Abstract
Pds5 is required for sister chromatid cohesion, and somewhat paradoxically, to remove cohesin from chromosomes. We found that Pds5 plays a critical role during DNA replication that is distinct from its previously known functions. Loss of Pds5 hinders replication fork progression in unperturbed human and mouse cells. Inhibition of MRE11 nuclease activity restores fork progression, suggesting that Pds5 protects forks from MRE11-activity. Loss of Pds5 also leads to double-strand breaks, which are again reduced by MRE11 inhibition. The replication function of Pds5 is independent of its previously reported interaction with BRCA2. Unlike Pds5, BRCA2 protects forks from nucleolytic degradation only in the presence of genotoxic stress. Moreover, our iPOND analysis shows that the loading of Pds5 and other cohesion factors on replication forks is not affected by the BRCA2 status. Pds5 role in DNA replication is shared by the other cohesin-removal factor Wapl, but not by the cohesin complex component Rad21. Interestingly, depletion of Rad21 in a Pds5-deficient background rescues the phenotype observed upon Pds5 depletion alone. These findings support a model where loss of either component of the cohesin releasin complex perturbs cohesin dynamics on replication forks, hindering fork progression and promoting MRE11-dependent fork slowing.
INTRODUCTION
Accurate DNA replication is essential to protect genome integrity and for the high-fidelity transmission of genomic information to daughter cells. Agents that arise from either normal metabolism or exposure to natural or artificial products in the environment constantly challenge progressing DNA replication forks. In addition to DNA lesions, increasing evidence suggests that intrinsic replication fork obstacles such as transcribing RNA polymerases, unusual DNA structures, tightly-bound protein–DNA complexes and oncogene activation may also perturb fork progression (1,2).
The ring-shaped cohesin complex tethers sister chromatids together during DNA replication and consists of two structural maintenance of chromosome (SMC) proteins, Smc1 and Smc3, and two non-SMC components, Scc1 (also referred as Rad21, Mcd1 or Kleisen) and Scc3 (also referred as SA or STAG) (3–5). Whereas a single Scc3 is present in yeast, two SA proteins, SA1 and SA2, are found in higher eukaryotes that form two distinct cohesin complexes in somatic cells: cohesin-SA1 and cohesin-SA2 (6,7). The cohesin ring complex is loaded onto chromosomes by the Nipped-B/NIPBL kollerin complex (Scc2 or delangin) and its partner MAU-2 (Scc4), and is removed by the releasin complex consisting of the Pds5 and Wapl proteins (8,9). The main function of cohesin is to hold newly replicated chromatids together in order to mediate sister chromatid cohesion and ensure accurate chromosome segregation during mitosis (10–12). However, increasing evidence suggests that cohesin plays crucial roles in multiple cellular processes including chromosome condensation, gene regulation and DNA damage repair (11,13). The fact that cohesion is established during S-phase, when new sister DNAs are synthesized by the replication complex, points to an intimate relationship between DNA replication and cohesion establishment. Indeed, cohesin has been shown to interact with several replication factors including PCNA and the alternative clamp loader formed by the Ctf18-Replication Factor C complex, as well as the CtF4 homotrimers. All these interactions contribute to proper cohesion establishment (14–20). However, our current knowledge of how the replication machinery regulates cohesin and vice versa is limited.
The Precocious Dissociation of Sister protein Pds5 was initially discovered as an essential factor for holding sister chromatids together during mitosis in Saccharomyces cerevisiae (21,22). The role of Pds5 in cohesion has been highly debated because Pds5 is required for sister chromatid cohesion, and somewhat paradoxically, also to remove cohesin from chromosomes (23,24). Recent studies suggest that Pds5 acts as a scaffold protein to recruit different factors to the cohesin ring (25–27). Depending on its binding partner, Pds5 may stabilize the cohesin ring on DNA or promote its unloading. As previously mentioned, Pds5 associates with Wapl and together they unload cohesin. Soronin competes with Wapl for binding to Pds5 (28). De-phosphorylation of Sororin by the Shugoshin and Protein Phosphatase 2A (SGO1-PP2A) complex strengthens Sororin binding to Pds5, stabilizing the cohesin complex at centromeres until metaphase (29). On the other hand, the mitotic factors Aurora B and CdK1 phosphorylate Soronin destabilizing its interaction with Pds5 and allowing Pds5 to re-associate with Wapl, leading to cohesin removal from chromosome arms (29,30).
Two Pds5 variants are expressed in mammalian cells, Pds5A and Pds5B (also known as APRIN). Both variants are required for cohesion establishment by facilitating Smc3 acetylation and recruiting Soronin and Wapl to the complex (26,31). However, Pds5B appears to be specifically important to ensure centromeric cohesion (31). Interestingly, recent reports point to a previously unappreciated function of Pds5 in DNA damage repair and homologous recombination (HR) (32–34). These studies have been sparked by the discovery that Pds5B interacts with the breast cancer susceptibility protein BRCA2, a protein well-known for its roles in HR and replication fork stability (35,36). Pds5B loss perturbs HR and sensitizes cells to DNA damaging agents (32), and recent mechanistic studies suggest that Pds5B stimulates Rad51-mediated strand invasion through its interaction with BRCA2 (33). A role of Pds5B in replication and DNA damage repair is also supported by observations that downregulation of Pds5B sensitizes breast cancer cells to DNA-damaging chemotherapy and that low Pds5B expression correlates with better breast and ovarian patient survival (32). While the cohesin regulatory and HR function of Pds5 have been relatively well-characterized, its role in DNA replication is poorly defined. Moreover, the contribution of the Pds5A variant to the replication and repair function of Pds5 remains unclear.
Here, we set out to investigate the function of the two isoforms of Pdsd5 in DNA replication. Our findings reveal that both Pds5 isoforms play an important role during replication fork progression in unperturbed cells. Loss of Pds5 leads to MRE11-dependent fork slowing and DSB accumulation. This function is uncoupled from the interaction with BRCA2 and seems to be related to the cohesin releasin activity of the Pds5 and Wapl proteins, suggesting that cohesin accumulation on replication forks may represent a new important obstacle to replication fork progression and genome duplication.
MATERIALS AND METHODS
Cell lines, cell culture, and gene silencing
Human Osteosarcoma (U-2 OS) cells (ATCC® HTB-96™) and hTERT Retinal Pigment Epithelial-1 (RPE) cells (ATCC® CRL-4000™) were transfected by siRNA technology to deplete Pds5B (Dharmacon ON-TARGETplus Human Pds5B siRNA Cat. No. L-010362-00-005, 50 nM for 72 h) (32); Pds5A (Dharmacon sequence: 5′-TTCTTCCTCAGGAACCC-CATT-3′, 50 nM for 48 h) (37); Wapl (Dharmacon sequence: 5′-CGGACTACCCTTAGCACAA-3′, 50 nM for 48 h) (37); Rad21 (Dharmacon sequence: 5′-AUAC-CUUCUUGCAG-ACUGU-3′, 50 nM for 48 h) (38); BRCA2 (Dharmacon ON-TARGETplus BRCA2, Cat. No. L-003462-00, 25 nM for 48 h) (39,40); MRE11 A (Dharmacon ON-TARGETplus, Cat. No. L-009271-00, 50 nM for 48 h) (41); Nbs1 (Dharmacon sequence: 5′-CCAACUAAAUUGCCAAGUAUU-3′, 50 nM for 48 h) (42) and Rad50 (Dharmacon sequence: 5′-CTGCGACTTGCTCCAGATAAA-3′, 50 nM for 48 h) (43), Rad51 (Ambion, Cat. No. 4390827, sequence- 5′-GUGCUGCAGCCUAAUGATT-3′, 50 nM, 48 h) (40), Smarcal1 (Dharmacon, Cat. No. D-013058-04-002, sequence- 5′-GAAUCUCACUUCCUCAAAA-3′, 20 nM, 72 h) (44). Silencer select negative control siRNA (Ambion, Cat. No. 4390843, 50 nM for 48 h) and Luciferase Duplex (Dharmacon, Cat. No. P-002099-01-20, 50 nM for 48 h) were used for the control experiments. RNAi transfection was carried out using RNAiMAX (ThermoFisher Scientific Cat. No. 13778030) following the manufacturer's instructions. U-2 OS and RPE cells were cultured in Dulbecco's modified Eagle's medium (DMEM Sigma, Poole, UK) supplemented with 10% Fetal Bovine Serum (FBS) and 100 U/ml penicillin and 100 μg/ml streptomycin (1%) at 37°C, 5% CO2.
One clone each of Pds5A knock-out (KO), Pds5B KO and Wild Type (WT) Mouse Embryonic Fibroblasts (MEFs) (31) that had been immortalized using SV40 large T antigen (iMEFs) were kindly provided by Dr Ana Losada (CINO Madrid Spain). iMEFs were cultured in Dulbecco's modified Eagle's medium (DMEM Sigma, Poole, UK) supplemented with 20% Fetal Bovine Serum (FBS) and 1% penicillin–streptomycin at 37°C, 5% CO2.
pRetroX Tet-ON Advanced HEK293T cells (Takara Bio Inc.) that stably express the doxycycline-regulated transactivator protein were transiently transfected with pRetroX-Tight.puro myc-BirA*Pds5B (APRIN) with Lipofectamine 2000 (Invitrogen; (45)). Cells were incubated with 0.5 μg/ml puromycin 48 h after transfection. Upon colony formation, single clones were isolated and screened for myc-BirA-Pds5B protein after doxycycline induction (1 μg/ml doxycycline for 24 h).
Antibodies for IP and western blot analysis
Levels of intracellular Pds5B, Pds5A, BRCA2, Rad21, Wapl, MRE11, Nbs1, Rad50, Rad51, Smarcal1, Chk-1, pChk1, H2AX, pRPA 32 (S33), γ-H2AX, PCNA, GAPDH, Histone H3 and Tubulin were determined by western blot analysis of cell extracts. Anti-Pds5B (APRIN, Novus, Cat. No.100-755; 1:1000); anti-Pds5A (Novus, Cat. No. A300-088A; 1:1000), anti-Wapl A-7 (Santa Cruz Biotechnology, Cat. No. sc-36518; 1:500), anti-BRCA2 (Calbiochem, Cat. No. OP-95; 1:1000), anti-Rad21 (Bethyl, Cat. No. A300-080A; 1:1000), anti- MRE11-A rabbit polyclonal (Sigma-Aldrich, Cat. No. M6193: 1:1000); anti-p95 Nbs1 (Abcam, Cat. No. ab32074; 1:1000) anti-Rad50 (Abcam, Cat. No. ab181602; 1:100); anti-PCNA (F-2) (Santa Cruz, Cat. No, sc-25280; 1:500); anti-Histone H3 (Abcam, Cat. No. ab1791; 1:30 000); anti-GAPDH (EPR16891) (Abcam, Cat. No. ab181602; 1:75 000); anti-rabbit (Thermo Fisher Scientific, Cat. No. 31460; 1:10 000); anti-mouse (Sigma-Aldrich, Cat. No. A3562; 1:10 000); anti-tubulin mouse monoclonal, (Sigma-Aldrich, Cat. No. T5168; 1:5000); anti-RAD51 rabbit polyclonal (Sigma-Aldrich, Cat. No. 05-0530-I; 1:1000); anti-Smarcal1 mouse monoclonal (Santa Cruz Biotechnology, Cat. No. E-12; 1:500); anti-Chk1 mouse monoclonal (Santa Cruz Biotechnology, Cat. No. sc-8408; 1:1000); anti-pChk1 (S345) rabbit monoclonal (Cell Signaling Technology, Cat. No. 2348; 1:1000); anti-phospho-H2AX mouse monoclonal (Abcam, Cat. No. ab26350; 1:1000); anti-CENPF rabbit polyclonal (Abcam, Cat. No. ab5; 1:300); anti-53BP1 mouse monoclonal (BD Biosciences Cat. No. 612522; 1:1000); anti-phospho RPA 32 (S33) rabbit polyclonal (Bethyl, Cat. No. A300-246A, 1:1000); Hoechst 33342 (Sigma-Aldrich. Cat. No. B2261).
