Abstract
X-inactive-specific transcript (Xist) is a long noncoding RNA (lncRNA) essential for inactivating one of the two X chromosomes in mammalian females. Random X chromosome inactivation is mediated by Xist RNA expressed from the inactive X chromosome. We found that Xist RNA is unspliced in naïve embryonic stem (ES) cells. Upon differentiation, Xist splicing becomes efficient across all exons independent of transcription, suggesting interdependent or coordinated removal of Xist introns. In female cells with mutated polypyrimidine tract binding protein 1 (Ptbp1), differentiation fails to substantially upregulate mature Xist RNA because of a defect in Xist splicing. We further found both Xist129 and XistCAS RNA are unspliced in Mus musculus 129SvJ/Mus castaneous (CAS) hybrid female ES cells. Upon differentiation, Xist129 exhibits a higher splicing efficiency than XistCAS, likely contributing to preferential inhibition of the X129 chromosome. Single cell analysis shows that the allelic choice of Xist splicing is linked to the inactive X chromosome. We conclude post-transcriptional control of Xist RNA splicing is an essential regulatory step of Xist induction. Our studies shed light on the developmental roles of splicing for nuclear-retained Xist lncRNA and suggest inefficient Xist splicing is an additional fail-safe mechanism to prevent Xist activity in ES cells.
INTRODUCTION
Female placental mammals transcriptionally silence one of the two X chromosomes to ensure a roughly equal gene dosage between males and females. This chromosome-wide silencing process, or X chromosome inactivation (XCI), is initiated early in development. Mouse XCI happens in two phases: imprinted and random XCI. Around the two- to four-cell stage, the paternal X chromosome is exclusively inactivated (1). This imprinted XCI is reverted in the inner cell mass of blastula (2). After implantation, either the paternal or maternal X chromosome is stochastically chosen to be inactivated in epiblast (3). Random XCI persists throughout the life.
Random XCI is believed to be triggered by upregulation of female-specific X-inactive-specific transcript (Xist) RNA from the future inactive X (Xi) chromosome (4–7). Overwhelming evidence shows Xist RNA acts in cis to coat the Xi chromosome and recruit epigenetic silencing factors. In both humans and mice, X chromosome does not initiate XCI without Xist RNA expression. Therefore, the consensus view is that Xist induction is necessary to initiate random XCI during development.
Differentiation of female mouse embryonic stem (ES) cells is the most favored model system to investigate Xist induction (8). Undifferentiated female ES cells derived from the inner cell mass exhibit two active X chromosomes. Mimicking embryo development, ES cell differentiation up-regulates Xist RNA from the future Xi chromosome, and goes through several stages to fully establish XCI (4,5,9). The initiation stage involves counting and selection of the future silenced X chromosome and induction of Xist RNA. With the embryoid bodies method, 48 h after induction of differentiation, Xist RNA spreads throughout and coats the Xi chromosome in cis (10). Then, high-order chromatin packaging and silencing of X-linked genes begin. Much later during differentiation (e.g. day 12 of differentiation), chromatin is further modified as XCI becomes fully established and irreversible. Maintenance of XCI appears independent of Xist RNA (11–13). Multiple studies have described transcriptional controls of the initial Xist up-regulation (4–7). The contribution of post-transcriptional regulation is still not well characterized.
The role of intron or splicing for long noncoding RNA like Xist is a mystery. Messenger RNA splicing generally promotes nuclear export and provides a mechanism to allow one protein-coding gene to generate multiple functional variants (14,15), but neither applies to nuclear-retained Xist RNA. By UCSC annotation, Xist precursor RNA contains eight exons. Retaining and deleting the last intron generates a long and short Xist isoform, respectively. The long isoform is the major isoform, while the short isoform is expressed lowly and only in certain differentiated tissues (16,17). Nevertheless, the two spliced variants were indistinguishable in mediating X-chromosome inactivation (17). Therefore, it is unclear why the Xist gene contains introns.
Notably, human and mouse Xist cDNA are only 47% identical but share a similar exon-intron structure, suggesting selection pressure to maintain Xist splicing (18). We asked whether Xist RNA splicing could be a regulatory checkpoint of Xist biogenesis. Surprisingly, we found that differentiation drastically increased Xist RNA splicing efficiency in C57BL/6 ES cells (or BL6 ES cells) and F1 2-1 ES cells (the hybrid of Mus musculus 129SvJaeJ and Mus musculus castaneus EiJ, which has been widely used to study allelic expression of Xist RNA and XCI). We and others previously discovered that RNA binding protein PTBP1 binds to Xist RNA (19–21). PTBP1 was also identified in a forward genetic screen as impairing Xist-dependent chromosome silencing (22). Since PTBP1 is well known to regulate splicing (23–26), we tested whether PTBP1 could be involved in differentiation-induced Xist splicing.
MATERIALS AND METHODS
Cell culture
Feeder dependent female WT and Ptbp1−/− BL6 mouse ES cells were expanded on inactive male murine embryonic fibroblast (MEF) feeders on 0.1% gelatinized tissue culture plates in DMEM media (Gibco cat. no. 10313039) supplemented with 15% ESC grade FBS (Gibco cat. no. 10439024), 1% nucleosides (EMD Millipore cat. no. ES008D), 1% Glutamax (Gibco cat. no. 35050061), 0.1 mM β-mercaptoethanol (Acros Organics cat. no. 125472500), and 1000 U/ml mLIF (EMD Millipore cat. no. ESG1106). Feeder independent female WT and Ptbp1−/− BL6 mouse ES cells, and F1 2-1 mouse ES cells were expanded on 0.1% gelatinized tissue culture plates in 2i culture media containing 50% Neurobasal (Gibco cat. no. 21103049) and 50% DMEM/F12 (Gibco cat. no. 11320082) supplemented with 1% B27 + RA (Gibco cat. no. 17504044), 1% N2 Supplement (R&D System cat.no. AR009), 1% Glutamax, 7.4 mM B27 Fraction V (Gibco cat. no. 15260037), 1% Penicillin-Streptomycin (GE Healthcare Life Sciences cat. no. SV30010), 3 μM CHIR99021 (Sigma cat. no. SML1046), 1 μM PD0325901 (Selleckchem cat. no. S1036), 150 μM 1-thioglycerol (Sigma cat. no. M6145) and 1000 U/ml mLIF (Gemini Bio-Products cat. no. 400–495). To assist initial attachment, 2i medium was supplemented with 2% ESC grade FBS for the first 24 h before switching to serum free.
Differentiation of embryonic stem cells
For monolayer differentiation, cells were plated feeder-free at 200–300 × 103 per six-well and were grown in LIF conditions for 24 h and then LIF was removed and cells were grown in differentiation media for 24 h (DMEM supplemented with 15% ESC FBS, 1% nucleosides, 1% l-glutamine, 0.1 mM β-mercaptoethanol and 1 μM retinoic acid (Sigma cat. no. R2625)). To generate embryoid bodies, feeder independent ES cells were plated at a density of 2.5 × 106 cells on 0.5% Agarose coated polystyrene dishes (Grenier Bio-One cat. no. 633102) and maintained in DMEM supplemented with 10% FBS, 1% Glutamax, 1% non-essential amino acids (GE Healthcare, cat. no. SH3023801), 1% sodium pyruvate (Lonza cat. no. 13-115E), and 0.1 mM betamercaptoethanol without mLIF. For western blots, WT and Ptbp1–/– mESCs were plated at 300 × 103 on 0.5% agarose coated six-well dishes (VWR, cat. no. 10062-892) to generate embryoid bodies. To minimize cell death, media was replenished every 2 days.
