Abstract
Polypyrimidine tract-binding protein 1 (PTBP1) is a highly conserved RNA-binding protein that is a well-known regulator of alternative splicing. Testicular tissue is one of the richest tissues with respect to the number of alternative splicing mRNA isoforms, but the molecular role(s) of PTBP1 in the regulation of these isoforms during spermatogenesis is still unclear. Here, we developed a germ cell–specific Ptbp1 conditional knockout (cKO) mouse model by using the Cre-loxP system to investigate the role of PTBP1 in spermatogenesis. Testis weight in Ptbp1 cKO mice was comparable to that in age-matched controls until 3 weeks of age; at ≥ 2 months old, testis weight was significantly lighter in cKO mice than in age-matched controls. Sperm count in Ptbp1 cKO mice at 2 months old was comparable to that in controls, whereas sperm count significantly decreased at 6 months old. Seminiferous tubules that exhibited degeneration in spermatogenic function were more evident in the 2-month-old Ptbp1 cKO mice than in controls. In addition, the early neonatal proliferation of spermatogonia, during postnatal days 1–5, was significantly retarded in Ptbp1 cKO mice compared with that in controls. An in vitro spermatogonia culture model (germline stem cells) revealed that hydroxytamoxifen-induced deletion of PTBP1 from germline stem cells caused severe proliferation arrest accompanied by an increase of apoptotic cell death. These data suggest that PTBP1 contributes to spermatogenesis through regulation of spermatogonia proliferation.
Keywords: Alternative splicing, Polypyrimidine track-binding protein, RNA-binding protein, Spermatogenesis, Spermatogonia proliferation
Spermatogenesis is a highly organized process to ultimately produce mature sperm sustainably throughout life. In the mouse, spermatogenesis occurs soon after birth when mitotically inactive differentiating germ cells, termed gonocytes [1, 2] or prospermatogonia [3], give rise to spermatogonia to initiate proliferation. Within the spermatogonial cell population, a small fraction of undifferentiated, unipotential spermatogonia, i.e., spermatogonial stem cells, self-renew to maintain their population [4, 5]. In addition, a subset of spermatogonial progenitor cells differentiate into meiotic spermatocytes followed by haploid spermatids [6] . This complex process of spermatogenesis is strictly regulated via various molecular mechanisms to orchestrate and maintain homeostasis during spermatogenesis.
Alternative splicing (AS), a mechanism to produce multiple gene products from a single gene locus, plays an important role in transcriptome and proteome complexity and facilitates proper tissue development [7, 8]. In humans, more than 90% of genes have splicing isoforms [9, 10]. The contribution of AS in cell lineage specification and differentiation is well investigated in neural development, especially brain formation [11,12,13]. Recently, high-throughput analysis of microarrays [14] and RNA sequencing [15,16,17] detected numerous gene isoforms resulting from AS within spermatogenic cell types in the testis, with numbers second only to the brain. Despite the high levels of AS that occur in the testis, the functional importance of AS for spermatogenesis is not well understood.
RNA-binding proteins (RBPs) play a central role in AS regulation by interacting with nascent transcripts to alter their mature isoforms via common forms of AS such as exon retention or skipping, 5′ or 3′ alternative exon, or mutually exclusive exon [12, 18]. The polypyrimidine tract-binding protein (PTBP) family includes highly conserved RBPs from Caenorhabditis elegans to mammals. They regulate AS, mainly via exon skipping [19], by binding to the polypyrimidine-rich intronic region of nascent RNA. Previous studies showed that genetic mutation in Drosophila PTBP (dmPTB or hephaestus), a male germline–specific mRNA isoform, causes infertility and a deficiency in spermatid individualization [20, 21].
In mice, three orthologous genes coding PTBPs are known: Ptbp1, Ptbp2 (also known as nPtb), and Ptbp3 (also known as Rod1). According to the public FANTOM5 mouse promoterome database (http://fantom.gsc.riken.jp/zenbu/), expression of Ptbp3 is high in embryonic stem cells and hematopoietic lineage cells but less evident in cells of the adult testis. By contrast, Ptbp2 is highly expressed by neurons and testis [22]. Recent studies using a mouse knockout (KO) model showed that germ cell–specific loss of function of Ptbp2 causes a severe abnormality in spermatogenesis, i.e., spermatid elongation was severely compromised in the conditional KO (cKO) model [23, 24], suggesting the necessary role of PTBPs in spermatogenesis via genes that are highly conserved.
Ptbp1, the focal gene of this study, regulates differentiation of many types of cells in mice including cardiomyocytes [25], neural cells [11, 26], and B lymphocytes [27]. PTBP1 seems to be widely expressed in various tissues, including testis (see FANTOM5 mouse promoterome database). However, our previous study [28] and another [29] reported that conventional Ptbp1 KO mice showed a complete embryonic lethal phenotype soon after implantation; thus the role(s) of PTBP1 in spermatogenesis is still unclear.
Here, we investigated the role(s) of PTBP1 in spermatogenesis in mice by using a germ cell–specific gene KO model for escaping the embryonic lethal phenotype observed in the conventional KO model. Although sperm counts in the Ptbp1 cKO mice were comparable to those in control mice at 2 months old, significantly more seminiferous tubules showed degeneration in spermatogenic function in the Ptbp1 cKO mice at that age. We also observed that early neonatal proliferation of spermatogonia was significantly retarded in the Ptbp1 cKO mice. Furthermore, hydroxytamoxifen-induced deletion of PTBP1 from cultured spermatogonia caused severe proliferation arrest accompanied by an increase in apoptotic cell death. These findings suggest that PTBP1 contributed to proper spermatogenesis through regulation of cell proliferation.
Materials and Methods
Animals and ethics
C57BL/6J mice were purchased from Japan SLC (Shizuoka, Japan). Ptbp1flox/flox mice were established in our laboratory by a conventional targeting strategy using 129-strain embryonic stem cells as described previously [26, 28] and backcrossed at least ten times to C57BL/6J mice. Ngn3-Cre transgenic mice [30] were introduced from RIKEN BioResource Research Center (Wakō, Japan) and backcrossed at least eight times to C57BL/6J mice. Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (mTmG) mice were purchased from The Jackson Laboratory (Bar Harber, ME, USA). CAG-CreMER mice were developed in our laboratory [31]. Mice were maintained under pathogen-free conditions in the experimental animal facility at the Institute of Medical Science, the University of Tokyo (Tokyo, Japan). All research protocols were conducted under guidelines approved by the Institutional Animal Care and Use Committee of the University of Tokyo (approval no. PA10-59).
