Abstract
The nearly conserved U54 of tRNA is mostly converted to a version of ribothymidine (T) in Bacteria and eukaryotes and to a version of pseudouridine (Ψ) in Archaea. Conserved U55 is nearly always modified to Ψ55 in all organisms. Orthologs of TrmA and TruB that produce T54 and Ψ55, respectively, in Bacteria and eukaryotes are absent in Archaea. Pus10 produces both Ψ54 and Ψ55 in Archaea. Pus10 orthologs are found in nearly all sequenced archaeal and most eukaryal genomes, but not in yeast and bacteria. This coincides with the presence of Ψ54 in most archaeal tRNAs and some animal tRNAs, but its absence from yeast and bacteria. Moreover, Ψ54 is found in several tRNAs that function as primers for retroviral DNA synthesis. Previously, no eukaryotic tRNA Ψ54 synthase had been identified. We show here that human Pus10 can produce Ψ54 in select tRNAs, including tRNALys3, the primer for HIV reverse transcriptase. This synthase activity of Pus10 is restricted to the cytoplasm and is distinct from nuclear Pus10, which is known to be involved in apoptosis. The sequence GUUCAm1AAUC (m1A is 1-methyladenosine) at position 53–61 of tRNA along with a stable acceptor stem results in maximum Ψ54 synthase activity. This recognition sequence is unique for a Ψ synthase in that it contains another modification. In addition to Ψ54, SF9 cells-derived recombinant human Pus10 can also generate Ψ55, even in tRNAs that do not contain the Ψ54 synthase recognition sequence. This activity may be redundant with that of TruB.
Keywords: tRNA modification, pseudouridine synthase, TruB, retroviral primer, mRNA modification, 1-methyladenosine
INTRODUCTION
A typical tRNA contains a 17-base stem–loop structure called the “common arm” or “TΨC arm” at position 49–65. Most tRNAs contain a consensus GUUCRANYC sequence at position 53–61 in this arm; the UUCRANY part of which forms the loop (Gupta 1985; Jühling et al. 2009). TΨC includes T (ribothymidine or 5-methyluridine) and Ψ (pseudouridine) modifications of the two U's at positions 54 and 55 in the consensus sequence, followed by a C at position 56. The U54 is commonly modified to T54 in most Bacteria and Eukarya, but rarely in Archaea. The U55 is converted to Ψ55 in nearly all tRNAs (Jühling et al. 2009). Most archaeal tRNAs, especially those of Euryarchaeota, contain Ψ or m1Ψ (1-methylpseudouridine) at position 54 instead of T (Gupta 1984, 1985, 1986; Jühling et al. 2009; Blaby et al. 2011; Chatterjee et al. 2012).
Bacterial tRNA methyltransferase, TrmA and tRNA Ψ synthase, TruB (and their eukaryal orthologs Trm2 and Pus4) produce T54 and Ψ55, respectively (Ny and Björk 1980; Nurse et al. 1995; Becker et al. 1997; Nordlund et al. 2000). Archaea lack orthologs of these enzymes. Archaea and Eukarya contain Cbf5, a box H/ACA guide RNA-dependent Ψ synthase, which structurally belongs to the TruB family of Ψ synthases (Koonin 1996; Watanabe and Gray 2000; Mueller and Ferre-D'Amare 2009). In vitro, archaeal Cbf5 can also produce Ψ55 in tRNA and Ψ at some positions in rRNA in a guide RNA-independent manner (Roovers et al. 2006; Gurha et al. 2007; Muller et al. 2007, 2008; Kamalampeta and Kothe 2012), but this activity of Cbf5 has only been shown in vivo for rRNA (Fujikane et al. 2018). On the other hand, archaeal Pus10 (PsuX), a Ψ synthase distinct from the TruB family (Watanabe and Gray 2000; Mueller and Ferre-D'Amare 2009; Rintala-Dempsey and Kothe 2017), can produce both Ψ54 and Ψ55 in tRNA, both in vitro and in vivo (Gurha and Gupta 2008; Joardar et al. 2013). Instead of a bacterial TrmA ortholog, an ortholog of bacterial RumA, an rRNA-methyltransferase, produces T54 in the tRNAs of some Archaea (Urbonavicius et al. 2008). Pus10 produces only Ψ55 in these T54-containing archaeal tRNAs (Roovers et al. 2006; Gurha and Gupta 2008; Joardar et al. 2013).