DNA fiber analysis
Asynchronously growing cells were labeled with two thymidine analogues: 20 μM 5-iodo-2′-deoxyuridine (IdU; Sigma-Aldrich) followed by 200 μM 5-chloro-2′-deoxyuridine (CldU; Sigma-Aldrich) for the indicated times. For the fork restart experiments, cells were washed twice with PBS after the first pulse and treated with 4 mM HU for 5 h before adding the second thymidine analog. To inhibit MRE11 activity, cells were treated with MRE11 inhibitor Mirin (Sigma Aldrich Cat. No. M9948). Cells were then harvested, lysed and the DNA was spread on a positive coated slide as described (46). Briefly, cells were harvested and resuspended in PBS at 100 000 cells/ml. 2 μl of this cell solution was mixed with 8 μl of lysis buffer (200 mM Tris–HCl pH 7.5; 50 mM EDTA; 0.5% SDS) on a glass slide. After 5 min, the slides were tilted at a 20–45° angle, and the resulting DNA spreads were air dried, fixed in 3:1 methanol/acetic acid and stored at 4°C. The DNA fibers were denatured with 2.5 M HCl for 1 h, washed with PBS, and blocked with 5% BSA in PBS for 1 h. DNA immunostaining was performed with rat anti-BrdU antibody (1:50; AbCys SA, Cat. No. ABC117 7513) for CldU and mouse anti-BrdU antibody (1:50; Becton Dickson, Cat. No. 347580) for IdU in a humid chamber at room temperature for 1 h. The following secondary antibodies were used: anti-rat Alexa 488 (1:100; Molecular Probes, Cat. No. A21470) and anti-mouse Alexa 546 (1:100; Molecular Probes, Cat. No. A21123) at room temperature for 45 min. The slides were air dried and mounted with Prolong Gold Antifade reagent (Invitrogen). Images were sequentially acquired (for double-label) with LAS AF software using TCS SP5 confocal microscope (Leica). A 63×/1.4 oil immersion objective was used. The DNA tract lengths were measured using ImageJ and the values were converted into micrometers using the scale bars created by the microscope. Statistical analysis was done using GraphPad Prism Software, (San Diego, CA, USA). Statistical significance was assessed using unpaired non-parametric Mann–Whitney compared ranks t-test. More than 300 fibers were measured for each data set. The statistics for all these experiments measuring changes in the size of the IdU or CldU tracts were calculated on the total number of DNA tracts measured in each given sample (usually n ≥ 300). For the fork restart experiments, the percentage of stalled forks was calculated on the basis of at least 1000 tracts counted in each independent experiment. All DNA fiber experiments were performed in duplicate or triplicate, as indicated in the figure legends. Additional information on the minimal number of tracts that should be measured for a reliable estimation of changes in fork speed within a given sample can be found in (47,48).
Affinity capture of biotinylated proteins
Cells were incubated for 24 h in complete media supplemented with 1 μg/ml doxycycline. Five hours before the end of the incubation period, biotin 50 μM was added to the media, with or without 4 mM hydroxyurea (HU). Non-induced cells (–DOX) were treated with 50 uM biotin. After three cold PBS washes, cells were lysed for 30 min at 4°C in a 1:4 ratio (g of cells:ml of buffer) using RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 0.1% SDS) supplemented with 250 U of benzonase, 0.5% sodium deoxycholate and 1× complete protease inhibitor (Roche). Lysates were then sonicated as followed: 3 × 10 s at 30% amplitude on ice and centrifuged at 16 000g for 30 min. Equal volumes of supernatants were incubated with 35 μl of sepharose beads (Streptavidin-Sepharose High Performance; GE Healthcare) for 3 h at 4°C under rotation. Beads were collected and washed five times in RIPA buffer. Bound proteins were eluted with 35 μl of Laemmli SDS-sample buffer and heated at 95°C for 10 min. Sample were loaded on NuPAGE™ 3–8% Tris-Acetate protein gels (Invitrogen) in Tris-Acetate running buffer according to the manufacturer's instructions and transferred on Nitrocellulose membrane (Amersham) using XCell II Blot Module (Invitrogen) in 20% methanol transfer buffer. Immunoblot were performed using anti-BRCA2-OP95 (1:250, Millipore), anti-Pds5 (1:5000, Bethyl Laboratories A300-538A), anti-GFP (1:1000, Roche).
GFP-trap pull-down
HEK293T were plated to 80% confluency and transfected with peGFP-C1, or peGFPC1-Pds5B vector using TransIT®-293 Transfection Reagent (Mirus Bio LLC) according to the manufacturer's protocol. Forty eight hours after transfection, cells were treated with 4 mM HU for 5 h, washed twice with cold PBS, and lysed in lysis buffer (50 mM Tris–Cl, pH 7.5, 150 mM NaCl, 0.5% NP40, PMSF, aprotinin, leupeptin, NaF, Na2VO4) supplemented with Benzonase (25 U) and 2.5 mM MgCl2 for 30 min at 4°C. Lysates were sonicated (3 × 10 s at 30% amplitude on ice) and centrifuged at 16 000g, 30 min, 4°C. Proteins were incubated with GFP-beads for 1 h at 4°C. Beads were washed four times with lysis buffer and bound proteins were eluted in 50 μl of Laemmli SDS-sample buffer and heated at 95°C for 10 min. Proteins were detected using the same approach described for the affinity capture of biotinylated proteins.
Chromatin fractionation
Chromatin fractionation experiments were performed following a previously described procedure (49). Briefly, proteins bound to chromatin were isolated using a two-step fractionation procedure, in which cytosolic proteins were extracted using a buffer containing 10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT 0.1% Triton X-100 and protease inhibitor cocktail. The pellet was then further fractionated releasing chromatin bound proteins using a buffer containing 3 mM EDTA, 0.2 mM EGTA, 1 mM DTT and protease inhibitor cocktail. Cytosolic fraction and chromatin bound proteins were run on a western blot to assess the levels of proteins bound to chromatin.
Neutral comet assay for DSB detection
Neutral comet assay was performed as previously described (40). Briefly, 700 cells were resuspended in 70 μl 0.5% low melting point agarose (Trevigen, Cat. No. 4250-050-02) and spread on a comet slide (Trevigen, Cat. No. 4250-200-03). Cells were lysed in a cold lysis solution (Trevigen, Cat. No. 4250-050-01) at 4°C for 30 min. DNA migration was performed in TBE buffer at 1 V/cm for 30 min. Slides were washed in milliQ water, fixed with ethanol 70% for 30 min and dried at room temperature. Comets were labeled with SYBR® Gold Nucleic Acid Gel Stain (ThermoFisher) for 30 min. Images were acquired with a fluorescence microscope (LEICA DMU 4000B; 20×/0.4 CORR) coupled to the LEICA DFC345FX camera. The images were analyzed using the OpenComet plug-in from ImageJ. At least 100 comets were scored per sample in each experiment. Olive moment measures DSBs as a product of the amount of DNA present (intensity) times the size of the broken DNA pieces (length) in the tail.
Chromosome spreads
Cells were treated with 10 μM nocodazole for 4 h. Cells were collected, washed and resuspended in 10 ml of warmed hypotonic solution (10 mM KCl, 10% FBS) for 10 min at 37°C. Cells were fixed by adding 500 μl of cold fixation buffer (acetic acid:ethanol 1:3). Cell pellets were washed four times with the cold fixation buffer and stored in this buffer at 4°C overnight. The nuclei were spread on cold slides. The slides were air dried overnight and mounted with Prolong Gold Antifade (Invitrogen) with DAPI. Images were acquired with a fluorescence microscope (LEICA DMU 4000B; 63×/1.40–0.60 NA oil) coupled to the LEICA DFC345FX camera. The images were analyzed with ImageJ. At least 50 metaphases per sample were scored in each experiment. Statistical significance was assessed using unpaired non-parametric Man–Whitney compared ranks t-test.
aniPOND
Samples for aniPOND were prepared as described (50). Briefly, U-2 OS cells were depleted for Pds5A and Pds5B using siRNA, or treated with negative siRNA as control. Five 90% confluent 15 cm plates of siRNA-depleted cells were required for each condition; two conditions for each depletion (± thymidine chase). Five additional plates of the siCrtl-depleted cells were prepared for the no-click control. Cells were labeled with 10 μM EdU for 20 min. For the thymidine chase experiments, cells were first washed with 1× PBS and then treated with fresh media containing 10 μM Thymidine for 1 h. After labeling and treatment, cells were washed with 1× PBS and nuclei were harvested using 8 ml of nuclei extraction buffer (New England Biolabs). Cells were scraped and the five plates used for each condition were pooled in 50 ml tubes. Samples were then treated with 10 ml of click reaction containing 25 μM biotin-azide, 10 mM (+)-sodium l-ascorbate and 2 mM CuSO4 and rotated at 4°C for 1 h. The no click control was treated with DMSO instead of biotin-azide. Samples were then centrifuged to pellet the nuclei; supernatant was removed and replaced with 1 ml Buffer 1 (B1) containing 25 mM NaCl, 2 mM EDTA, 50 mM Tris–HCl pH 8.0, 1% IGEPAL and protease inhibitor and rotated again at 4°C for 30 min. This step was repeated twice. Samples were centrifuged to pellet the nuclei; supernatant was removed and replaced with 500 μl of B1 and sonicated six times for 10 s on and 10 s off at 20 amplitude. Samples were centrifuged and supernatant was transferred to fresh tubes. To adjust the NaCl concentration to physiological levels 500 μl of Buffer 2 (B2) containing 150 mM NaCl, 2 mM EDTA, 50 mM Tris–HCl pH 8.0, 0.5% IGEPAL and protease inhibitor was added to each sample. 20 μl of each sample were removed and transferred to fresh tubes contained 20 μl of Laemli buffer and labeled as ‘input’. The remaining sample (labeled as ‘capture’) was incubated overnight (16–20 h) with 100 μl of streptavidin-coated beads for the streptavidin biotin capture step. Proteins were eluted with 30 μl of Laemli buffer and boiled at 95°C for 15 min. Western blots were performed to monitor the amount of Rad21, Pds5A and Pds5B at the replication forks (– thymidine chase) and behind the replication forks (+ thymidine chase).
iPOND-SILAC mass spectrometry
Samples for iPOND-SILAC MS were prepared as previously described (51,52). Briefly, VC8 cells complemented with human BRCA2 were cultured in heavy isotope media and compared to VC8 cells lacking BRCA2 cultured in light isotope media. After labeling with EdU and treating with hydroxyurea, the cells were harvested and mixed prior to performing the iPOND purifications. Mass spectrometry data was analyzed by MaxQuant (53). At least two peptides were required for protein identification. Quantitation is reported as the log2 of the normalized heavy/light ratios.