Immunohistochemistry and measurement of seminiferous tubule area
Mice were euthanized, and then their testes were dissected. The testes were fixed with phosphate-buffered saline (PBS) containing 4% (w/v) paraformaldehyde (Nacalai Tesque, Kyoto, Japan) at 4°C overnight with gentle shaking. After fixation, testes were dehydrated in a graded ascending series of ethanol (25–100%, v/v), embedded in paraffin, and sectioned at a thickness of 5 μm. Sections were deparaffinized in Lemosol A (FUJIFILM Wako Pure Chemical, Osaka, Japan) and rehydrated by serial incubations in ethanol (100% to 70%, v/v) followed by washing in deionized, distilled water twice. For antigen retrieval, rehydrated sections were heated in sodium citrate buffer [10 mM sodium citrate containing 0.05% (v/v) Tween 20, pH 6.0] or EDTA buffer [10 mM Tris, 1 mM EDTA containing 0.05% (v/v) Tween 20, pH 9.0] for 20 min at 120°C in an autoclave. Sections were then permeabilized with 0.3% (v/v) or 0.025% Triton X diluted in PBS or Tris-buffered saline (TBS), respectively and blocked using bovine serum albumin (Nacalai Tesque) at room temperature for 1 h. Slides were incubated with primary antibodies diluted in SignalStain antibody diluent (Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight with gentle shaking. Then, sections were incubated with Alexa Fluor 488–, 555–, or 647–conjugated host species–specific secondary antibodies (all from Thermo Fisher Scientific, Tokyo, Japan) for 1 h at room temperature in the dark. After the antibody treatment, sections were incubated with 0.1% (w/v) Sudan black B (Sigma-Aldrich, St. Louis, MO, USA) solution for quenching autofluorescent signals [32, 33]. The nucleus was counterstained with 4,6-diamidino-2-phenylindole and mounted with a coverslip with anti-fade mountant solution (PermaFluor Aqueous Mounting Medium or ProLong Glass, Thermo Fisher). Fluorescent signals were detected using a fluorescent microscope (BZ-9000 or BZ-X700, Keyence, Osaka, Japan). The primary antibodies used were as follows: rabbit anti-PLZF (sc-22839, Santa Cruz Biotechnology Inc., Dallas, TX, USA), goat anti-PTBP1 (sc-16547, Santa Cruz Biotechnology), rabbit anti-WT1 (sc-192, Santa Cruz Biotechnology), rabbit anti-MVH (ab13840, Abcam, Cambridge, MA, USA), rat anti-germ cell–specific nuclear antigen (clone TRA98, BioAcademia, Osaka, Japan), rabbit anti-cleaved caspase-3 (CC3) antibody (9664, Cell Signaling Technology), rat anti-haploid sperm cell–specific antigen (clone TRA54, ab92286, Abcam), goat anti-SCP3 (sc-20845, Santa Cruz Biotechnology), and goat anti-GATA4 (sc-1237, Santa Cruz Biotechnology). For measurement of area of seminiferous tubules, tubules of which major axis length was less than 2-fold of minor axis were selected, and area was analyzed using ImageJ software (https://imagej.nih.gov/ij/).
Hematoxylin and eosin (HE) staining
Testes were fixed with Bouin’s solution (FUJIFILM Wako Pure Chemical) at 4°C overnight with gentle shaking. Fixed testes were washed in PBS and then dehydrated by serial incubations in ethanol (70–100%, v/v). Dehydrated testes were embedded in paraffin, sectioned, or rehydrated as described under Immunohistochemistry. For histological analysis, sections were stained with hematoxylin and eosin (HE) by a standard HE staining procedure. Stained sections were dehydrated by serial incubations in ethanol (50–100%, v/v) followed by permeation by incubation twice in Lemosol A (FUJIFILM Wako Pure Chemical). Sections were mounted with a coverslip with Mount-Quick (Daido Sangyo, Saitama, Japan) and observed under a fluorescence microscope (BZ-9000, Keyence).
Germline stem (GS) cell development
A mouse having mTmG; CAG-CreMER; Ptbp1flox/flox (Ptbp1-FF) or mTmG; CAG-CreMER; Ptbp1flox/wt (Ptbp1-FW) was used for germline stem (GS) cell development. Testes of 7–10-day-old postpartum neonates were collected and digested into single cells enzymatically as described previously [34]. Single cells were washed twice with PBS containing 2 mM EDTA and 0.5% (w/v) bovine serum albumin. THY1.2-positive cells, the fraction in which undifferentiated spermatogonia are highly enriched [35, 36], were sorted by magnetic-activated cell sorting technology (CD90.2 MicroBeads, Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. The isolated cell fraction was seeded onto mitotically inactivated feeder cells (X-ray–irradiated or Mitomycin C–treated mouse embryonic fibroblasts) to develop into stably self-renewing GS cells. GS cells were cultured as described previously [37] at 37°C in a humid atmosphere of 5% CO2 and 95% air. GS cells were passaged every 7 to 10 days at a density of 1–2 × 105 cells/ml. For initiation of tamoxifen-induced recombination of the flox allele, 1 μM 4-hydroxytamoxifen (4OHT; LKT Laboratories, St. Paul, MN, USA) was added to the culture medium.
RNA extraction, cDNA synthesis, and quantitative polymerase chain reaction (qPCR)
Total RNA from mouse tissues, except brain, was extracted using Sepasol-RNA I Super G solution (Nacalai Tesque) according to the manufacturer’s instructions and then treated with DNase I (Takara, Shiga, Japan) to digest potentially contaminating genomic DNA. For extracting RNA from brain, a NucleoSpin RNAII system (Macherey-Nagel, Düren, Germany) was used in which DNase for digestion of contaminated genomic DNA was included. cDNA was reverse transcribed from RNA using a SuperScript VILO cDNA synthesis kit (Thermo Fisher). Synthesized cDNA was used for quantitative polymerase chain reaction (qPCR) in a PCR reaction mixture of THUNDERBIRD SYBR qPCR Mix (Toyobo, Osaka, Japan) using the StepOne system (Thermo Fisher). The fold difference was calculated using the ΔΔCt method [38] with Gapdh as the reference. Primer sequences were as follows: for Ptbp1, forward GGTCTCTTCCGTGTGCCATG and reverse CTGCGCTCCTGTTGTCACCT, and for Gapdh, forward ATGAATACGGCTACAGCAACAGG and reverse CTCTTGCTCAGTGTCCTTGCTG.
