperature (Tm) of 59.6°C as evidenced by UV and CD melting experiments in the presence of 70 mM KCl and 20 mM KPi (Supplementary Figure S5). Moreover, within the transcription factor binding timescale of few milliseconds to hundred seconds (45,46), this G-quadruplex structure largely remains a monomer under physiological conditions and evolves into aggregated higher-order structures only over time.
A small sequence change facilitated NMR structure determination
Variants of the TP3 sequence were examined, where each non-tetrad forming guanine residue (G2, G6 and G14) was replaced by a thymine (G-to-T substitution). Imino proton spectra of mutants show that single thymine mutations at G2 and G14 result in the co-existence of multiple G-quadruplexes (Supplementary Figure S6), while G-to-T substitution at position 6 (TP3-T6) shows a NMR spectrum very similar to that of TP3 (Figure 3A), indicating the formation of the same G-quadruplex fold. Furthermore, the TP3-T6 G-quadruplex structure displays improved temporal stability upon exposure to room temperature, enabling a detailed NMR structural study. CD spectra of TP3-T6, measured at concentrations of 6 and 250 μM, displays two positive peaks at 265 and 290 nm along with a trough at 240 nm (Supplementary Figure S7), indicative of a (3+1) hybrid G-quadruplex, as observed for TP3.
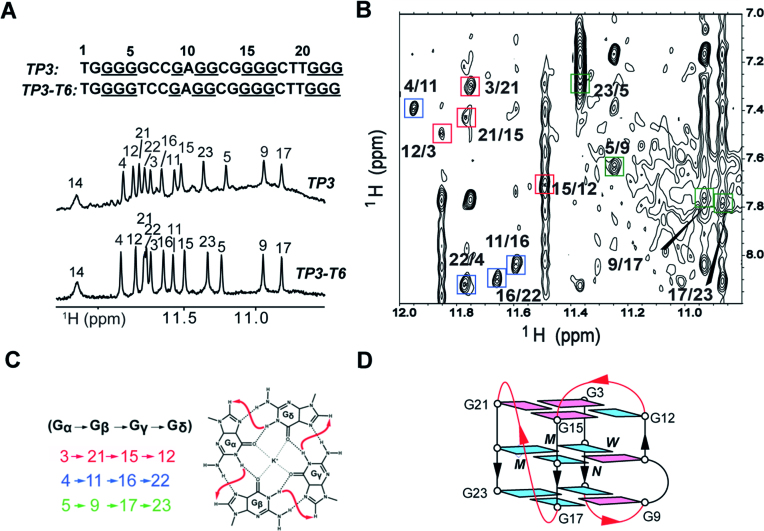
Figure 3.
(A) Imino proton spectra of TP3 and the mutated TP3-T6 sequence with peak assignments on top. (B) NOESY spectrum (mixing time, 250 ms) showing the H1/H8 connectivity of G-tetrads. Guanine H1/H8 cross-peaks for G-tetrads are framed and labeled. The residues in different G-tetrads are indicated by different colors. (C) Specific H1-H8 connectivity pattern of the three tetrads. (D) Schematic of the folding topology of TP3-T6. Syn and anti guanines are colored as magenta and cyan respectively. W, M and N represent wide, medium and narrow groove widths respectively.