Orthologs of archaeal Pus10 are present in most eukaryotes, but not in bacteria and dikaryon fungi including yeast (Watanabe and Gray 2000; Roovers et al. 2006; Gurha and Gupta 2008; Fitzek et al. 2018). This coincides with the presence of Ψ54 in certain tRNAs of animals, and its absence from the tRNAs of bacteria and yeast (Jühling et al. 2009). Certain isoacceptors of mammalian tRNAs for Gln, Trp, Pro, Thr, and Arg and of chicken tRNAs for Pro and Trp contain Ψ, instead of T, at position 54 (Harada et al. 1975, 1979, 1989; Fournier et al. 1978; Harada and Nishimura 1980; Yang et al. 1983; Keith 1984; Kuchino et al. 1987; Harada 1989; Jühling et al. 2009).
Several Ψ54-containing tRNAs are reported to associate with retroviruses because these function as primers in retroviral DNA synthesis or were isolated from leukemia cells. These are tRNAs for Trp, Pro, Arg, Thr, and Gln (Harada et al. 1975, 1979; Harada and Nishimura 1980; Hu and Dahlberg 1983; Colicelli and Goff 1986; Harada 1989; Marquet et al. 1995; Mak and Kleiman 1997). The amount of a particular Ψ54-containing tRNAGln increases in Moloney murine leukemia virus-infected mouse cells and in Ehrlich ascites cells (Kuchino et al. 1987). This tRNA has a UmUG (Um is 2′-O-methyluridine) anticodon, which is suggested to read-through the UAG stop codon between the gag and pol genes to produce a viral protease. Similarly, mammalian tRNALys2, which normally contains Tm (2′-O-methylribothymidine) at position 54, increases in SV40-transformed mouse fibroblast (BALB/3T3) cells and other dividing and proliferating tissues (Raba et al. 1979). This tRNA shows a mixture of U, T, Tm, and Ψ at position 54 in a 3:30:60:10 ratio (Raba et al. 1979). The tRNA Ψ54 synthase activity has also been identified in Xenopus laevis oocytes (Nishikura and De Robertis 1981; Grosjean et al. 1996).
So far, no protein responsible for the tRNA Ψ54 synthase activity has been identified in eukaryotes. Previously done multiple sequence alignments (McCleverty et al. 2007; Fitzek et al. 2018) have shown that though eukaryotic Pus10 proteins are much larger than archaeal Pus10, they do contain the full set of five residues (catalytic Asp, an Arg/Lys, a Tyr/Phe, a hydrophobic residue and a Leu) that are conserved within the active sites of all Ψ synthases (McCleverty et al. 2007; Mueller and Ferre-D'Amare 2009). In addition, both archaeal and eukaryotic Pus10 share two more conserved residues with several Ψ synthases: an Arg, two residues N-terminal, and an Asp, two residues C-terminal to the catalytic Asp, which have been shown to be important for the activity of archaeal Pus10 (Joardar et al. 2013; Kamalampeta et al. 2013; Fitzek et al. 2018). The crystal structure of human Pus10 suggests that it is a bona fide Ψ synthase (McCleverty et al. 2007). Structural alignment of human Pus10 and Methanocaldococcus jannaschii Pus10 showed nearly superimposable catalytic domains (Joardar et al. 2013). Although, the Ψ synthase activity of human Pus10 has not been demonstrated, it has been shown that the version of this protein located in the nucleus is involved in TRAIL-induced apoptosis (Aza-Blanc et al. 2003; Jana et al. 2017).