Flow cytometry
Flow cytometry was used to detect intracellular proteins or to analyze phases of cell cycle. For protein detection, cells were fixed with 1% (v/v) paraformaldehyde prepared in PBS for 10 min at room temperature and then permeabilized using 90% (v/v) ice-cold methanol for 30 min. Permeabilized cells were incubated with rabbit anti-CC3 antibody (9664, Cell Signaling Technology) or goat anti-PTBP1 (sc-16547, Santa Cruz Biotechnology) at 4°C for 1 h and then exposed to Alexa Fluor 647–conjugated host species–specific secondary antibodies (Thermo Fisher). For cell cycle analysis, cells were fixed and permeabilized in ethanol (90% (v/v) prepared in PBS) overnight at –20°C. Thereafter, DNA was stained with propidium iodide (50 µg/ml, Sigma-Aldrich) and endogenous RNA was simultaneously digested with RNase A (200 µg/ml, Merck Millipore) for 20 min at 37°C. Stained cells were analyzed using a FACSCalibur or FACSVerse flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed using FlowJo software (FlowJo, LLC; https://www.flowjo.com).
Statistical analysis
All experiments were replicated at least three times. The data are presented as means ± standard error of the mean. Differences between genotypes or ages were tested using Student’s t-test except GS cell proliferation assay. Differences of GS cell proliferation were tested by one- way ANOVA followed by Tukey's multiple comparison tests. P values less than 0.05 were considered significant.
Results
PTBP1 is highly expressed by spermatogonia and spermatocyte in testicular germ cells
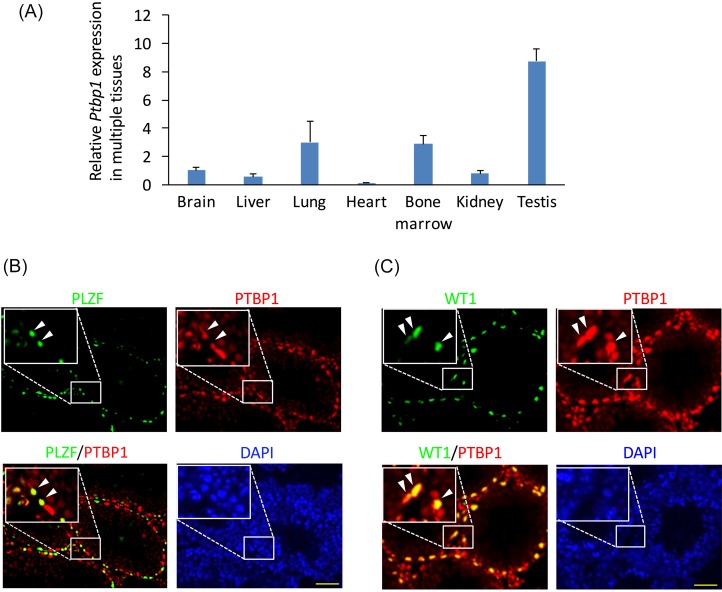
To compare expression levels of Ptbp1 mRNA in various tissues (e.g., brain, liver, lung, heart, bone marrow, kidney, and testis), these tissues were collected and used for qPCR analysis. All of the tissues analyzed expressed Ptbp1, but the level was the highest in testis (Fig. 1A). The testis consist of germ cells at the various stages of differentiation (i.e., undifferentiated or differentiating spermatogonia, meiotic spermatocytes, round or elongated haploid spermatids) as well as somatic cells such as Sertoli cells and Leydig cells that provide structural and nutritional support for germ cell maintenance and differentiation. PTBP1 protein localization in the testes was determined by immunohistochemistry using adult wild type mice. The expression of PTBP1 was strong in PLZF-positive undifferentiated spermatogonia (Fig. 1B) as well as in Wilms tumor gene product (WT1)-positive Sertoli cells (Fig. 1C), but it became less evident in cells located on the apical side of seminiferous tubules (Fig. 1B and 1C). These results indicated that PTBP1 expression in testicular germ cells is strong in mitotic spermatogonia and becomes weaker after entering meiosis.
Fig. 1.
PTBP1 is highly expressed by spermatogonia and spermatocytes in testicular germ cells. (A) Quantitative polymerase chain reaction of Ptbp1 expression in various tissues. Gene expression was standardized using Gapdh, and the expression level of the brain was designated as 1-fold [n = 3 in each tissue, except testis (n = 2)]. (B and C) Immunohistochemical analysis of PTBP1 expression in the wild-type adult testis. Sections were stained with (B) anti-PLZF (a marker of spermatogonia, arrowhead) or (C) anti-WT1 (a marker of Sertoli cells, arrowhead) as well as with anti-PTBP1. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Scale bar denotes 50 µm.
