Abstract
DNA Topoisomerase I (TopoI) in eubacteria is the principle DNA relaxase, belonging to Type 1A group. The enzyme from Mycobacterium smegmatis is essential for cell survival and distinct from other eubacteria in having several unusual characteristics. To understand genome-wide TopoI engagements in vivo, functional sites were mapped by employing a poisonous variant of the enzyme and a newly discovered inhibitor, both of which arrest the enzyme activity after the first transestrification reaction, thereby leading to the accumulation of protein-DNA covalent complexes. The cleavage sites are subsets of TopoI binding sites, implying that TopoI recruitment does not necessarily lead to DNA cleavage in vivo. The cleavage protection conferred by nucleoid associated proteins in vitro suggest a similar possibility in vivo. Co-localization of binding and cleavage sites of the enzyme on transcription units, implying that both TopoI recruitment and function are associated with active transcription. Attenuation of the cleavage upon Rifampicin treatment confirms the close connection between transcription and TopoI action. Notably, TopoI is inactive upstream of the Transcription start site (TSS) and activated following transcription initiation. The binding of TopoI at the Ter region, and the DNA cleavage at the Ter indicates TopoI involvement in chromosome segregation, substantiated by its catenation and decatenation activities.
INTRODUCTION
Topoisomerases maintain topological homeostasis during a variety of cellular processes. For example, during transcription, the movement of the RNA polymerase generates positive supercoils ahead and negative supercoils behind the transcribing machinery (1). The torsional stress generated during the process is relieved by the action of different classes of topoisomerases. Their catalytic action involves DNA binding, cleavage, strand passage and religation. The Escherichia coli genome encodes four topoisomerases—two of each type (Type 1 and Type II) (2–6). Previous work carried out in the E. coli to elucidate the cellular function of the topoisomerases have revealed that positive supercoils that are generated ahead of the RNA polymerase are removed by DNA gyrase and Topoisomerase IV (TopoIV) while the negative supercoils are acted upon by TopoI and TopoIV. TopoIV and to a lesser extent Topoisomerase III (TopoIII), separate the sister chromatids during chromosome segregation (1,3,7–11). However, not all bacteria have the full complement of topoisomerases. For example, mycobacterial genomes code for a limited repertoire of topoisomerases, a departure from a number of eubacterial genomes that have a luxury of having more than one topoisomerase of each type. The Mycobacterium tuberculosis (Mtb) genome encodes only DNA gyrase (Type II) and TopoI (Type 1) (12–15). Similarly, TopoI and DNA gyrase are the major topoisomerases in M. smegmatis, and both are essential for cell survival (16,17). The absence of TopoIII and TopoIV in mycobacteria, implies that the mycobacterial topoisomerases could have evolved to carry out additional functions. Indeed, mycobacterial DNA gyrase is a dual-function enzyme, catalysing robust decatenation in addition to supercoiling (18–20). However, additional roles if any, for TopoI are yet to be investigated.
Given the key role of topoisomerases during DNA replication, transcription, recombination and chromosome segregation, understanding how these enzymes act in vivo is of interest. A few studies have carried out genome-wide binding of topoisomerases (21–25). While these investigations have provided insights into the process of topoisomerase recruitment at various sites where topoisomerases need to be engaged, their catalytic action in vivo is less understood. In one of the studies, TopoIV activity sites were mapped in the E. coli by trapping fluoroquinolone induced enzyme-DNA complexes (21). Similarly, camptothecin (CPT) induced cleavage has been studied with a mammalian topoisomerase I, TOP1, a Type 1B enzyme (23). However, functional sites of other topoisomerases are yet to be mapped, which is important given the diverse cellular roles played by different topoisomerases. Notably, no such studies have been carried out with Type 1A topoisomerases, which are found in all eubacteria and eukaryotes. In this context, understanding in vivo action of TopoI in mycobacteria is of high relevance given its essentiality and the principal role played in DNA relaxation.
Here, we have employed multiple approaches to elucidate the genome-wide distribution of TopoI binding versus catalytic engagements. High throughput sequencing was used to map the genome-wide occupancy as well as action sites determined by genetic and chemical interventions, which allowed trapping of covalent adducts following the first transestrification reaction. Importantly, steady state TopoI action could also be visualized by capturing reaction intermediates without any perturbation for the first time in any system. Our study revealed that the action landscape of TopoI in the genome is also driven by transcription induced supercoiling. Notably, all the binding sites were not accessible at a given time in vivo for TopoI activity and the basis for this inaccessibility of the sites was investigated. We also demonstrated the recruitment of TopoI at the Ter region of the chromosome to carry out decatenation function, indicating multiple crucial roles played by the sole Type 1A enzyme present in the cell.
MATERIALS AND METHODS
Bacterial strains and culture conditions
Mycobacterium smegmatis TopoI D108A, a toprim mutant of TopoI (28) was sub-cloned in pMIND vector (26) to attain MsTopoI D108A clone. MsTopoI D108A harbouring M. smegmatis (mc2 155) cells were grown at 37°C in 7H9 medium (Difco) enriched with 0.05% Tween 80 and 0.2% glycerol or on solid Middlebrook 7H11 medium (Difco) supplemented with 0.5% glycerol. Appropriate antibiotics were added wherever required.
Trapping enzyme–DNA covalent complexes
Mycobacterium smegmatis competent cells were transformed with MsTopoI D108A containing plasmid. The cells were grown till OD600 nm of 0.4–0.6 followed by induction with 25 ng/ml of tetracycline for 4 h. The harvested cell pellets were resuspended in 200 μl of RLT Plus buffer (Qiagen) and incubated at room temperature for 10 min for lysis. The samples were centrifuged at 13 000 rpm for 10 min at 4°C and supernatants were collected. Protein–DNA covalent complexes were ethanol precipitated and resuspended in 200 μl of 8 mM NaOH. DNA was quantified using a nano drop spectrophotometer (Thermo Scientific) and spotted on PVDF membrane by Bio-Dot apparatus (Bio Rad) as described (27). The complexes were probed for TopoI using specific monoclonal antibodies (mAb) (28). Similarly, for imipramine dot-blot, exponentially growing M. smegmatis cells were treated with 10× MIC of imipramine for 12 h, covalent complexes isolated and dot-blot assays were carried out as described above. Dot-blots were performed several times and similar results were obtained.
RNA isolation and quantitative PCR
RNA was isolated from exponentially growing M. smegmatis harbouring MsTopoI D108A and imipramine treated cells by using RNAZol-RT reagent (Sigma). RNA concentration and purity was checked spectrophotometrically and on 1.2% formaldehyde gel respectively. cDNA was synthesized using Applied Bioscience cDNA synthesis kit following manufacturer's instructions. Quantitative PCR (qPCR) was carried out using Bio Rad CFX96 Touch Real Time PCR Machine. 16S rRNA was used as internal control for normalization. The data represented is mean±SD from the three independent experiments.