Here we show that human Pus10 can produce Ψ54 in select tRNAs, including tRNALys3, the primer for HIV DNA synthesis. It shows maximum activity when the TΨC arm consensus sequence is restricted to GUUCAm1AAUC (m1A is 1-methyladenosine and the underlined U is modified to Ψ54). U55 and A58 are essential in this sequence. So far, the involvement of another modification in Ψ formation has not been reported. Destabilization of the acceptor stem of the tRNA by non-Watson–Crick pairs considerably reduces this activity. Recombinant human Pus10 can also selectively produce Ψ54 in certain tRNAs and tRNA-fragments. In addition, recombinant Pus10 shows tRNA Ψ55 synthase activity in tRNAs that normally contain Ψ54 as well as those that do not contain Ψ54. This Ψ55 activity of Pus10 may be redundant with that of TruB inside the cell. However, the tRNA Ψ54 synthase activity of Pus10 is unique.
RESULTS
Ψ is present at position 54 in certain mammalian tRNAs
The presence of Ψ54 in mammalian tRNAs was mostly determined by sequencing the individually isolated tRNA species. Mammalian genomes contain multiple tRNA genes that are transcribed to produce a tRNA pool of several isoacceptors and isodecoders for each amino acid. Therefore, we determined whether a significant population of tRNAs for certain amino acids in HeLa and mouse liver cells contained Ψ54. We used primer extension after 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT) treatment of tRNA-enriched total RNA to determine the presence or absence of Ψ54 in the human and mouse tRNA pools of some amino acids (Fig. 1A,B). As expected, all tRNAs contain Ψ at position 55. Ψ is also present at position 54 in HeLa tRNAs for Gln, Trp, and Thr, but not Phe and Asp, and mouse tRNAs for Gln and Arg, but not Phe. The presence of Ψ54, if any, in Lys tRNAs could not be determined because Tm (2′-O-methylribothymidine) commonly present at position 54 in these tRNAs inhibits primer extension for unknown reasons. This has also been observed by others (Muthuswami et al. 2002). Most mammalian tRNAs contain m1A at position 58, which strongly inhibits primer extension. However, it does allow some extension after misincorporation during reverse transcription (Safra et al. 2017b). AlkB-mediated demethylation of m1A58 before CMCT treatment reduces the stops (dark bands) at position 58 and improves the resolution of Ψ54 and Ψ55 in some cases (Fig. 1C). Nucleotide sequences of some of these tRNAs are shown in Supplemental Figure S1A. Our data agree with the reported presence or absence of Ψ54 in these tRNAs (Jühling et al. 2009).
