Abstract
As a novel IGF system member, igf3 plays an important role in gonadal development of teleost fish. Although studies have reported the unusual expression of igf3 in fish gonad, whether the igf3 affects the expression of long noncoding RNAs (lncRNAs) in gonad remains unknown. In this study, an igf3 knockdown common carp (Cyprinus carpio) model was established by RNA interference. Then RNA sequencing of C. carpio gonad after igf3 knockdown was performed. A total of 327,169,410 and 306,305,018 clean reads were identified from control and igf3-dsRNA interference group, respectively. After a stringent filtering, RNA-seq yielded 14199 lncRNA and 106932 mRNA transcripts with 124 and 353 differentially expressed lncRNAs and mRNAs. Our dataset provides an extensive resource for understanding the potential regulatory molecular mechanism of igf3 in early stage of gonadal development in C. carpio.
Subject terms: RNA sequencing, RNAi, Long non-coding RNAs
Background & Summary
Insulin-like growth factors (IGFs) are key factors that regulate the growth and reproduction axis. As a new ligand of IGFs, igf3 plays an important role in the development and maturation of the gonads in fish 1–3 , especially in the maturation of oocytes and the process of spermatogenesis 4,5 . We reported previously that the igf3 mRNA of common carp (Cyprinus carpio) was highly expressed in the gonads and blood, and Igf3 protein was localized in the ovary granulosa cells and testis spermatogonium and spermatids 3 . In addition, igf3 also serves as a mediator of luteinizing hormone (LH) action in zebrafish (Danio rerio) ovulation 6 , and stimulate spermatogonial differentiation by activating β-catenin signaling or through some estrogens 7,8 . To better understand the regulatory pathway of igf3 in gonadal development of fish, we are interested in the transcriptional regulation of lncRNAs and mRNAs expression profiles following igf3 knockdown in common carp.
The molecular mechanisms of gonadal development are complex processes that involve in many germ cell-specific gene products in testis or ovary that undergo strict developmental regulations 9,10 . LncRNAs are a novel class of RNAs with the length over 200 bp and without the potential of protein-coding 11 . By now, there are tens of thousands of lncRNAs have been discovered across human, mouse, nematode, zebrafish etc. 12 . For human, 58,648 of genes were classified as lncRNAs 13 . The existing studies show that lncRNAs play a key role in epigenetic, transcriptional and post-transcriptional control of gene expression and maintenance of stem cell, signal transduction, cell proliferation and differentiation, metabolism, and individual development 14–17 . In testis, lncRNAs affect germ cell development and spermatogenesis, such as tsx, mrh1, dmr and hongES2 18–21 . In ovary, many lncRNAs that regulate ovarian development have been identified 22–25 . LncRNA A-30-P01019163 may affect ovarian cell cycle and proliferation by regulating p2rx7 expression in the ovary 26 . All of these observations indicate that lncRNAs play important roles in gonadal development.
RNA interference technology is an effective method in studying the gene function of organisms. In this study, we first established a living model of igf3 knockdown C. carpio via double-stranded RNA (dsRNA) injection, and then a high-throughput RNA sequencing (RNA-seq) was employed to profile the primordial gonad transcriptome of igf3 knockdown and control C. carpio. The aims of the experiment were to discover and characterize lncRNAs in carp gonad tissue and identify key genes, lncRNAs, and pathways that are associated with igf3. This is the first study to describe the expression profiles of lncRNAs and mRNA in carp gonad with igf3 knockdown. Our data will provide a very useful resource for studying the functional role of igf3 in gonadal development.
Methods
Animals
The experimental common carp (Cyprinus carpio) is a new aquaculture variety derived from the original parents of Jian carp (C. carpio var. jian), Huanghe carp (C. carpio haematopterus) and Heilongjiang common carp (C. carpio haermatopterus) after five generations combined selection by FFRC, CAFS. This newly improved strain is designated as FFRC No. 2 strain common carp. It is one of the dominant aquaculture species in China 27 . The C. carpio were obtained from the Qiting Pilot Research Station (Yixing, China), which is affiliated with the FFRC. Juvenile fishes at 8 weeks post hatching (10.92 ± 0.14 g) were maintained in the aquaculture experimental facilities during the acclimation and experimental periods under 12-h light/dark cycles at 28 ± 1 °C, and fed twice daily with compound feed (Tech-bank Co., Ltd., Ningbo, China).
RNA interference (RNAi)
This description of RNA interference is extended from the protocol described in the related research manuscript 28 . The dsRNA of igf3 was synthesized in vitro using TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. The template for igf3-dsRNA synthesis was prepared by amplifying gonad cDNA with the primers igf3 iF and igf3 iR (Table 1 (available online only)). The concentration of dsRNA was measured at 260 nm with a BioPhotometer (Eppendorf, Hamburg, Germany), purity and integrity were examined by 1% agarose gel electrophoresis, and then stored at −20 °C until used.