Actinomycin D treatments
Cells were plated at 100 × 103 per six-well for 24 h prior to actinomycin D treatment. For stability experiments, cells were treated with 5 μg/ml actinomycin-D (Sigma, cat. no. A1410) or an equal volume of DMSO vehicle (Sigma, cat. no. D2438) in growth media for 8 h. Percent Xist remaining was calculated as the ratio of Xist transcripts at the time of collection relative to time 0. For transcription inhibition prior to the inflection point of enhanced splicing, cells were differentiated for 6 h in media lacking LIF and supplemented with 1 μM retinoic acid. After 6 h of differentiation, cells were treated with 5 μg/ml actinomycin D for 4 h added directly to growth media. Total RNA was collected from undifferentiated cells (0 h), after 6 h of differentiation, and after 2 or 4 h of actinomycin D post treatment (8 and 10 h).
Generation of Ptbp1−/− ES cells
Mouse Ptbp1loxp/loxp ES cells were generated following a previously published protocol (27). In brief, healthy E3.5 blastocysts were collected from Ptbp1loxp/loxp intercrosses and individually cultured in KSOM medium (DMEM high glucose with 15% (vol/vol) KO serum, 2 mM GlutaMAX,1 mM sodium pyruvate, 0.1 mM MEM NEAA, 0.1 mM β-mercaptoethanol, 103 U/ml of LIF, 1 μM PD0325901 and 3 μM CHIR99021. After overnight culture, blastocysts were transferred to defined serum-free medium (1:1 mixture of DMEM-F12/N2 and Neurobasal/B27, 1 mM GlutaMAX, 0.5 mM sodium pyruvate, 0.1 mM MEM NEAA, 0.1 mM β-Mercaptoethanol) plus 2i/LIF to allow attachment to the feeder layer. After outgrowth from blastocysts, cells were disaggregated and cultured in the serum-free medium plus 2i/LIF together with MEF. Cells were monitored daily for colony formation. Cells exhibiting differentiation or atypical ES cell morphology were discarded and the healthy ES cell lines were maintained in the serum-free medium plus 2i/LIF with MEF. Ptbp1−/− mES cells were generated via expressing Cre recombinase in Ptbp1loxp/loxp ES cells. Ptbp1 homozygous knockout ES cell clones were identified by PCR genotyping and confirmed by PTBP1 Western blot.
RNA Isolation, cDNA synthesis and RT-qPCR
Trizol isolation of RNA was performed following manufacturer's protocol (Life Technologies). Isolated RNA was treated with 4 units of Turbo DNase (Ambion) at 37°C for 35 min followed by Phenol-Chloroform extraction (pH 4.5, VWR cat. no. 97064-744). First-strand cDNA was generated using 1 μg RNA and 200 units of M-MLV reverse transcriptase (Promega cat. no. M1705) with a RT primer in a 20 μl reaction (28). For analyzing Xist expression and splicing, the strand-specific RT primer (50μM) was a Xist gene specific reverse primer (E6-E7 reverse) targeting exon 7 unless specified otherwise. For X-linked and differentiation genes, random hexamers (30 μM) were used. For polyadenylated (poly(A)+) RNA, oligodT primers (50μM) were used for cDNA generation. The RT-qPCRs were conducted using a Quantstudio 6 Flex Real-Time PCR instrument with 2x Syber Master Mix (Life Technologies) (29,30). Three technical replicates of all samples were performed for each biological replicate. An additional no-template control was included in every experiment. Outliers were defined by a Ct variance higher than 0.5. Analysis was done with Quanstudio 6 software and Microsoft Excel. Statistics test were based on ΔCt (for Xist with the same RNA input) or ΔΔCt values (for other genes).
Splicing assay
Splicing assays were performed essentially as previously described (31). Standard conditions for Taq polymerase from New England Biolabs (cat. no. M0267E) were used. For amplification of spliced regions of Xist spanning exons 1–2, exons 2–3 and exons 2–6, exon specific primers for corresponding exons were used (Supplementary Table). The PCRs were performed at 60°C (Tm) for 29–35 cycles. PCR products >1 kb were resolved in 1.5% TAE Agarose gel. Smaller products were resolved with capillary electrophoresis (QIAxcel Advanced System) or on a 2% TBE Agarose gel (32). Relative quantities of spliced and unspliced products were measured on the QIAxcel software (Qiagen) or ImageLab (Bio-Rad) and the splicing ratio was calculated as the molar ratio of spliced to unspliced PCR products. The electropherograms of capillary electrophoresis were converted to virtual gel images by QIAxcel for presentation.
Allele specific splicing assay and restriction fragment length polymorphism analysis
PCR reactions were performed under standard conditions for Taq polymerase (New England Biolabs) on RT products derived from F1 2-1 cells. For Xist 30–35 cycles was used. PCR products were digested with 2.5 units of Tfi1 (New England Biolabs cat. no. R0546L) for 2 h at 65°C. For single cell studies, PCR products were digested with 2.0 units of Tfi at 37°C overnight. For X-linked genes G6pdx and Ogt, 35–40 cycles were used. PCR products were digested with Ava1 (NEB, cat. no. R0152S) or ScrF1 (NEB, cat. no. R0110S) to cut Ogt or G6pdx respectively. Samples were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter cat. no. A63881). Eluted PCR products were diluted 1:3–1:5 when necessary before analysis on the QIAxcel Capillary Electrophoresis system (Qiagen) using the QIAxcel DNA Screening Kit (Qiagen, cat. no. 929004). The QIAxcel Alignment Marker QX 15-600bp (Qiagen, cat. no. 929530) and the Qiaxcel 25–500 bp size marker (Qiagen, cat. no. 929560) were used in the analysis. For confirmation of allele specific digestion by Tfi1, genomic DNA collected from the brain of wildtype Mus musculus castaneus EIJ and Mus musculus domesticus 129S4/SvJaeJ mice were used.
Single cell sorting and cDNA preparation
WT F1 2-1 ES cells were differentiated into embryoid bodies for 6–8 days. Cells were washed two times with 1× PBS and stained with propidium iodide (PI) (1:1500) (BD Pharmigen, cat. no. 556547). We used three stringent gating steps to select single cells: (i) negative staining of propidium iodide as an indicator of healthy cells; (ii) typical FSC and SSC settings to filter out cell debris and cell chunks; (iii) linearity between SSC height and SSC area to ensure single cell (singlet in sorting). Single PI-negative WT F1 2-1 cells from embryoid bodies were then sorted into PCR strips using a MoFlo Astrios EQ (Beckman Coulter) according to manufacturers' protocol. The cDNA was prepared using SMART-Seq v4 Ultra Low Input Kit (Clontech) according to manufacturer's protocol.
Immunoblotting
For primary antibodies, we used antibodies to PTBP1-NT (1:1000), PTBP1-CT (1:200), GAPDH (1:1000, Ambion, AM4300). For secondary antibodies, we used Alexa Fluor 488 donkey anti rabbit (Invitrogen, A21206) and AF 647 donkey anti mouse (Invitrogen, A31571).