Chromatin immunoprecipitation
Exponentially growing MsTopoI D108A M. smegmatis cultures were harvested and pellets were re-suspended in immune-precipitation buffer and samples were processed as described earlier (22). TopoI bound DNA was pulled down using anti-TopoI antibodies (28). Mock-IP reactions were without TopoI antibodies. DNA was extracted twice with phenol-chloroform, precipitated and re-suspended in 50 μl of water. For imipramine ChIP, exponentially growing cells were treated with 10× MIC of imipramine for 12 h and ChIP was carried out. To identify TopoI binding, formaldehyde crosslinked ChIP was performed (22). TopoI enrichment was checked on TopoI targeted genes (16) using qPCR. The data represented is mean±SD from the three independent experiments.
ChIP-Seq library construction, sequencing and data analysis
For library construction DNA fragments of 150– 250 bp were selected. Sequencing was performed on the Illumina Genome Analyzer IIx using the Paired-Read Cluster Generation Kit v4. The data were processed using the Illumina Pipeline Software v1.60. The sequences with adapters and low quality reads (Phred Score < 30) were removed using Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). These reads were aligned to the M. smegmatis genome (Assembly accession: GCA_000767705.1 [ASM76770v1] http://bacteria.ensembl.org/Mycobacterium_smegmatis/Info/Index ) using Bowtie2 (version 2.2.6) (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). The peak calling was performed using MACS2 (version 2.1.1.20160309) (https://github.com/taoliu/MACS). Target prediction was performed using ChIP seeker (https://www.ncbi.nlm.nih.gov/pubmed/25765347) and annotations from Ensembl Bacteria for M.smegmatis (ftp://ftp.ensemblgenomes.org/pub/bacteria/release38/gff3/bacteria_7_collection/mycobacterium_smegmatis_str_mc2_155/)). For visualization, alignment files were converted to BedGraph format and uploaded to UCSC genome browser. The data have been uploaded to NCBI (SRA accession number-PRJNA503689).
Primer extension
Mycobacterium smegmatis cells harboring MsTopoI D108A (OD600nm = 0.4–0.6) were induced with 25 ng/ml of tetracycline. Another set of cells were treated with Rifampicin for 10 min prior to induction. Genomic DNA was isolated from all the cultures using CTAB method (29). Primer extensions were carried out with 2 pmol of P32 labeled specific primers (Supplementary Table DS1) and 1 U of Taq DNA polymerase in a reaction mixture containing 20 mM Tris–HCl (pH-8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4 and 1% Triton X-100 and dNTPs. The samples were subjected to 29 and 10 cycles of 95°C for 1 min, 95°C for 30 s, 58°C for 50 s and 72°C for 1.5 min. The products were ethanol precipitated and resolved on 8 M urea 12% polyacrylamide gel. Similar primer extension analysis was carried out with M. smegmatis cells treated with imipramine. The experiments were performed multiple times and similar trends were observed.
Catenation and decatenation assays
Catenation of M13 single stranded DNA was carried out in a 20 μl reaction volume containing 20 mM Tris–HCl (pH 8.0), 20 mM KCl, 6 mM MgCl2, 5 mM spermidine, 0.5 mM DTT, 50 μg/ml BSA, 30% glycerol (v/v) or 10% PEG-400 or 10% PEG-8000, 100 ng of DNA substrate and 0.2 μM MsTopoI. Reaction mixtures were incubated at 52°C for 1 h and terminated by adding 1% SDS and 1mM EDTA. The products were ethanol precipitated and resolved on 0.8% agarose gel. Gels were stained with SYBR™ Gold (Invitrogen) and image was documented by Biorad gel documentation system. For decatenation assay, catenated DNA (kDNA, Inspiralis) was incubated with increasing concentration of MsTopoI at 37°C for 30 min in the assay buffer containing 40 mM Tris–HCl (pH 8.0), 20 mM NaCl, 1 mM EDTA, 5 mM MgCl2 and 5% glycerol and terminated by adding 1% SDS and 1mM EDTA. The products were resolved on 0.8% agarose gel and stained with EtBr. The experiments were performed multiple times and similar results were obtained.
Oligonucleotide cleavage assays
Thirty two mer oligonucleotides were designed from the cleavage and binding peaks (Supplementary Table DS1). Cleavage assays were carried with MsTopoI and 5′ end-labelled 32-mer oligonucleotides and in assay buffer (40 mM Tris–HCl (pH 8.0), 20 mM NaCl and 1 mM EDTA) at 37°C for 30 min. The products were resolved on 8 M urea and 12% polyacrylamide gel once reaction was terminated with heat inactivation at 95°C for 2 min in a buffer containing 45% formamide. Cleavage assays were also performed in the presence of HU or Lsr2 and the resultant products were resolved on denaturing polyacrylamide gels as described above. The experiments were performed multiple times and similar results were obtained.
RESULTS
Experimental design and validation
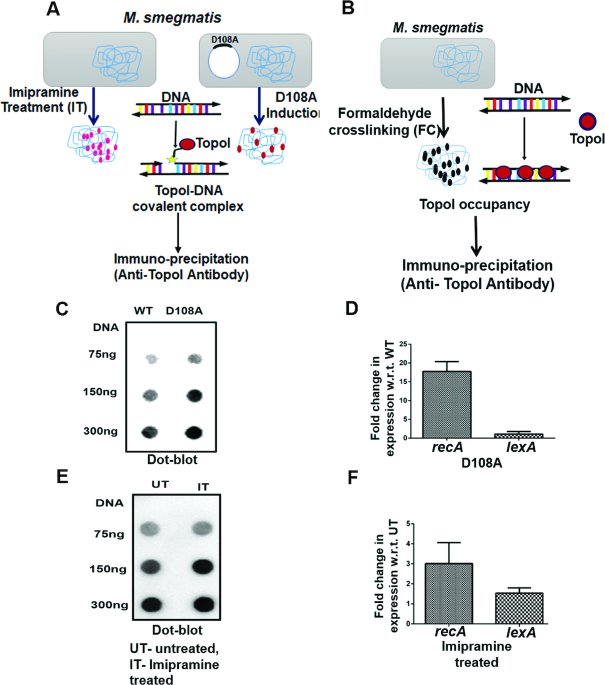
Although genome-wide occupancy studies with topoisomerases would reveal the interaction of the enzyme across the genome, such analysis do not pin-point the sites of catalytical activity. In order to map functional sites of TopoI action, two strategies were employed (Figure 1A). In the first approach, a poisonous variant of TopoI is used, which generates covalent adducts between the enzyme and DNA, arresting the reaction at that stage (30). We have shown previously that an alanine substituted mutant of the first conserved amino acid in the DXDXE motif in the toprim region of MsTopoI (D108A) leads to the accumulation of covalent complexes in vitro (30). Instead of completing the two sequential transesterification reactions, in the mutant enzyme catalysed reaction, the covalent complexes are accumulated even in the presence of Mg2+, which otherwise promotes religation (30). That the mutant enzyme arrests the enzyme reaction in vivo can be shown in an assay where covalent complexes are detected using TopoI specific antibodies. In the second approach, imipramine, a newly discovered inhibitor of mycobacterial TopoI, is employed (31). This tricyclic antidepressant drug specifically inhibits mycobacterial TopoI, generating cleaved complexes (31). Thus, treatment with imipramine leads to the arrest of the TopoI reaction after the first transesterification (31) in a manner analogous to fluoroquinolone arrested ternary complexes of DNA–DNA gyrase and DNA–TopoIV (21,32,33). These complexes can also be conveniently captured by the antibodies against MsTopoI without requiring formaldehyde crosslinking used for conventional ChIP. However, formaldehyde crosslinking would allow mapping of genome-wide binding of the enzyme (Figure 1B).