Table 1. Primer sequences used in this study.
| Gene ID | Primer sequnces (5' → 3') | Product size | Application | ||
|---|---|---|---|---|---|
| igf3 | igf3 iF | F | TAATACGACTCACTATAGGGTGTACTGCGTCCTGATCCTG | 482 bp | RNA interference |
| igf3 iR | R | TAATACGACTCACTATAGGGGAAGGTTGCTGCTGTGTTGA | |||
| igf3 | F | AGCAGCGATACCAGAAGCAT | 91 bp | qRT-PCR | |
| igf3 | R | GAGTTCCACCGGTAAAGCGT | |||
| lncRNA | MSTRG.42108 | F | GCTCCAGCTTGATCCTCATC | 81 bp | |
| R | GAGACGAGCTGAAGCCTTTC | ||||
| MSTRG.13209 | F | CATTCTCCATGTAGGGCTCC | 143 bp | ||
| R | ACTGGAGGAGCTGAGAACCA | ||||
| MSTRG.10998 | F | ACCTTTAAACAAACGGCACG | 150 bp | ||
| R | CCAACTGGGATCCTGAAGAA | ||||
| MSTRG.43997 | F | GACACCTTGGGAGGAACTGA | 198 bp | ||
| R | CTTTGCTTTTCACCTCAGGG | ||||
| MSTRG.7759 | F | GCAAGCCATCTCTCGACTCT | 95 bp | ||
| R | AGCAGCATCTCCTTCAGCTC | ||||
| MSTRG.43046 | F | AGGAGTGAAGTCCTGAGGCA | 148 bp | ||
| R | GATCCCCGTTCACTGTCTGT | ||||
| MSTRG.19541 | F | AGCCGACAATAGAGCGGACT | 166 bp | ||
| R | CAGAATCGCTCCCGTTTGAAG | ||||
| MSTRG.36412 | F | AAGGCCACGAGATGAAGAGA | 127 bp | ||
| R | GCCCAGCAATAAAAGGAACA | ||||
| MSTRG.29920 | F | AAGGGGGAGGTCAACTTGGT | 103 bp | ||
| R | ACAGAGCCTGGAGAGCCATT | ||||
| MSTRG.16585 | F | GTCTTAGGAGGACGGGACGA | 125 bp | ||
| R | CCGTGTCACGTGAGTTCTCC | ||||
| MSTRG.48206 | F | GCCCCTTGTGTTTTTCTCAA | 169 bp | ||
| R | TCATTTTGGAGGGTGGAAAT | ||||
| MSTRG.22976 | F | GCTCCAGTGGACCGTAAAGA | 92 bp | ||
| R | CAGATGGAGGTCCTGGAGAA | ||||
| MSTRG.2504 | F | ACAAGCTTTGGTCTGGACATTT | 99 bp | ||
| R | GCTCTTCTGTTCATTTCGACCT | ||||
| MSTRG.40241 | F | AACGCCATCTTCATTCGTGT | 144 bp | ||
| R | AAAACTCCTGCTGAAGCGAC | ||||
| MSTRG.36182 | F | GCGAAGAAGAACAGTTTGCC | 155 bp | ||
| R | ACAATCCTTCCCGAGTCAGA | ||||
| MSTRG.23436 | F | TTTTGTAACCCCCAGCTTTG | 128 bp | ||
| R | TTTACAGACCCCCTTTCCCT | ||||
| MSTRG.27815 | F | TCAGGGGAAGTGTTGCTTCT | 95 bp | ||
| R | GCTGAGCCGTTTCAGAAAGT | ||||
| MSTRG.7538 | F | AATGGCTGGGTGTGTGTGTA | 181 bp | ||
| R | TGTCGAATATCTGCGGGAAT | ||||
| MSTRG.51187 | F | AGACGGGGGCGAGAATACAA | 120 bp | ||
| R | CACTAATCCTGGCCCTGGGA | ||||
| MSTRG.18820 | F | GGAGACAGCAGGGAACACAT | 178 bp | ||
| R | GTCCGTGTTTTGCTCCTCAT | ||||
| mRNA | CAFS_CommonCarp_T_00220460 | F | CCCGTGTGTTTCATTGCTGA | 200 bp | |
| R | GTGCAGGCAGGATCATTGAG | ||||
| CAFS_CommonCarp_T_00106217 | F | TGCTATCGAGTGGCAAACTG | 221 bp | ||
| R | TTAAGGGGAGGAGAGCCATT | ||||
| CAFS_CommonCarp_T_00276083 | F | CTGGACCAAGAGTGGTGGTT | 137 bp | ||
| R | CACCAAACTCCCTCCTGAAA | ||||
| CAFS_CommonCarp_T_00055524 | F | CTGGGAGGTGGACAAGTCAT | 228 bp | ||
| R | TTCTGGAGGTGGCGTTAGTT | ||||
| CAFS_CommonCarp_T_00253405 | F | ACACCCCACCTCACAGAGAC | 143 bp | ||
| R | GGTCACCAACCTGTTCTCGT | ||||
| CAFS_CommonCarp_T_00128677 | F | AAACCTACACGTGTGGAGCC | 194 bp | ||
| R | CGGTACGTGGTTCCTCTGTT | ||||