RESULTS
ES cell differentiation induces Xist RNA splicing
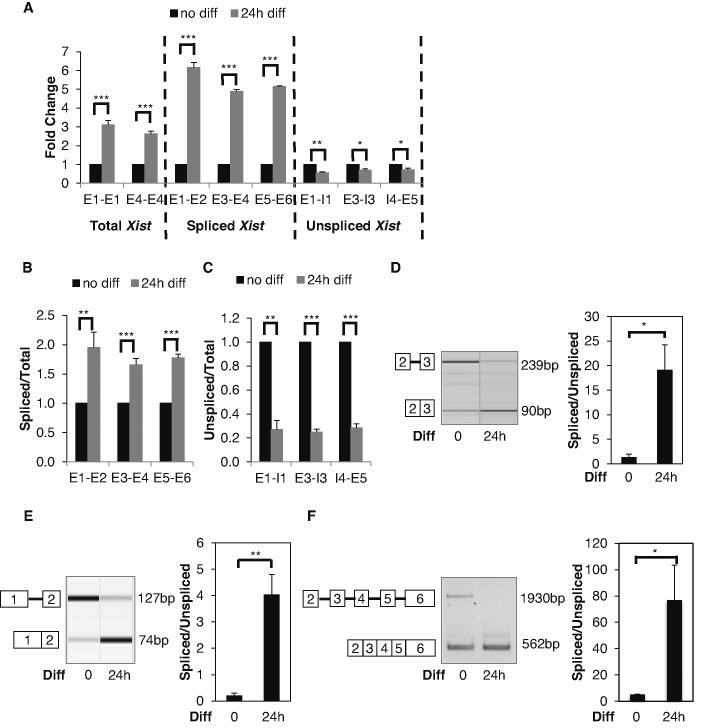
To systemically investigate the initial phase of Xist induction, we designed qPCR-quality primer pairs to measure total, spliced, and unspliced Xist transcripts. For total Xist, the primer pair targets the same exon and is indifferent to spliced vs. unspliced transcripts. For spliced Xist, the forward and reverse primers anneal to different exons separated by a large intron and are unable to amplify unspliced Xist. For unspliced Xist, each primer pair flanks a specific splice site hybridizing to the intron and exon respectively. We used multiple qualified primer pairs for each Xist species to increase the robustness of the analysis.
We performed strand-specific RT-qPCR and found total Xist was upregulated in F1 2-1 cells 24 h after differentiation in a monolayer culture (Figure 1A). We observed a much higher increase in the spliced Xist transcript, which in theory reflected the aggregated effect of splicing and RNA decay. By contrast, we found a modest but consistent decrease in the unspliced Xist transcript (Figure 1A), which should be caused by both transcription and splicing.
Figure 1.
Differentiation Induces Xist Splicing. (A) RT-qPCR analysis of total, spliced, and unspliced Xist transcripts. Adherent F1 2-1 cells were differentiated by LIF withdrawal and addition of RA. (B) Ratios of spliced/total Xist transcripts quantified by RT-qPCR. (C) Ratios of unspliced/total Xist transcripts quantified by RT-qPCR. (D) Splicing assay examines intron 2 splicing before and 24 h after differentiation. Representative virtual gel image of intron 2 splicing (left panel). Quantified intron 2 splicing ratio (right panel) (E) Splicing assay examines intron 1 splicing before and 24 h after differentiation. Representative virtual gel image of intron 1 splicing (left panel). Quantified intron 1 splicing ratio (right panel). (F) Splicing assay examines splicing from exon 2 to exon 6 before and 24 h after differentiation. Representative gel image (left panel). PCR cycles were 33 and 29 for undifferentiated and differentiated cells respectively. For differentiated cells, PCR amplification for 33 cycles did not produce the unspliced product and the spliced product was over saturated, so amplification of 29 cycles were used for the presentation. Quantified splicing ratio (right panel). Error bars indicate S.E.M. of three biological replicates. *P-value<0.05; **P-value<0.01; ***P-value<0.001 Student's t-test (one-tailed, unpaired).
A reasonable explanation for these observations is that differentiation activates Xist transcription and splicing. To mask the transcription effect, we normalized the spliced and unspliced Xist to total Xist. After this normalization, the spliced/total Xist ratio was up-regulated by ∼2-fold (Figure 1B), whereas the unspliced/total Xist ratio down-regulated by ∼3-fold (Figure 1C).
Enhanced splicing is a parsimonious explanation for a decrease in the unspliced transcript and an increase in the spliced transcript. To directly test whether differentiation affects Xist RNA splicing, we designed a primer pair flanking intron 2 (the smallest intron of Xist) to simultaneously detect unspliced and spliced transcripts (Figure 1D). Intron 2 is 149 nt, so unspliced Xist can be reasonably detected alongside spliced Xist and allows calculation of a splicing ratio (spliced/unspliced) as a measure of splicing efficiency. We found differentiation clearly induced intron splicing (Figure 1D).
To test whether differentiation-induced splicing occurs for other introns, we focused on intron 1. We were unable to identify an effective primer pair flanking intron 1 to amplify the unspliced transcript, probably due to the size of intron 1 (2794 nt). Therefore, we designed an intronic forward primer upstream from the intron 1−exon 2 boundary and a reverse primer on exon 2 to detect the intron 1-containing unspliced variant. A multiplex RT-PCR reaction containing this primer pair and another forward primer on exon 1 effectively amplifies both the spliced (74 bp) and unspliced (127 bp) Xist transcripts. Consistently, we observed that splicing was significantly enhanced after differentiation (Figure 1E).
These data suggest a low splicing efficiency of Xist RNA in undifferentiated ES cells. An alternative possibility is higher stability of the spliced transcript in differentiated ES cells. To test this, we measured the half-lives of the spliced Xist transcript by measuring percentage of remaining transcripts upon actinomycin D treatment. We found no change in the stability of the spliced Xist transcript before or after differentiation (Supplementary Figure S1).
We next asked whether differentiation-induced Xist splicing was regulated individually per intron or coordinated across all introns. We reasoned that uncoordinated regulation of intron splicing would lead to isoforms whose introns were not 100% removed. In contrast, coordinated regulation would result in either fully unspliced or spliced transcripts (i.e. all-in or all-out). It was also possible that some, but not all, intron splicing was coordinated. To distinguish these possibilities, we used primers flanking multiple exons (2–6) to detect Xist transcripts.
Our results were consistent with the hypothesis that differentiation-induced Xist splicing is coordinated across introns: only completely spliced and unspliced transcripts were detected by RT-PCR (Figure 1F). Because PCR efficiency is skewed toward the spliced product and the unspliced product is much larger, spliced Xist was over-represented in this analysis. A recent study using long-read sequencing showed that most nascent RNAs are either fully spliced or fully unspliced in fission yeast (33). Our result suggests that this might also be happening in mammals.
Taken together, these data are consistent with the following model for Xist biogenesis: naïve ES cells express Xist transcripts, which remain largely unspliced due to splicing deficiency; differentiation enhances splicing efficiency while transcription ramps up. Enhancing splicing competence is thus likely one of the regulatory events of development-induced Xist expression.