Loss of PTBP1 causes an increase in degeneration of seminiferous tubules
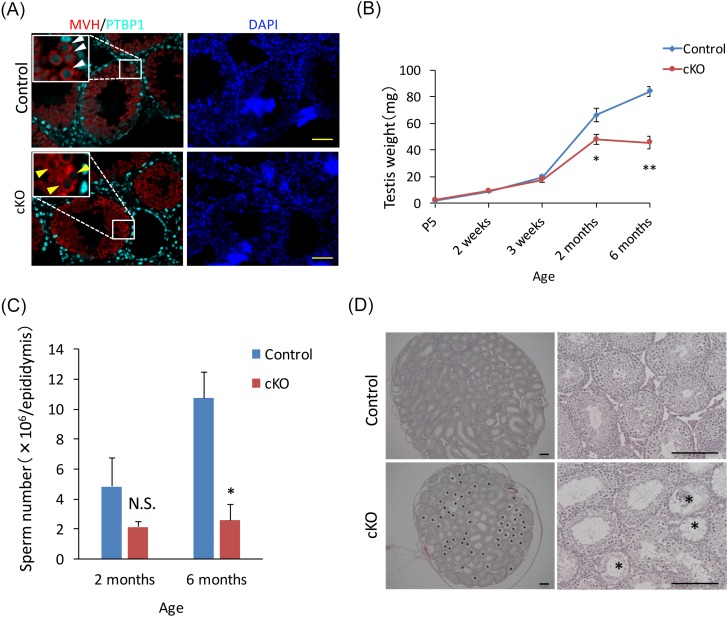
Our previous study [28] and another study [29] revealed that the conventional Ptbp1 KO mouse exhibits embryonic lethality soon after implantation. To escape this lethal phenotype, we developed a germ cell–specific Ptbp1 cKO mouse to explore the role of PTBP1 in spermatogenesis. First, we checked the gene KO efficiency of our Ptbp1 cKO model by immunohistochemistry. The results clearly showed that PTBP1 expression was lost only in MVH-positive germ cells in the Ptbp1 cKO testis at high efficiency (Fig. 2A, arrowhead). Second, we compared the growth of the testis during postnatal development. Testis weight was comparable between control and cKO mice until 3 weeks of age, but the cKO testis weight was significantly lighter than that of the control at 2 months old (47.9 ± 3.8 and 66.2 ± 5.2 mg, respectively; P < 0.05) and 6 months old (45.5 ± 4.6 and 83.9 ± 3.8 mg, respectively; P < 0.01) (Fig. 2B). Although sperm counts from the cauda epididymis of 2-month-old mice did not differ significantly between the genotypes, this count was significantly lower in 6-month-old cKO mice than in age-matched controls (0.26 ± 0.10 × 107 and 1.1 ± 0.2 × 107, respectively; [) (Fig. 2C). Finally, we compared the histology of the testis between control and Ptbp1 cKO mice. HE staining of testes from 2-month-old Ptbp1 cKO mice exhibited an increase in seminiferous tubules undergoing degeneration of spermatogenesis compared with the control testes (Fig. 2D, asterisk). These results suggest that PTBP1 plays an important role(s) in spermatogenesis, although it is not a necessary protein for spermatogenesis.
Fig. 2.
Loss of PTBP1 causes an increase in seminiferous tubules showing degeneration of spermatogenesis. (A) Immunohistochemical analysis of PTBP1 expression in control (top) and the Ptbp1 conditional knockout (cKO) (bottom). Sections were stained with anti-MVH (mouse VASA homolog, a marker of germ cells) and anti-PTBP1. The white arrowhead indicates germ cells expressing PTBP1 in the control mouse, and the yellow arrowhead indicates germ cells without PTBP1 expression in the Ptbp1 cKO mouse. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Scale bar denotes 50 µm. (B) Fluctuation of the weight of testis during development in Ptbp1 cKO and control mice. The asterisk depicts a significant difference (* P < 0.05, ** P < 0.01; n ≥ 3). (C) Sperm counts from cauda epididymis of a 2- or 6-month-old Ptbp1 cKO mouse and control. The asterisk depicts a significant difference (P < 0.05; n ≥ 3). N.S., not significant. (D) Hematoxylin and eosin staining of testis from a 2-month-old Ptbp1 cKO mouse (bottom) and age-matched control (top). The asterisk depicts seminiferous tubules exhibiting degeneration of spermatogenesis. Scale bar denotes 200 µm.
Loss of PTBP1 reduces proliferation of spermatogonia
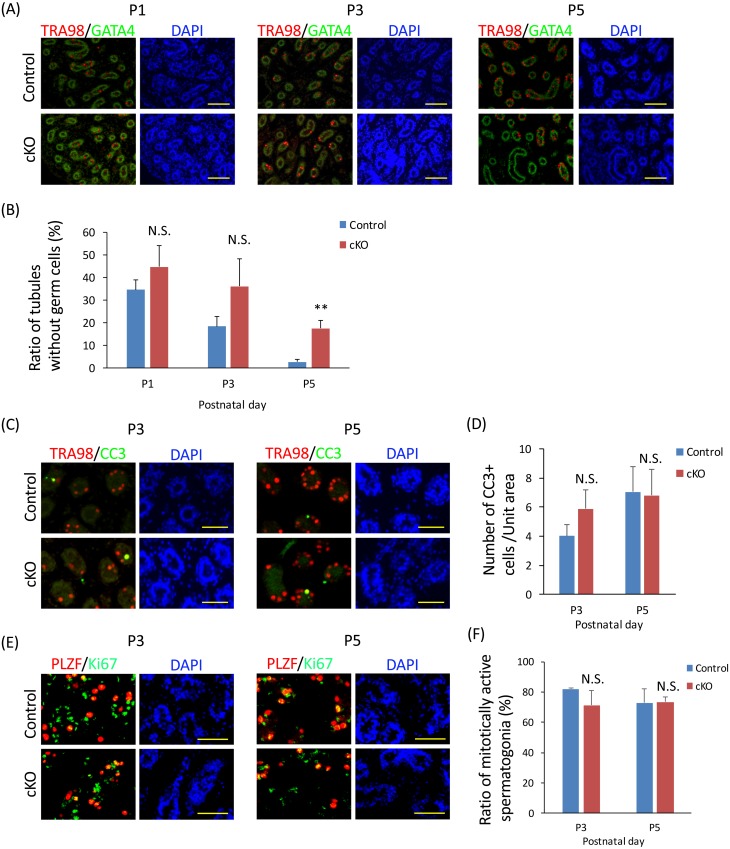
To determine how the loss of PTBP1 causes defects in spermatogenesis, testes were examined using immunohistochemistry methods. During the neonatal period, the number of seminiferous tubules without germ cells gradually decreased in the control, whereas this number remained high in the Ptbp1 cKO mice (Fig. 3A); the difference was significant at postnatal day 5 (Fig. 3B). We also checked progression of first spermatogenesis by immunohistochemistry. The result revealed that ratio of tubules having TRA54-positive haploid spermatid in the Ptbp1 cKO did not differ compared to the control (Supplementary Fig. 1: online only), suggesting that progression of spermatogenesis in the Ptbp1 cKO was, at least until this point, comparable to the control. We next checked whether this retarded expansion of spermatogonia observed in the neonate Ptbp1 cKO was attributed to an increase of cell death or reduced cell proliferation. Interestingly, neither number of CC3-positive apoptotic cells (Fig. 3C and 3D) nor ratio of Ki67-positive cycling cells (Fig. 3E and 3F) differed between the control and the Ptbp1 cKO. Since anti-Ki67 stains cells which enter cell cycle at any phases, these results suggest that retarded expansion of spermatogonia shown in the Ptbp1 cKO neonate was attributed to prolonged cell cycle and slower proliferation of spermatogonia but not an increase of apoptotic cell death.