| CAFS_CommonCarp_T_00047108 | F | ATTCGGGCGTAGCACAATAC | 81 bp | ||
| R | ATGTCACTTTCTGGGATCGG | ||||
| CAFS_CommonCarp_T_00071301 | F | CTCAGGGGGAAAAACAAACA | 123 bp | ||
| R | AAACTCCGTGCAATACGACC | ||||
| CAFS_CommonCarp_T_00083670 | F | AGACGAAGGAGGGAGGAGAG | 179 bp | ||
| R | CCGAGGATGATCTCACCAGT | ||||
| CAFS_CommonCarp_T_00206701 | F | CTATGACAAAGCCTTCCCCA | 131 bp | ||
| R | GCTGACGTTCTTTTTCGAGG | ||||
| CAFS_CommonCarp_T_00037501 | F | TGCATCAGAGTTTCACTGCC | 117 bp | ||
| R | GACGGACTGACAGCAGAACA | ||||
| CAFS_CommonCarp_T_00190084 | F | CACCGGTCAGAAACCGTACT | 165 bp | ||
| R | ATCCGCTTGTGGATTTTCAG | ||||
| CAFS_CommonCarp_T_00208822 | F | GCAAAAGAGTGTGTGCAGGA | 166 bp | ||
| R | AGAGTTTAAGCGGCTCCACA | ||||
| CAFS_CommonCarp_T_00088409 | F | CAGCATTCTGAAAGGAAGGC | 167 bp | ||
| R | ATCCTGTTTTCGCCAACATC | ||||
| CAFS_CommonCarp_T_00213038 | F | GAGACCCAAAATCAAGCCAA | 125 bp | ||
| R | TACGGTGTGGTGTTGAGCAT | ||||
| CAFS_CommonCarp_T_00239240 | F | CTGAAGCAGGAGTACCAGCC | 87 bp | ||
| R | GTCAGCACCTTTACCCAGGA | ||||
| CAFS_CommonCarp_T_00112860 | F | GCCAAGCACAACACTGCTTA | 152 bp | ||
| R | GGACAACTTGGGGAGTTTGA | ||||
| CAFS_CommonCarp_T_00038343 | F | TCAGAACGTGGTGTCTGAGC | 181 bp | ||
| R | TTGTACGAGGAGCTGCAATG | ||||
| CAFS_CommonCarp_T_00052776 | F | CGAAAACCCAGCAGTTCTGT | 149 bp | ||
| R | TGCTGCCCTCTCGATAACTT | ||||
| CAFS_CommonCarp_T_00128308 | F | GTGATCAGCGCTGTGATGTT | 122 bp | ||
| R | CCTGTCACGAGAATGCAGAA | ||||
| β-actin | β-actin | F | GCTATGTGGCTCTTGACTTCGA | 89 bp | |
| R | CCGTCAGGCAGCTCATAGCT |
Preliminary experiment showed that the optimum interfering effect was observed at the 24th hour after the intraperitoneal injection of igf3-dsRNA with the dose of 5 μg/g body weight. In this experiment, juvenile C. carpio were randomly divided into two groups with 15 fishes per group. The experimental group was injected intraperitoneally of igf3-dsRNA and the control group was injected intraperitoneally of H2O at an equal dose based on body weight. Gonad samples were obtained at 24 h after injection, and 9 fish samples were obtained per group. Gonad samples were snap-frozen in liquid nitrogen and stored at −80 °C until processed. The expression level of igf3 was tested by qRT-PCR, primers used as shown in Table 1 (available online only).
Total RNA isolation and qualification
Total RNA was extracted from gonad tissues using TRIZOL (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, genomic DNA was removed from RNA sample using DNase I (New England Biolabs). The concentration and integrity of RNA was estimated using the NanoDrop 2000 (Thermo Scientific, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), respectively. And only high quality samples with an RNA Integrity Number (RIN) value greater than or equal to 6.8 were used to construct the sequencing library.