Enhanced Xist splicing is independent of Xist transcription
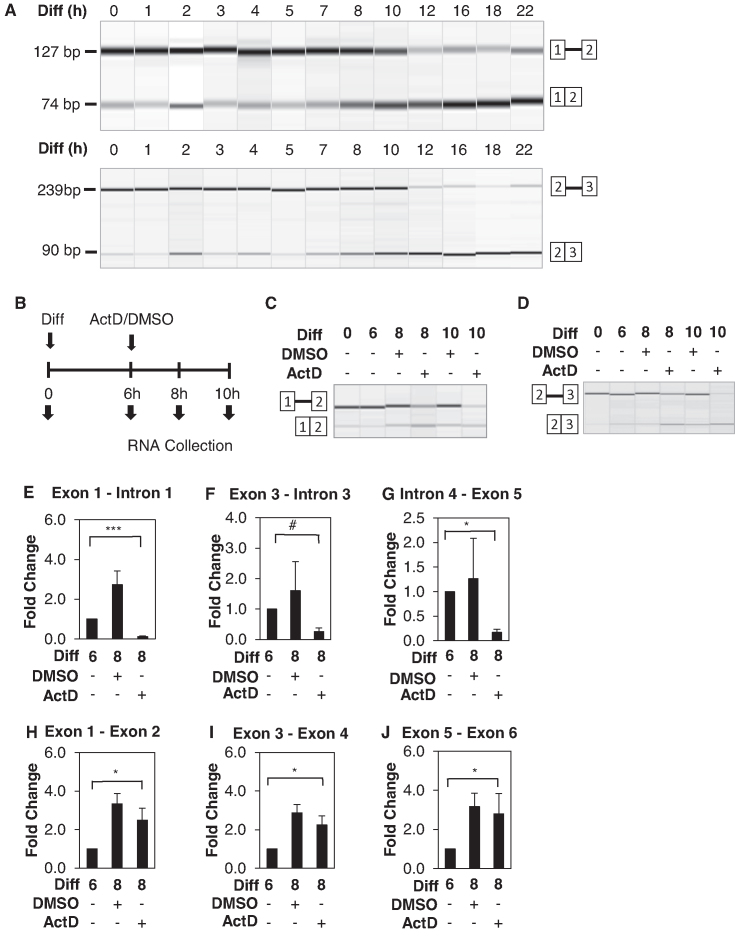
One outstanding question is whether pre-existing Xist becomes spliced upon differentiation or if only newly synthesized Xist is spliced. This can be tested by inhibiting Xist transcription before splicing activation and examining whether the pre-existing Xist transcripts become spliced. To do this, we first determined the timepoint of the splicing change. Xist splicing does not immediately increase upon differentiation with retinoic acid but begins to change around 7–8 h post differentiation (Figure 2A). After that Xist splicing is gradually enhanced until 18 h post differentiation. We therefore define 7 h post differentiation as the inflection point for splicing enhancement.
Figure 2.
Enhanced Xist splicing is independent of Xist transcription. (A) Representative RT-PCR virtual gel images of intron 1 splicing (top panel) and intron 2 splicing (bottom panel) in adherent F1 2-1 cells between 0–22 h post differentiation (RA induction and LIF withdrawal). (B) Schematic diagram of experimental procedures. Cells were differentiated for 6 hours before treatment with 5ug/ml Actinomycin D or control DMSO. RNA was collected before differentiation (time 0), for 6 h post differentiation induction (time 6), for 2 and 4 h post treatment (time 8 and time 10). Representative RT-PCR virtual gel image of (C) intron 1 and (D) intron 2splicing following the aforementioned experimental procedures. (E–G) RT-qPCR analysis of unspliced Xist transcripts using primers detecting various introns. (H–J) RT-qPCR of spliced Xist transcripts using primers for various exon–exon junctions. Error bars indicate S.E.M. of three biological replicates. #P-value = 0.06; *P-value < 0.05; ***P-value < 0.001 Student's t-test (one-tailed, unpaired).
We inhibited transcription with actinomycin D at 6 h post differentiation and analyzed Xist splicing at 8 and 10 h post differentiation (Figure 2B). In scenario 1, if only newly synthesized Xist is spliced, actinomycin D treatment will prevent the production of spliced Xist. In scenario 2, if pre-existing unspliced Xist RNA becomes spliced after the inflection point, spliced Xist transcripts will still be generated even with actinomycin D treatment. As a result, spliced Xist transcripts would appear abundant relative to unspliced Xist transcripts because unspliced Xist transcripts are lost to splicing without being supplied from transcription.
We found that the production of spliced Xist transcripts was largely unaffected by actinomycin D treatment (Figure 2C and D). In contrast, unspliced Xist transcripts were clearly diminished by actinomycin D, consistent with transcription inhibition. This was observed with two different primer pairs examining splicing of two different introns. These data support scenario 2 showing that the pre-existing unspliced Xist RNA becomes spliced after the inflection point.
To quantitatively measure magnitudes of the expression changes in the spliced and unspliced Xist RNA individually, we performed RT-qPCR. Compared to before the inflection point (i.e. 6 h post differentiation), unspliced Xist stayed relatively flat after DMSO treatment but decreased by about 4-fold as early as 2 h after actinomycin D treatment (Figure 2E–G). In contrast, spliced Xist remained up-regulated after actinomycin D treatment (Figure 2H–J). Therefore, differentiation-induced splicing continues in actinomycin D-treated cells.
The above results indicate that Xist splicing can occur post-transcriptionally, because most pre-existing unspliced Xist transcripts likely have completed their transcription. Also supporting post-transcriptional splicing of Xist was that data concerning splicing of the upstream (e.g. intron 1) and downstream introns (e.g. introns 3, 4 and 5) were consistent with one another (Figure 2E–J). Xist transcripts are polyadenylated (34). To further test whether Xist splicing is post-transcriptional, we analyzed Xist splicing in polyadenylated (poly(A)+) RNA fractions. We found most polyadenylated Xist transcripts were unspliced in ES cells and the splicing of these poly(A)+ Xist RNA was enhanced by differentiation (Supplementary Figure S2A-B).
Taken together, these data show that Xist splicing can occur post-transcriptionally. We could not rule out co-transcriptional Xist splicing. Co-transcriptional splicing could be tested with nascent chromatin-associated RNA fractions under the assumption that chromatin-associated RNAs undergo transcription and post-transcriptional RNAs are detached from chromatin. However, Xist RNA is tethered to the X chromosome, so an analysis of chromatin-associated RNA would not be informative.
Allelic difference in Xist RNA splicing efficiency is associated with nonrandom Xist induction
Although largely random, differentiation-induced X chromosome inactivation can be skewed (35). For example, in 129/Castaneus hybrid mice, the 129 allele is preferentially chosen for XCI. This stochastic allelic difference is associated with preferential expression of Xist from X129 rather than from XCAS (36). The exact mechanism of nonrandom Xist expression is still unclear, but likely involves a myriad of coordinated events. We hypothesized that nonrandom Xist induction was associated with allelic differences in Xist RNA splicing.