Immunofluorescence microscopy
Twenty four hours following siRNA treatment, U-2 OS cells were seeded onto microscope coverslips and incubated at 37°C for an additional 40 h. The cells were extracted with 1× PBS containing 0.2% Triton X-100 and protease inhibitors (Pierce) for 10 min on ice and fixed with 3.2% paraformaldehyde in 1× PBS. The cells were then washed extensively with IF Wash Buffer (1× PBS, 0.5% NP-40 and 0.02% NaN3), then blocked with IF Blocking Buffer (IF Wash Buffer plus 10% FBS) for at least 30 min. Cells were incubated with primary antibodies diluted in IF Blocking Buffer overnight at 4°C. Cells were stained with secondary antibodies and mounted using Prolong Gold mounting medium (Invitrogen). Epifluorescence microscopy was performed on an Olympus fluorescence microscope (BX-53) using an UPlanS-Apo 60×/1.35 oil immersion lens. Images were obtained using an ORCA-Flash4.0 LT digital camera and cellSens Dimension software. Raw images were exported into Adobe Photoshop, and for any adjustments in image contrast or brightness, the levels function was applied. Nuclear Rad21 mean intensity levels were measured using the histogram function in Photoshop. The number of 53BP1 nuclear bodies in CENP-F negative (G1) cells was quantified manually. At least 200 cells were analyzed for each experiment.
RESULTS
Pds5 controls replication fork progression
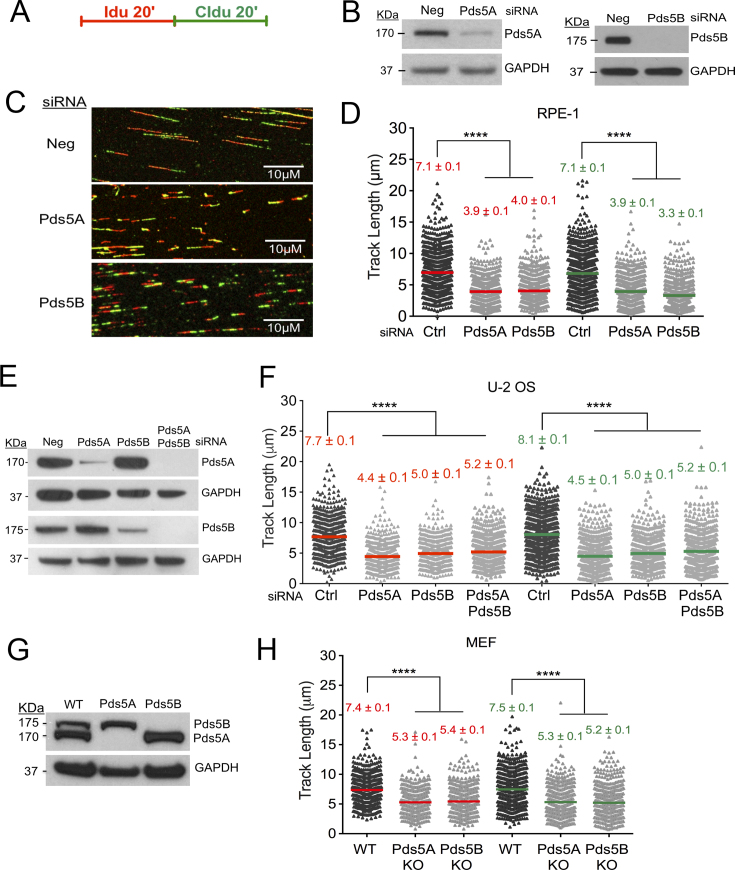
We sought to investigate the contribution of the two isoforms of Pds5 in replication fork progression by genome-wide single-molecule DNA replication assays. We knocked down Pds5A and Pds5B by siRNA in two different human cells lines commonly used to study perturbations in replication fork dynamics, the human retinal pigment epithelial (RPE-1) and osteosarcoma U-2 OS cells. Replication fork progression was monitored by pulse-labeling cells with the first thymidine analog 5-Iodo-2′-deoxyuridine (IdU, red label) for 20 min followed by treatment with the second thymidine analog 5-chloro-2′-deoxyuridine (CIdU, green label) for the same amount of time in the absence of genotoxic stress (Figure 1A). Shortening of the first and second tract for each fork was measured as a readout of a defect in fork progression only on forks characterized by contiguous IdU-CldU signals (and not on forks that have only one label) to ensure that the shortening phenotype is indeed due to a defect in fork progression and not to premature termination events. Pds5B loss in RPE-1 cells caused a marked reduction in the IdU and CldU tract (approximately 44%) length (Figure 1A–D). A similar reduction was observed upon Pds5A depletion (Figure 1A–D). Moreover, the CldU/IdU ratio was very close to 1 (Supplementary Figure S1A) indicating that the replication fork defects associated with Pds5A or Pds5B loss equally perturbs both tracts. Similar results were obtained with the U-2 OS cells (Figure 1E, F and Supplementary Figure S1B). Co-depleting both variants in U-2 OS cells did not further increase tract shortening, suggesting that the two proteins are acting in the same pathway (Figure 1F). The same results were also confirmed using Pds5A and Pds5B knockout immortalized mouse embryonic fibroblasts (Pds5A KO and Pds5B KO iMEFs) ruling out the possibility that the phenotype observed with the siRNA depleted cells might be associated to an off-target effect of the selected siRNAs (Figure 1G, H and Supplementary Figure S1C). Collectively, these results suggest that both Pds5 isoforms play an important and previously unappreciated role in replication fork progression in unperturbed cells.
Figure 1.
Pds5 controls replication fork progression. (A) Single-molecule DNA fiber-labeling scheme. (B) Expression of Pds5A and Pds5B after siRNA knockdown in RPE-1 cells. (C) Representative DNA fiber images of RPE-1 cells transfected with control siRNA (Ctrl), Pds5A, or Pds5B siRNA. Scale bar 10 μm. (D) Size distribution of IdU and CIdU tract length in Pds5A and Pds5B depleted RPE-1 cells. Cells were transfected with control siRNA (Ctrl), Pds5A, or Pds5B siRNA before IdU and CldU labeling, as indicated. Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (E) Expression of Pds5A and Pds5B after siRNA knockdown in U-2 OS cells. (F) Size distribution of IdU and CIdU tract length in Pds5A and Pds5B depleted U-2 OS cells. Cells were transfected with control siRNA (Ctrl), Pds5A, Pds5B and Pds5A/Pds5B siRNA before IdU and CldU labeling, as indicated. Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (G) Expression of Pds5A and Pds5B in the Pds5B and Pds5A knockout MEFs. (H) Size distribution of IdU and CIdU tract length in WT, Pds5B and Pds5A knockout MEF cells. Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001.
Pds5 role in replication fork progression is uncoupled from BRCA2
Pds5B was previously reported to interact with BRCA2 (32,34). First, we confirmed the Pds5B–BRCA2 interaction using HEK293T cells expressing a myc-BirA-tagged version of Pds5B (Supplementary Figure S2A). We also confirmed the same results using a GFP-tagged version of the protein and showed that the interaction persists under conditions of replication stress (Supplementary Figure S2B). Next, we utilized a recently improved iPOND (isolation of Proteins on Nascent DNA) protocol coupled to SILAC mass spectrometry (51,52) to study whether Pds5 loads on nascent DNA and whether its loading its affected by BRCA2 status. Using iPOND, we compared the association of Pds5 as well as other sister chromatid cohesion factors with nascent DNA in the BRCA2 mutant VC8 hamster cell line and its complemented derivative expressing wild-type BRCA2 (36,54,55). We found that most of sister chromatid cohesion components, including the cohesin-ring components Rad21, SA1 and SA2, the cohesion loading complex subunit MAU2, and the cohesin unloading factors Pds5A, Pds5B, and Wapl are present on nascent DNA (Table 1). Robustness of the method was evident from the identification of all six components of the replicative helicase MCM2-7, as well as of PCNA and RPA. The main components of the histone core complex (H2A, H2B, H3 and H4) were also present consistent with their rapid assembly onto newly synthesized DNA. Moreover, the log2 ratios of amount of proteins detected in BRCA2-proficient versus deficient cells showed that the association of all the cohesin components as well as of the core replication factors was not significantly affected by BRCA2 status (Table 1, column ‘No treatment ± BRCA2’). These results suggest that the reported BRCA2–Pds5B interaction does not affect the ability of Pds5 as well as other sister chromatid cohesion factors to interact with newly synthesized DNA.
Table 1.