Library preparation for lncRNA sequencing
In this study, 3 μg total RNA from each gonad sample was used as the input material for library preparation. Six cDNA sequencing libraries, designated control (control-1, control-2 and control-3) and treatment (igf3-dsRNA-1, igf3-dsRNA-2, and igf3-dsRNA-3), were constructed using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) after removal of ribosomal RNA by the Epicentre Ribo-zero rRNA Removal Kit (Epicentre, Madison, WI, USA). Then, first strand cDNA was synthesized using random hexamer primers and M-MuLV reverse transcriptase; second strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H and reaction buffer, dUTP replaced dTTP in the reaction buffer used in second strand cDNA synthesis. The resulting double-stranded cDNA was ligated to adaptors after being end-repaired and A-tailed. Length of 350–400 bp cDNA fragments were preferred and isolated after purifying using the AMPure XP system (Bechman Coulter, Beverly, USA). PCR amplification was performed to enrich cDNA libraries. Finally, PCR products were purified and library quality was assessed on the Agilent Bioanalyzer 2100 system. The libraries were then sequenced on the Illumina HiSeq X platform, generating 150 bp paired-end reads.
Transcriptome assembly and lncRNA identification
Quality control and reads statistics were determined by FastQC 29 . At the same time, Q20, Q30 and GC contents of the clean reads were calculated, and all the downstream analyses were based on clean reads with high quality. The final clean reads were then mapped to the C. carpio reference genome (version 3.0) using Hisat2 aligner. We used reference annotation file of C. carpio (version 3.0, provided by CAFS, http://www.fishbrowser.org/database/Commoncarp_genome) to guide the transcripts assembly of each sample with StringTie program (v1.3.3b) 30 . Then the output GTF files were merged into a single unified transcript using stringTie merge function. The merged transcripts were compared to the reference annotation using gffcompare program (v0.10.1, https://ccb.jhu.edu/software/stringtie/gffcompare.shtml). To identify potential lncRNAs, transcripts which were longer than 200 bp and belonged to special class code (“j”, at least one splice junction is shared with a reference transcript; “o”, generic exonic overlap with a reference transcript; “i”, a transfrag falling entirely within a reference intron; “u”, unknown, intergenic transcript; “x”, exonic overlap with reference on the opposite strand) were retained. Furthermore, TransDecoder (version rel16JAN2014) was used to identify transcripts with open-reading frame (ORF), while NONCODE and NR database were conducted to discover known lncRNAs and protein-coding transcript homologs. Additionally, we used Coding Potential Calculator (CPC, version 0.9-r2) to evaluate the coding potential of transcripts. Combined with above computational approaches, the predicted novel lncRNAs had to meet the following requirements: 1)transcripts with an FPKM (fragments per kilobase per million mapped reads) value < 0.1 were removed; 2) transcripts which had ORF or be homologous to known lncRNAs transcripts or protein-coding transcripts were discarded; 3) transcripts with CPC score larger than 0 were abandoned. The remained transcripts from the intersection results were considered as non-coding.
Differential expression of lncRNA and mRNA
The TPM (Transcripts Per Million) was used to estimate the expression levels of lncRNAs and mRNAs in every sample. Differential expression analyses in the control and treatment groups were performed using the R package (edgeR 3.22.3), an absolute value of |log2 (fold change)| > 1 and an adjusted P-value of < 0.05 were set as the filter criteria for significant differential expression. The differential cluster analysis of genes and a volcano plot of differentially expressed lncRNAs and mRNAs were drawn by the E Charts platform (http://www.ehbio.com/ImageGP/index.php/Home/Index/index.html).
Quantitative real-time-PCR (qRT-PCR) validation
RNA samples from the gonad of 6 individuals used for the RNA-seq were analyzed by qRT-PCR. First-strand cDNA was synthesized using PrimeScript RT Master Mix (Takara, Japan). qRT-PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using SYBR Premix Ex Taq II (Takara) according to the manufacturer’s protocol. Specific primers of genes and lncRNAs as shown in Table 1 (available online only) were designed using the Primer Premier 5. Each 25 μL reaction volume contained 12.5 μL 2 × SYBR Premix Ex Taq II, 1.0 μL diluted cDNA template (100 ng RNA), 9.5 μL PCR-grade water, and 1.0 μL of each 10 μM primer. The qRT-PCR conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 63 °C for 30 s. The relative lncRNA and mRNA expression levels were normalized to β-actin and calculated using the 2−ΔΔCt method 31 .