Figure 1.
Experimental approaches and validation. (A) Schematic of experimental approaches to map TopoI functional sites. Imipramine treated (IT) and D108A mutant mediated protein-DNA covalent adducts were immune-precipated using anti-TopoI antibodies and sequenced. (B) Overall occupancy of TopoI is determined by formaldehyde crosslinking of TopoI and DNA (FC). TopoI linked to DNA was immune-precipitated using anti-TopoI antibodies. (C) Dot-blots showing Protein-DNA covalent complexes isolated from MsTopoI D108A overexpressing M. smegmatis (mc2 155) cells and (E) imipramine treated M. smegmatis cells probed with TopoI antibodies. (D) qPCR showing increased recA expression in MsTopoI D108A overexpressing M. smegmatis cells and (F) imipramine treated M. smegmatis cells. The data represented is mean ± SD from the three independent experiments. UT = untreated, IT = imipramine treated.
To validate the two approaches, MsTopoI D108A was overexpressed in M. smegmatis (Supplementary Figure S1), TopoI–DNA covalent adducts were isolated and detected using dot-blot. The TopoI signals increased with increasing DNA concentration (Figure 1C) indicating the accumulation of DNA-protein covalent adducts. DNA damage brought about by DNA breakage was confirmed by the qPCR of recA expression. The increase in recA expression indicated the induction of SOS response upon protein-DNA covalent complex formation (Figure 1D). As in the case of D108A, after imipramine treatment, protein-DNA covalent complexes were isolated, which were detected by immunoblots (Figure 1E). Higher recA expression indicated DNA damage due to DNA cleavage after the drug treatment (Figure 1F). Thus, with both the approaches, probing the TopoI action in vivo resulted in accumulation of reaction intermediates, which are captured by the antibodies and the cleavage sites in DNA determined by sequencing. Our previous studies indicated that fabG, aldolase and groS genes were down regulated in MsTopoI knockdown strain (16). TopoI enrichment on these TopoI targeted genes was confirmed using ChIP-qPCR for D108A ChIP (Supplementary Figure S2A), formaldehyde crosslinked ChIP (FC ChIP, Supplementary Figure S2B) and imipramine treated ChIP samples (IT ChIP, Supplementary Figure S2C) prior to the genome-wide analysis.
Analyses of genome-wide TopoI recruitment and activity
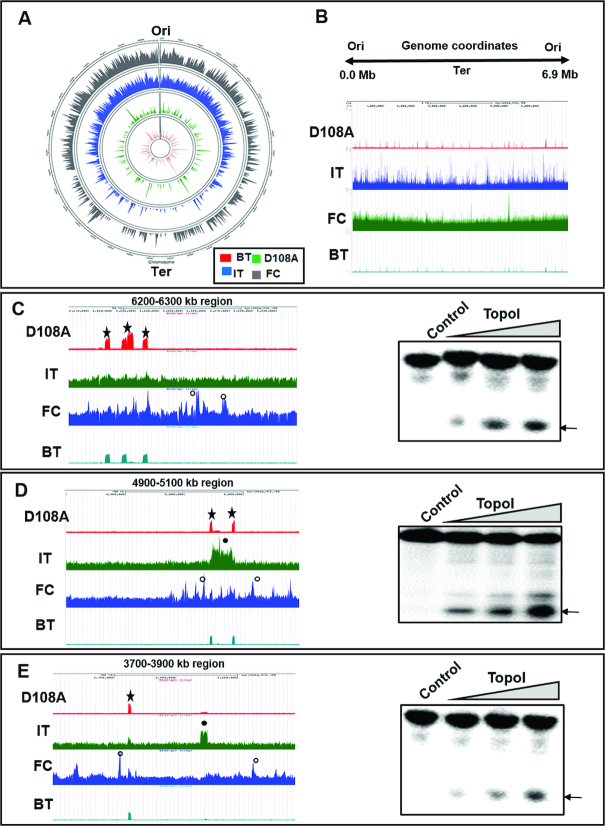
The genome wide TopoI occupancy and the action sites are represented on the circos plot using circlize tool (https://cran.rstudio.com/web/packages/circlize/circlize.pdf) (Figure 2A). The grey circle depicts TopoI occupancy on the M. smegmatis genome, whereas blue and green circles represent TopoI action sites mapped by IT ChIP-seq and D108A ChIP seq respectively. While the enzyme recruitment is found throughout the genomic landscape, the major cleavage sites appear to be localized to specific regions across the genome (Figure 2A and Supplementary Figure S3 A-G). To verify that the cleavage peaks are indeed obtained by TopoI activity, 32mer oligonucleotides were designed from three different genomic coordinates—6200–6300 kb, 4900–5100 kb and 3700–3900 kb (Figure 2C–E, left panel, respectively). Cleavage assays with these three sets of oligonucleotides and TopoI indicated that in vivo cleavage peaks obtained by sequencing are genuine cleavage sites (Figure 2C–E, right panel). Figure 2B depicts the linear UCSC browser snapshot where Ori is represented at both the extremities and the Ter is located in the middle. High density of the TopoI peaks were found near Ori (Supplementary Figure S4A and B) for all the samples (D108A, IT and FC) used for the study. These patterns, were retained even after copy number and sequencing depth normalization. Similar higher abundance of peaks for binding of topoisomerases near Ori were seen in earlier studies with the Escherichia coli and Mtb (21,22,25). Genomic coordinates for TopoI functional sites were further analysed (Supplementary Figure S3A–G) and three of the highly-enriched sites are expanded to obtain better insights into TopoI in vivo action (Figure 2C–E). Analysis of the zoomed regions showed more cleavage peaks for IT ChIP compared to D108A cleavage peaks, indicating drug induced additional cleavage of DNA (Figure 2E, closed circle). Similarly, in studies with the E. coli TopoIV, hundreds of cleavage sites were obtained in norfloxacin ChIP (21). Remarkably, the examination of circos plot (Figure 2A) revealed that in the experiments conducted in the absence of imipramine treatment or without MsTopoI D108A expression, the basal TopoI activity was captured during the natural reaction of the enzyme (BT- ChIP). These sites were at identical locations (0.65 Pearson correlation coefficient with D108A, Supplementary Figure S5 A) to the cleavage sites mapped using TopoI poisonous variant D108A (Figure 2C–E, star) but are subsets of mutant induced cleavage peaks (∼150 peaks for BT ChIP and ∼270 peaks for D108A). Thus, the different approaches used to assess TopoI functional sites show remarkably similar patterns, indicating the robustness of the methods employed (Pearson correlation coefficient for D108A and IT = 0.61, FC and IT = 0.59 and D108 and FC = 0.48, Supplementary Figure S5 B-D).
Figure 2.