Table summarizing the relative abundance of selected proteins obtained by iPOND-mass spectrometry. Values are log2 of the ratio observed with and without BRCA2. Positive values indicate proteins that are more abundant in cells containing BRCA2. Peptide numbers are the number of peptides with quantifiable Heavy/Light ratios in that experiment. Values highlighted in bold are significant using the ‘Significance A’ test in Perseus after applying Benjamini Hochberg FDR correction. See Cox and Mann 2008 Nature Biotechnology for details of the statistical test (53)
| No treatment ± BRCA2 | Hydroxurea Replicate 1 ± BRCA2 | Hydroxurea Replicate 2 ± BRCA2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Uniprot ID | Protein name | log2 ratio | % variability | # peptide ratios | log2 ratio | % variability | # peptide ratios | log2 ratio | % variability | # peptide ratios |
| Histones | ||||||||||
| G3H154 | Histone H2A | 0.15 | 5.08 | 7 | 0.40 | 9.85 | 10 | –0.13 | 32.13 | 8 |
| G3HDT6 | Histone H2A type 1 | –0.19 | 15.97 | 148 | –0.18 | 17.40 | 73 | –0.33 | 21.56 | 68 |
| G3I968 | Histone H2A | 0.21 | 3.99 | 8 | 0.35 | 4.89 | 7 | 0.03 | 8.40 | 7 |
| G3HDT7 | Histone H2B | NO | 11.39 | 29 | 0.06 | 24.93 | 57 | –0.32 | 21.69 | 72 |
| G3H2T7 | Histone H3 | –0.07 | 9.51 | 122 | 0.00 | 18.26 | 50 | –0.22 | 30.15 | 52 |
| G3HHM2 | Histone H3.1t | –0.06 | 24.16 | 98 | 0.17 | N/A | 1 | –0.17 | N/A | 1 |
| G3HDT9 | Histone H4 | –0.04 | 29.12 | 6 | –0.62 | 118.58 | 4 | –0.45 | 15.90 | 5 |
| G3HPV7 | Histone H4 | –0.07 | 21.56 | 264 | 0.03 | 22.53 | 179 | –0.13 | 24.47 | 196 |
| Replicative Helicase | ||||||||||
| G3H7V9 | DNA helicase MCM2 | –0.01 | 14.26 | 67 | 0.07 | 23.19 | 49 | 0.23 | 26.90 | 38 |
| G3I1H0 | DNA helicase MCM3 | 0.04 | 13.26 | 116 | 0.05 | 16.32 | 56 | 0.27 | 15.65 | 54 |
| G3I2I1 | DNA helicase MCM4 | –0.02 | 23.90 | 60 | 0.07 | 34.40 | 53 | 0.22 | 27.95 | 45 |
| A0A061IB94 | DNA helicase MCM5 | 0.00 | 30.46 | 81 | 0.06 | 18.41 | 67 | 0.20 | 17.03 | 47 |
| G3GZQ9 | DNA helicase MCM6 | 0.00 | 16.55 | 70 | 0.04 | 13.92 | 31 | 0.28 | 20.82 | 28 |
| A0A061I6D0 | DNA helicase MCM7 | –0.05 | 21.72 | 87 | 0.09 | 32.17 | 62 | 0.26 | 14.10 | 54 |
| PCNA | ||||||||||
| P57761 | Proliferating cell nuclear antigen; PCNA | 0.16 | 18.19 | 169 | 0.25 | 8.91 | 39 | 0.72 | 25.95 | 45 |
| RAD51 | ||||||||||
| P70099 | DNA repair protein RAD51 | NO | N/A | N/A | 2.26 | 40.85 | 2 | 3.17 | 108.35 | 7 |
| Replication Protein A | ||||||||||
| G3HJH9 | Replication protein A 14 kDa subunit; RPA3 | NO | N/A | N/A | 0.49 | 10.30 | 8 | 0.56 | 7.00 | 33 |
| G3H201 | Replication protein A 32 kDa subunit; RPA2 | 0.21 | 11.05 | 13 | 0.54 | 9.30 | 45 | 0.68 | 19.40 | 37 |
| G3IP86 | Replication protein A 70 kDa subunit; RPA1 | 0.23 | 20.21 | 32 | 0.55 | 13.64 | 23 | 0.78 | 28.57 | 29 |
| Cohesin Subunits | ||||||||||
| G3IBF6 | Sister chromatid cohesion protein PDS5-like A | 0.13 | 20.60 | 35 | –0.19 | 16.90 | 16 | 0.02 | 22.34 | 14 |
| G3HM82 | Sister chromatid cohesion protein PDS5-like B | 0.07 | 25.80 | 54 | 0.10 | 15.83 | 47 | –0.09 | 19.24 | 45 |
| G3H0A3 | Double-strand-break repair protein rad21-like | 0.07 | 16.43 | 14 | 0.14 | 45.91 | 14 | 0.05 | 16.23 | 13 |
| A0A061I2V2 | Cohesin subunit SA-1 isoform 1 | –0.02 | 13.97 | 11 | 0.01 | 12.70 | 11 | –0.15 | 17.13 | 12 |
| G3HY68 | Cohesin subunit SA-2 | 0.07 | 33.00 | 33 | 0.23 | 10.32 | 21 | 0.09 | 19.47 | 16 |
| G3H467 | Cohesin loading complex subunit SCC4-like | 0.02 | N/A | 1 | 0.05 | 12.01 | 2 | NO | N/A | N/A |
| A0A061IQY6 | Putative MAU2 chromatid cohesin protein | 0.08 | 17.13 | 30 | 0.12 | 16.23 | 11 | –0.15 | 20.45 | 12 |
| N/A | WAPL cohesin release factor | 0.03 | 6.74 | 8 | 0.22 | 19.07 | 5 | –0.13 | 20.81 | 7 |
*NO = not observed.
Aside from its well-established role in HR, BRCA2 has recently emerged as an important factor during replication stress response (35,36,40,44,56–59). Thereby, we also sought to investigate whether the interaction of sister chromatid cohesion factors with replication forks might be specifically affected by BRCA2 status under conditions of replication stress. To this purpose, we repeated the iPOND experiments treating cells with 4 mM HU for 2 hours and compared the log2 ratios of amount of proteins detected in BRCA2-proficient versus deficient cells in the presence of HU (Table 1, columns ‘Hydroxyurea ± BRCA2’). In this case, we used the Rad51 protein as a positive control for a factor that should dissociate from replication forks upon BRCA2 loss (56,60,61). Indeed, we found that the Rad51 association was significantly decreased in the BRCA2 mutant cells. However, we did not find any significant change in the association of the sister chromatid cohesion factors and the core replisome components with replication forks suggesting that the function of BRCA2 in replication stress response is uncoupled from Pds5B (Table 1, columns ‘Hydroxyurea ± BRCA2’).
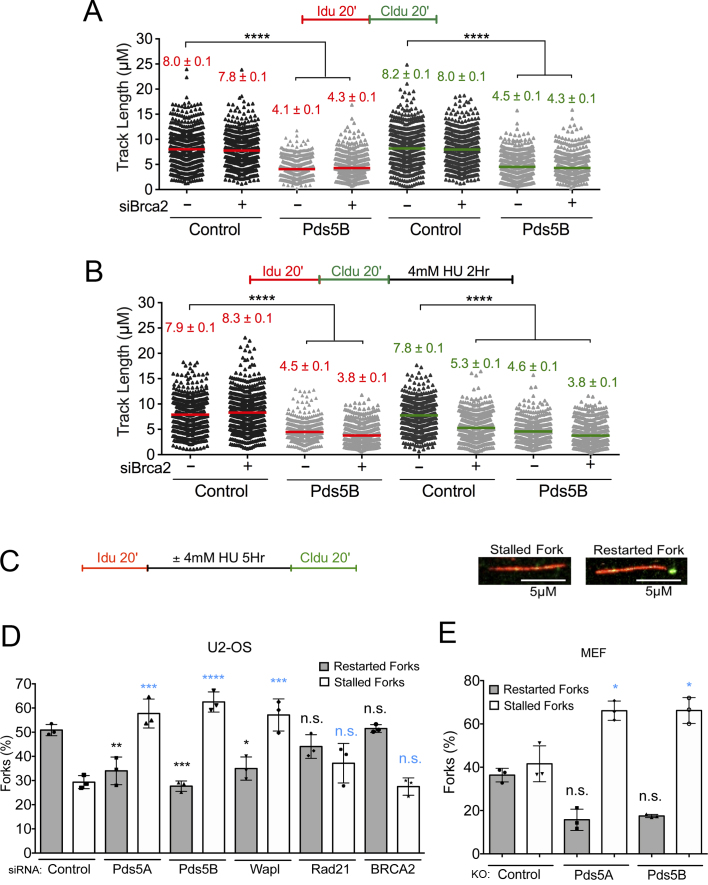
In agreement with the iPOND results, we found that BRCA2 loss did not lead to the same fork progression defect observed upon Pds5A and Pds5B loss in unperturbed U-2 OS cells, suggesting that the Pds5B–BRCA2 interaction is not relevant for the fork progression function of Pds5B (Figure 2A and Supplementary Figure S1D). Moreover, co-depletion of Pds5B and BRCA2 did not further exacerbate the fork progression defect observed in Pds5B depleted cells, confirming that BRCA2 does not play a role during normal replication, as previously reported (36). To study fork progression under conditions of genotoxic stress, we pulsed-labeled replication events with the two thymidine analogs, followed by treatment with 4 mM hydroxyurea (HU) for 2 h (Figure 2B). Previous studies suggested that BRCA2 is required to protect stalled replication forks from nucleolytic degradation following genotoxic stress induction (36,58,59). In agreement with these observations, we found that BRCA2 loss led to a significant shortening of the CldU tract in HU-treated cells, whereas the length of the IdU tracts was not significantly affected (Figure 2B). These results are consistent with notion that BRCA2 loss leads to an extensive nucleolytic degradation of the newly synthesized DNA, with a consequent loss of the signal originating from the thymidine analog that was incorporated immediately before HU treatment. Pds5B loss behaved differently from BRCA2 because it promoted a significant shortening of both IdU (red) and CldU (green) tracts in the presence and absence of HU (Figures 2A and B). Specifically, treatment with HU did not further exacerbate the fork progression defect observed in untreated Pds5B depleted cells. Moreover, combined depletion of Pds5B and BRCA2 yielded to a minor further decrease in the IdU and CldU tracts. However, a proper analysis of the double-depletion experiments is complicated by the fact that the tracts were already very short upon Pds5B loss. Taken together these results suggest that Pds5B is acting independently of BRCA2 at the replication forks and that, unlike BRCA2, is important for replication fork progression both in perturbed and unperturbed conditions.
Figure 2.
The role of Pds5 in replication fork progression is uncoupled from BRCA2 and compromises fork restart upon prolonged HU treatment. (A, B) Size distribution of IdU and CldU tract length in Pds5B, BRCA2 and Pds5B/BRCA2 siRNA depleted U-2 OS Cells. Untreated cells (A) and HU treated cells (B). Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (C) Single-molecule DNA fiber labeling scheme used for the fork restart experiments and representative images of stalled and restarting forks. Scale bar 5 μm. (D) Quantification of stalled/terminated forks and restarting forks after HU treatment in control, Pds5A, Pds5B, Wapl and Rad21, BRCA2 siRNA depleted U-2 OS cells. Out of three repeats, the percentage is established on at least 1000 tracts scored for each data set. Data are represented as mean ± SD. Statistics: two-way ANOVA. P values, restarted forks: siControl versus siPds5A **P = 0.0088; siControl versus siPds5B ***P = 0.0010; siControl versus siWapl *P = 0.0127; siControl versus siRad21 ns, P = 0.3881; siControl versus siBRCA2 ns, P = 0.9998. Stalled forks: siControl versus siPds5A ***P = 0.0002; siControl versus siPds5B ****P < 0.0001; siControl versus siWapl ***P = 0.0002; siControl versus siRad21 ns, P = 0.2757; siControl versus siBRCA2 ns, P = 0.9878. ns, not significant. (E) Quantification of stalled/terminated forks and restarting forks in WT, Pds5A, and Pds5B knockout MEF cells. Out of three repeats, the percentage is established on at least 1000 tracts scored for each data set. Data are represented as mean ± SD. Statistics: two-way ANOVA. P values, restarted forks: siControl versus siPds5A *P = 0.0474; siControl versus siPds5B ns, P = 0.0617. Stalled forks: siControl versus siPds5A *P = 0.0273; siControl versus siPds5B *P = 0.0269. ns, not significant.
Pds5 is required for fork restart after prolonged HU treatment
Given that Pds5 loss affects replication forks progression, we sought to investigate whether it also affects the ability of replication forks to restart after HU removal. To this purpose, we pulse-labeled Pds5A- and Pds5B-depleted U-2 OS cells with IdU for 20 minutes, followed by treatment with HU for five hours and labeling with the second thymidine analog CldU for 20 min after drug removal (Figure 2C). Using this labeling scheme, we can calculate the percentage of forks that have restarted by quantifying tracts that are red followed by green (restarting forks) and those that are only red (representing replication forks that were unable to restart following genotoxic stress induction) (Figure 2C). We found that, unlike BRCA2, loss of Pds5A or Pds5B lead to a significant increase in the percentage of stalled replication forks after prolonged exposure to HU (Figure 2D). Concomitantly, we also observed a parallel decrease in the percentage of restarting forks. The same results were obtained using the Pds5A and Pds5B KO MEF cells (Figure 2E).