Statistical analysis
All data are presented as the mean ± standard error of the mean (SEM) and were analyzed using the statistical software SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). Comparisons between the two groups were performed using Student’s t-test. Statistical significance was determined at P < 0.05.
Code availability
The code used in this manuscript of R package (edgeR 3.22.3) is available at bioconductor (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html). The gffcompare program (v0.10.1) is available at GffCompare (https://ccb.jhu.edu/software/stringtie/gffcompare.shtml). The TransDecoder (rel16JAN2014) is available at sourceforge (https://sourceforge.net/projects/transdecoder/). The Coding Potential Calculator (0.9-r2) is available at mybiosoftware (http://www.mybiosoftware.com/cpc-0-9r2-assess-protein-coding-potential-transcripts.html).
Data records
The raw fastq files for the RNA-seq data have been deposited in the NCBI Sequence Read Archive (NCBI-SRA) under accession numbers SRR7944794-SRR7944799 (Data Citation 1), and sample metadata expression estimates can be found on the NCBI Gene Expression Omnibus (Data Citation 2). The information of lncRNAs and mRNAs, information of lncRNA overlap with protein coding genes and annotations of both lncRNA and protein coding genes can be found on the Figshare (Data Citation 3).
Technical Validation
Efficiency of igf3 knockdown in juvenile C. carpio
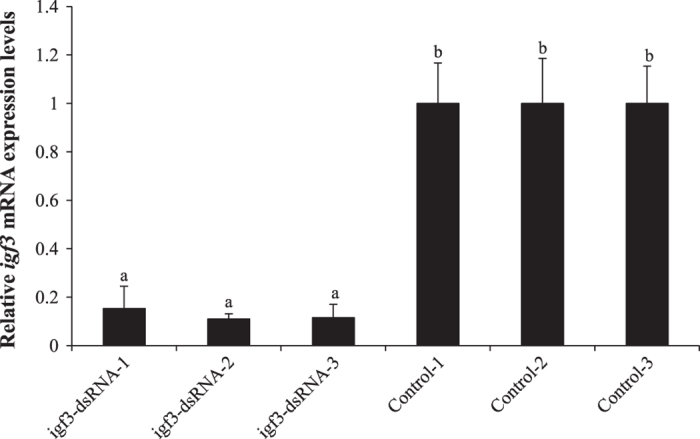
The mRNA expression level of igf3 was examined by qRT-PCR after RNAi with igf3-dsRNA injection. The qRT-PCR results of the igf3 RNAi were given in Fig. 1, injection of igf3-dsRNA resulted in significant down-regulation of igf3 mRNA expression compared with the control group (P < 0.05), and it reached an 87.4% decrease at the 24th hour.
Figure 1. The effect of igf3-dsRNA injection analysis by qRT-PCR.
Overview of RNA-sequencing
In this study, 6 cDNA libraries were constructed using total gonadal RNA from 3 control groups and 3 treatment groups. In total, 663,158,182 raw reads of 150 bp were produced from the 6 cDNA libraries, and 633,474,428 clean reads amounted to 95.02 Gb. More than 80% of the clean reads were mapped to the C. carpio reference genome. The average number of sequences that had unique and multiple positions mapped to the reference sequence was 68.17 and 13.41%, respectively. Approximately 34% of the reads substantially mapped to the positive and negative chains of the genome (Table 2).
Table 2. Alignment of statistical results of reads.