Analyses of TopoI binding and cleavage sites distribution in the genome. (A) Circos plot for whole genome TopoI functional sites. Innermost red circle represents basal TopoI cleavage (BT), green circle depicts D108A induced cleavage and blue circle is drug imipramine induced (IT) cleavage. The grey circle depicts TopoI occupancy by formaldehyde crosslinked ChIP (FC) and outermost circle represents genomic coordinates of M. smegmatis (mc2 155). (B) UCSC genome browser view of TopoI binding and cleavage peaks across the M. smegmatis genome (0.0–6.9 Mb). (C–E) UCSC browser higher magnification view of different genomic coordinates. D108A (star), IT (closed circle) and TopoI FC (open circle) are marked. Additional drug induced IT cleavage peaks are seen compared to D108A induced cleavage. In all the three genomic coordinates chosen basal cleavage (BT) at same location as D108A. Right panels show in vitro DNA cleavage by MsTopoI. Oligonucleotides designed from one of the peaks of genomic coordinates 6200–6300, 4900–5100 and 3700–3900 kb respectively were analysed for cleavage as described in Materials and Methods. The resultant products were analysed on 8M urea 12% polyacrylamide gel. Arrow indicates the cleavage product.
Insights into TopoI sequence specific interaction in vivo
Previous studies revealed that unlike most other topoisomerases, both M. smegmatis and Mtb TopoI interact with DNA in sequence specific fashion. They recognised the sequence CG/TCTTC/G, termed as strong topoisomerase site (STS) and cleaved DNA between the two T residues (34–37). Earlier studies also revealed that the enzyme requires a longer length of at least 20 nt DNA for binding and cleavage (38) unlike the E. coli TopoI, which can cleave oligonucleotides of 7–8 nt length (39). Further, foot printing studies revealed a large protected DNA region due to MsTopoI binding, which also encompassed upstream region of STS (37). Thus, it is probable that the enzyme would recognise similar sequences in vivo, although DNA cleavage sites in vivo have not been mapped for any Type 1A enzyme. Hence, major cleavage peaks were analysed to identify common recognition motif. The consensus motif was generated (Supplementary Figure S6A) (P value = 1e–4) for the enriched cleavage sites using MEME suite (40). The motif derived was similar to the motif obtained from in vitro studies (34,35), which indicated that the mycobacterial TopoI exhibits sequence preference in vivo, in genomic context as well. To further validate, oligonucleotides designed from the derived motif from one of the regions where both binding and cleavage is seen was assessed for TopoI cleavage. With the increasing enzyme concentrations, the cleavage of the oligonucleotides was increased (Supplementary Figure S6B). The oligonucleotides with one or two nucleotide variation from the consensus sequence showed reduced cleavage activity indicating the sequence specific interaction of the enzyme (Supplementary Figure S6C–E). Notably, when oligonucleotides from the genome sequence where peaks were not found or totally non-specific sequences were taken, cleavage was not observed (Supplementary Figure S6F and G).
Some binding sites are recalcitrant to cleavage by TopoI
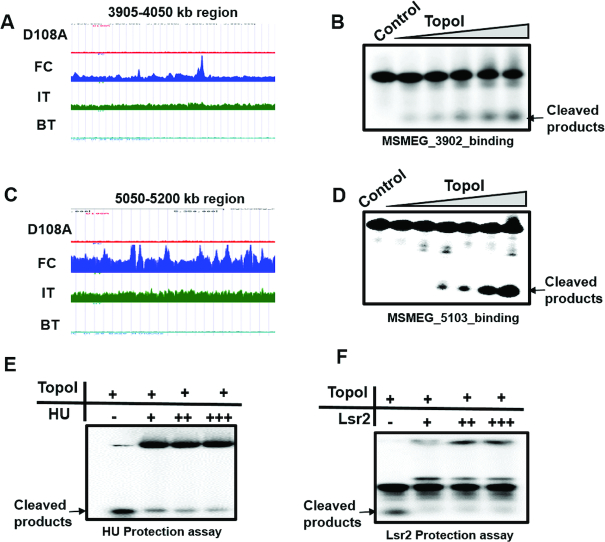
Comparison of TopoI binding and cleavage data showed that cleavage peaks were absent in a few locations where corresponding binding peaks were seen (Figure 2C and D, open circle). These results indicated that although TopoI is recruited, the sites may be inaccessible for cleavage. For example-from Figure 2C–E, the Supplementary figure S3 A-G and Figure 3A and C, it is evident that TopoI mediated DNA cleavage sites are not seen at several locations where peaks for recruitment are observed. Thus, although TopoI sites are distributed throughout the genome, not all the sites where it is recruited are accessible for TopoI action in vivo. In order to understand the basis for the inaccessibility of the enzyme for the substrate, oligonucleotides were designed from these regions and in vitro cleavage reactions with MsTopoI were carried out. Cleavage of these oligonucleotides at comparable efficiency to that of any other sites indicated that these sites are actually genuine TopoI sites with respect to enzyme recognition and cleavage (Figure 3B and D). This inaccessibility of the sites in vivo for the enzyme action could be due to the masking of the sites by the binding of other DNA binding proteins or topological constraints rendering the sites refractory to cleavage.
Figure 3.
Protection of cleavage sites by NAPs. (A and C) UCSC browser view in higher magnification for 3905–4050 and 5050–5200 kb genomic coordinates respectively. (B and D) Cleavage assay with DNA having TopoI recognition motif but cleavage not seen in vivo. Oligonucleotides (32mer) from DNA binding peaks were treated with increasing concentration of MsTopoI (0.01–0.16 μM). The resultant products were analysed on 8M urea 12% polyacrylamide gel. (E and F) Protection assay with NAPs. 32mer oligonucleotides were incubated with increasing concentration of HU (E) or Lsr2 (F) and cleavage assays were carried out with TopoI (0.2 μM). The resultant products were analysed on 8 M urea and 12% polyacrylamide gel.
NAPs, by virtue of their DNA binding, compaction, silencing and genome organisation roles, are the most likely candidates, which hinder the TopoI action at these sites in vivo (41–43). To address whether binding of NAPs to the DNA leads to protection from TopoI action, two of the well-studied NAPs of mycobacteria—HU and Lsr2, were chosen for protection analysis. HU is an abundant NAP, conserved among all the eubacterial species and is considered the archetypal bacterial counterpart of eukaryotic histones (44). The Lsr2 protein is specific to mycobacteria but has properties similar to H-NS from the E. coli (45,46). One of the major roles of H-NS is to repress gene transcription by binding to AT-rich sequences in a sequence independent fashion (47,48). In addition, H-NS is also a key player in silencing genomic regions (49). In order to test whether HU and Lsr2 protect the binding sites from TopoI mediated cleavage, the oligonucleotides were incubated with HU or Lsr2 and then TopoI cleavage assays were carried out. Both the NAPs inhibited TopoI mediated cleavage (Figure 3E and F), implying NAPs mediated protection could play a role in the inaccessibility of the sites in vivo.