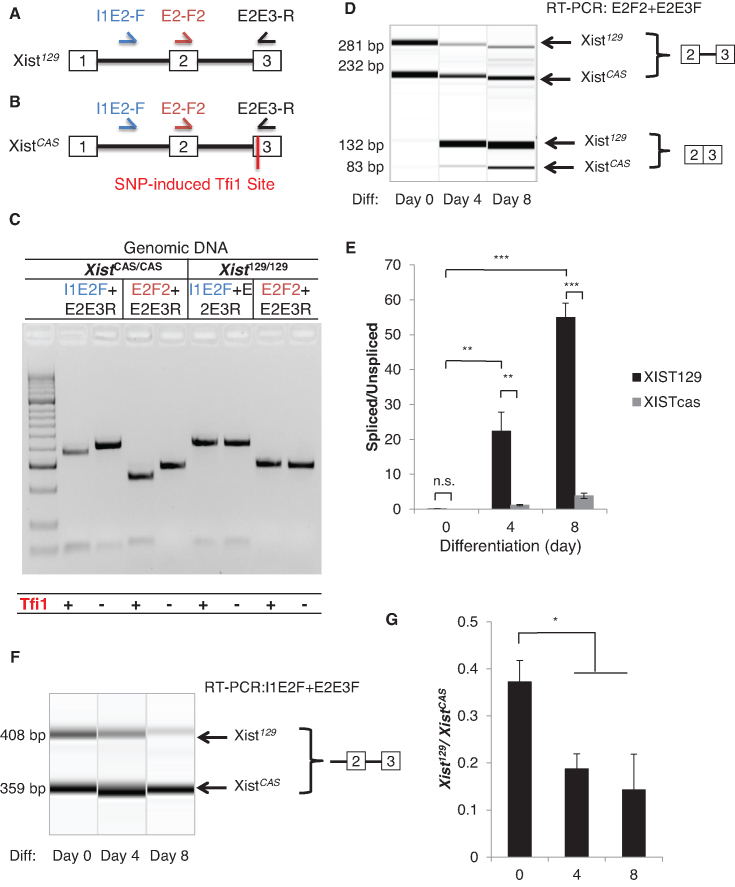
To simultaneously measure spliced and unspliced Xist variants from 129 and CAS alleles, an exonic polymorphic site near the exon-intron boundary and compatible with splicing assay is needed. We cloned and sequenced all exon–intron boundaries using genomic DNA from M. musculus domesticus 129SvJaeJ and M. musculus castaneus EiJ. We found a new single nucleotide polymorphism (SNP) in exon 3 close to the 3′ splice site, which generates a Tfi1 enzyme recognition site specifically in the CAS allele (Figure 3A and B).
Figure 3.
Allelic difference in Xist splicing is associated with non-random Xist induction. (A, B) Schematics of the primer locations and allele-specific restriction enzyme digestion for distinguishing the unspliced and spliced Xist transcripts from the Xist129 and XistCAS alleles. SNP induced Tfi1 cut site is present in exon 3 of the XistCAS allele. (C) A representative gel showing Tfi1 specifically cuts exon 3 of XistCAS/CAS genomic DNA but not Xist129/129 DNA. Genomic DNA was amplified using the I1E2F or the E2F2 forward primer and the E2E3R primer before Tfi1 treatment. (D) Splicing assay detects allele-specific expression of unspliced and spliced transcripts from the Xist129 and XistCAS alleles before and after differentiation. (E) Quantification of intron 2 splicing ratios of XistCAS and Xist129 transcripts, respectively. (F) A representative RT-PCR virtual gel image shows the expression of unspliced XistCAS and unspliced Xist129 transcripts using I1E2-F and E2E3-R primers. (G) Ratios of unspliced Xist129 transcript relative to unspliced XistCAS transcript upon differentiation. Error bars indicate S.E.M. of three biological replicates. ns, P-value > 0.05; *P-value < 0.05; **P-value< 0.01; ***P-value < 0.001.
To test the new SNP, we PCR amplified the sequences around the SNP locus from M. musculus domesticus 129SvJaeJ and M. musculus castaneus EiJ genomic DNA. For both Xist129 and XistCAS, E2-F2 plus E2E3-R primers yielded a 281bp PCR product, and I1E2-F plus E2E3-R primers yielded a 408 bp product (Figure 3C). While the 129 amplicons were resistant to Tfi1 restriction enzyme digestion, the CAS amplicons were completely digested resulting in faster-migrating products (232 bp and 49 bp from the E2F2 amplicon, 359 bp and 49 bp from the I1E2-F amplicon). Therefore, Tfi1 digestion of RT-PCR products should be appropriate to distinguish Xist129 and XistCAS RNA.
To compare the splicing efficiency of Xist129 and XistCAS RNA, we performed RT-PCR followed by restriction fragment length polymorphism (RFLP) for allele-specific quantification based on the Tfi1 site. In undifferentiated F1 2-1 ES cells, we found neither Xist129 nor XistCAS were efficiently spliced (Figure 3D). Spliced Xist129 and XistCAS RNA were clearly up-regulated after differentiation. The majority of the spliced Xist transcript originated from Xist129. Even though both Xist129 and XistCAS RNA became more efficiently spliced, the Xist129 splicing ratio (spliced/unspliced) was significantly larger than that of XistCAS, suggesting higher splicing efficiency for Xist129 (Figure 3E). These data indicated efficient splicing of the Xist129allele is associated with preferential expression of Xist129 RNA.
To further test the differential splicing efficiency of Xist129 and XistCAS RNA, we directly measured premature unspliced Xist129 and XistCAS RNA transcripts and calculated Xist129/XistCAS before and after differentiation (Figure 3F and G). I1E2-F and E2E3-R primers amplified only the unspliced Xist transcript. Even with presumed higher Xist129than XistCAS transcription, unspliced Xist129precursor RNA was consistently lower than unspliced XistCAS RNA, showing that Xist129is intrinsically spliced more effectively. Furthermore, the Xist129/XistCAS ratio decreased as cells differentiated (Figure 3G). These data suggest that Xist RNA splicing might contribute to the choice mechanism selecting the future inactive X chromosome.
Allele specific Xist splicing is linked to the choice of inactive X chromosome
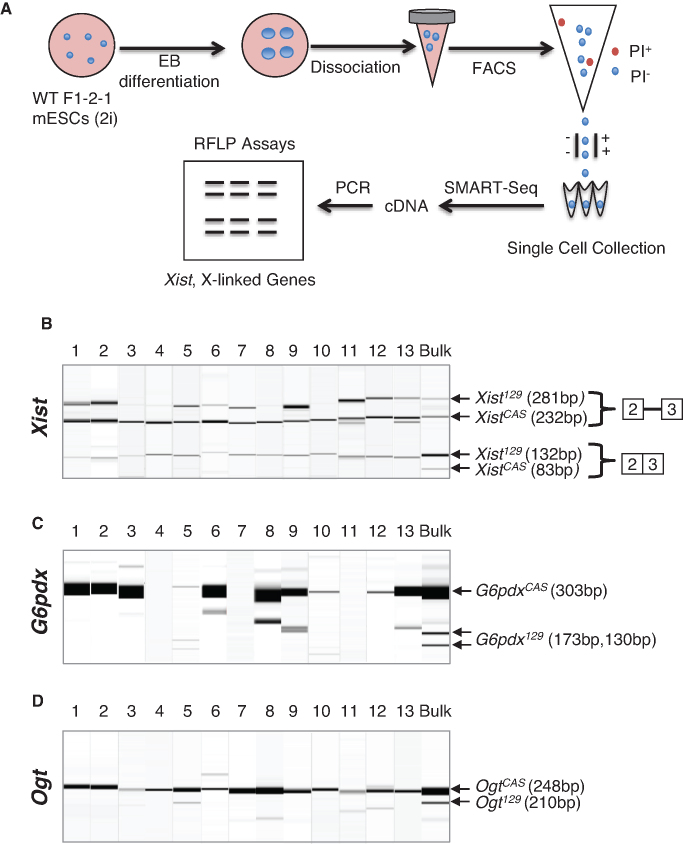
To test whether Xist RNA splicing is linked to the choice of inactive X chromosome, we compared allele-specific Xist splicing and allele-specific X-linked gene expression at the single cell level. We differentiated F1 2-1 cells and isolated single differentiating cells from embryoid bodies by FACS. We generated cDNA directly from single intact cells using SMART-seq. We then performed restriction fragment length polymorphism (RFLP) analysis to determine allele-specific Xist splicing and allele-specific expression of X-linked genes in single cells. Two X-linked genes (G6pdx and Ogt) were assessed to increase the rigor of the analysis and they were correlated when both were detected. For cells exhibiting dropouts (no detectable expression) for G6pdx or Ogt1, we relied on one detected gene.