| Sample name | Control-1 | Control-2 | Control-3 | igf3-dsRNA-1 | igf3-dsRNA-2 | igf3-dsRNA-3 |
|---|---|---|---|---|---|---|
| Notes: Raw reads: The number of total reads. Clean reads: The number of clean reads. Clean base (G): The data of clean reads. Total mapped reads: The number of sequences matched to the genome. Multiple mapped reads: The number of sequences that had multiple positions mapped to the reference sequence. Uniquely mapped reads: The number of sequences that had unique positions mapped to the reference sequence. Read-1, read-2 mapped: The number of read-1 and read-2 that locate on the genome; the statistical proportion of the two parts should be substantially the same. Reads map to '+': The number of reads mapped to the positive strand of the genome. Reads map to '-': The number of reads mapped to the negative strand of the genome. | ||||||
| Raw reads | 103838052 | 129702320 | 109996624 | 98107722 | 130263708 | 91249756 |
| Clean reads | 99410868 | 123563848 | 104194694 | 94321406 | 122675946 | 89307666 |
| Clean base (G) | 14.91 | 18.53 | 15.63 | 14.15 | 18.4 | 13.4 |
| Total mapped reads | 81530347(82.01%) | 99404340(80.45%) | 85870625(82.41%) | 77049685(81.69%) | 99493319(81.10%) | 73064096(81.81%) |
| Multiple mapped | 12853963(12.93%) | 18112363(14.66%) | 12915566(12.40%) | 15084557(15.99%) | 15762693(12.85%) | 10390489(11.63%) |
| Uniquely mapped | 68676384(69.08%) | 81291977(65.79%) | 72955059(70.02%) | 61965128(65.70%) | 83730626(68.25%) | 62673607(70.18%) |
| Read-1 mapped | 34516969(34.72%) | 40477251(32.76%) | 36673789(35.20%) | 31095154(32.97%) | 41919370(34.17%) | 31496714(35.27%) |
| Read-2 mapped | 34159415(34.36%) | 40814726(33.03%) | 36281270(34.82%) | 30869974(32.73%) | 41811256(34.08%) | 31176893(34.91%) |
| Reads map to ‵+′ | 34383536(34.59%) | 40550020(32.82%) | 36580679(35.11%) | 31056772(32.93%) | 41822537(34.09%) | 31350486(35.10%) |
| Reads map to ‵−′ | 34292848(34.50%) | 40741957(32.97%) | 36374380(34.91%) | 30908356(32.77%) | 41908089(34.16%) | 31323121(35.07%) |
Identification of lncRNAs and mRNAs
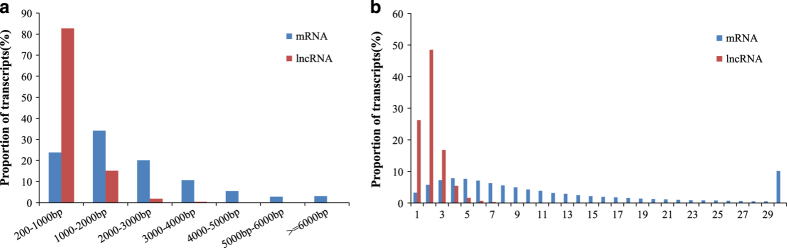
RNA-seq yielded 14199 lncRNA and 106932 mRNA transcripts after a stringent filtering process to discard transcripts without the characteristics of lncRNAs and mRNAs. These transcripts were randomly distributed into 50 chromosomes of C. carpio, quite notably, the most number of lncRNAs and mRNAs were located in the chromosome 31 and 38 with the percentage of 1.79 and 1.65%, respectively (Information of carp lncRNAs and mRNAs (as surfaced at figshare), Data Citation 3). The length of the lncRNAs ranged from 200 to 5759 bp, with 82.69 and 15.07% of lncRNAs having a length of 200–1000 bp and 1000–2000 bp, respectively, whereas the mRNA transcripts were mainly longer than 1000 bp (Fig. 2a), and the lncRNAs had an average length of 643 bp and 1.6 exons, were less than that of the mRNA (Fig. 2b). In addition, the types of lncRNA overlapping with protein coding genes are ‘j’, ‘o’ and ‘x’ (The information of lncRNA overlap with protein coding genes (as surfaced at figshare), Data Citation 3), and the annotations of both lncRNA and protein coding genes can be found in Data Citation 3 (Annotations of lncRNA and protein coding genes (as surfaced at figshare), Data Citation 3).
Figure 2. Identification of lncRNAs and mRNAs in C. carpio gonad.
(a) Length of lncRNAs and mRNAs. (b) Exonic content of lncRNAs and mRNAs.
Differential expression of lncRNA and mRNA
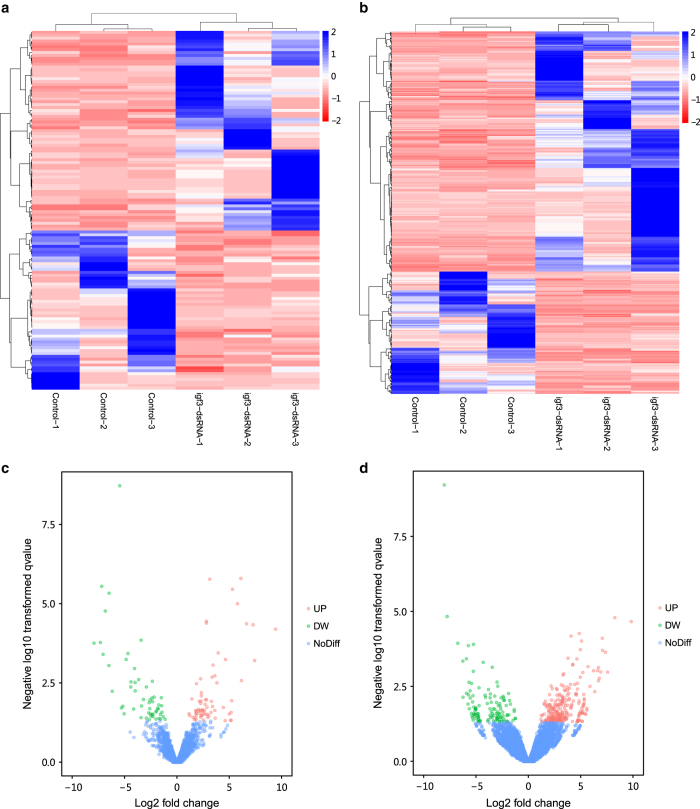
We investigated the differential expression levels of lncRNAs and mRNAs between control group and treatment group. As shown in Fig. 3a,b, hierarchical clustering revealed the significant differential expression (absolute log2 (fold change) > 1, P < 0.05) of 124 lncRNAs and 353 mRNAs. In differentially expressed lncRNAs, 69 lncRNAs were up-regulated, whereas 55 lncRNAs were down-regulated in the treatment group (Fig. 3c). Furthermore, among the differentially expressed mRNAs, 234 mRNAs were up-regulated, whereas 119 mRNAs were down-regulated (Fig. 3d). Among these differently expressed genes, there were some key genes associated with gonadal development, such as fibroblast growth factor receptor 2 (FGFR2), fibronectin-like, serine/threonine-protein phosphatase 4, serine-rich adhesin.