Unlike Pds5, BRCA2 depletion did not significantly affect the ability of the stalled forks to restart upon drug removal (Figure 2D), in agreement with recent findings (36,40,58). Of note, other studies pointed to a potential role for BRCA2 in fork restart suggesting that its effect might vary as a function of the cell type or the particular conditions used to stall replication forks (62,63). The different fork restart phenotypes observed upon Pds5 and BRCA2 depletion in our cell lines further confirm that the Pds5 and BRCA2 play distinct functions during replication stress. Next, we investigated whether the fork restart defect associated with Pds5 loss was shared by other cohesion factors. Depletion of Rad21, one of the core components of the cohesin ring, did not significantly affect the ability of replication forks to restart upon drug removal (Figure 2D). On the other hand, depletion of the other cohesin unloading factor, Wapl, caused a significant increase in the percentage of stalled replication forks, mimicking the effect of Pds5A or Pds5B depletion (Figure 2D). These results suggest cohesin removal function of Pds5A/Pds5B and Wapl is important for normal replication fork progression and restart.
MRE11 activity is the cause of the fork progression defect associated with Pds5 loss
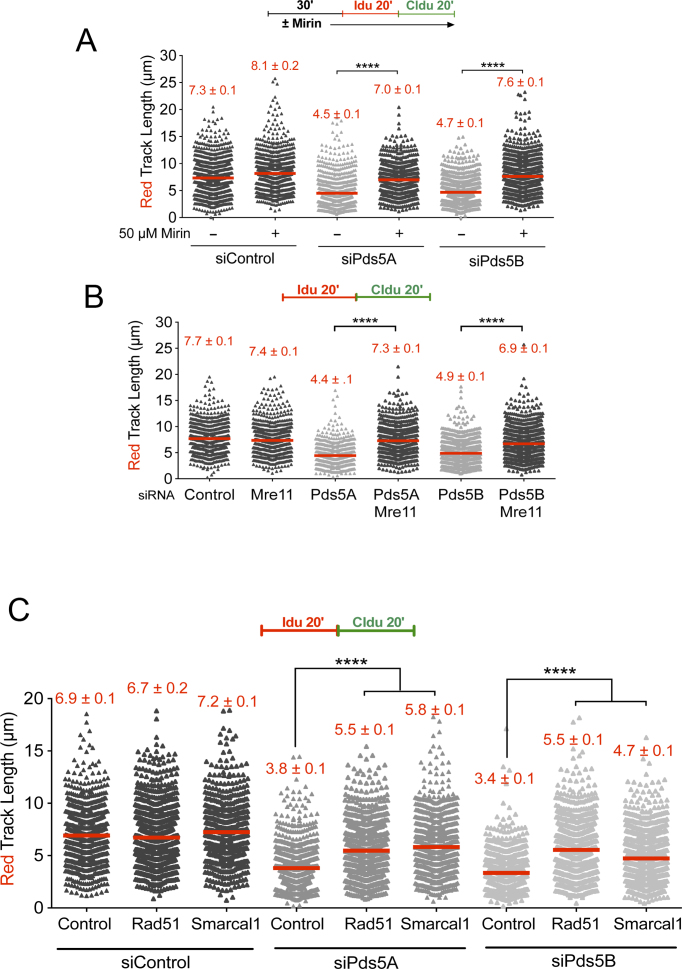
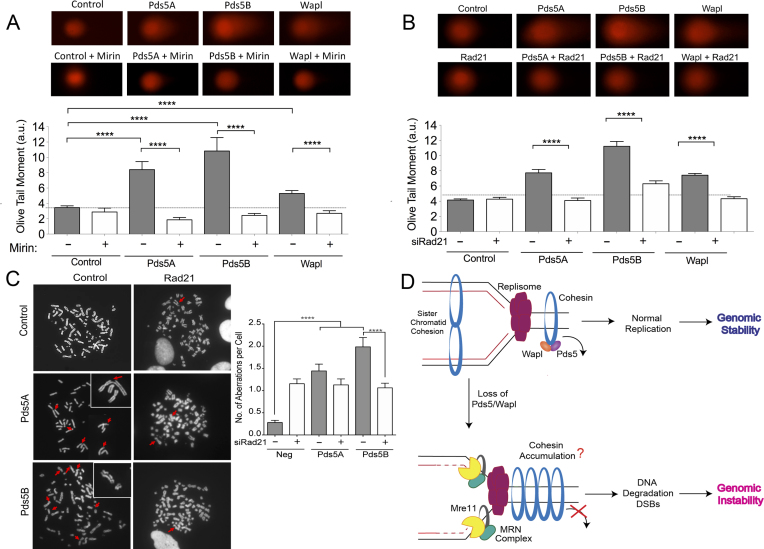
The MRE11 nuclease forms a trimeric complex comprising MRE11, Rad50 and Nbs1 (MRN) and it is best known for its role in DSB resection (64,65). Recent studies suggest that MRE11-dependent resection is also crucial for stalled replication fork processing through a tightly regulated process that must ensure a limited degradation of nascent DNA strands required for efficient fork processing and restart (66–68). However, MRE11-dependent resection can also lead to extensive and uncontrolled degradation of stalled replication forks in the absence of selected Fanconi Anemia (FA) and Homologous Recombination (HR) factors, including the Fanconi Anemia Complementation Group D2 (FANCD2) factor, BRCA1 and BRCA2 (36,58,59). To investigate whether the fork slowing phenotype associated with Pds5 loss is linked to nucleolytic degradation, we inhibited MRE11 activity with the Mirin inhibitor before pulse-labeling cells with the thymidine analogs (Figure 3A). We found that MRE11 inhibition rescued the fork slowing phenotype of Pds5A or Pds5B-depleted U-2 OS (Figure 3A) and RPE-1 (Supplementary Figure S3A) cells, suggesting that Pds5A and Pds5B play an important role in regulating MRE11 activity. MRE11 knockdown in Pds5A or Pds5B depleted U-2 OS, or in Pds5B knockout MEFs, led to very similar results ruling out any possible off-target effect of the MRE11 inhibitor (Figure 3B and Supplementary Figure S3B). Furthermore, the same results were obtained by knocking down the other two subunits of the MRN complex, Nbs1 and Rad50 (Supplementary Figure S3C–F). These results suggest that the MRN complex is responsible for fork slowing phenotype observed in Pds5 depleted cells.
Figure 3.
MRE11 degrades reversed replication forks in the absence of Pds5. (A) Single-molecule DNA fiber labeling scheme (top). Size distribution of IdU tract length in Pds5A and Pds5B siRNA depleted U-2 OS cells treated with 50 μM Mirin. Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (B) Size distribution of IdU tract length in Pds5A, Pds5B, MRE11, Pds5A/MRE11 and Pds5B/MRE11 siRNA depleted U-2 OS cells. Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (C) Size distribution of IdU length in Rad51 or Smarcal1 siRNA depleted U-2 OS cells in the presence and absence of Pds5A or Pds5B. Bars represent the mean. Out of three repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001.
MRE11-dependent fork slowing could arise as a consequence of the MRE11-dependent degradation of replication forks, as already discussed, or an inhibitory effect of MRE11 on fork movement independent of fork degradation. Recent studies suggest that the regressed arms of reversed replication forks are the entry point for MRE11 degradation in the absence of central HR factors (40,44,56,57). Thus, we sought to investigate whether depletion of factors required fork replication fork reversal abrogates MRE11-dependent fork slowing. Indeed, we found that loss of the central recombinase Rad51 (69) or the Smarcal1 translocase (44,56,70), which are both required for replication fork reversal, rescues the MRE11-dependent fork slowing phenotype associated with loss of Pds5A or Pds5B (Figure 3C and Supplementary Figure S3G and H). Collectively, these results suggest that the MRE11 nuclease might target reversed replication forks that accumulate upon Pds5 loss.
Loss of Rad21 rescues fork progression upon Pds5 depletion
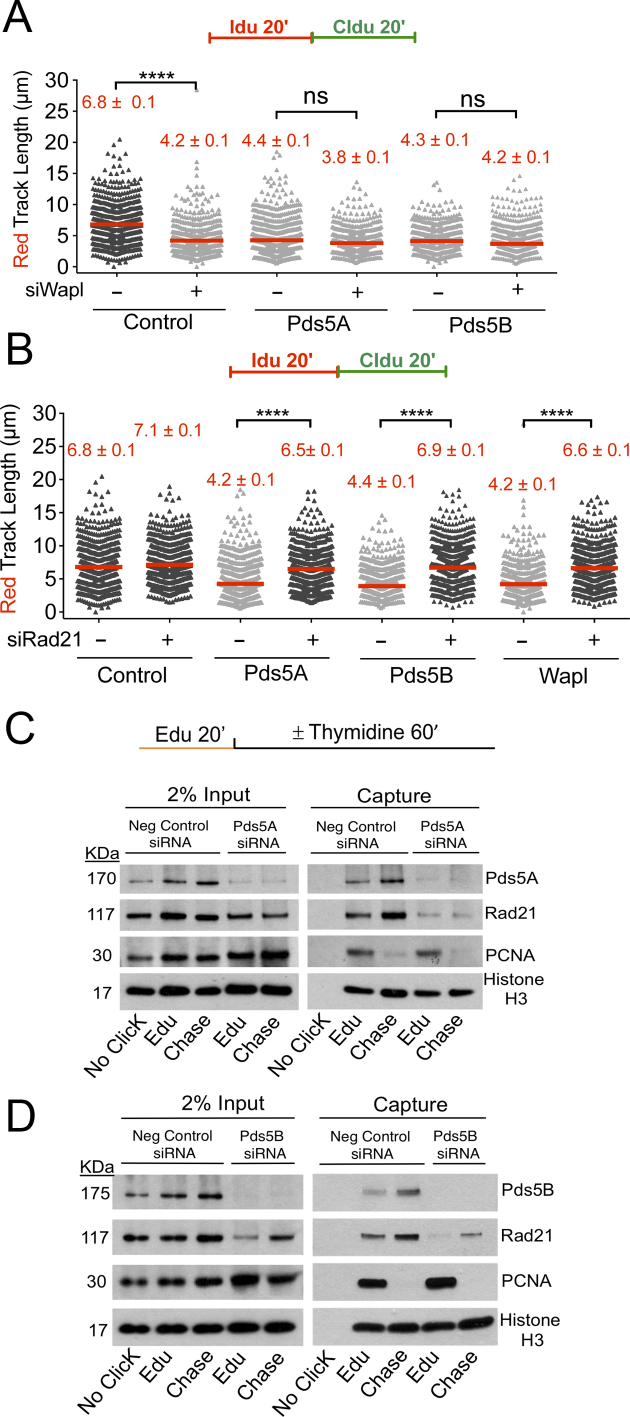
Next, we sought to investigate whether the fork progression defect observed upon Pds5A or Pds5B depletion could be recapitulated by knocking down other sister chromatid cohesion factors. We found that depletion of Wapl, which together with Pds5 is needed to release cohesin from DNA, lead to the same shortening of the IdU tracts observed upon Pds5A or Pds5B loss (Figure 4A and Supplementary Figure S4A). Moreover, co-depletion of Pds5A or Pds5B and Wapl did not lead to a further decrease in the size of the IdU tracts pointing to an epistatic role for Pds5 and Wapl in replication fork protection. In contrast to Pds5 and Wapl, depletion of core cohesin-ring component Rad21 did not significantly affect fork progression (Figure 4B and Supplementary Figure S4B–D). Surprisingly, co-depletion of Pds5A or Pds5B and Rad21 completely rescued the fork slowing phenotype observed upon Pds5A or Pds5B depletion alone. Similarly, co-depletion of Wapl and Rad21 rescued the fork slowing phenotype of Wapl depleted cells (Figure 4B). These results suggest that the fork slowing phenotype observed upon Pds5 or Wapl depletion is linked to an aberrant loading of cohesin on replication forks and can be rescued by cohesin removal.