Fig. 3.
Loss of Ptbp1 reduces proliferation of spermatogonia during neonatal period. (A) Immunohistochemical analysis of germ cell distribution in control (top) and a Ptbp1 conditional knockout (cKO) (bottom) mouse during the neonatal period. Sections were stained with anti-TRA98 (a marker of germ cells) and anti-GATA4 (a marker of the Sertoli cells and Leydig cells). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Scale bar denotes 100 µm. (B) Ratio of tubules without germ cells. The asterisk depicts a significant difference (** P < 0.01; n ≥ 3). (C) Immunohistochemical analysis of testis sections from neonatal Ptbp1 cKO and control for TRA98 and cleaved caspase-3 (CC3). Nuclei were stained with DAPI. Scale bar denotes 50 µm. (D) Number of CC3-positive apoptotic cells (P > 0.05; n ≥ 3). (E) Immunohistochemical analysis of testis sections from neonatal Ptbp1 cKO and control for PLZF and Ki67. Nuclei were stained with DAPI. Scale bar denotes 50 µm. (F) Ratio of mitotically active spermatogonia (P > 0.05; n = 3).
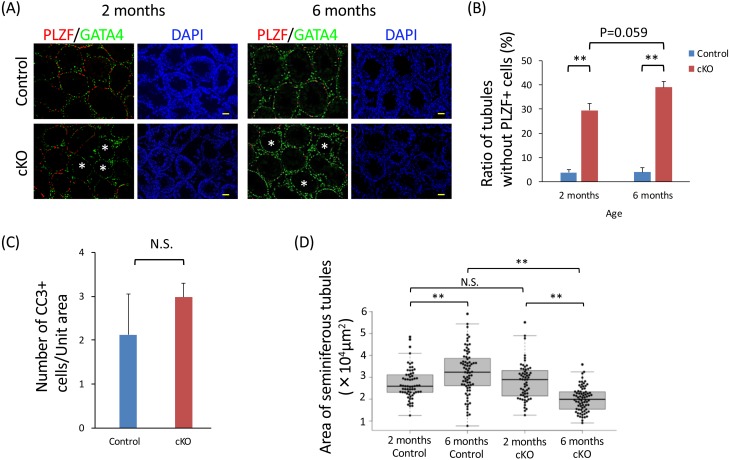
Because the difference in sperm number and testis weight between the Ptbp1 cKO and control mice became more evident in the 6-month-old mice compared with the younger 2-month-old mice, we next analyzed adult testes from 2- and 6-month-old mice by immunohistochemistry methods. The results revealed that seminiferous tubules without PLZF-positive spermatogonia were significantly more abundant in the Ptbp1 cKO mice than in the control mice, both at 2 and 6 months of age (Fig. 4A, asterisk), and the ratio tended to become larger (P = 0.059) in the Ptbp1 cKO mice with age (Fig. 4B). On the other hand, number of apoptotic cells in the Ptbp1 cKO at 6 months of age did not differ compared to the age matched control (Fig. 4C). Furthermore, the area of seminiferous tubules, representing the activity level of spermatogenesis [39,40,41], became larger in the 6-month-old testes than in the 2-month-old testes in the control mice, but became smaller in the Ptbp1 cKO mice with age (Fig. 4D). Taken together, these results suggest that PTBP1 assists proliferation of spermatogonia during both neonatal and adult periods.
Fig. 4.
Loss of Ptbp1 reduces proliferation of spermatogonia during adulthood. (A) Immunohistochemical analysis of testis from 2- and 6-month-old Ptbp1 cKO (bottom) mice and age-matched controls (top). Sections were stained with anti-PLZF (a marker of spermatogonia) and anti-GATA4 (a marker of the Sertoli cells and Leydig cells). Nuclei were stained with DAPI. Scale bar denotes 50 µm. (B) Ratio of tubules without PLZF-positive spermatogonia. The asterisk depicts a significant difference (** P < 0.01; n = 3). (C) Number of CC3-positive apoptotic cells in the testis from 6-month-old Ptbp1 cKO mice and age-matched controls (P > 0.05; n = 3). (D) Area of seminiferous tubules from 2- and 6-month-old Ptbp1 cKO mice and age-matched controls. The asterisk depicts a significant difference (data were collected from three individual mice in each genotype and age; ** P < 0.01; n = 3).