Figure 3. Differentially expressed lncRNAs and mRNAs.
(a and b) are hierarchical clustering of the differentially expressed lncRNAs and mRNAs, blue indicates relative high expression, and red indicates relative low expression. (c and d) are volcano plots of differentially expressed lncRNAs and mRNAs. X-axis represents fold change (log 2) and Y-axis represents P (−log 10). Red points indicate up-regulated (UP, X-axis > 0) transcripts; green points indicate down-regulated (DW, X-axis < 0) transcripts; blue points indicate no significant difference (NoDiff).
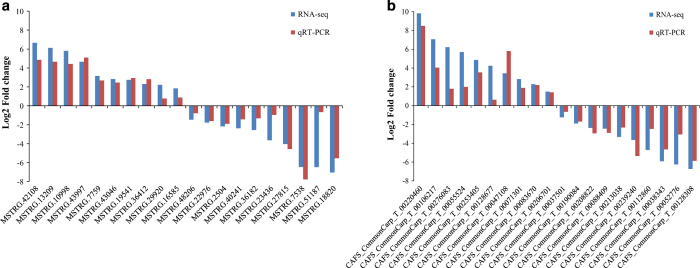
To further evaluate the reliability of RNA sequencing, 20 differentially expressed lncRNAs (10 up-regulated and 10 down-regulated) and 20 differentially expressed mRNAs (10 up-regulated and 10 down-regulated) were randomly selected to validate the relative expression levels in the gonad of the control and treatment group using qRT-PCR. As shown in Fig 4, the trend of differential expression of these 40 randomly selected genes (lncRNAs, Fig. 4a, mRNAs, Fig. 4b) is 100% consistent with our RNA-seq data, indicating that the RNA-seq data was reliable.
Figure 4. Illustrating of qRT-PCR confirmation for RNA-seq.
Validation of 40 selected differentially expressed lncRNAs (a) and mRNAs (b). The lncRNAs and mRNAs were expressed as log2 (fold change) values (RNA-seq) and −ΔΔCt values (qRT-PCR).
Additional Information
How to cite this article: Song, F. et al. Long noncoding RNA and mRNA expression profiles following igf3 knockdown in common carp, Cyprinus carpio. Sci. Data. 6:190024 https://doi.org/10.1038/sdata.2019.24 (2019).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This research was funded by the Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2018HY-XKQ02-02), the Chinese Earmarked Fund for Modern Agro-Industry Technology Research System (CARS-45) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX17_0651).
Footnotes
The authors declare no competing interests.