Figure 4.
Cohesin depletion rescues the fork progression defects associated with the loss of Pds5 or Wapl. (A) Single-molecule DNA fiber labeling scheme (top). Size distribution of IdU tract length in Pds5A, Pds5B, Wapl, Pds5A/Wapl and Pds5B/Wapl siRNA depleted U-2 OS cells (bottom). Bars represent the mean. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (B) Size distribution of IdU tract length in Pds5A, Pds5B, Wapl, Pds5A/Rad21, Pds5B/Rad21 and Wapl/Rad21 siRNA depleted U-2 OS cells. Bars represent the median. Out of two repeats; n ≥ 300 tracts scored for each data set. Statistics: Mann–Whitney; ****P < 0.0001. (C) AniPOND of Pds5A siRNA depleted U-2 OS cells. (D) AniPOND of Pds5B siRNA depleted U-2 OS cells. No-Click control: no biotin was added. Edu: cells treated with EdU for 20 minutes and then harvested. Chase: cells treated with Edu for 20 min, washed and treated with a thymidine chase for 1 h and then harvested. Proteins present at the replication forks were recovered using streptavidin beads. Proteins enriched at the fork were detected by western blot.
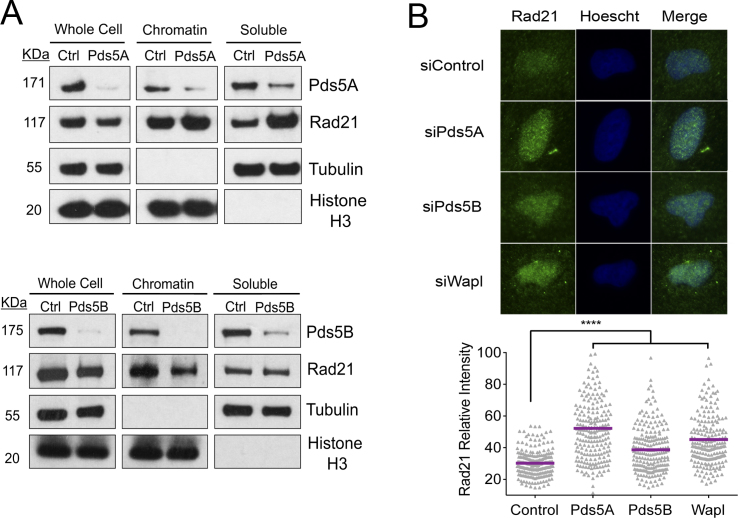
Loss of Pds5 decreases cohesin levels behind replication forks
On the basis of our observation that Rad21 depletion rescues the fork progression defect associated with Pds5 loss, we sought to investigate whether loss of Pds5A or Pds5B affects cohesin loading on replication forks. To this end, we studied the recruitment of Rad21 to the replication forks using a slightly modified version of the iPOND protocol, called aniPOND (50). Moreover, we can test whether the same proteins are recruited at some distance behind forks using a thymidine chase. Histone 3 (H3) was used as a loading control. PCNA was used as a positive control for proteins present at the fork and as a negative control in the chase, as PCNA travels with replication forks and would not be captured by the EdU label behind the fork. We found that Pds5 and Rad21 were associated with the newly synthesized tracts (EdU) (Figure 4C and D), in agreement with the iPOND-mass spectrometry results (Table 1). Moreover, the thymidine chase experiments showed that both proteins were also present behind the fork, in agreement with the notion that cohesin relocates behind replication forks to ensure sister chromatid cohesion. On the other hand, loss of Pds5A and Pds5B decreased Rad21 levels both at forks and behind the forks (Figure 4C and D). These data suggest that Pds5 depletion leads to a significant reduction of the cohesin levels on replication forks and behind them. Next, we sought to investigate whether the overall levels of chromatin bound Rad21 are affected by loss of Pds5 by chromatin fractionation and immunofluorescence experiments (Figure 5). We found the levels of chromatin bound Rad21 remain unchanged upon Pds5 depletion. These experiments, combined with the iPOND experiments described above, suggest the cohesin rings bound ahead of replication forks might not be efficiently relocated behind the forks in the absence of Pds5 leading to a reduction of cohesin levels behind forks without a significant change in the overall cohesin levels bound to DNA.
Figure 5.
Loss of Pds5 does not alter cohesin loading on chromatin. (A) Levels of Rad21 bound to chromatin after depletion of Pds5A (top) or Pds5B (bottom). Chromatin Fractionation of Pds5 depleted U-2 OS cells. Fractions separated included whole cell, chromatin bound and soluble cytoplasmic proteins. Samples were probed with Pds5A, Pds5B, Rad21, Tubulin and H3 antibodies. (B) Relative intensity of Rad21 bound to chromatin after immunofluorescence staining of Pds5A or Pds5B depleted cells. Cells were pre-extracted to remove soluble proteins, leaving chromatin bound proteins intact. Quantification of the Rad21 relative intensity signal. At least 100 cells were scored for each data set. n = 2. Data are represented as mean ± SEM. Statistics: Mann–Whitney; ****P < 0.0001.
Pds5 or Wapl loss leads to increased DSB accumulation, which is rescued by Rad21 depletion
To investigate the functional consequences of the fork progression defect observed upon Pds5 or Wapl depletion, we monitored whether loss of these essential cohesin removal factors leads to increased double-strand breaks (DSBs) accumulation and chromosomal aberrations. Using neutral comet assays, we found that depletion of Pds5A, Pds5B or Wapl lead to a significant increase in DSBs in unperturbed U-2 OS cells (Figure 6A). The same results were recapitulated using the Pds5A and Pds5B knockout MEFs and was further exacerbated by HU treatment (Supplementary Figure S4E). As an additional readout of DSBs accumulation, we also found that loss of Pds5A or Pds5B leads to a marked increase in γ-H2AX and 53BP1 nuclear bodies, even though it did not significantly affect the cellular levels of p-Chk1 or p-RPA (Supplementary Figure S5). In agreement with the results of the DNA fiber experiments, we also found that inhibition of MRE11 activity rescues the DSBs detected in the absence of Pds5A, Pds5B, or Wapl suggesting that the DSBs accumulation caused by the loss the releasin factors is a consequence of the MRE11-dependent processing of the nascent DNA strands (Figure 6A). Moreover, we found that loss of Rad21 also rescued the observed increase in DSBs supporting the notion that these DSBs are associated to the replication defects linked to Pds5 or Wapl depletion (Figure 6B).
Figure 6.
Loss of Pds5 leads to increased DSBs and chromosomal aberrations. (A) Neutral Comet assay monitoring DSB formation in in Pds5A, Pds5B or Wapl siRNA depleted U-2 OS cells treated with 50 μM Mirin or left untreated. Representative images of comets of Pds5A, Pds5B, Rad21 or Wapl siRNA depleted cells with or without 50 μM Mirin (top). Out of three repeats; n ≥ 100 comets scored for each data set. Data are represented as mean ± SEM. Statistics: Mann–Whitney; ****P < 0.0001 (bottom). (B) Neutral Comet assay monitoring DSB formation in Pds5A, Pds5B, Wapl, Pds5A/Rad21, Pds5B/Rad21 and Wapl/Rad21 siRNA depleted U-2 OS cells. Representative images of comets of Pds5A, Pds5B, Rad21, Wapl, Pds5A/Rad21, Pds5B/Rad21 and Wapl/Rad21 siRNA depleted cells (top). Out of three repeats; n ≥ 100 comets scored for each data set. Data are represented as mean ± SEM Statistics: Mann–Whitney; ****P < 0.0001 (bottom). (C) Accumulation of DSB as measured by chromosome spread. Representative images of metaphases of Pds5A, Pds5B, Rad21, Pds5A/Rad21 and Pds5B/Rad21 siRNA depleted cells (left). Red arrows point to DSBs. Bar graph, distribution of number of chromosomal abnormalities per metaphase. At least 50 metaphases were counted from three independent experiments. Mean shown, n = 3. Statistics: unpaired t-test; ****P < 0.0001. (D) Proposed model. Cohesin is recruited to origins in early S-Phase. Pds5/Wapl unload cohesin ahead of the fork to allow fork passage. Cohesin is promptly re-assembled behind the replication fork to achieve sister chromatid cohesion. Whether the two sister chromatids are embraced by two interacting cohesin complexes, as shown in the figure, or by a single-complex is still controversial in the field (84–87). In the absence of Pds5 or Wapl, there is a reduction in cohesin loading behind the forks suggesting that there might be a consequent accumulation of cohesin rings ahead of replication forks. This aberrant re-distribution of the cohesin rings might cause MRE11-dependent degradation of the newly synthesized DNA strands, leading tract shortening, DSB accumulation and chromosomal instability. Tract shortening could also be due to an inhibitory effect of MRE11 on fork movement independent of fork degradation as discussed in the main text.
Chromosome spread analysis showed that Pds5A and Pds5B depletion also led to increased chromosomal instability in unperturbed U-2 OS cells (Figure 6C). This effect was again partially rescued by Rad21 depletion. However, Rad21 depletion alone also led to a significant increase in chromosomal breaks and alterations in cell morphology (Figure 6C and Supplementary Figure S4F), in agreement with the notion that loss of cohesion leads to improper chromosomal segregation and increased genomic instability (11,71). Collectively, these results suggest that the fork slowing phenotype associated with the Pds5 loss leads to consequent accumulation of DSBs and chromosomal aberrations. This phenotype can be partially rescued by Rad21 depletion suggesting that it may be associated with aberrant accumulation of cohesin rings on replication forks, as also suggested by our aniPOND experiments.