Induced KO of Ptbp1 from GS cells exhibits severe growth arrest and an increase in apoptotic cell death in vitro
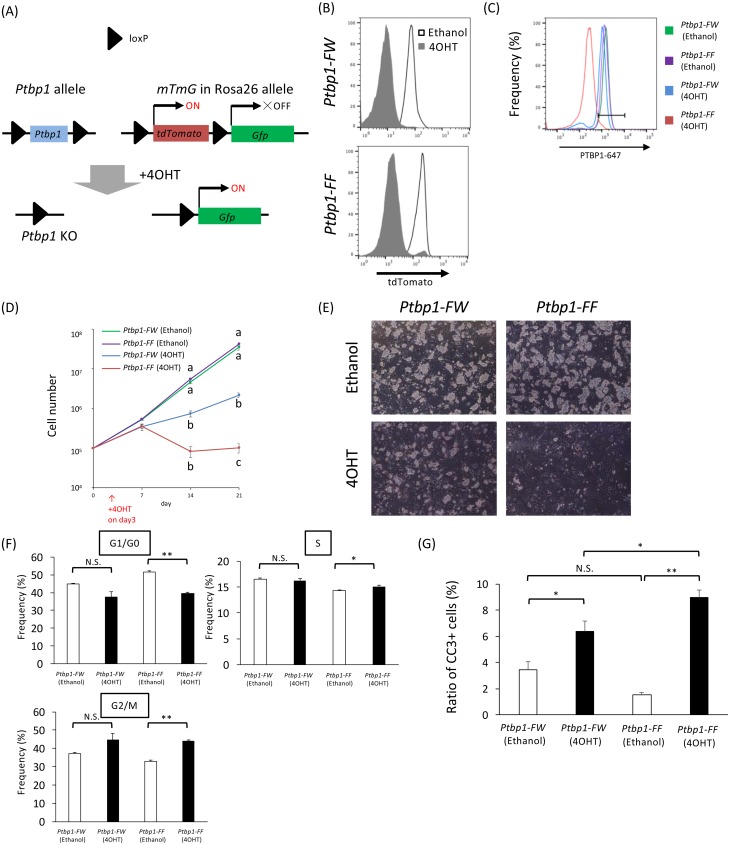
Undifferentiated spermatogonia can proliferate on mouse embryonic fibroblasts in vitro under stimulation with fibroblast growth factor 2 and glial cell line–derived neurotrophic factor, while retaining spermatogonial characteristics [34, 36, 42]. To evaluate the role of PTBP1 in spermatogonia in vitro, we developed GS cells from a mouse having mTmG, CAG-CreMER, and Ptbp1 flox transgenic loci, in which Cre-mediated loxP recombination is induced by 4OHT (Fig. 5A). Administration of 4OHT to the culture medium effectively recombined the mTmG locus (Fig. 5B), and PTBP1 protein clearly disappeared only in the 4OHT-administered Ptbp1-FF GS cells (Fig. 5C). Using this culture model, we next determined the proliferation activity of GS cells. The results clearly showed that proliferation was severely suppressed in the Ptbp1-FF GS cells treated with 4OHT, although 4OHT-administered Ptbp1-FW also showed significant reduction of cell proliferation compared to the placebo (ethanol)-administered Ptbp1-FW or Ptbp1-FF (Fig. 5D and 5E). Next, we checked cell cycle of GS cells by flow cytometry. Ratio of GS cells at G1/G0, S, or G2/M phase did not differ significantly between ethanol- and 4OHT-administered Ptbp1-FW, whereas ratio of GS cells at G2/M phase became significantly more, and at G1/G0 phase became significantly less in the 4OHT-administered Ptbp1-FF GS cells compared to the ethanol-administered Ptbp1-FF GS cells (Fig. 5F). In addition, the number of CC3–positive GS cells, which underwent apoptotic cell death, was significantly (P < 0.05) more in the Ptbp1-FF GS cells than in the Ptbp1-FW GS cells after 4OHT administration (Fig. 5G). These results suggest that PTBP1 contributes to cell cycle regulation and suppresses apoptotic cell death in spermatogonia in vitro.
Fig. 5.
Ptbp1 cKO germline stem (GS) cells exhibit severe growth arrest and an increase in apoptotic cell death in vitro. (A) Schematic of allele recombination mediated by the Cre-loxP system. (B) Flow cytometry to analyze tdTomato-positive GS cells in the Ptbp1-FW (top) and Ptbp1-FF (bottom). 4-Hydroxytamoxifen (4OHT) (induced recombination) or ethanol (placebo) was added to the culture medium. (C) Flow cytometry to analyze PTBP1 expression in Ptbp1-FW GS cells and Ptbp1-FF GS cells treated with 4OHT or ethanol. (D) Proliferation of Ptbp1-FW GS cells and Ptbp1-FF GS cells treated with 4OHT or ethanol. Different superscripts depict significant differences within same day (** P < 0.01; n = 3). (E) Representative morphology of Ptbp1-FW GS cells and Ptbp1-FF GS cells treated with 4OHT or ethanol. (F) Percentages of cells in G0/G1, S, and G2/M phases. The asterisk depicts a significant difference (* P < 0.05, ** P < 0.01; n = 3). (G) Ratio of CC3 positive GS cells. Ptbp1-FW GS cells and Ptbp1-FF GS cells were treated with 4OHT or ethanol, fixed, stained with an anti-CC3 antibody, and analyzed by flow cytometry. The asterisk depicts a significant difference (* P < 0.05, ** P < 0.01; n ≥ 4). Ptbp1-FW, mTmG; CAG-CreMER; Ptbp1flox/wt; Ptbp1-FF, mTmG; CAG-CreMER; Ptbp1flox/flox.
Discussion
We investigated the role of PTBP1 during spermatogenesis by using a germ cell–specific gene KO model for escaping its complete lethality in conventional KO mice [28]. Our results demonstrated that loss of PTBP1 from testicular germ cells caused proliferation arrest of early neonatal spermatogonia as well as an increase in seminiferous tubules exhibiting degeneration of spermatogenesis in the postpubertal male. In addition, the cross-sectional area of seminiferous tubules, an indicator of spermatogenesis activity [39,40,41], became larger in the control mice, whereas significantly smaller in the Ptbp1 cKO mice with age (2 months old vs. 6 months old). An in vitro GS cell culture model showed that loss of PTBP1 caused severe growth arrest accompanied by an increase in apoptotic cell death. Together, these data indicate that PTBP1 supports spermatogenesis through regulation of cell proliferation activity in spermatogonia.
PTBPs are highly conserved RBPs among species. PTBP1 controls cellular differentiation or specification, especially during neural cell development [13, 26, 43]. For example, PTBP1 governs transition of neural progenitor cells to neurons by regulating AS of neuron-specific exons [13]. During germ cell development, loss of function of the germ cell–specific isoform of PTBP homolog dmPTB in Drosophila results in severe disruption of cones of the spermatid individualization complex and causes male-only infertility [21]. Failure of spermatogenesis caused by lack of PTBPs is also observed in the mouse. Germ cell–specific KO of Ptbp2, one of the PTBP orthologs in mice, shows comparable activity in spermatogonial mitosis, or entry into meiosis, where it causes loss of mature spermatozoa due to severe arrest of spermatid elongation, and results in a massive increase in multinucleated giant cell formation in seminiferous tubules [23]. The present study also revealed abnormal spermatogenesis caused by lack of PTBP1, although the abnormal phenotype was mainly observed in spermatogonial proliferation, not in spermatid maturation as shown in the Ptbp2 cKO in mice. PTBP1 is strongly expressed by spermatogonia and becomes less evident in the differentiating spermatocytes or spermatids (Fig. 1B and 1C). By contrast, PTBP2 expression is weak in spermatogonia but becomes stronger at the onset of meiosis entry, maintaining this high level until the round spermatid stage [15]. The different phenotypes between Ptbp1 cKO and Ptbp2 cKO mice may be attributed to the different spatiotemporal expression patterns of PTBP1 and PTBP2 in the testis. Another possibility is that, although Ptbp1 and Ptbp2 are orthologous genes, and show binding to similar exons, a certain amount of AS is regulated differentially by PTBP1 and PTBP2, as observed in neural cells [12]. Therefore, target RNAs of PTBP1 or PTBP2 in the testicular germ cells may be distinct, at least to some extent, and result in a different phenotype in the KO models.