Transcription modulates TopoI activity
During transcription, the transcribing RNAP would induce the formation of both positive and negative supercoils ahead and behind it, respectively, facilitating the recruitment of topoisomerases to transcription units. Indeed a few recent studies show such recruitment of topoisomerases (21–25). Co-localization of TopoI and DNA gyrase along with RNAP in transcription units (TUs) has been described in Mtb indicating a close connection between topology and transcription (22). In a study in mammalian cell lines, the TOP1 activity was activated by RNA polymerase II (RNAPII) phosphorylation. The inactive TOP1 bound at the upstream of TSS becomes fully active after the transcription pause release triggered by phosphorylation of C-terminal domain of RNAPII (23). Thus, to examine such a close connection between transcription and topoisomerase function, both TopoI occupancy and activity were analysed on highly and lowly expressed genes located across the genome. The occupancy and activity of the enzyme were found more on highly expressed genes compared to lowly expressed genes (not shown). To compare the binding verses activity pattern of the enzyme during transcription, both binding (FC ChIP) and activity (D108A ChIP) of TopoI were plotted along with RNAP ChIP data. The initiating and elongating RNAP were distinguished by holoenzyme ChIP (RNAP sigma ChIP) verses core enzyme ChIP (RNAP beta ChIP) respectively (50). DNA-bound TopoI was co-localized with RNAP from 1.5 kb upstream of TSS to 1.5 kb downstream (Figure 4A and B). Although TopoI was bound upstream of TSS along with initiating RNAP (RNAP sigma) (Figure 4A), the cleavage sites were co-localised with elongating RNAP (RNAP beta) in the downstream 1.5 kb region (Figure 4B) showing that TopoI activity is associated with active transcription. That the transcription drives TopoI recruitment and function is confirmed by treating the cells with Rifampicin, which inhibits transcription. The primer extension reactions with the genomic DNA isolated from the MsTopoI D108A overexpressing strain for 4900–5100 and 3700–3900 kb genomic coordinates (Figure 2D and E) showed reduction in cleavage after treatment with Rifampicin, indicating the requirement of active transcription for TopoI activity (Figure 4C and D, lanes 2 and 4). Similar results were obtained with DNA cleavage of imipramine treated cells. Rifampicin treatment attenuated in vivo TopoI cleavage in case of imipramine mediated cleavage as well (Figure 4C and D, lane 3 and 5).
Figure 4.
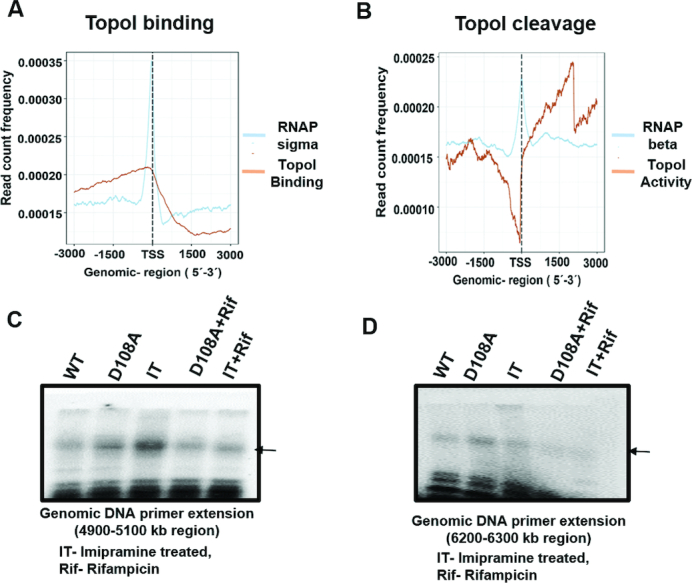
Transcription modulates TopoI activity. (A) The M. smegmatis genome is divided into the windows of 6kb (−3 to +3 kb) around the TSS. The reads falling in each overlap window were normalized to get read count frequency (y-axis) for TopoI binding (FC) and RNAP binding (RNAP sigma). (B) Similarly normalized read count frequency TopoI activity (D108A) and elongating RNAP (RNAP beta) were plotted. (C and D) Genomic DNA primer extension showing attenuation of in vivo TopoI mediated DNA cleavage by Rifampicin. Genomic DNA was isolated from MsTopoI D108A, MsTopoI D108A treated with Rifampicin (Rif) before induction of MsTopoI D108A and WT M. smegmatis (WT, control), imipramine treated cells (IT) and Rifampicin+imipramine (Rif+IT) treated cells. Primer extension was performed with specific forward primers (Ms_murA_4934_99bp_F and Ms_topA_6160_F) for 4900–5100 kb (C) and 6200–6300 kb (D) respectively with Taq DNA polymerase. The resulting products were analysed on 8M urea 12% polyacrylamide gel. Arrow indicates the cleavage product.
TopoI action near Ter region
Unexpectedly, ChIP-seq analysis revealed TopoI binding and cleavage peaks near the Ter region in M. smegmatis genome suggesting that the enzyme is catalytically engaged at these sites (Figure 5A). That the cleavage sites found at the Ter region are due to genuine TopoI activity was confirmed by primer extension of the genomic DNA isolated from M. smegmatis harbouring MsTopoI D108A. Appearance of cleavage signals in case of D108A confirmed the in vivo activity of TopoI at the Ter region (Figure 5B). Towards the completion of DNA replication, daughter DNA molecules form precatenane and catenane at the Ter region. These interlinked DNA duplex molecules are resolved by the decatenation activity of TopoIV in a majority of eubacteria examined, where TopoIV is a dedicated decatenase (51,52). In addition to TopoIV, TopoIII, a Type 1A enzyme also participates in the removal of precatenane in the E. coli (53,54). However, in the absence of both TopoIII and TopoIV in genus Mycobacterium (14,17), TopoI appears to be carrying out the role of TopoIII. To verify, catenation and decatenation activities of TopoI were analysed. Using phage M13 ssDNA, we show that TopoI has catenation activity (Figure 5C) confirming our earlier observations (34). Importantly, its decatenation activity is visualized with kDNA as a substrate (Figure 5D). Thus, mycobacterial TopoI is a dual-function enzyme, the relaxase of the cell also having decatenation activity likely to be functioning in the removal of precatenane/catenane generated during replication.
Figure 5.
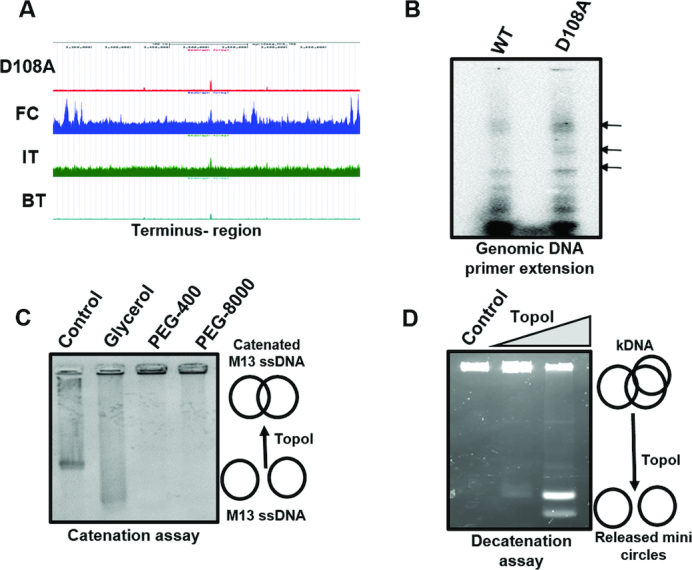
TopoI catalysed catenation and decatenation. (A) UCSC browser view for Ter-region in higher magnification indicating the specific recruitment (FC) and action peaks (D108A and IT) for TopoI. (B) Primer extension of genomic DNA to confirm D108A induced cleavage at Ter region. Genomic DNA was isolated from MsTopoI D108A overexpressing M. smegmatis cells. Primer extension was carried out using forward primer (Ms_3434_3432_F) with Taq DNA polymerase and analysed as described in the Materials and Methods and legends for figure 5. (C) Catenation assay with single-stranded M13 DNA. 0.2 μM of TopoI was incubated with 100 ng of M13 ssDNA in a buffer containing 20 mM Tris (pH 8.0), 20 mM KCl, 6 mM MgCl2, 5 mM spermidine, 50 μg/ml BSA and 0.5 mM DTT in the presence of 30% glycerol or 10% PEG-400 or 10% PEG-8000. The formation of catenated products were analysed on 0.8% agarose gels. (D) Decatenation assay with kDNA. 0.2 uM of TopoI was incubated with 200 ng of kDNA in a buffer containing 20 mM Tris, 20 mM NaCl, 5 mM MgCl2. The mini circles released by the action of TopoI were analysed on 0.7% agarose gels. Control: reaction mixture without TopoI.