DISCUSSION
Replication fork progression is constantly challenged by DNA lesions and intrinsic replication fork obstacles. Cohesin is loaded on the DNA in late G1 phase and is present on the DNA throughout S phase while DNA is replicated (17,72). Here, we found that the Pds5 protein plays a crucial role in replication fork progression by regulating a proper re-distribution of the cohesin rings on the ongoing replication forks. In its absence, replication forks are targeted by the MRE11 nuclease and fail to promptly restart when challenged with HU. This function of Pds5 is uncoupled to its previously reported interaction with BRCA2 and is shared by the other component of the cohesin releasin complex, Wapl. On the basis of these results, we propose that perturbing cohesin dynamics on replication forks poses an important and previously unappreciated form of replication challenge that must be properly controlled to ensure replication fork progression.
Pds5B has been shown to interact with several HR factors, including Rad51, Palb2 and BRCA2 upon DNA damage induction (32–34). Recent studies showed that BRCA2 and Rad51 have a HR-independent role in replication fork stability upon genotoxic stress induction (40,44,56,57). We found that, unlike BRCA2, Pds5B is required for replication fork progression in unperturbed conditions suggesting that the role of Pds5 during DNA replication is distinct from the previously reported function of BRCA2 and Rad51 in replication fork stability. We also found that Pds5B depletion increases fork stalling under conditions of replication stress. This phenotype is again different from that of BRCA2-deficient cells because loss of BRCA2 does not significantly affect the ability of replication forks to restart upon drug removal (36,40,58). Of note, loss of the other Pds5 variant, Pds5A, which does not interact with BRCA2, mimics the phenotype observed upon Pds5B depletion further supporting the idea that the function of the two Pds5 variants during replication is uncoupled from BRCA2. Along the same lines, our iPOND data show that the loading of Pds5, as well as the core component of the cohesin complex, is not affected by BRCA2 loss both in the presence and absence of replication stress. On the basis of these results, we propose the role of Pds5 during replication is independent from its interaction with HR factors, and is may be connected to its function in cohesin removal.
On the other hand, the interaction of Pds5B with HR factors might become important under conditions that lead to replication fork breakage and DSB accumulation. In this context, Pds5B seems to be important for the proper localization of BRCA2 and Rad51 at damaged sites during HR-mediated repair (32–34). In agreement with this conclusion, Pds5B was shown to interact with BRCA2 in response to agents that stall replication forks under conditions of prolonged drug treatment, which cause significant DSB accumulation (32). This conclusion is also supported by our data showing that the interaction between Pds5B–BRCA2 persists upon prolonged exposure to HU treatment pointing to a potential role of this complex following replication fork breakage.
We found that chemical inhibition of MRE11 activity using Mirin or reduction in protein levels by siRNA or shRNA rescued the pronounced shortening phenotype associated with Pds5A or Pds5B loss both in human and mouse cells. Furthermore, we were able to rescue this resection phenotype by depleting the two other components of the MRN complex, Nbs1 and Rad50. Specifically, Nbs1 depletion prevents MRE11 and Rad50 from being transported into the nucleus (73), whereas loss of Rad50 destabilizes the whole MRN complex (74). On the basis of these results, we propose that the MRE11 nuclease is responsible for the pronounced slowing phenotype associated with Pds5A or Pds5B loss. MRE11 travels with the replication machinery and the limited MRE11-dependent resection of nascent strands is required for efficient fork processing and restart (66–68). However, MRE11 activity can also lead to an extensive and uncontrolled degradation of replication intermediates in specific genetic backgrounds (75). The reason for the MRE11-dependent fork slowing phenotype associated with Pds5 loss could be either due to the MRE11-dependent degradation of the replication forks or an inhibitory effect of MRE11 on fork movement independent of fork degradation. In this regard, recent studies suggest that the regressed arms of reversed replication forks are the entry point for MRE11 in the absence of central HR factors (40,44,56,57). Indeed, we found that suppression of replication fork reversal by depletion of the central recombinase Rad51 (69) or the Smarcal1 translocase (44,56,70) rescues the MRE11-dependent fork slowing phenotype associated with loss of Pds5A or Pds5B. These data suggest that reversed forks might be the structures targeted by MRE11 in the absence of Pds5 and that the DSBs detected upon Pds5 loss might arise form nucleolytic cleavage of reversed forks. Interestingly, reversed forks were previously shown to be degraded by MRE11 only upon HU treatment under specific genetic backgrounds, such as in BRCA1- or BRCA2-deficient cells (40,44,56,57). In particular, these studies suggested that BRCA proteins load on the reversed forks to protect the regressed arms from degradation only under conditions of replication stress. We posit that the reversed forks that form upon Pds5 loss in the absence of HU might not trigger BRCA recruitment, thus remaining unprotected and exposed to nucleolytic degradation. However, accurate electron microscopy studies would be needed to fully validate this conclusion.
Structural studies previously reported that the inner diameter of the cohesin ring is approximately 50 nm (5,76). Advances in single molecule technology have helped uncover the flexibility of the cohesin ring loaded on DNA. In particular, recent work suggests that the cohesin ring may be loaded on the DNA in a collapsed form making the diameter of the ring much smaller than initially predicted (approximately 10–19 nm) (77). Moreover, single-molecule experiments have shown that cohesin is loaded topologically onto the DNA where it can translocate along the strand if pushed by DNA motor proteins (77,78). The replisome is a multi-protein macromolecular machinery, much larger than the inner diameter of the cohesin ring pore (79,80). Therefore, cohesin molecules present ahead of replication forks need to be promptly removed by the releasin complex to allow passage of the replication forks unless forks are somehow capable of traversing through the rings (81). Cohesin will then need to be reloaded behind the fork to hold the newly synthesized strands together and ensure proper sister chromatid cohesion. In this regard, a recent study showed that cohesin ubiquitination facilitates cohesin dynamics on replication forks (82). The exact mechanism by which this process occurs is still much debated in the field. Our data show that disruption of the releasin complex, by Pds5 or Wapl depletion, perturbs replication fork progression, leading to replication tract shortening. This phenotype is rescued by depletion of the core cohesin-ring component Rad21. Together, these findings support a model whereby loss of either component of the releasin complex compromises the ability of the cohesin rings to be removed from DNA and the aberrant re-distribution of cohesin rings on replication forks lead to the observed replication defects. Consistently, our aniPOND experiments show that there is a decreased amount of cohesin loaded behind the forks following Pds5A and Pds5B knockdown suggesting that the absence of Pds5 or Wapl perturbs cohesin dynamics on replication forks. Unfortunately, the iPOND approach is not designed to detect proteins ahead of replication forks and future studies should aim to test this possibility by using alternative techniques. Moreover, our aniPOND data do not rule out the alternative possibility that there is simply an overall reduction in cohesin levels bound to DNA upon Pds5 and Wapl knockdown. To rule out this alternative possibility, we performed chromatin fractionation and immunofluorescence experiments showing that overall levels of Rad21 remain largely unchanged upon Pds5 depletion. These experiments, coupled with our observation that Pds5 loss decreases the Rad21 levels behind forks, support a model where Pds5 loss compromises the proper relocation of cohesin rings on replication forks.
As discussed above, MRE11 travels with the replication machinery (66–68). A possible interpretation for why perturbing cohesin dynamics on replication forks leads to MRE11-dependent degradation could be simply that the inefficient relocalization of cohesin rings slows down replication forks and gives more time for MRE11 to act on the forks, leading to extensive nascent strand degradation (Figure 6D). Alternatively, aberrant loading of cohesin might somehow affect the distribution or activity of the MRN complex in the ongoing replication forks thereby leading to uncontrolled nascent strand DNA resection. However, future biochemical studies would need to be performed to define the mechanistic link between aberrant cohesin loading and MRE11 activity.
In summary, our data suggest that altering cohesin ring dynamics by Pds5 or Wapl loss perturbs normal replication fork progression and leads to uncontrolled MRE11-dependent degradation of the nascent DNA strands. This degradation leads to increased DSB accumulation and genomic instability (Figure 6D). Our findings provide a new rationale for the recently identified tumor suppressor function of Pds5B (83) and for the previously reported correlation between Pds5B expression levels and breast cancer patient survival following treatment with DNA-damaging chemotherapy (32).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Ana Losada (Spanish National Cancer Research Centre (CNIO), Madrid, Spain) for providing the Pds5A and Pds5 knockout immortalized mouse embryonic fibroblasts and to Maria Jasin (Memorial Sloan Kettering Cancer Center) providing the VC8 and VC8+BRCA2 cells. We would like to thank Laura Sesma and Anthony Couturier in J.-Y.M. laboratory for technical help.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM108648 to A.V.]; Department of Defense BRCP Breakthrough Award [BC151728 to A.V.]; National Institutes of Health [R01 CA193318 to N.M.], Alvin Siteman Cancer Research Fund (to N.M.); Siteman Investment Program (to N.M.); A.K.B. is supported by the National Institutes of Health Cell and Molecular Biology Training Grant [T32 GM007067-40]; Canadian Institutes of Health Research (top J.-Y.M); FRQS fellowship (to L.G.-S.); FRQS research chair in Genome Stability (to J.-Y.M.). Funding for open access charge: NIH grant [GM108648].
Conflict of interest statement. None declared.