FIGURE 1.
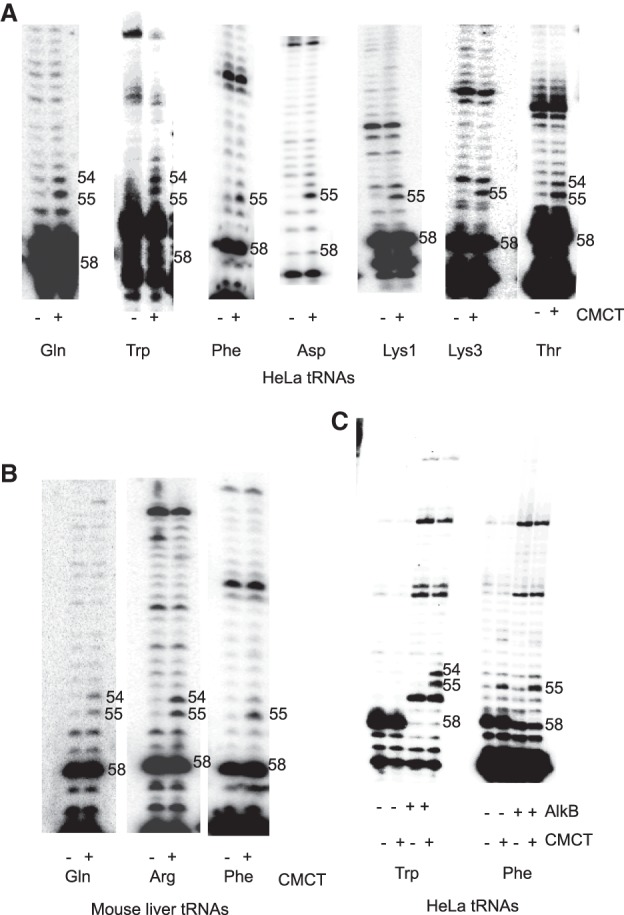
CMCT-primer extension analyses to determine the presence of Ψ54 in selected mammalian tRNAs. (A) HeLa cell and (B) mouse liver tRNA-enriched total RNAs were either not treated (−) or treated with CMCT (+) for 20 min, followed by alkali treatment before primer extension reactions. The sequence of each primer (Supplemental Table S1) for the tRNA of a particular amino acid corresponds to the 15 bases at position 62–76 of one isoacceptor for that amino acid (indicated below each panel). A dark band in the CMCT lane but not in the untreated lane indicates a Ψ. Dark bands in both lanes at the same position are caused by inhibition of primer extension by a modified residue at that position or due to a strong secondary and/or tertiary structure in that region. Numbers 54 and 55 on the side indicate the presence of Ψ at these positions. T54 of a tRNA does not produce a dark band in either lane. Very dark bands at position 58 (marked) in both lanes in most tRNAs are due to the presence of m1A, which significantly inhibits primer extension. Tm at position 54 also inhibits primer extension, which causes dark bands in both lanes in Lys tRNAs. (C) HeLa cell tRNA-enriched total RNAs were treated with AlkB to demethylate m1A58, and then CMCT-primer extension was done as in A. Treatment (+) or no-treatment (−) by AlkB and CMCT is indicated below each lane. For unknown reasons, there is a strong stop at position 56 in tRNATrp, which becomes prominent after nearly complete demethylation of m1A58 by AlkB (compare with tRNATrp in A). Demethylation of m1A58 in the same AlkB reaction mixture of tRNA-enriched total RNA is complete in tRNATrp and only partial in tRNAPhe.
Human tRNA Ψ54 synthase activity is selective toward certain tRNAs
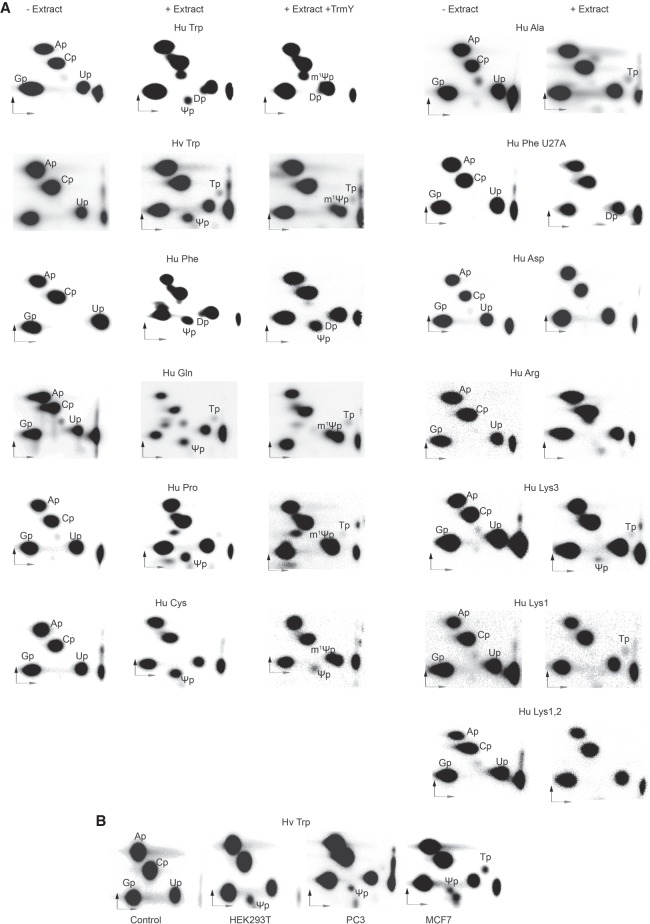
HeLa cell extract was used to treat [α-32P]UTP-labeled transcripts of human tRNATrp. After RNase T2 digestion and separation of the products by two dimensional (2D) thin layer chromatography (TLC), radioactive spots were seen for Ψ and dihydrouridine (D), in addition to those for A, C, G, and U (Fig. 2A). RNase T2 digestion of RNA produces ribonucleoside-3′-monophospates (Np). Consequently, in this nearest-neighbor analysis, labeled 5′ phosphate, originally derived from the [α-32P]-labeled NTP used to produce the transcript, is transferred to the 3′ side of the preceding residue. Therefore, labeled U (or modified U) spots in the RNase T2 digest of [α-32P]UTP-labeled RNA would be derived from the first of two adjacent U's in the RNA sequence. Such adjacent pairs of U in human tRNATrp are present at positions 47–48 and 54–55 (Supplemental Fig. S1A). We believe that the HeLa cell extract converted U at positions 47 and 54 to D and Ψ, respectively, because although the modification status of human tRNATrp is not known, bovine tRNATrp has D47 and Ψ54 (Jühling et al. 2009). Ψ at position 54 was confirmed by conversion of this Ψ to 1-methyl-Ψ (m1Ψ) by M. jannaschii TrmY, which converts the Ψ54 of tRNA to m1Ψ (Chatterjee et al. 2012). We further confirmed that this Ψ is derived from U54 by using the tRNATrp of Haloferax volcanii, an archaeon (Fig. 2A). This tRNA only has two adjacent U's, at positions 54–55 and the nucleotide sequences of these two tRNATrp are nearly identical at positions 28–42 in the anticodon arm and 53–61 in the TΨC arm (Supplemental Fig. S1A). As expected, TLC analysis of H. volcanii tRNATrp showed a Ψ but no D (Fig. 2A). This Ψ could also be converted to m1Ψ. We interchangeably used these two Trp tRNAs, since they provided similar results.
FIGURE 2.
Human tRNA Ψ54 synthase activity is selective. (A) Several [α-32P]UTP-labeled human (Hu) tRNA and H. volcanii tRNATrp (Hv Trp) transcripts (indicated above panels) were incubated with HeLa cell extracts. See sequences of transcripts in Supplemental Figure S1A. Purified RNA products were further treated with recombinant M. jannaschii TrmY and SAM, to convert Ψ54 to m1Ψ54, and the RNA products were further purified. RNase T2 digests of untreated (panels in the first column), extract-treated (panels in the second column), and extract and TrmY-treated RNAs (panels in the third column, for some transcripts) were separated by 2D-TLC using solvents I and II in the first and second dimensions (denoted by dark and light arrows), respectively. Representative autoradiograms of repeat experiments are shown. The labels indicate the 3′-monophospates of corresponding ribonucleosides. Trace amounts of Tp are observed in some reactions that are produced by conversion of U54 to T54 in the presence of residual SAM in the cell extracts. Sometimes Dp and m1Ψp merge into Up. (B) Representative figures of 2D-TLC analyses of reactions similar to those in A using [α-32P]UTP-labeled H. volcanii tRNATrp transcripts and extracts of HEK293T, PC3, and MCF7 cells.