DISCUSSION
Supercoiling is an indispensable component and inevitable consequence of DNA and RNA metabolism (3). The excess of supercoiling, both positive and negative, generated during replication and transcription have to be removed by the activity of topoisomerases to manage normal cellular function (1–3,55). We recently provided experimental demonstration for the co-localization of DNA gyrase and TopoI with RNAP by analysing the genome-wide binding profile of DNA gyrase, TopoI and RNAP in Mtb as a validation of the twin supercoiled domain model (22). The distribution profile of DNA gyrase and TopoI correlated well with that of RNAP indicating the functional interplay between these enzymes and RNAP (22). Such an interaction of the topoisomerases with transcription apparatus, following their recruitment is necessary for further continuation of the transcription elongation. Although these studies implied the function of topoisomerases during transcription, their actual catalytical activity per se was not examined. By applying multiple approaches, we provide the first direct experimental verification of genome-wide action of a prokaryotic TopoI during transcription and also uncover its hitherto unknown role in chromosome segregation. To identify MsTopoI functional sites, the site specific cleavage characteristic and the processivity attributed to the enzyme have been considered while exploiting a poisonous mutant of TopoI and an inhibitor, both of which arrest the reaction after DNA cleavage, creating imbalance in cleavage-religation equilibrium.
It is imperative that negative supercoiling generated during transcription in the vicinity of the RNAP has to be resolved for the continuation of the process. Indeed from the data presented in the Figure 4A and B, we are able to visualize the close linkage between transcription elongation and TopoI activity. The cleavage activity of the enzyme is high throughout the ORF (open reading frame) and low at upstream promoter region and at the ends of the genes. Although, TopoI binding is seen at promoter proximal upstream region as well as at TSS, the absence of cleavage indicates the inactivity of the enzyme at the promoter region. Thus, although both RNAP and TopoI would co-exist at and in the vicinity of the TSS, TopoI is not functionally active prior to the initiation of the transcription process. In the hind sight, this is not surprising given that the negative supercoils generated at the promoter by RNAP binding to –10 and –35 elements would be needed to retain promoter melted state by the enzyme (56–58). Much of the underwound DNA sequence at the promoter region is wrapped around RNAP to favour transcription initiation and hence DNA will not be available for TopoI action (59,60). However, during transcription elongation, continuously generated negative supercoils must be resolved by the action of TopoI throughout gene body as seen in the present study. In this context, the observation that TopoI physically interacts with RNAP gains significance (61). The authors hypothesized that the enzyme may migrate with the transcription elongation complex to prevent the accumulation of supercoiling (61). Our results provide experimental support for the co-migration model of TopoI and RNAP during transcription elongation. The data presented also imply that TopoI recruited at the promoter has to be retained in an inactive state prior to transcription initiation. This is reminiscent of the behaviour of eukaryotic TOP1 (a Type 1B enzyme), which is recruited to the promoter but remains inactive until activated (23). The enzyme is activated after the activation of CTD of RNAPII by phosphorylation within a zone of ∼1.5 kb downstream of the transcription pause site (23). Transcription by RNAPII is resumed once the opposing torque is removed by the relaxation activity of activated TOP1 (23). Key factors (BRD4 and PTEFb) participate in this TOP1-RNAPII interplay to overcome the transcriptional pause and to commence the elongation process. It is a moot point whether the bacterial TopoI would also require such elaborate activation process involving complex interplay. Alternatively, the activation of the enzyme may simply be brought about by the mechanical relay of the topological stress created by RNAP movement. Nevertheless, there appears to be a parallel in the involvement of distinct types of topoisomerases during initiation and elongation of transcription in prokaryotes and eukaryotes, although these two enzymes function differently. Being a Type 1B topoisomerase, TOP1 in eukaryotes acts ahead of transcription machinery to remove positive supercoils whereas bacterial TopoI (Type 1A) acts behind transcription machinery to remove negative supercoils.
Our analysis of TopoI cleavage sites in vivo revealed several interesting features. For the first time in any system, steady state reaction intermediates of a topoisomerase action were captured possibly reflecting the high quality of the antibodies used, which arrest enzyme-DNA covalent adducts without drug treatment. That these are genuine reaction intermediates and not the artefacts is evident given that the same peaks were seen when the poisonous variant MsTopoI D108A was induced in the cells and in the drug imipramine treated samples, which arrest the reaction intermediates after the first transestrification reaction. Moreover, TopoI mediated genomic cleavage was verified by PCR analysis of isolated genomic DNA. It is rather obvious why the steady state arrested cleavage peaks detected were fewer in number, as the reaction would have gone to completion in most sites where enzyme is engaged in the process. However, when the TopoI function is perturbed by the addition of imipramine, a large number ie ∼1000 cleavage peaks were generated. The number of peaks with the poisonous variant (MsTopoI D108A) is lower than drug treated samples because of the fact that both MsTopoI WT and MsTopoI D108A would compete for the sites. The peaks seen with D108A induction are thus due to its dominant negative effect.
Another notable feature is that while a majority of the sites were cleaved in the genome by the enzyme a few were resistant. The refractoriness of the sites could be due to chromosome compaction mediated by several factors. The bacterial chromosome is packed into a condensed structure termed nucleoid, which displays higher order organisation (41–43). Protein mediated packaging involving NAPs and topological maintenance brought about by topoisomerases join together to manage higher order genome organisation (62). By virtue of their multiple binding modes viz looping, bridging and coating, NAPs such as H-NS have the ability to occlude the binding of other proteins and/or silence gene expression (41). Similarly HU, a major NAP in all eubacteria, has multiple roles in the cell (41,63). The protection of TopoI sites from cleavage by HU and Lsr2 (H-NS homologue in mycobacteria), two of the NAPs tested, probably hints at dynamic interplay in vivo. We showed previously that HU and TopoI physically and functionally interact in mycobacteria (64). At higher concentrations of HU, it inhibited the TopoI activity (64). Similarly, occlusion of TopoIV binding and cleavage by H-NS binding is also documented (21). Nucleoid dynamics at various stages of growth has been well documented (41). Under different experimental conditions and different growth phases some TopoI sites would become inaccessible due to the alteration in chromatin architecture with changes in steady state levels of various NAPs.