We proceeded to study the G-quadruplex structure of the TP3-T6 sequence in more detail. Spectral assignment of TP3-T6 was carried out by the site-specific (4%) 15N-enrichment approach. Guanine imino (H1) and aromatic (H8) protons were unambiguously assigned for TP3-T6 (Supplementary Figure S8) and some of the assignments were also extrapolated from TP3 (Supplementary Figures S2 and S3) and vice versa as proton chemical shifts and spectral patterns in both sequences are extremely similar. The imino proton at ∼12.2 ppm in TP3-T6 was assigned to G14 by site-specific 15N-enrichment experiments (Supplementary Figure S8). The imino proton of G2 was identified at ∼10.8 ppm by 15N-enrichment experiments at 10°C (Supplementary Figure S9). Assignments of thymine residues were determined via through-bond correlation experiments as well as T-to-U substitutions (Supplementary Figure S10). The folding topology was determined using NOESY experiments (Figure 3 and Supplementary Figures S11 and S12). Cyclic NOE connectivities between the imino (H1) proton of a guanine and the aromatic (H8) proton of the neighboring guanine in the same tetrad (Figure 3B) established G-tetrad alignments for TP3-T6: G3·G21·G15·G12 (red), G4·G11·G16·G22 (blue) and G5·G9·G17·G23 (green) (Figure 3C). High-intensity intra-residue H8-H1′ NOE cross peaks were observed for five guanine residues G3, G9, G11, G15 and G21 (Supplementary Figure S11), indicating the syn glycosylic conformation adopted by them, whereas other guanines in the tetrad core adopt an anti glycosylic conformation. NOE sequential connectivity between residues H1′(n)-H8(n+1) strongly depends on their glycosylic conformation (39). Syn-to-anti inter-residue connections, are supported by the observation of specific rectangular NOE patterns of H1′(n)–H8(n+1) and H1′(n+1)–H8(n) cross peaks (Supplementary Figure S11). Additionally, syn·syn·syn·anti alignment of the top and anti·anti·anti·syn alignment of the two bottom G-tetrads are confirmed by guanine H1–H1 and H8–H8 NOE peaks (Supplementary Figure S12). Thus, the folding topology of TP3-T6 reveals a (3+1) hybrid G-quadruplex scaffold, in which three strands (involving the G3–G5, G15–G17 and G21–G23 tracts) are oriented in one direction and the fourth (G9–G12 tract) in the opposite direction connected by two edgewise loops and one propeller loop (Figure 3D). Strand orientations in TP3-T6 result in two medium grooves, one narrow groove and a wide groove being present. The first edgewise loop (TCC) connects across the wide groove at the bottom, the second edgewise loop (CG) connects across the narrow groove at the top, while the third propeller loop (CTT) traverses across the medium groove connecting two parallel stands.
G4, G11, G16 and G22 form the central tetrad layer. This is independently confirmed in solvent exchange experiments, whereby the imino protons of these residues are among the most protected after 20 minutes (Supplementary Figure S13). Adenine interruption (A10) in the second guanine tract is incorporated as a bulge between two tetrads. The close proximity of G11 to the A10 bulge might account for the relatively shorter exchange time of the G4 imino proton, which is hydrogen-bonded to G11(N7), as compared to other central guanines.
Solution structure of the TP3-T6 quadruplex
The solution structure of TP3-T6 was computed based on NMR restraints (Table 1) extracted from NOE cross-peaks in NOESY experiments.
Table 1.
Statistics of the computed structures of TP3-T6a
| A. NMR restraints | |
| distance restraints | |
| Intraresidue | |
| Exchangeable | 0 |
| Non-exchangeable | 312 |
| Interresidue | |
| Exchangeable | 45 |
| Non-exchangeable | 75 |
| other restraints | |
| hydrogen-bond restraints | 53 |
| dihedral restraints | 13 |
| planarity restraints | 4 |
| repulsion restraints | 2 |
| B. Structure Statistics | |
| NOE violations | |
| number (>0.2 Å) | 0.2 ± 0.4 |
| deviations from the ideal covalent geometry | |
| bond lengths (Å) | 0.003 ± 0.000 |
| bond angles (deg) | 0.687 ± 0.006 |
| impropers (deg) | 0.339 ± 0.005 |
| pairwise all heavy atom rmsd values (Å) | |
| G-tetrad core | 0.402 ± 0.122 |
| all residues | 0.601 ± 0.142 |
aPDB ID 6AC7.