Mitotically inactive gonocytes begin proliferation soon after birth and form a population of spermatogonia [1, 2]. This study showed that seminiferous tubules without germ cells became fewer according to neonatal development in the control, whereas they remained high in the Ptbp1 cKO mice (Fig. 3B), suggesting that proliferation might be downregulated as a result of the loss of PTBP1. Cell proliferation arrest was also observed in vitro (Fig. 5D). It would note that 4OHT-administered Ptbp1-FW also showed retarded proliferation compared to the ethanol-administered Ptbp1-FF or Ptbp1-FW. It is unclear whether this deleterious effect was attributed to toxicity of 4OHT or heterogeneous deletion of Ptbp1. On the other hand, 4OHT-administerd Ptbp1-FF GS cells represented far severe phenotype than 4OHT-administerd Ptbp1-FW GS cells , suggesting that PTBP1 contributes to spermatogonial cell proliferation. In addition, a previous study [29] revealed that 5-bromo-2[prime]-deoxyuridine–incorporating activity, which represents DNA synthesis and cell proliferation, is significantly lower in Ptbp1 null embryos. These results indicated that PTBP1 positively controls cell proliferation in many types of cells. By contrast, we could not detect any significant changes of Ki67-positive cycling cell numbers in spermatogonia in vivo between Ptbp1 cKO mice and controls either during neonatal stage (Fig. 3E and 3F). Ki67 is a common marker for cell proliferation and is able to distinguish a group of cells in G0 phase (Ki67 negative) or in G1/S/G2/M phase (Ki67 positive). By contrast, Ki67 staining does not necessarily relate to the speed of cell proliferation. Indeed, embryonic stem cells lacking PTBP1 had a slower proliferation speed than cells with PTBP1; this retarded proliferation was caused by an increase in M-phase arrest, but not of G0-phase arrest [28]. We also observed the similar phenomena that ratio of G2/M phase became more in the Ptbp1-FF GS cells after 4OHT administration. In addition, our in vivo study showed that the area of seminiferous tubules became smaller in the Ptbp1 cKO mice according to age, whereas this area increased in the controls (Fig. 4D), suggesting a lower activity of spermatogenesis in the Ptbp1 cKO mice. Therefore, it is possible that PTBP1 controls spermatogonial cell proliferation via regulating the speed of cell division. Further study to investigate the speed of spermatogonial proliferation in the Ptbp1 cKO mice would be useful, perhaps by using 5-bromo-2’-deoxyuridine incorporation labeling or CreERT/loxP-mediated pulse reporter labeling followed by lineage tracing.
Loss of PTBP1 in cultured GS cells causes an increase in apoptotic cell death in GS cells in vitro (Fig. 5G); however, immunohistochemistry did not show a significant increase of cleaved caspase-3 (CC3)–positive cells in vivo either during neonatal or adult stages (Fig. 3D and 4C). Several studies reported opposing results of PTBP1-mediated induction of apoptosis. For example, knockdown of PTBP1 in human colon cancer cell lines resulted in an increase in apoptosis [44]. The conventional Ptbp1 KO mouse also showed an increase in apoptotic cell death in the fetus soon after implantation (embryonic day 7.5) [29]. By contrast, drug-induced apoptosis occurs more frequently in the PTBP1-high A127 cell line compared with the PTBP1-low LN18 cell line, cell lines from human glioblastoma [45], and knockdown of PTBP1 in the human chronic myeloid leukemia K562 cell line strengthens tolerance against drug-induced apoptosis [46]. Therefore, apoptotic response after Ptbp1 manipulation might vary among types of cells or tissues.
The next step is to identify PTBP1 target RNAs and their networks to regulate homeostasis of spermatogonia. Thus, further study is needed via RNA immunoprecipitation followed by sequencing, or RNA sequencing, to clarify PTBP1 target RNA, transcriptome, or AS changes in spermatogonia for mining detailed molecular functions regulated by PTBP1.
Supplementary
Acknowledgments
Authors thank Dr Shosei Yoshida for providing a Ngn3-Cre mouse, and Dr Hirotake Ichise, Dr Taeko Ichise, Dr Takahiko Chimura, Dr Hiroki Sasanuma, and Dr Yasuhiro Yamada for their valuable advice and discussions for carrying out the research. This work was supported in part by KAKENHI grant (17K07132) from the Ministry of Education, Culture, Sports, and Technology (MEXT), Japan to MO, and KAKENHI grants (JP17H01394 and JP 25112007), Japan Agency for Medical Research and Development grant (JP18gm5010001), Takeda Science Foundation, and The Tokyo Biochemical Research Foundation to MI.
