Recently, we received the letter to the editor [1] in response to our recently published paper titled “Systematic Review and Meta-Analysis of Diagnostic Accuracy of miRNAs in Patients with Pancreatic Cancer [2].” We appreciate the interest and comments from Jayaraj et al. regarding our work. However, there are some issues in the comments that need to be addressed.
Regarding the differences between this study and previous publications [3–5], we wish to elaborate on the following: (1) 109 studies are included in this meta-analysis, which was more than the 9 studies in Wan et al. [3], the 52 studies in Ding et al. [4], and the 36 studies in Pei et al. [5]; (2) More subgroup analyses than in those articles [3–5] were performed, including race, source of control, miRNA profiling, and the combination of miR-21. In particular, subgroup analyses by the combination of miR-21 were not involved in the previous three articles. We found that miR-21s as biomarkers for the early diagnosis of PaC were more valuable than other miRNAs; (3) Our review conducted metaregression analyses to explore potential sources of heterogeneity and to confirm the results of subgroup analyses, but Wan et al. [3] and Pei et al. [5] did not. A large sample and more suitable statistical analysis method allow our study to provide more accurate and reliable information for future studies.
The diagnostic cut-off point plays an important role in disease diagnosis. There is not a singular diagnostic cut-off point for different miRNA profiling and source of miRNA. We originally prepared our subgroup analysis and sensitivity analysis based on different miRNA profiling and source of miRNA, but half of the included studies did not report the testing threshold. Therefore, such subgroup analysis and sensitivity analysis cannot be performed, which may have an impact on the results. It was recommended that diagnostic test articles should report thresholds to provide raw information for future meta-analysis.
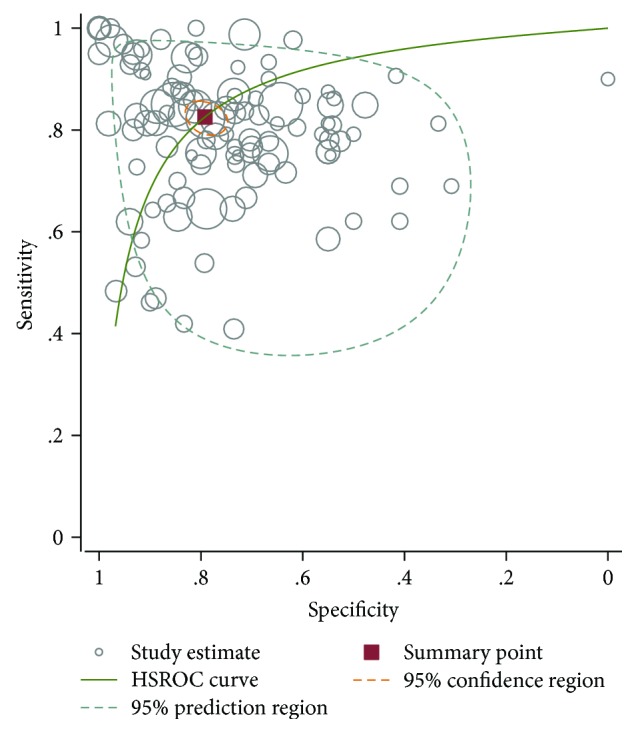
The analysis of diagnostic threshold was performed in our meta-analysis. Spearman's correlation coefficient was 0.147 (P = 0.127), suggesting that there is no threshold effect. Then an I-square parameter was adopted to estimate the heterogeneity between the studies. However, we agree with Jayaraj et al.'s suggestion that a Tau-squared statistical parameter might be suitable for being the estimated variation of heterogeneity between the effects for test accuracy observed in different studies. For the meta-analysis of diagnostic test accuracy (DTA), the hierarchical summary receiver operating characteristic (HSROC) allows both the existence of threshold effects and the heterogeneity between studies. We subsequently conducted HSROC and found that the pooled SEN was 0.82 (95% CI, 0.79–0.85) and the pooled SPE was 0.79 (95% CI, 0.75–0.82) (Figure 1) and basically consistent with the results of the original study, which also shows that our results are reliable and credible.
Figure 1.
The Cochrane Handbook for Systematic Reviews explicitly mentions not to use methods like the Begg or Egger tests for publication bias of DTA and argues that it is best to use the test proposed by Deeks [6]. Applying such tests for funnel plot asymmetry in systematic reviews of DTA may often cause publication bias being incorrect [7]. Deeks' tests should be preferred to assess publication bias in DTA meta-analyses [8].
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Jayaraj R., Kumarasamy C., Royam M. M., et al. Comment on “Systematic review and meta-analysis of diagnostic accuracy of miRNAs in patients with pancreatic cancer”. Disease Markers. 2018;2018:2. doi: 10.1155/2018/6904569.6904569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun X., Zhou X. Systematic review and meta-analysis of diagnostic accuracy of miRNAs in patients with pancreatic cancer. Disease Markers. 2018;2018:13. doi: 10.1155/2018/6292396.6292396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan C., Shen Y., Yang T., Wang T., Chen L., Wen F. Diagnostic value of microRNA for pancreatic cancer: a meta-analysis. Archives of Medical Science. 2012;5(5):749–755. doi: 10.5114/aoms.2012.31609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Z., Wu H., Zhang J., Huang G., Ji D. MicroRNAs as novel biomarkers for pancreatic cancer diagnosis: a meta-analysis based on 18 articles. Tumor Biology. 2014;35(9):8837–8848. doi: 10.1007/s13277-014-2133-4. [DOI] [PubMed] [Google Scholar]
- 5.Pei Z., Liu S. M., Huang J. T., et al. Clinically relevant circulating microRNA profiling studies in pancreatic cancer using meta-analysis. Oncotarget. 2017;8(14):22616–22624. doi: 10.18632/oncotarget.15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macaskill P., Gatsonis C., Deeks J. J., Harbord R. M., Takwoingi Y. Chapter 10: Analysing and Presenting Results. The Cochrane Collaboration; 2010. Cochrane handbook for systematic reviews of diagnostic test accuracy; pp. 1–61. [Google Scholar]
- 7.Deeks J. J., Macaskill P., Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of Clinical Epidemiology. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 8.van Enst W. A., Ochodo E., Scholten R. J. P. M., Hooft L., Leeflang M. M. Investigation of publication bias in meta-analyses of diagnostic test accuracy: a meta-epidemiological study. BMC Medical Research Methodology. 2014;14(1):p. 70. doi: 10.1186/1471-2288-14-70. [DOI] [PMC free article] [PubMed] [Google Scholar]