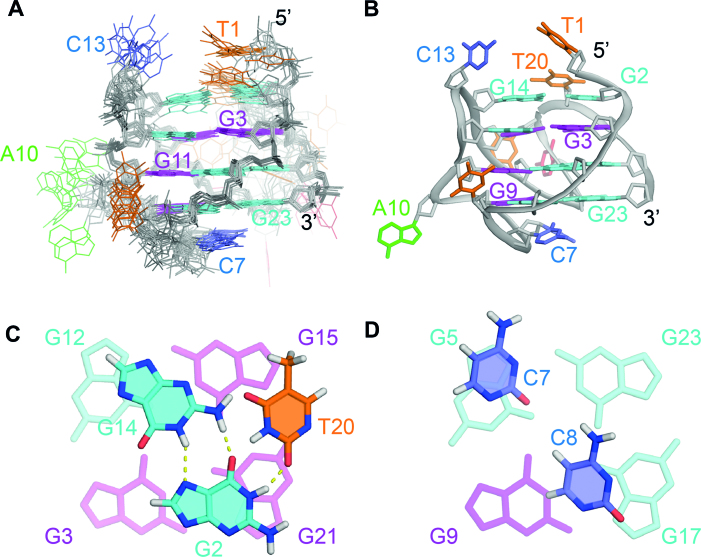
The ten lowest-energy structures are presented in Figure 4A. The G-tetrad core displays a greater convergence compared to the loops. Stacking interactions are observed between cytosine residues C7 and C8 in the TCC edgewise loop and guanine residues in the bottom tetrad (Figures 4).
Figure 4.
NMR solution structure of the TP3-T6 G-quadruplex (PDB ID: 6AC7). (A) Ten lowest energy superimposed refined structures. (B) Ribbon view of a representative structure. Anti guanine residues are colored cyan, syn guanine residues are colored magenta, thymine residues are colored orange, cytosine residues are colored red and adenine residues are colored green. (C) Close-up view of the G2·G14·T20 base triple capping the top of the structure. (D) Close-up view of the C7 and C8 bases at the bottom of the structure.
Formation of a base triple capping structure
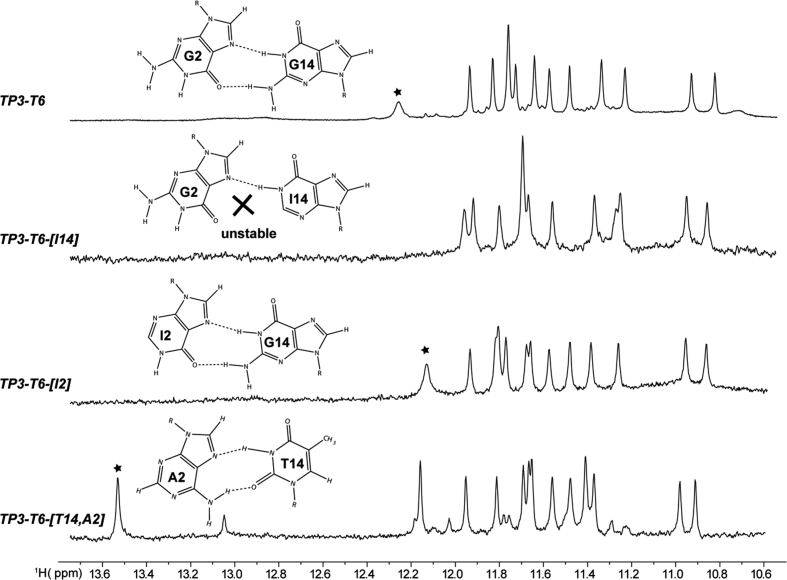
In addition to the three G-tetrad layers, the structure also possesses a G2·G14·T20 base triple (Figure 4C), where the three bases come from distinct strands. G2 and G14 interact via reverse Hoogsteen bonding, with H21 and H1 of G14 hydrogen bonding with O6 and N7 of G2 respectively. This is further supported by the observation of a NOE peak between the aromatic (H8) proton of G2 with the imino (H1) proton of G14 (Supplementary Figure S14). Inosine-substitution experiments at G2 and G14 positions also confirm the reverse Hoogsteen hydrogen bonding between G2 and G14, where the G14 imino proton peak vanishes for I14 substitution but remains for the I2 substitution (Figure 5). We also observed that the reversed Hoogsteen base pair G14·G2 could be substituted by an alternative Hoogsteen base pair T14·A2 (Figure 5).