References
- 1.Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res C Embryo Today 2009; 87: 1–26. [DOI] [PubMed] [Google Scholar]
- 2.Pui HP, Saga Y. NANOS2 acts as an intrinsic regulator of gonocytes-to-spermatogonia transition in the murine testes. Mech Dev 2018; 149: 27–40. [DOI] [PubMed] [Google Scholar]
- 3.McCarrey JR. Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biol Reprod 2013; 89: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 2010; 328: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hara K, Nakagawa T, Enomoto H, Suzuki M, Yamamoto M, Simons BD, Yoshida S. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 2014; 14: 658–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21: 776–798. [PubMed] [Google Scholar]
- 7.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 2008; 40: 225–231. [DOI] [PubMed] [Google Scholar]
- 8.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- 9.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 2012; 338: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Zheng S, Han A, Lin CH, Stoilov P, Fu XD, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife 2014; 3: e01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuong JK, Lin CH, Zhang M, Chen L, Black DL, Zheng S. PTBP1 and PTBP2 Serve Both Specific and Redundant Functions in Neuronal Pre-mRNA Splicing. Cell Reports 2016; 17: 2766–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, Black DL, Kharchenko PV, Sharp PA, Walsh CA. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 2016; 166: 1147–1162.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan Z, Garrett-Engele PW, Johnson JM, Castle JC. Evolutionarily conserved and diverged alternative splicing events show different expression and functional profiles. Nucleic Acids Res 2005; 33: 5659–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid R, Grellscheid SN, Ehrmann I, Dalgliesh C, Danilenko M, Paronetto MP, Pedrotti S, Grellscheid D, Dixon RJ, Sette C, Eperon IC, Elliott DJ. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res 2013; 41: 10170–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, Dym M, de Massy B, Mikkelsen TS, Kaessmann H. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Reports 2013; 3: 2179–2190. [DOI] [PubMed] [Google Scholar]
- 17.Margolin G, Khil PP, Kim J, Bellani MA, Camerini-Otero RD. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 2014; 15: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Reports 2013; 5: 357–366. [DOI] [PubMed] [Google Scholar]
- 19.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans 2008; 36: 641–647. [DOI] [PubMed] [Google Scholar]
- 20.Robida MD, Singh R. Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J 2003; 22: 2924–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robida M, Sridharan V, Morgan S, Rao T, Singh R. Drosophila polypyrimidine tract-binding protein is necessary for spermatid individualization. Proc Natl Acad Sci USA 2010; 107: 12570–12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu M, Hecht NB. Polypyrimidine tract-binding protein 2 binds to selective, intronic messenger RNA and microRNA targets in the mouse testis. Biol Reprod 2011; 84: 435–439. [DOI] [PubMed] [Google Scholar]
- 23.Zagore LL, Grabinski SE, Sweet TJ, Hannigan MM, Sramkoski RM, Li Q, Licatalosi DD. RNA binding protein Ptbp2 is essential for male germ cell development. Mol Cell Biol 2015; 35: 4030–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannigan MM, Zagore LL, Licatalosi DD. Ptbp2 controls an alternative splicing network required for cell communication during spermatogenesis. Cell Reports 2017; 19: 2598–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Wang L, Welch JD, Ma H, Zhou Y, Vaseghi HR, Yu S, Wall JB, Alimohamadi S, Zheng M, Yin C, Shen W, Prins JF, Liu J, Qian L. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature 2017; 551: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibasaki T, Tokunaga A, Sakamoto R, Sagara H, Noguchi S, Sasaoka T, Yoshida N. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cereb Cortex 2013; 23: 1824–1835. [DOI] [PubMed] [Google Scholar]
- 27.Monzón-Casanova E, Screen M, Díaz-Muñoz MD, Coulson RMR, Bell SE, Lamers G, Solimena M, Smith CWJ, Turner M. The RNA-binding protein PTBP1 is necessary for B cell selection in germinal centers. Nat Immunol 2018; 19: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibayama M, Ohno S, Osaka T, Sakamoto R, Tokunaga A, Nakatake Y, Sato M, Yoshida N. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS J 2009; 276: 6658–6668. [DOI] [PubMed] [Google Scholar]
- 29.Suckale J, Wendling O, Masjkur J, Jäger M, Münster C, Anastassiadis K, Stewart AF, Solimena M. PTBP1 is required for embryonic development before gastrulation. PLoS One 2011; 6: e16992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, Suda T, Nabeshima Y. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol 2004; 269: 447–458. [DOI] [PubMed] [Google Scholar]
- 31.Ichise H, Hori A, Shiozawa S, Kondo S, Kanegae Y, Saito I, Ichise T, Yoshida N. Establishment of a tamoxifen-inducible Cre-driver mouse strain for widespread and temporal genetic modification in adult mice. Exp Anim 2016; 65: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erben T, Ossig R, Naim HY, Schnekenburger J. What to do with high autofluorescence background in pancreatic tissues - an efficient Sudan black B quenching method for specific immunofluorescence labelling. Histopathology 2016; 69: 406–422. [DOI] [PubMed] [Google Scholar]
- 33.Kajimura J, Ito R, Manley NR, Hale LP. Optimization of single- and dual-color immunofluorescence protocols for formalin-fixed, paraffin-embedded archival tissues. J Histochem Cytochem 2016; 64: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003; 69: 612–616. [DOI] [PubMed] [Google Scholar]
- 35.Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 2009; 136: 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 2010; 142: 468–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozawa M, Fukuda T, Sakamoto R, Honda H, Yoshida N. The histone demethylase FBXL10 regulates the proliferation of spermatogonia and ensures long-term sustainable spermatogenesis in mice. Biol Reprod 2016; 94: 92. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 39.Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 1999; 99: 649–659. [DOI] [PubMed] [Google Scholar]
- 40.Wu X, Peppi M, Vengalil MJ, Maheras KJ, Southwood CM, Bradley M, Gow A. Transgene-mediated rescue of spermatogenesis in Cldn11-null mice. Biol Reprod 2012; 86: 139–: 1–11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Y, Zhang L, Bai Y, Zhou HM, Campbell AM, Chen H, Yong W, Zhang W, Zeng Q, Shou W, Zhang ZY. Phosphatase of regenerating liver 2 (PRL2) deficiency impairs Kit signaling and spermatogenesis. J Biol Chem 2014; 289: 3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2004; 101: 16489–16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife 2016; 5: e11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniguchi K, Sugito N, Kumazaki M, Shinohara H, Yamada N, Nakagawa Y, Ito Y, Otsuki Y, Uno B, Uchiyama K, Akao Y. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett 2015; 363: 17–27. [DOI] [PubMed] [Google Scholar]
- 45.Bielli P, Bordi M, Di Biasio V, Sette C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res 2014; 42: 12070–12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juan WC, Roca X, Ong ST. Identification of cis-acting elements and splicing factors involved in the regulation of BIM Pre-mRNA splicing. PLoS One 2014; 9: e95210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.