We found some cells expressed Xist from both X chromosomes (Figure 4) and some did not (Supplementary Figure S3). Between the onset of Xist propagation and the full establishment of XCI is an intermediate state when Xist transcription continues from both chromosomes while (most) X-lined genes are being silenced from the inactive X chromosome (37,38). When XCI is fully established, Xist is expressed only from the Xi chromosome and is turned off in the active chromosome (4,5,9). Therefore, those expressing Xist from both X chromosomes likely reflected the intermediate state, whereas those expressing Xist from a single X chromosome probably entered the irreversible state of XCI. We therefore named them intermediate cells and mature cells regarding XCI stages, respectively. In mature cells, allelic preference of Xist splicing could not be fairly assessed, and any data interpretation would be confounded by probable allelic bias of Xist transcription. In intermediate cells, allelic preference of Xist splicing can be more fairly examined with bi-allelic transcription.
Figure 4.
Allele specific Xist splicing is linked to the choice of inactive X chromosome. (A) Schematic diagram of the experimental procedure using embryoid body differentiation, FACS, and SMART-Seq for single cell analysis of allele-specific Xist splicing and allele-specific X-linked gene expression in F1 2-1 cells. (B) Restriction fragment length polymorphism analysis combined with splicing assay detects allelic specific expression of unspliced and spliced Xist transcripts from the X129 and XCAS chromosomes in intermediate cells. (C and D) Restriction fragment length polymorphism analysis detects allelic expression of X-linked genes (C) G6pdx and (D) Ogt from the X129 and XCAS chromosomes. Bulk means RNA samples from bulk cells.
We found all the intermediate cells predominantly spliced Xist129 RNA and simultaneously expressed the X-linked genes predominantly from the XCAS chromosome (Figure 4). In 12 out of 13 cells (all except cell #5), spliced XistCAS was not detectable, demonstrating strong selection for allele-specific Xist splicing. Consequently, these cells expressed X-linked genes exclusively from the XCAS chromosome. Cell #5 had a weak signal for the spliced XistCAS transcript and, interestingly, appeared to express G6pdx about equally from both X chromosomes. However, its Ogt expression from XCAS was much stronger than from X129. Cell #5 was likely at an earlier differentiation stage than the other intermediate cells, meaning it was on its way to splice Xist129 exclusively and silence X129 chromosome completely.
Under the null hypothesis that Xist splicing is not linked to the choice of X-inactivation, the probability of observing 12 out of 13 individual intermediate cells that exclusively splice Xist129 and inhibit X129 chromosome is close to 0 (or 0.0017 based on binomial distribution). In all mature cells, the spliced Xist and X-linked genes originated from different X chromosomes (Supplementary Figure S3). Therefore, we conclude that allele-specific Xist splicing is intrinsically linked to the choice of inactive X chromosome. This is also consistent with the notion that Xist splicing is required for Xist’s activity to induce XCI (see Disucssion).
Xist RNA splicing is impaired in Ptbp1 mutant cells
To test whether PTBP1 influences Xist splicing, we generated female Ptbp1−/− ES cells from a Ptbp1loxp allele via homologous recombination. This Ptbp1loxp allele contains two loxp sites flanking Ptbp1 exon 2, deletion of which eliminates functional PTBP1 proteins (Supplementary Figure S4A). The female Ptbp1−/− ES cell line was identified by genotyping PCR (Supplementary Figure S4B) and confirmed by western blot using an antibody recognizing the N terminus of PTBP1 protein (Supplementary Figure S4C). When using an antibody reactive to the C-terminus of PTBP1, we detected a smaller and substantially less abundant protein (Supplementary Figure S4D), which might correspond to an N-terminally truncated PTBP1 protein isoform.
We next asked whether Xist splicing was affected in Ptbp1−/− cells. EB differentiation for 2 or 4 days in WT BL6 ES cells dramatically enhanced Xist splicing (Supplementary Figure S4E-F). Ptbp1−/− cells displayed lower splicing than WT cells. An alternative explanation for the shortage of spliced Xist transcripts in female Ptbp1−/− cells is decreased stability of the spliced transcript, such that any differentiation-enhanced splicing ratios would be attenuated by loss of the spliced transcript. However, we found no change in the stability of the spliced Xist transcript before or after differentiation (Supplementary Figure S4G). Nonetheless, PTBP1 depletion did not entirely block differentiation-induced Xist splicing at 2 or 4 days of differentiation.
We then asked whether enhanced Xist splicing correlated with any change in PTBP1 protein levels during EB differentiation but found no such correlation (Supplementary Figure S4H). PTBP1 was unchanged in the first day of differentiation and decreased in the second day of differentiation. Xist splicing is enhanced during the first day of differentiation while PTBP1 level has not changed. Second, differentiation reveals a negative relationship between PTBP1 level and Xist splicing: decrease in PTBP1 and increased in Xist splicing. Xist splicing efficiency is therefore not correlative with PTBP1 expression level but only requires a certain level of PTBP1 protein.
One possibility for the observed phenotype was that Ptbp1−/− ES cells differentiated more slowly than WT cells. To test this, we measured the expression of pluripotency marker Nanog, epiblast marker Fgf5, and neuroectoderm marker Zfp521. In WT cells, we found the expected decrease in Nanog expression upon differentiation (Supplementary Figure S4I-K). Nanog appeared to be slightly re-activated around day 4, as previously noted by Trott et al. (39). Fgf5 was induced as early as day 2 of EB differentiation and Zfp521 induced at day 4, consistent with sequential manifestation of development stages. We observed similar trends in Ptbp1−/− cell differentiation except that PTBP1 knockout appeared to accelerate the differentiation (Supplementary Figure S4I–K). If PTBP1 controlled Xist splicing through its influence on differentiation, Ptbp1−/− cells should have showed stronger induction of Xist splicing, which is not the case. Therefore, impaired Xist splicing in Ptbp1−/− cells cannot be explained by slow differentiation. We conclude PTBP1 affects Xist splicing independent of PTBP1’s interactions with cellular differentiation (Supplementary Figure S4L).
DISCUSSION
Our studies provide comprehensive evidence for splicing as an essential regulatory step in Xist induction. First, naïve stem cells expressed mostly unspliced Xist. Second, after normalizing to total Xist RNA levels, spliced Xist was upregulated and unspliced Xist was reduced during the early stage of differentiation. Third, differentiation-induced Xist splicing was significantly impaired (though not completely arrested) in Ptbp1 mutant cells, suggesting involvement of trans-factors in its splicing regulation. Fourth, in hybrid Xist129/XistCAS cells, Xist129 and XistCAS RNA were differentially spliced, indicating the existence of splicing cis-elements responsive to differentiation. Since the intrinsic bias for expressing Xist from the 129 allele has been ascribed to effects of the X-controlling element (Xce) (35), future studies should focus on Xce to identify splicing-controlling elements. Finally, allele-specific Xist splicing is intrinsically linked to the choice of inactive X chromosome.
Xist splicing probably has been overlooked historically because: (i) Xist was first cloned as spliced cDNA from somatic cells; (ii) the spliced Xist cDNA transgene is sufficient for XCI; (iii) methods for Xist detection have been designed to detect the spliced Xist transcript and (iv) Xist splicing was thought to occur quickly and passively.