Figure 5.
Imino proton spectra of TP3-T6 and sequences mutated at the indicated positions. An asterisk indicates the imino proton in the Hoogsteen base pair between residues 2 and 14.
The capping structure confers additional stability to TP3-T6 and a higher thermal stability is observed for TP3-T6 as compared to TP3-T6-[I14] in which the capping structure is disrupted (Supplementary Figure S15 and Table 2). Such hydrogen-bond interactions between edgewise loop residues and 5′ terminal residues were previously reported in the literature for (3+1) hybrid G-quadruplexes, where they have been found to play a critical role in the stabilization of one conformation over the other by providing additional stability to the structure (43,47–52).
Table 2.
Thermal stability of TP3-T6 and mutated sequences
| Name | Sequence (5′ → 3′) | T m (°C)a |
|---|---|---|
| TP3-T6 | TGGGGTCCGAGGCGGGGCTTGGG | 60.2 ± 0.9 |
| TP3-T6-[I14] | TGGGGTCCGAGGCIGGGCTTGGG | 57.3 ± 0.6 |
| TP3-T6-[I2] | TIGGGTCCGAGGCGGGGCTTGGG | 60.2 ± 0.7 |
| TP3-T6-[T14, A2] | TAGGGTCCGAGGCTGGGCTTGGG | 63.6 ± 0.8 |
aMelting temperature (Tm) was measured in 70 mM KCl and 20 mM KPi (pH 7).
Addtionally, we detected a sharp imino proton resonance of G2 at 10°C (Supplementary Figure S9), indicative of the hydrogen bonding present between G2 and T20 (Figure 4C and D). Therefore, the overall base triple capping structure in TP3-T6 comprises of the 5′ terminal G2 residue which is centrally positioned and forms hydrogen bonding with G14 in the edgewise loop and T20 in the propeller loop. The resulting G·G·T capping structure formed on the top tetrad serves as a protective layer, limiting the exposure of guanine residues of the top tetrad to solvent, which is in agreement with solvent exchange experiments whereby the imino protons of G12 and G15 located in the top tetrad are protected from D2O solvent exchange after 20 minutes and the imino proton of G15 remains protected even after 2 h (Supplementary Figure S13). Similar base triple capping structures stacked on the top and bottom tetrad, such as A·A·A, T·G·A, T·A·T, T·T·T and protonated T18·A20+·G5 base triples have been previously reported to contribute to the overall stability of the quadruplex or favor one particular conformation over another (47–55).
Bulge-containing (3+1) hybrid G-quadruplex scaffold for drug targeting
Research over the past 25 years on G-quadruplexes has resulted in the discovery of several G-quadruplexes with a (3+1) G-tetrad core, in which three strands are oriented in one direction and the fourth in the opposite direction. The (3+1) hybrid folding topology was first observed for a G-quadruplex formed by the Tetrahymena telomeric sequence d(T2G4)4 in Na+ solution, whereby a three-layered intramolecular G-quadruplex with a syn·syn·syn·anti G-tetrad and two anti·anti·anti·syn G-tetrads was reported (56). A dimeric, asymmetric quadruplex in Na+ solution formed by the human telomeric sequence d(GGGTTAGGGTTAGGGT), as well as intramolecular G-quadruplexes in K+ solution formed by the human telomeric sequence d[TAGGG(TTAGGG)3] and many variants, were also later found to adopt a (3+1) hybrid folding topology (43,49–52,57). Besides the telomeric region, (3+1) hybrid G-quadruplexes have also been found to be present in several gene promoters such as hTERT, BCL2 and VEGFR-2, highlighting a possible role for such scaffolds in transcriptional regulation (47,58,59). In this study, we have revealed a (3+1) hybrid G-quadruplex in the promoter of PARP1—a gene with a very important role in DNA repair and of significant therapeutic relevance.