One of the prevailing views is that the main role of bacterial TopoI is in the removal of R-loops to reduce genome instability (65,66). This idea has gathered support from the studies with the E. coli TopoI and TopoIII. The E. coli has the luxury of having three relaxases (TopoI, TopoIII and TopoIV) to carry out DNA relaxation. As a result, backup relaxation activity is provided by one or the other enzyme when the activity of individual enzyme is compromised (3,7,9,11). However, in genus Mycobacterium and in a number of other bacteria where there is single Type 1A topoisomerase and where there is no backup relaxase, the primary role of TopoI has to be the removal of negative supercoils generated during active transcription and replication. The data presented in the present study reinforce such a thesis. The negative supercoils removal by the enzyme would also resolve the problem of excessive R-loops.
A major finding in the present manuscript is the detection of TopoI activity at the Ter region. A distinct cleavage peak is seen with D108A ChIP, IT ChIP and even in basal cleavage, indicating that TopoI is active at the Ter site. The ability of the enzyme to catenate and decatenate the DNA together with the action at the Ter site imply a role for it in chromosome segregation. Normally, in well studied eubacteria such as the E. coli, entangled daughter DNA molecules are segregated by TopoIV activity. In no small measure, TopoIII, which also exhibits decatenase activity, participates in the removal of replication precatenanes. The absence of both the enzymes in mycobacteria necessitates that these microorganisms evolve strategies to solve DNA segregation problem towards the end of replication. Indeed, previous studies have revealed that in addition to its supercoiling function, DNA gyrase from both Mtb and M. smegmatis is a strong decatenase (18–20). Its recruitment to the Ter site indicated a role for the enzyme in resolution of daughter chromosome catenanes (22). The present analysis provide evidence for the involvement of TopoI in chromosome segregation. Thus, it appears that both DNA gyrase and TopoI participate in chromosome segregation by virtue of their decatenation properties, akin to the role played by TopoIV and TopoIII in the E. coli. Participation of both DNA gyrase and TopoI in chromosome segregation, however, is without any precedent. Moreover, TopoI in mycobacteria appears to have acquired properties of TopoIII. Dual-function of both DNA gyrase and TopoI in mycobacteria thus seem to compensate for the underrepresentation of topoisomerases in the genus.
To conclude, the genome-wide analysis of TopoI has provided the insights into its indispensable functions during transcription and additional responsibility in removing catenated products at replication terminus. The enzyme's recruitment at promoter in an inactive state and activated role during transcription process is akin to what is seen in eukaryotes during transcription elongation. The approaches used to map TopoI activity in M. smegmatis have the potential (poisonous variant, newly discovered drug) to be readily applied for understanding the in vivo functional role of other topoisomerases in both prokaryotes and eukaryotes.
DATA AVAILABILITY
The data have been uploaded to NCBI (SRA accession number-PRJNA503689).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Adwait Godbole and Wareed Ahmed for critical inputs in various experiments, Olivier Espeli for valuable suggestions, S. Chandrashekaran for providing M13 ssDNA and members of V.N. laboratory for discussions. Bhavna Padmanabhan and Sandhya Rao are acknowledged for their technical assistance. P.R. is a Senior Research Fellow of University of Grant Commission, Government of India. V.N. is a recipient of J.C. Boss fellowship of Department of Science and Technology, Government of India.
Author contributions: The author(s) have made the following declarations about their contributions: conceived and designed the experiments: V.N. and P.R. performed the experiments: P.R. analysed the data: V.N. and P.R. wrote the paper: V.N. and P.R.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology, Government of India [MCB/VNR/DBT/496] and Life science research, education and training at JNCSAR [BT/INF/22/SP27679/2018]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu L.F., Wang J.C.. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001; 70:369–413. [DOI] [PubMed] [Google Scholar]
- 3. Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. [DOI] [PubMed] [Google Scholar]
- 4. Espeli O., Marians K.J.. Untangling intracellular DNA topology. Mol. Microbiol. 2004; 52:925–931. [DOI] [PubMed] [Google Scholar]
- 5. Zechiedrich E.L., Khodursky A.B., Bachellier S., Schneider R., Chen D., Lilley D.M., Cozzarelli N.R.. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000; 275:8103–8113. [DOI] [PubMed] [Google Scholar]
- 6. Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M.. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011; 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ullsperger C., Cozzarelli N.R.. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996; 271:31549–31555. [DOI] [PubMed] [Google Scholar]
- 8. Drolet M., Wu H.Y., Liu L.F.. Roles of DNA topoisomerases in transcription. Adv. Pharmacol. 1994; 29a:135–146. [DOI] [PubMed] [Google Scholar]
- 9. Seol Y., Hardin A.H., Strub M.P., Charvin G., Neuman K.C.. Comparison of DNA decatenation by Escherichia coli topoisomerase IV and topoisomerase III: implications for non-equilibrium topology simplification. Nucleic Acids Res. 2013; 41:4640–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J.C. Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol. 1971; 55:523–533. [DOI] [PubMed] [Google Scholar]
- 11. Cozzarelli N.R. DNA gyrase and the supercoiling of DNA. Science. 1980; 207:953–960. [DOI] [PubMed] [Google Scholar]
- 12. Gadelle D., Filee J., Buhler C., Forterre P.. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003; 25:232–242. [DOI] [PubMed] [Google Scholar]
- 13. Lew J.M., Kapopoulou A., Jones L.M., Cole S.T.. TubercuList–10 years after. Tuberculosis. 2011; 91:1–7. [DOI] [PubMed] [Google Scholar]
- 14. Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E. 3rd et al. . Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 393:537–544. [DOI] [PubMed] [Google Scholar]
- 15. Jain P., Nagaraja V.. An atypical type II topoisomerase from Mycobacterium smegmatis with positive supercoiling activity. Mol. Microbiol. 2005; 58:1392–1405. [DOI] [PubMed] [Google Scholar]
- 16. Ahmed W., Menon S., Karthik P.V., Nagaraja V.. Reduction in DNA topoisomerase I level affects growth, phenotype and nucleoid architecture of Mycobacterium smegmatis. Microbiology. 2015; 161:341–353. [DOI] [PubMed] [Google Scholar]
- 17. Kapopoulou A., Lew J.M., Cole S.T.. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis. 2011; 91:8–13. [DOI] [PubMed] [Google Scholar]
- 18. Kumar R., Riley J.E., Parry D., Bates A.D., Nagaraja V.. Binding of two DNA molecules by type II topoisomerases for decatenation. Nucleic Acids Res. 2012; 40:10904–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manjunatha U.H., Dalal M., Chatterji M., Radha D.R., Visweswariah S.S., Nagaraja V.. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 2002; 30:2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aubry A., Fisher L.M., Jarlier V., Cambau E.. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2006; 348:158–165. [DOI] [PubMed] [Google Scholar]