To precisely measure splicing, the amplicon sizes for the spliced and unspliced transcripts should be comparable. PCR efficiency is skewed toward smaller amplicons (or spliced transcripts). The larger the difference between the amplicons, the greater the bias. Our splicing assay examining intron 2 yielded amplicon sizes of 239 and 90 bp (Figure 1D). For analyzing the largest Xist intron (i.e. intron 1), multiplex PCR was used to produce amplicons of similar sizes (127 bp versus 74 bp, Figure 1E). We also used a primer pair targeting exon 2 and 6, which yielded a unspliced product (1930 bp) and a spliced product (562 bp, Figure 1F). In this case, the unspliced Xist has become difficult to detect even in ES cells. When we assayed the splicing of intron 1, intron 2 and introns 2–5 with the same RNA samples, the spliced/unspliced ratios increased in that order, which correlates with the difference in amplicon sizes.
Another required consideration for detecting unspliced Xist transcripts is the pluripotency state of ES cells. An intrinsic relationship between pluripotency and X chromosome inactivation has been reported and the underlying mechanism through regulating Xist expression has been suggested (40). Xist splicing may be negatively influenced by the pluripotency state, and spontaneous ES cell differentiation would hinder revealing the true splicing status of Xist in ES cells.
Although Xist splicing has been below the radar, its functional importance is hinted at in previous publications. The most convincing sufficiency test for Xist’s direct role in chromosome inactivation was based on doxycycline-inducible Xist transgene lines (10). These lines were transduced with a 15kb mouse spliced Xist cDNA. Upregulation of this ectopic spliced Xist by doxycycline was sufficient to trigger inactivation of the chromosome containing the transgene in the undifferentiated ES cells. The result suggests that all factors except spliced Xist required for XCI initiation are present in ES cells. Just as our research shows that naïve ES cells express unspliced Xist, Wutz and Jaenisch's results imply that spliced Xist rather than unspliced Xist is essential to initiate XCI.
Two additional studies, though not specifically investigating Xist splicing, also show a strong relationship between the Xist splicing status and Xist’s ability to inactivate chromosomes. The mutant cells in these two studies exhibited a gain and a loss of Xist splicing. Heard et al. introduced a 480-kb YAC containing full human Xist gene (exons + introns) into mouse ES cells (41,42). Interestingly, even before in vitro differentiation the undifferentiated mouse ES cells tended to express the spliced Xist RNA from the human transgene, which functionally coated the host chromosomes. This unexpected result was not explored further in their publications but, similar to results from Wutz et al., implied the functional importance of Xist splicing.
In the second study, Royce-Tolland et al. deleted the A repeat of the Xist gene on the allele 129 of 129/CAS hybrid cells (43). The A repeat sequence resides within exon 1 and is necessary for XCI (44). These differentiated mutant cells did not inactivate the X129 chromosome. Interestingly, the mutant cells also failed to splice Xist RNA from allele 129. These results are consistent with the notion that Xist splicing correlates with functional XCI.
Our single cell analyses further strengthen the notion that Xist splicing is intrinsically linked to the choice of inactive chromosome. The striking association between allele specific Xist splicing and allele specific X-linked gene expression at the single cell level show that Xist splicing would be a critical regulatory step. Some studies suggested that differential transcription induction might not be the selection mechanism for allele-specific Xist expression. For example, Panning's group reported that the choice of Xist allele for induction was not initiated by unequal distribution of histone modification marks (e.g. H3K27me3 and H3K4me2) (43). Demethylation of the Xist locus due to deficient DNA methyltransferase activity activated Xist expression in somatic but not undifferentiated ES cells, suggesting DNA methylation is inessential for Xist repression in ES cells (45). These data raise the possibility of a role for post-transcriptional regulation in Xist allele induction.
We have not isolated cells that splice XistCAS. In bulk-cell analysis of allele-specific expression, the ratio of spliced Xist129 to spliced XistCAS is about 10:1. Therefore, the chance of isolating cells expressing spliced XistCAS is low to begin with. The ratio of spliced Xist129 cells to spliced XistCAS cells may be further skewed by FACS. We used three stringent gating steps to select single cells (Methods and Materials). Only ∼1% of total FACS events (cells) passed these stringent criteria. It is possible that some idiosyncratic paraments disproportionally led to the discard of the spliced XistCAS cells. We also note that allelic expression levels vary among cells partly due to heterogeneity in differentiation stages (or cell states) and intrinsic technical variance and bias in this kind of single cell analysis (46,47).
Xist splicing might be essential for Xist to form high-order structures to spread throughout and coat X chromosome, which is an activity observed only for the spliced Xist in literature. Xist introns could serve as a structural hindrance by themselves or as binding sites for factors blocking Xist’s ability to tether to, spread, or coat X chromosome. Alternatively, exon-exon junctions could create new binding sites for factors that act positively in mediating XCI.
Our data provides an explanation to reconcile some previous observations. While some believed transcription upregulation was the mechanism of inducing Xist, others disagreed with transcriptional activation when using an intron 1 probe to measure Xist transcription (37). We observed a significant increase in total Xist, and an even larger increase in spliced Xist, but a modest decrease in unspliced Xist. We propose that differentiation up-regulates Xist transcription and splicing. Increased splicing efficiency occludes up-regulation of the unspliced Xist transcripts (measured by the intron 1 probe). Therefore, our data is consistent with previous publications and, at the same time, provides novel insights.
Repression of Xist splicing presumably provides a fail-safe mechanism to inhibit Xist activity in ES cells. The repression of Xist in ES cells has been attributed to pluripotency factors, because de-repression of pluripotency factors up-regulate Xist expression in female cells (40,48). We found Xist transcription is detectable in ES cells, but its splicing is repressed, meaning Xist splicing is not the default. Splicing may be a context-dependent regulatory checkpoint for Xist expression, thereby contributing to regulation of Xist function. Determining the source of Xist splicing repression and activation could reveal novel splicing regulatory mechanisms.