HeLa cell extract treatment of human tRNAAla did not generate any Ψ (Fig. 2A). However, similar treatment of human tRNAPhe did show a Ψ (Fig. 2A). This Ψ is not derived from position 54 because TrmY does not convert it to m1Ψ. Human tRNAPhe contains two consecutive U's at three positions: 16–17, 27–28, and 54–55 (Supplemental Fig. S1A). Native human tRNAPhe contains D, Ψ, and T at positions 16, 27, and 54, respectively. The Ψ in our TLC is derived from Ψ27, which is known to be produced by Pus1 (Rintala-Dempsey and Kothe 2017). We confirmed this by using a mutant tRNAPhe, where U27 was changed to an A. No Ψ was produced in this case (Fig. 2A).
Several other human tRNA transcripts were also treated with HeLa cell extracts and treated with TrmY, as needed, to determine the production of Ψ54 (Fig. 2A). Ψ54 was produced in tRNAGln and tRNAPro, but not in tRNAAsp and tRNAArg. The extracts did produce Ψ54 in tRNALys3, but not in tRNALys1, and (mixed) tRNALys1,2 (Fig. 2A). We also used extracts of HEK293T, PC3, and MCF7 cells to show that tRNA Ψ54 synthase activity is present in these cells too (Fig. 2B).
Overall these data suggest that human cell extracts can produce Ψ54 in some isoacceptors of certain amino acids, but not of others.
Human tRNA Ψ54 synthase activity is in the cytoplasm
We treated [α-32P]UTP-labeled H. volcanii tRNATrp with nuclear and cytoplasmic extracts of HeLa cells. Ψ was observed only when the cytoplasmic extract was used (Fig. 3A), suggesting that tRNA Ψ54 synthase activity is restricted to the cytoplasm and not present in the nucleus. This was not due to lack of overall Ψ synthase activity in the nuclear extracts, as observed by the presence of Ψ in 1D-TLC of nuclease P1 digests of [α-32P]UTP-labeled H. volcanii tRNATrp treated with such extracts (Fig. 3B). Nuclease P1 generates labeled modified or unmodified versions of all U's in [α-32P]UTP-labeled RNA. This Ψ could come from any other position of tRNA, because several Ψ synthases are present in the nucleus (Rintala-Dempsey and Kothe 2017). Our nuclear and cytoplasmic extracts did not show any significant cross-contamination (Fig. 3C). Although tRNA Ψ54 synthase activity was observed in the cytoplasm and not in the nucleus (Fig. 3A), immunoblots using anti-Pus10 antibody detected Pus10 in the nucleus and not in the cytoplasm (Fig. 3C). This inconsistency is discussed later. Addition of nuclear extract to cytoplasmic extract did not increase or decrease Ψ54 synthesis. m1A58 in the transcripts enhances Ψ54 production (see later). However, nuclear extracts did not produce Ψ54 even in m1A58-containing tRNAs.
FIGURE 3.
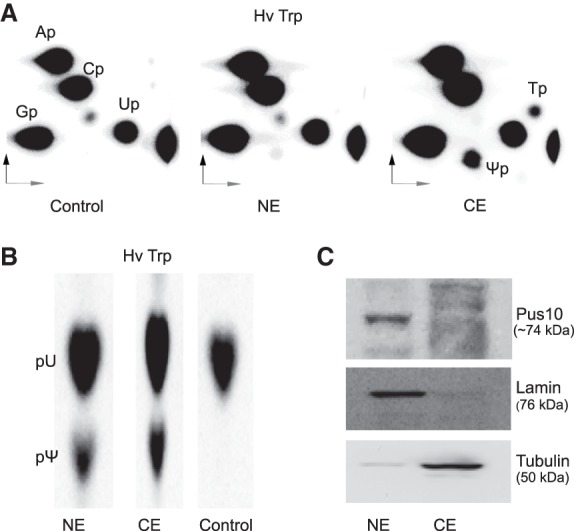
Human tRNA Ψ54 synthase activity is present in the cytoplasm. 2D-TLC of RNase T2 (A) and 1D-TLC (in solvent I) of nuclease P1 (B) digests of HeLa nuclear (NE) and cytoplasmic (CE) extract-treated [α-32P]UTP-labeled H. volcanii tRNATrp (Hv Trp). Relevant modified residues are labeled. “p” before and after a nucleoside letter in (A,B) indicates the 5′ and 3′ phosphate of that nucleoside. (C) Immunoblots of nuclear and cytoplasmic extracts using anti-Pus10 (HPA049582, Sigma) antibody. Purity of extracts was determined by using anti-lamin A (ab26300, Abcam) and anti-tubulin (ab6046, Abcam) antibodies as nuclear and cytoplasmic markers, respectively.