Data Citations
References
- Wang D. S et al. Discovery of a gonad-specific IGF subtype in teleost. Biochem. Biophys. Res. Commun. 367, 336–341 (2008). [DOI] [PubMed] [Google Scholar]
- Li J., Liu Z., Wang D. & Cheng C. H. Insulin-Like Growth Factor 3 Is Involved in Oocyte Maturation in Zebrafish. Biol. Reprod. 84, 476–486 (2011). [DOI] [PubMed] [Google Scholar]
- Song F. et al. A Novel igf3 Gene in Common Carp (Cyprinus carpio): Evidence for Its Role in Regulating Gonadal Development. PLoS One. 11, e0168874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chu L., Sun X., Liu Y. & Cheng C. H. IGFs mediate the action of LH on oocyte maturation in zebrafish. Mol. Endocrinol. 29, 373–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nã³Brega R. H. et al. Fsh Stimulates Spermatogonial Proliferation and Differentiation in Zebrafish via Igf3. Endocrinology 156, 3804–3817 (2015). [DOI] [PubMed] [Google Scholar]
- Li J., Niu C. & Cheng C. H. K. Igf3 serves as a mediator of LH in zebrafish ovulation. Biol. Reprod. 6, 1235–1243 (2018). [DOI] [PubMed] [Google Scholar]
- Safian D., van der Kant H. J. G., Crespo D., Bogerd J. & Schulz R. W. Follicle-Stimulating Hormone Regulates igfbp Gene Expression Directly or via Downstream Effectors to Modulate Igf3 Effects on Zebrafish Spermatogenesis. Front. Endocrinol. 8, 328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safian D., Bogerd J. & Schulz R. W. Igf3 activates β-catenin signaling to stimulate spermatogonial differentiation in zebrafish. J. Endocrinol. 238, 245–257 (2018). [DOI] [PubMed] [Google Scholar]
- Chocu S., Calvel P., Rolland A. D. & Pineau C. Spermatogenesis in mammals: proteomic insights. Syst. Biol. Reprod. Med. 58, 179–190 (2012). [DOI] [PubMed] [Google Scholar]
- Shen Z. G., Yao H., Guo L., Li X. X. & Wang H. P. Ribosome RNA Profiling to Quantify Ovarian Development and Identify Sex in Fish. Sci. Rep. 7, 4196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova I. V., Hennelly S. P., Tung C. S. & Sanbonmatsu K. Y. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J. Mol. Biol. 425, 3731–3746 (2013). [DOI] [PubMed] [Google Scholar]
- Yi Z. et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic. Acids. Res. 44, D203–D208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Lu X. & Yuan L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta. 1839, 1097–1109 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang R. & Ying K. Long non‑coding RNAs: Novel links in respiratory diseases (Review). Mol. Med. Rep. 11, 4025–4031 (2015). [DOI] [PubMed] [Google Scholar]
- Larsson L., Castilho R. M. & Giannobile W. V. Epigenetics and its role in periodontal diseases: a state-of-the-art review. J. Periodontol. 86, 556–568 (2015). [DOI] [PubMed] [Google Scholar]
- Dianatpour A. & Ghafouri-Fard S. Long Non Coding RNA Expression Intersecting Cancer and Spermatogenesis: A Systematic Review. Asian. Pac. J. Cancer Prev. 18, 2601–2610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera M. C. et al. Tsx, Produces a Long Noncoding RNA and Has General Functions in the Germline, Stem Cells, and Brain. PLoS Genet. 7, e1002248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G., Akhade V. S. & Donakonda S. mrhl RNA, a Long Noncoding RNA, Negatively Regulates Wnt Signaling through Its Protein Partner Ddx5/p68 in Mouse Spermatogonial Cells. Mol. Cell Biol. 32, 3140–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lu H., Xin D., Cheng H. & Zhou R. A novel ncRNA gene from mouse chromosome 5 trans-splices with Dmrt1 on chromosome 19. Biochem. Biophys Res. Commun. 400, 696–700 (2010). [DOI] [PubMed] [Google Scholar]
- Ni M. J. et al. Identification and Characterization of a Novel Non-Coding RNA Involved in Sperm Maturation. PLoS One 6, e26053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y. H. & Macdonald P. M. RNA sequences required for the noncoding function of oskar RNA also mediate regulation of Oskar protein expression by Bicoid Stability Factor. Dev. Biol. 407, 211–223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi G. M. et al. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol. Hum. Reprod. 20, 719–735 (2014). [DOI] [PubMed] [Google Scholar]
- Mandal A. C. et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141, 4618–4627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara S., Kurihara M. & Kimura A. P. A long non-coding RNA transcribed from conserved non-coding sequences contributes to the mouse prolyl oligopeptidase gene activation. J. Biochem. 155, 243–256 (2014). [DOI] [PubMed] [Google Scholar]
- Gao Q. et al. Long non-coding RNAs regulate effects of β-crystallin B2 on mouse ovary development. Mol. Med. Rep. 14, 4223–4231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Nguyen N. H. & Zhu W. Genetic evaluation of a selective breeding program for common carp Cyprinus carpio conducted from 2004 to 2014. BMC Genet. 16, 94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H. et al. Characterization, expression, and function analysis of gonad-inhibiting hormone in Oriental River prawn, Macrobrachium nipponense, and its induced expression by temperature. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 185, 1–8 (2015). [DOI] [PubMed] [Google Scholar]
- Schmieder R. & Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.