REFERENCES
- 1. Berti M., Vindigni A.. Replication stress: getting back on track. Nat. Struct. Mol. Biol. 2016; 23:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gligoris T., Lowe J.. Structural insights into ring formation of cohesin and related Smc complexes. Trends Cell Biol. 2016; 26:680–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gruber S., Haering C.H., Nasmyth K.. Chromosomal cohesin forms a ring. Cell. 2003; 112:765–777. [DOI] [PubMed] [Google Scholar]
- 5. Peters J.M., Tedeschi A., Schmitz J.. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008; 22:3089–3114. [DOI] [PubMed] [Google Scholar]
- 6. Losada A., Yokochi T., Kobayashi R., Hirano T.. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 2000; 150:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sumara I., Vorlaufer E., Gieffers C., Peters B.H., Peters J.M.. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 2000; 151:749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murayama Y., Uhlmann F.. DNA Entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell. 2015; 163:1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ouyang Z., Zheng G., Tomchick D.R., Luo X., Yu H.. Structural basis and IP6 requirement for Pds5-Dependent cohesin dynamics. Mol. Cell. 2016; 62:248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorsett D., Strom L.. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr. Biol.: CB. 2012; 22:R240–R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mehta G.D., Rizvi S.M., Ghosh S.K.. Cohesin: a guardian of genome integrity. Biochim. Biophys. Acta. 2012; 1823:1324–1342. [DOI] [PubMed] [Google Scholar]
- 12. Nasmyth K., Haering C.H.. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009; 43:525–558. [DOI] [PubMed] [Google Scholar]
- 13. Gelot C., Guirouilh-Barbat J., Le Guen T., Dardillac E., Chailleux C., Canitrot Y., Lopez B.S.. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol. Cell. 2016; 61:15–26. [DOI] [PubMed] [Google Scholar]
- 14. Bermudez V.P., Maniwa Y., Tappin I., Ozato K., Yokomori K., Hurwitz J.. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:10237–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borges V., Smith D.J., Whitehouse I., Uhlmann F.. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma. 2013; 122:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guillou E., Ibarra A., Coulon V., Casado-Vela J., Rico D., Casal I., Schwob E., Losada A., Mendez J.. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 2010; 24:2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moldovan G.L., Pfander B., Jentsch S.. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006; 23:723–732. [DOI] [PubMed] [Google Scholar]
- 18. Samora C.P., Saksouk J., Goswami P., Wade B.O., Singleton M.R., Bates P.A., Lengronne A., Costa A., Uhlmann F.. Ctf4 links DNA replication with sister chromatid cohesion establishment by recruiting the Chl1 helicase to the replisome. Mol. Cell. 2016; 63:371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sherwood R., Takahashi T.S., Jallepalli P.V.. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 2010; 24:2723–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fumasoni M., Zwicky K., Vanoli F., Lopes M., Branzei D.. Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polalpha/Primase/Ctf4 complex. Mol. Cell. 2015; 57:812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartman T., Stead K., Koshland D., Guacci V.. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 2000; 151:613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panizza S., Tanaka T., Hochwagen A., Eisenhaber F., Nasmyth K.. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol.: CB. 2000; 10:1557–1564. [DOI] [PubMed] [Google Scholar]
- 23. Gause M., Misulovin Z., Bilyeu A., Dorsett D.. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol. Cell. Biol. 2010; 30:4940–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Losada A., Yokochi T., Hirano T.. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 2005; 118:2133–2141. [DOI] [PubMed] [Google Scholar]
- 25. Goto Y., Yamagishi Y., Shintomi-Kawamura M., Abe M., Tanno Y., Watanabe Y.. Pds5 Regulates Sister-Chromatid cohesion and chromosome Bi-orientation through a conserved protein interaction module. Curr. Biol.: CB. 2017; 27:1005–1012. [DOI] [PubMed] [Google Scholar]
- 26. Muir K.W., Kschonsak M., Li Y., Metz J., Haering C.H., Panne D.. Structure of the Pds5-Scc1 complex and implications for cohesin function. Cell Rep. 2016; 14:2116–2126. [DOI] [PubMed] [Google Scholar]
- 27. Zhou L., Liang C., Chen Q., Zhang Z., Zhang B., Yan H., Qi F., Zhang M., Yi Q., Guan Y. et al. . The N-terminal non-kinase-domain-mediated binding of haspin to Pds5B protects centromeric cohesion in mitosis. Curr. Biol.: CB. 2017; 27:992–1004. [DOI] [PubMed] [Google Scholar]
- 28. Nishiyama T., Ladurner R., Schmitz J., Kreidl E., Schleiffer A., Bhaskara V., Bando M., Shirahige K., Hyman A.A., Mechtler K. et al. . Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 2010; 143:737–749. [DOI] [PubMed] [Google Scholar]
- 29. Nishiyama T., Sykora M.M., Huis in ’t Veld P.J., Mechtler K., Peters J.M.. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13404–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanke M., Tahara E., Huis In’t Veld P.J., Nishiyama T.. Cohesin acetylation and Wapl-Pds5 oppositely regulate translocation of cohesin along DNA. EMBO J. 2016; 35:2686–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carretero M., Ruiz-Torres M., Rodriguez-Corsino M., Barthelemy I., Losada A.. Pds5B is required for cohesion establishment and Aurora B accumulation at centromeres. EMBO J. 2013; 32:2938–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brough R., Bajrami I., Vatcheva R., Natrajan R., Reis-Filho J.S., Lord C.J., Ashworth A.. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012; 31:1160–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Couturier A.M., Fleury H., Patenaude A.M., Bentley V.L., Rodrigue A., Coulombe Y., Niraj J., Pauty J., Berman J.N., Dellaire G. et al. . Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Res. 2016; 44:10879–10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kusch T. Brca2-Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J. Cell Sci. 2015; 128:717–727. [DOI] [PubMed] [Google Scholar]
- 35. Boulton S.J. Cellular functions of the BRCA tumour-suppressor proteins. Biochem. Soc. Trans. 2006; 34:633–645. [DOI] [PubMed] [Google Scholar]
- 36. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terret M.E., Sherwood R., Rahman S., Qin J., Jallepalli P.V.. Cohesin acetylation speeds the replication fork. Nature. 2009; 462:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Diaz-Martinez L.A., Gimenez-Abian J.F., Clarke D.J.. Cohesin is dispensable for centromere cohesion in human cells. PLoS One. 2007; 2:e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chun J., Buechelmaier E.S., Powell S.N.. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 2013; 33:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lemacon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N. et al. . MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017; 8:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ewald B., Sampath D., Plunkett W.. ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance. Cancer Res. 2008; 68:7947–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yuan J., Chen J.. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J. Biol. Chem. 2010; 285:1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roques C., Coulombe Y., Delannoy M., Vignard J., Grossi S., Brodeur I., Rodrigue A., Gautier J., Stasiak A.Z., Stasiak A. et al. . MRE11-RAD50-NBS1 is a critical regulator of FANCD2 stability and function during DNA double-strand break repair. EMBO J. 2009; 28:2400–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R. et al. . Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell. 2017; 68:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., Burke B.. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2194–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quinet A., Carvajal-Maldonado D., Lemacon D., Vindigni A.. DNA fiber analysis: mind the gap!. Methods Enzymol. 2017; 591:55–82. [DOI] [PubMed] [Google Scholar]
- 47. Techer H., Koundrioukoff S., Azar D., Wilhelm T., Carignon S., Brison O., Debatisse M., Le Tallec B.. Replication dynamics: biases and robustness of DNA fiber analysis. J. Mol. Biol. 2013; 425:4845–4855. [DOI] [PubMed] [Google Scholar]
- 48. Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A. et al. . DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015; 208:545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mendez J., Stillman B.. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000; 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leung K.H., Abou El Hassan M., Bremner R.. A rapid and efficient method to purify proteins at replication forks under native conditions. BioTechniques. 2013; 55:204–206. [DOI] [PubMed] [Google Scholar]
- 51. Dungrawala H., Cortez D.. Purification of proteins on newly synthesized DNA using iPOND. Methods Mol. Biol. 2015; 1228:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sirbu B.M., Couch F.B., Cortez D.. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012; 7:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 54. Kraakman-van der Zwet M., Overkamp W.J., van Lange R.E., Essers J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P.P., Errami A., Tan R.T. et al. . Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002; 22:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wiegant W.W., Overmeer R.M., Godthelp B.C., van Buul P.P., Zdzienicka M.Z.. Chinese hamster cell mutant, V-C8, a model for analysis of Brca2 function. Mutat. Res. 2006; 600:79–88. [DOI] [PubMed] [Google Scholar]
- 56. Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L. et al. . Smarcal1-Mediated fork reversal triggers Mre11-Dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 2017; 67:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P. et al. . Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017; 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J. et al. . Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016; 535:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ying S., Hamdy F.C., Helleday T.. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012; 72:2814–2821. [DOI] [PubMed] [Google Scholar]
- 60. Jensen R.B., Carreira A., Kowalczykowski S.C.. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010; 467:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Raghunandan M., Chaudhury I., Kelich S.L., Hanenberg H., Sobeck A.. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi anemia core complex. Cell cycle. 2015; 14:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rondinelli B., Gogola E., Yucel H., Duarte A.A., van de Ven M., van der Sluijs R., Konstantinopoulos P.A., Jonkers J., Ceccaldi R., Rottenberg S. et al. . EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017; 19:1371–1378. [DOI] [PubMed] [Google Scholar]
- 64. Williams R.S., Williams J.S., Tainer J.A.. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007; 85:509–520. [DOI] [PubMed] [Google Scholar]
- 65. Stracker T.H., Petrini J.H.. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell. Biol. 2011; 12:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Costanzo V. Brca2, Rad51 and Mre11: performing balancing acts on replication forks. DNA Repair. 2011; 10:1060–1065. [DOI] [PubMed] [Google Scholar]
- 67. Costanzo V., Robertson K., Bibikova M., Kim E., Grieco D., Gottesman M., Carroll D., Gautier J.. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 2001; 8:137–147. [DOI] [PubMed] [Google Scholar]
- 68. Hashimoto Y., Puddu F., Costanzo V.. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol. 2012; 19:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M.. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015; 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Poole L.A., Cortez D.. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 2017; 52:696–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat. Rev. Cancer. 2014; 14:389–393. [DOI] [PubMed] [Google Scholar]
- 72. Kenna M.A., Skibbens R.V.. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 2003; 23:2999–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R. 3rd, Hays L., Morgan W.F., Petrini J.H.. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998; 93:477–486. [DOI] [PubMed] [Google Scholar]
- 74. Paull T.T., Gellert M.. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999; 13:1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pasero P., Vindigni A.. Nucleases acting at stalled forks: how to reboot the replication program with a few shortcuts. Annu. Rev. Genet. 2017; 51:477–499. [DOI] [PubMed] [Google Scholar]
- 76. Haering C.H., Lowe J., Hochwagen A., Nasmyth K.. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002; 9:773–788. [DOI] [PubMed] [Google Scholar]
- 77. Stigler J., Camdere G.O., Koshland D.E., Greene E.C.. Single-Molecule imaging reveals a collapsed conformational state for DNA-Bound cohesin. Cell Rep. 2016; 15:988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Davidson I.F., Goetz D., Zaczek M.P., Molodtsov M.I., Huis In ’t Veld P.J., Weissmann F., Litos G., Cisneros D.A., Ocampo-Hafalla M., Ladurner R. et al. . Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 2016; 35:2671–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kulczyk A.W., Moeller A., Meyer P., Sliz P., Richardson C.C.. Cryo-EM structure of the replisome reveals multiple interactions coordinating DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E1848–E1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sun J., Shi Y., Georgescu R.E., Yuan Z., Chait B.T., Li H., O’Donnell M.E.. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015; 22:976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K.P., Shirahige K., Uhlmann F.. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006; 23:787–799. [DOI] [PubMed] [Google Scholar]
- 82. Frattini C., Villa-Hernandez S., Pellicano G., Jossen R., Katou Y., Shirahige K., Bermejo R.. Cohesin ubiquitylation and mobilization facilitate stalled replication fork dynamics. Mol. Cell. 2017; 68:758–772. [DOI] [PubMed] [Google Scholar]
- 83. Denes V., Pilichowska M., Makarovskiy A., Carpinito G., Geck P.. Loss of a cohesin-linked suppressor APRIN (Pds5b) disrupts stem cell programs in embryonal carcinoma: an emerging cohesin role in tumor suppression. Oncogene. 2010; 29:3446–3452. [DOI] [PubMed] [Google Scholar]
- 84. Eng T., Guacci V., Koshland D.. Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol. Biol. Cell. 2015; 26:4224–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Haering C.H., Farcas A.M., Arumugam P., Metson J., Nasmyth K.. The cohesin ring concatenates sister DNA molecules. Nature. 2008; 454:297–301. [DOI] [PubMed] [Google Scholar]
- 86. Morales C., Losada A.. Establishing and dissolving cohesion during the vertebrate cell cycle. Curr. Opin. Cell Biol. 2018; 52:51–57. [DOI] [PubMed] [Google Scholar]
- 87. Zhang N., Kuznetsov S.G., Sharan S.K., Li K., Rao P.H., Pati D.. A handcuff model for the cohesin complex. J. Cell Biol. 2008; 183:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.