PTBP1 significantly impairs but does not completely block differentiation-induced Xist splicing. The influence of differentiation and that of PTBP1 on Xist splicing are likely uncoupled, even though PTBP1 and differentiation are intrinsically linked (Supplementary Figure S4L). PTBP1’s effect on Xist splicing is not because PTBP1 restrains differentiation. Likewise, differentiation-induced Xist splicing is not due to differentiation-mediated downregulation of PTBP1. These suggest that PTBP1 plays a permissive rather than instructive role in regulating Xist splicing and PTBP1’s effect may be indirect. PTBP1 does interact with E repeat of Xist RNA transcripts, but it is unclear how this interaction could mediate splicing of all Xist introns (21). One possibility is that the E repeat serves as a ‘landing pad’ for PTBP1 to bring other splicing factors in proximity. Alternatively, PTBP1 might control the expression of splicing factors critical for Xist RNA splicing.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Xiang-dong Fu (University of California, San Diego) and Dr Douglas Black (University of California, Los Angeles) for providing Ptbp1loxp/loxp mice. We thank Dr Douglas Black for the PTBP1 antibodies. We thank Anastasia Schimmel and Anthony Linares for technical assistance on blastocyst culture and isolation of ES cell colonies. We thank Dr Barbara Panning (University of California, San Francisco), Leeanne Goodrich, and Karen Leung for providing F1 2-1 ES cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (NIH) [R01NS104041, R01MH116220]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Huynh K.D., Lee J.T.. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003; 426:857–862. [DOI] [PubMed] [Google Scholar]
- 2. Mak W., Nesterova T.B., de Napoles M., Appanah R., Yamanaka S., Otte A.P., Brockdorff N.. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004; 303:666–669. [DOI] [PubMed] [Google Scholar]
- 3. Monkhorst K., Jonkers I., Rentmeester E., Grosveld F., Gribnau J.. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008; 132:410–421. [DOI] [PubMed] [Google Scholar]
- 4. Lessing D., Anguera M.C., Lee J.T.. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet. 2013; 14:85–110. [DOI] [PubMed] [Google Scholar]
- 5. Augui S., Nora E.P., Heard E.. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011; 12:429–442. [DOI] [PubMed] [Google Scholar]
- 6. Plath K., Mlynarczyk-Evans S., Nusinow D.A., Panning B.. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002; 36:233–278. [DOI] [PubMed] [Google Scholar]
- 7. Senner C.E., Brockdorff N.. Xist gene regulation at the onset of X inactivation. Curr. Opin. Genet. Dev. 2009; 19:122–126. [DOI] [PubMed] [Google Scholar]
- 8. Chaumeil J., Okamoto I., Heard E.. X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 2004; 376:405–419. [DOI] [PubMed] [Google Scholar]
- 9. da Rocha S.T., Heard E.. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 2017; 24:197–204. [DOI] [PubMed] [Google Scholar]
- 10. Wutz A., Jaenisch R.. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 2000; 5:695–705. [DOI] [PubMed] [Google Scholar]
- 11. Brown C.J., Willard H.F.. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994; 368:154–156. [DOI] [PubMed] [Google Scholar]
- 12. Rack K.A., Chelly J., Gibbons R.J., Rider S., Benjamin D., Lafreniére R.G., Oscier D., Hendriks R.W., Craig I.W., Willard H.F.. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet. 1994; 3:1053–1059. [DOI] [PubMed] [Google Scholar]
- 13. Csankovszki G., Panning B., Bates B., Pehrson J.R., Jaenisch R.. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 1999; 22:323–324. [DOI] [PubMed] [Google Scholar]
- 14. Zheng S., Black D.L.. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013; 29:442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng S. Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression. Int. J. Dev. Neurosci. 2016; 55:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J.S., Choi H.W., Araúzo-Bravo M.J., Schöler H.R., Do J.T.. Reactivation of the inactive X chromosome and post-transcriptional reprogramming of Xist in iPSCs. J. Cell Sci. 2015; 128:81–87. [DOI] [PubMed] [Google Scholar]
- 17. Yue M., Ogawa Y.. CRISPR/Cas9-mediated modulation of splicing efficiency reveals short splicing isoform of Xist RNA is sufficient to induce X-chromosome inactivation. Nucleic Acids Res. 2018; 46:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hendrich B.D., Brown C.J., Willard H.F.. Evolutionary conservation of possible functional domains of the human and murine XIST genes. Hum. Mol. Genet. 1993; 2:663–672. [DOI] [PubMed] [Google Scholar]
- 19. Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y.. Systematic discovery of Xist RNA binding proteins. Cell. 2015; 161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McHugh C.A., Chen C.-K., Chow A., Surka C.F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015; 521:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vuong J.K., Lin C.-H., Zhang M., Chen L., Black D.L., Zheng S.. PTBP1 and PTBP2 serve both specific and redundant functions in neuronal Pre-mRNA splicing. Cell Rep. 2016; 17:2766–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., Schermelleh L., Nesterova T.B., Brockdorff N.. A pooled shRNA screen identifies Rbm15, spen, and wtap as factors required for Xist RNA-Mediated silencing. Cell Rep. 2015; 12:562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kafasla P., Mickleburgh I., Llorian M., Coelho M., Gooding C., Cherny D., Joshi A., Kotik-Kogan O., Curry S., Eperon I.C. et al. Defining the roles and interactions of PTB. Biochem. Soc. Trans. 2012; 40:815–820. [DOI] [PubMed] [Google Scholar]
- 24. Zheng S., Gray E.E., Chawla G., Porse B.T., O’Dell T.J., Black D.L.. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat. Neurosci. 2012; 15:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vuong C.K., Black D.L., Zheng S.. The neurogenetics of alternative splicing. Nat. Rev. Neurosci. 2016; 17:265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keppetipola N., Sharma S., Li Q., Black D.L.. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012; 47:360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Czechanski A., Byers C., Greenstein I., Schrode N., Donahue L.R., Hadjantonakis A.-K., Reinholdt L.G.. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 2014; 9:559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen L., Zheng S.. Identify alternative splicing events based on position-specific evolutionary conservation. PLoS One. 2008; 3:e2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z., Vuong J.K., Zhang M., Stork C., Zheng S.. Inhibition of nonsense-mediated RNA decay by ER stress. RNA. 2017; 23:378–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng S., Chen L.. A hierarchical Bayesian model for comparing transcriptomes at the individual transcript isoform level. Nucleic Acids Res. 2009; 37:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng S., Damoiseaux R., Chen L., Black D.L.. A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Res. 2013; 23:998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L., Zheng S.. Studying alternative splicing regulatory networks through partial correlation analysis. Genome Biol. 2009; 10:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herzel L., Straube K., Neugebauer K.M.. Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res. 2018; 28:1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S.. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992; 71:515–526. [DOI] [PubMed] [Google Scholar]
- 35. Cattanach B.M., Isaacson J.H.. Controlling elements in the mouse X chromosome. Genetics. 1967; 57:331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stavropoulos N., Lu N., Lee J.T.. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:10232–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panning B., Dausman J., Jaenisch R.. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997; 90:907–916. [DOI] [PubMed] [Google Scholar]
- 38. Sheardown S.A., Duthie S.M., Johnston C.M., Newall A.E., Formstone E.J., Arkell R.M., Nesterova T.B., Alghisi G.C., Rastan S., Brockdorff N.. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997; 91:99–107. [DOI] [PubMed] [Google Scholar]
- 39. Trott J., Martinez Arias A.. Single cell lineage analysis of mouse embryonic stem cells at the exit from pluripotency. Biol. Open. 2013; 2:1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Navarro P., Chambers I., Karwacki-Neisius V., Chureau C., Morey C., Rougeulle C., Avner P.. Molecular coupling of Xist regulation and pluripotency. Science. 2008; 321:1693–1695. [DOI] [PubMed] [Google Scholar]
- 41. Heard E., Mongelard F., Arnaud D., Avner P.. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol. Cell. Biol. 1999; 19:3156–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heard E., Mongelard F., Arnaud D., Chureau C., Vourc’h C., Avner P.. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:6841–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Royce-Tolland M.E., Andersen A.A., Koyfman H.R., Talbot D.J., Wutz A., Tonks I.D., Kay G.F., Panning B.. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nat. Struct. Mol. Biol. 2010; 17:948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoki Y., Kimura N., Kanbayashi M., Amakawa Y., Ohhata T., Sasaki H., Sado T.. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009; 136:139–146. [DOI] [PubMed] [Google Scholar]
- 45. Beard C., Li E., Jaenisch R.. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995; 9:2325–2334. [DOI] [PubMed] [Google Scholar]
- 46. Stegle O., Teichmann S.A., Marioni J.C.. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015; 16:133–145. [DOI] [PubMed] [Google Scholar]
- 47. Chen L., Zheng S.. BCseq: accurate single cell RNA-seq quantification with bias correction. Nucleic Acids Res. 2018; 46:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erwin J.A., del Rosario B., Payer B., Lee J.T.. An ex vivo model for imprinting: mutually exclusive binding of Cdx2 and Oct4 as a switch for imprinted and random X-inactivation. Genetics. 2012; 192:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.