Besides being characterized by loop and strand orientations, G-quadruplexes can also be characterized by unique and intrinsic structural features such as bulges, providing recognition elements that can, in principle, be exploited for specific small-molecule targeting. The bulge formation was observed in a variety of contexts with varying the type, the number and the positions of bulge residues, as well as the type of the G-quadruplex scaffold, suggesting a widespread existence of bulge-containing G-quadruplex-forming sequences (26). Several G-quadruplex forming sequences with residues capable of forming single-nucleotide or multi-nucleotide bulges upon folding into a G-quadruplex were identified by the G4-seq method via sequencing DNA from primary human B lymphocytes (11). Thymine bulges within parallel-stranded G-quadruplexes have been reported for LTR-IV, present in the long terminal repeat promoter region of HIV-1, 22RT, present within the KRAS nuclease-hypersensitive element (NHE) region, and AT11, an anti-proliferative oligonucleotide (27,28,60). A cytosine bulge has also been observed within the parallel-stranded RNA and DNA G-quadruplexes formed by the 5′-end sequence of hTERC (61,62). Although adenine bulges have also been previously reported to exist within G-quadruplex structures (26,59), we report here the solution structure of a G-quadruplex with an adenine bulge in a new structural context. The detailed structural characteristics of the (3+1) hybrid G-quadruplex TP3-T6 with an adenine bulge present an opportunity for PARP1 regulation by selective non-canonical G-quadruplex targeting.
CONCLUSION
We have revealed the (3+1) hybrid G-quadruplex folding topology adopted by the G-rich promoter sequence of the PARP1 gene. The interesting structural features of the PARP1 G-quadruplex are an adenine residue bulge and a unique G·G·T triple capping structure formed by the central edgewise loop, propeller loop and 5′ flanking terminal which could be exploited as distinct recognition elements to generate small-molecule ligands to target this G-quadruplex structure with specificity. This structure presents an opportunity to further explore the therapeutic potential of PARP1 inhibition via selective G-quadruplex DNA targeting.
DATA AVAILABILITY
SpinWorks and SPARKY are open source softwares available at http://home.cc.umanitoba.ca/∼wolowiec/spinworks/ and https://www.cgl.ucsf.edu/home/sparky/ respectively.
Atomic coordinates and structure factors for the reported NMR structures have been deposited with the Protein Data bank under accession number 6AC7.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brahim Heddi for his assistance with NMR experiments and Giovanni Marsico for bioinformatics analysis that led to the identification of the PARP1 G-quadruplex sequence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore National Research Foundation Investigatorship [NRF-NRFI2017-09]; Nanyang Technological University (NTU Singapore) (to A.T.P.); The Balasubramanian laboratory is core-funded by Cancer Research UK [C14303/A17197]; Cancer Research UK programme [C9681/A18618]; S.B. is a Welcome Trust Senior Investigator [099232/Z/12/Z]; The authors acknowledge the use of NMR facilities at the NTU Institute of Structural Biology. Funding for open access charge: Singapore National Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. D’Amours D., Desnoyers S., D'Silva I., Poirier G.G.. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999; 342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 2. Gibson B.A., Kraus W.L.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell. Biol. 2012; 13:411–424. [DOI] [PubMed] [Google Scholar]
- 3. Kraus W.L. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell. Biol. 2008; 20:294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo X., Kraus W.L.. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012; 26:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ray Chaudhuri A, Nussenzweig A.. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Livraghi L., Garber J.E.. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med. 2015; 13:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J. Clin. Oncol. 2008; 26:3785–3790. [DOI] [PubMed] [Google Scholar]
- 8. Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009; 361:123–134. [DOI] [PubMed] [Google Scholar]