- 21. El Sayyed H., Le Chat L., Lebailly E., Vickridge E.. Mapping Topoisomerase IV Binding and Activity Sites on the E. coli Genome. PLoS Genet. 2016; 12:e1006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ahmed W., Sala C., Hegde S.R., Jha R.K., Cole S.T., Nagaraja V.. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet. 2017; 13:e1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H. et al. . RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell. 2016; 165:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dalla Rosa I., Huang S.Y., Agama K., Khiati S., Zhang H., Pommier Y.. Mapping topoisomerase sites in mitochondrial DNA with a poisonous mitochondrial topoisomerase I (Top1mt). J. Biol. Chem. 2014; 289:18595–18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeong K.S., Ahn J., Khodursky A.B.. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 2004; 5:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blokpoel M.C., Murphy H.N., O’Toole R., Wiles S., Runn E.S., Stewart G.R., Young D.B., Robertson B.D.. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005; 33:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiianitsa K., Maizels N.. A rapid and sensitive assay for DNA-protein covalent complexes in living cells. Nucleic Acids Res. 2013; 41:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leelaram M.N., Bhat A.G., Godbole A.A., Bhat R.S., Manjunath R., Nagaraja V.. Type IA topoisomerase inhibition by clamp closure. FASEB J. 2013; 27:3030–3038. [DOI] [PubMed] [Google Scholar]
- 29. Wilson K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001; doi:10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 30. Bhat A.G., Leelaram M.N., Hegde S.M., Nagaraja V.. Deciphering the distinct role for the metal coordination motif in the catalytic activity of Mycobacterium smegmatis topoisomerase I. J. Mol. Biol. 2009; 393:788–802. [DOI] [PubMed] [Google Scholar]
- 31. Godbole A.A., Ahmed W., Bhat R.S., Bradley E.K., Ekins S., Nagaraja V.. Targeting Mycobacterium tuberculosis topoisomerase I by small-molecule inhibitors. Antimicrob. Agents Chemother. 2015; 59:1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Franco R.J., Drlica K.. DNA gyrase on the bacterial chromosome. Oxolinic acid-induced DNA cleavage in the dnaA-gyrB region. J. Mol. Biol. 1988; 201:229–233. [DOI] [PubMed] [Google Scholar]
- 33. Snyder M., Drlica K.. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 1979; 131:287–302. [DOI] [PubMed] [Google Scholar]
- 34. Bhaduri T., Bagui T.K., Sikder D., Nagaraja V.. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J. Biol. Chem. 1998; 273:13925–13932. [DOI] [PubMed] [Google Scholar]
- 35. Sikder D., Nagaraja V.. Determination of the recognition sequence of Mycobacterium smegmatis topoisomerase I on mycobacterial genomic sequences. Nucleic Acids Res. 2000; 28:1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Godbole A.A., Leelaram M.N., Bhat A.G., Jain P., Nagaraja V.. Characterization of DNA topoisomerase I from Mycobacterium tuberculosis: DNA cleavage and religation properties and inhibition of its activity. Arch. Biochem. Biophys. 2012; 528:197–203. [DOI] [PubMed] [Google Scholar]
- 37. Sikder D., Nagaraja V.. A novel bipartite mode of binding of M. smegmatis topoisomerase I to its recognition sequence. J. Mol. Biol. 2001; 312:347–357. [DOI] [PubMed] [Google Scholar]
- 38. Bhaduri T., Basak S., Sikder D., Nagaraja V.. Inhibition of Mycobacterium smegmatis topoisomerase I by specific oligonucleotides. FEBS Lett. 2000; 486:126–130. [DOI] [PubMed] [Google Scholar]
- 39. Tse-Dinh Y.C., McCarron B.G., Arentzen R., Chowdhry V.. Mechanistic study of E. coli DNA topoisomerase I: cleavage of oligonucleotides. Nucleic Acids Res. 1983; 11:8691–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 42. Fisher J.K., Bourniquel A., Witz G., Weiner B., Prentiss M., Kleckner N.. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013; 153:882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Umbarger M.A., Toro E., Wright M.A., Porreca G.J., Bau D., Hong S.H., Fero M.J., Zhu L.J., Marti-Renom M.A., McAdams H.H. et al. . The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell. 2011; 44:252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong J.T., New D.C., Wong J.C., Hung V.K.. Histone-like proteins of the dinoflagellate Crypthecodinium cohnii have homologies to bacterial DNA-binding proteins. Eukaryot. Cell. 2003; 2:646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen J.M., German G.J., Alexander D.C., Ren H., Tan T., Liu J.. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 2006; 188:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gordon B.R., Imperial R., Wang L., Navarre W.W., Liu J.. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008; 190:7052–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gordon B.R., Li Y., Wang L., Sintsova A., van Bakel H., Tian S., Navarre W.W., Xia B., Liu J.. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:5154–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dorman C.J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004; 2:391–400. [DOI] [PubMed] [Google Scholar]
- 49. Atlung T., Ingmer H.. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997; 24:7–17. [DOI] [PubMed] [Google Scholar]
- 50. Landick R., Krek A., Glickman M.S., Socci N.D., Stallings C.L.. Genome-Wide mapping of the distribution of CarD, RNAP sigma(A), and RNAP beta on the Mycobacterium smegmatis chromosome using chromatin immunoprecipitation sequencing. Genom. Data. 2014; 2:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Khodursky A.B., Peter B.J., Schmid M.B., DeRisi J., Botstein D., Brown P.O., Cozzarelli N.R.. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:9419–9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peter B.J., Ullsperger C., Hiasa H., Marians K.J., Cozzarelli N.R.. The structure of supercoiled intermediates in DNA replication. Cell. 1998; 94:819–827. [DOI] [PubMed] [Google Scholar]
- 53. DiGate R.J., Marians K.J.. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988; 263:13366–13373. [PubMed] [Google Scholar]
- 54. Hiasa H., DiGate R.J., Marians K.J.. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 1994; 269:2093–2099. [PubMed] [Google Scholar]
- 55. Seol Y., Neuman K.C.. The dynamic interplay between DNA topoisomerases and DNA topology. Biophys. Rev. 2016; 8:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D.. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013; 20:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N.. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013; 20:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 59. Darst S.A., Kubalek E.W., Kornberg R.D.. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature. 1989; 340:730. [DOI] [PubMed] [Google Scholar]
- 60. deHaseth P.L., Helmann J.D.. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol. Microbiol. 1995; 16:817–824. [DOI] [PubMed] [Google Scholar]
- 61. Banda S., Cao N., Tse-Dinh Y.C.. Distinct Mechanism evolved for Mycobacterial RNA polymerase and topoisomerase I Protein-Protein interaction. J. Mol. Biol. 2017; 429:2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thanbichler M., Wang S.C., Shapiro L.. The bacterial nucleoid: a highly organized and dynamic structure. J. Cell. Biochem. 2005; 96:506–521. [DOI] [PubMed] [Google Scholar]
- 63. Macvanin M., Adhya S.. Architectural organization in E. coli nucleoid. Biochim. Biophys. Acta Gene Regul. Mech. 2012; 1819:830–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ghosh S., Mallick B., Nagaraja V.. Direct regulation of topoisomerase activity by a nucleoid-associated protein. Nucleic Acids Res. 2014; 42:11156–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brochu J., Vlachos-Breton E., Sutherland S., Martel M., Drolet M.. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet. 2018; 14:e1007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006; 59:723–730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been uploaded to NCBI (SRA accession number-PRJNA503689).