- 9. Scott C.L., Swisher E.M., Kaufmann S.H.. Poly (ADP-ribose) polymerase inhibitors: recent advances and future development. J. Clin. Oncol. 2015; 33:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lord C.J., Ashworth A.. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017; 355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 12. Simonsson T., Pecinka P., Kubista M.. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998; 26:1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soldatenkov V.A., Vetcher A.A., Duka T., Ladame S.. First evidence of a functional interaction between DNA quadruplexes and poly(ADP-ribose) polymerase-1. ACS Chem. Biol. 2008; 3:214–219. [DOI] [PubMed] [Google Scholar]
- 16. Fekete A., Kenesi E., Hunyadi-Gulyas E., Durgo H., Berko B., Dunai Z.A., Bauer P.I.. The guanine-quadruplex structure in the human c-myc gene's promoter is converted into B-DNA form by the human poly(ADP-ribose)polymerase-1. PLoS One. 2012; 7:e42690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cogoi S., Paramasivam M., Membrino A., Yokoyama K.K., Xodo L.E.. The KRAS promoter responds to Myc-associated Zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010; 285:22003–22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salvati E., Scarsella M., Porru M., Rizzo A., Iachettini S., Tentori L., Graziani G., D’Incalci M., Stevens M.F.G., Orlandi A. et al. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene. 2010; 29:6280–6293. [DOI] [PubMed] [Google Scholar]
- 19. Gellert M., Lipsett M.N., Davies D.R.. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962; 48:2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 21. Davis J.T. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004; 43:668–698. [DOI] [PubMed] [Google Scholar]
- 22. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukundan V.T., Phan A.T.. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013; 135:5017–5028. [DOI] [PubMed] [Google Scholar]
- 27. De Nicola B., Lech C.J., Heddi B., Regmi S., Frasson I., Perrone R., Richter S.N., Phan A.T.. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 2016; 44:6442–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kerkour A., Marquevielle J., Ivashchenko S., Yatsunyk L.A., Mergny J.L., Salgado G.F.. High-resolution three-dimensional NMR structure of the KRAS proto-oncogene promoter reveals key features of a G-quadruplex involved in transcriptional regulation. J. Biol. Chem. 2017; 292:8082–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heddi B., Martin-Pintado N., Serimbetov Z., Kari T.M., Phan A.T.. G-quadruplexes with (4n - 1) guanines in the G-tetrad core: formation of a G-triad.water complex and implication for small-molecule binding. Nucleic Acids Res. 2016; 44:910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guedin A., Gros J., Alberti P., Mergny J.L.. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010; 38:7858–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amrane S., Adrian M., Heddi B., Serero A., Nicolas A., Mergny J.-L., Phan A.T.. Formation of pearl-necklace monomorphic G-quadruplexes in the human CEB25 minisatellite. J. Am. Chem. Soc. 2012; 134:5807–5816. [DOI] [PubMed] [Google Scholar]
- 32. Yu Z., Gaerig V., Cui Y., Kang H., Gokhale V., Zhao Y., Hurley L.H., Mao H.. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J. Am. Chem. Soc. 2012; 134:5157–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim K.W., Phan A.T.. Structural basis of DNA quadruplex–duplex junction formation. Angew. Chem. Int. Ed. Engl. 2013; 52:8566–8569. [DOI] [PubMed] [Google Scholar]
- 34. Lim K.W., Nguyen T.Q., Phan A.T.. Joining of multiple duplex stems at a single quadruplex loop. J. Am. Chem. Soc. 2014; 136:17969–17973. [DOI] [PubMed] [Google Scholar]
- 35. Greco M.L., Kotar A., Rigo R., Cristofari C., Plavec J., Sissi C.. Coexistence of two main folded G-quadruplexes within a single G-rich domain in the EGFR promoter. Nucleic Acids Res. 2017; 45:10132–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chung W.J., Heddi B., Schmitt E., Lim K.W., Mechulam Y., Phan A.T.. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kocman V., Plavec J.. Tetrahelical structural family adopted by AGCGA-rich regulatory DNA regions. Nat. Commun. 2017; 8:15355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M.. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003; 160:65–73. [DOI] [PubMed] [Google Scholar]
- 39. Adrian M., Heddi B., Phan A.T.. NMR spectroscopy of G-quadruplexes. Methods. 2012; 57:11–24. [DOI] [PubMed] [Google Scholar]
- 40. Karsisiotis A.I., Hessari N.M., Novellino E., Spada G.P., Randazzo A., Webba da Silva M.. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. Engl. 2011; 50:10645–10648. [DOI] [PubMed] [Google Scholar]
- 41. del Villar-Guerra R., Trent J.O., Chaires J.B.. G-quadruplex secondary structure from circular dichroism spectroscopy. Angew. Chem. Int. Ed. Engl. 2018; 57:7171–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vorlickova M., Kejnovska I., Sagi J., Renciuk D., Bednarova K., Motlova J., Kypr J.. Circular dichroism and guanine quadruplexes. Methods. 2012; 57:64–75. [DOI] [PubMed] [Google Scholar]
- 43. Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J.. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006; 128:9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Do N.Q., Phan A.T.. Monomer-dimer equilibrium for the 5′-5′ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chemistry (Easton). 2012; 18:14752–14759. [DOI] [PubMed] [Google Scholar]
- 45. Hager G.L., McNally J.G., Misteli T.. Transcription dynamics. Mol. Cell. 2009; 35:741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang A.Y., Balasubramanian S.. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012; 134:19297–19308. [DOI] [PubMed] [Google Scholar]
- 47. Lim K.W., Lacroix L., Yue D.J., Lim J.K., Lim J.M., Phan A.T.. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010; 132:12331–12342. [DOI] [PubMed] [Google Scholar]
- 48. Wirmer-Bartoschek J., Bendel L.E., Jonker H.R.A., Grun J.T., Papi F., Bazzicalupi C., Messori L., Gratteri P., Schwalbe H.. Solution NMR structure of a ligand/hybrid-2-G-quadruplex complex reveals rearrangements that affect ligand binding. Angew. Chem. Int. Ed. Engl. 2017; 56:7102–7106. [DOI] [PubMed] [Google Scholar]
- 49. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsugami A., Xu Y., Noguchi Y., Sugiyama H., Katahira M.. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007; 274:3545–3556. [DOI] [PubMed] [Google Scholar]
- 51. Dai J., Punchihewa C., Ambrus A., Chen D., Jones R.A., Yang D.. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007; 35:2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dai J., Carver M., Punchihewa C., Jones R.A., Yang D.. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007; 35:4927–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kuryavyi V., Patel D.J.. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010; 18:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kuryavyi V., Majumdar A., Shallop A., Chernichenko N., Skripkin E., Jones R., Patel D.J.. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) triad and a T-T-T triple. J. Mol. Biol. 2001; 310:181–194. [DOI] [PubMed] [Google Scholar]
- 55. Galer P., Wang B., Sket P., Plavec J.. Reversible pH switch of two-quartet G-quadruplexes formed by human telomere. Angew. Chem. Int. Ed. Engl. 2016; 55:1993–1997. [DOI] [PubMed] [Google Scholar]
- 56. Wang Y., Patel D.J.. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994; 2:1141–1156. [DOI] [PubMed] [Google Scholar]
- 57. Zhang N., Phan A.T., Patel D.J.. (3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005; 127:17277–17285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dai J., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D.. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006; 128:1096–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y., Lan W., Wang C., Cao C.. A putative G-quadruplex structure in the proximal promoter of VEGFR-2 has implications for drug design to inhibit tumor angiogenesis. J. Biol. Chem. 2018; 293:8947–8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Do N.Q., Chung W.J., Truong T.H.A., Heddi B., Phan A.T.. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017; 45:7487–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martadinata H., Phan A.T.. Formation of a stacked dimeric G-quadruplex containing bulges by the 5′-terminal region of human telomerase RNA (hTERC). Biochemistry. 2014; 53:1595–1600. [DOI] [PubMed] [Google Scholar]
- 62. Meier M., Moya-Torres A., Krahn N.J., McDougall M.D., Orriss G.L., McRae E.K.S., Booy E.P., McEleney K., Patel T.R., McKenna S.A. et al. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018; 46:5319–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SpinWorks and SPARKY are open source softwares available at http://home.cc.umanitoba.ca/∼wolowiec/spinworks/ and https://www.cgl.ucsf.edu/home/sparky/ respectively.
Atomic coordinates and structure factors for the reported NMR structures have been deposited with the Protein Data bank under accession number 6AC7.