Pus10 is the catalytic protein for tRNA Ψ54 formation in human cells
We treated [α-32P]UTP-labeled H. volcanii tRNATrp with extracts of two previously described Pus10-knockdown strains of PC3 cells (Jana et al. 2017) (along with a vector control) to determine any change in the amount of tRNA Ψ54 synthesis. Both strains showed a reduction in Ψ54 synthesis in comparison to controls, when measured relative to Pus1-mediated human tRNAPhe Ψ27 synthesis by the same extracts (Fig. 4A; Supplemental Fig. S2A). We prepared a similar Pus10-knockdown strain of HEK293T cells (Supplemental Fig. S2B). It also showed reduced Ψ54 synthesis (Fig. 4B; Supplemental Fig. S2C).
FIGURE 4.
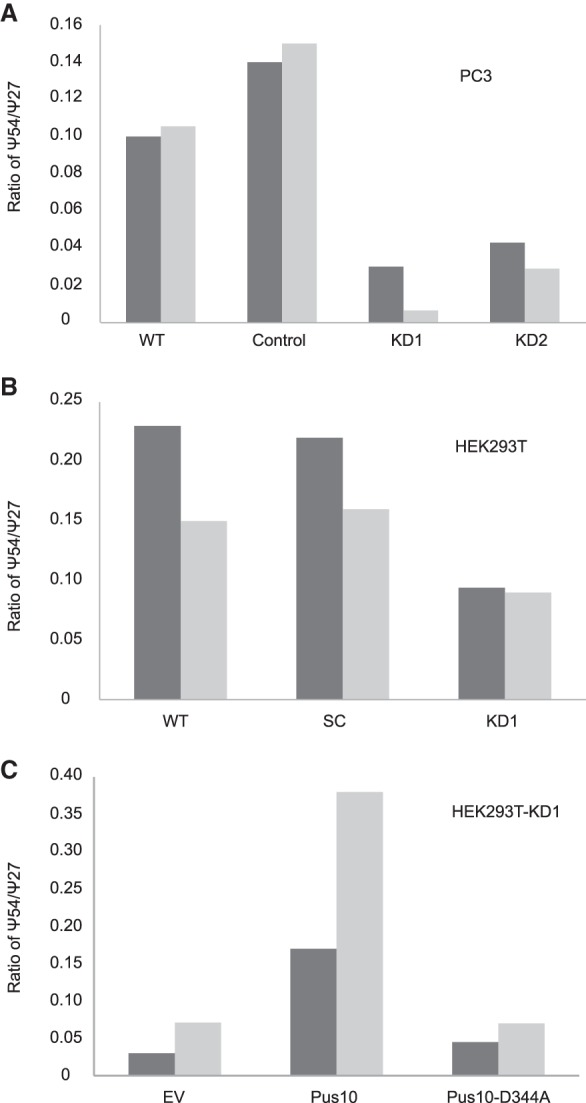
Human Pus10 is the catalytic protein for tRNA Ψ54 synthesis. (A) tRNA Ψ54 synthase activity (percent Ψ54 in tRNATrp /percent Ψ27 in tRNAPhe by the same extract) (see Supplemental Fig. S2A) by extracts of wild-type (WT), vector control (Control), and Pus10-knockdown (KD1 and KD2) strains of PC3 cells (dark bars—based on 2D-TLC analyses in Supplemental Fig. S2A; light bars—another independent replicate) showing reduction of Ψ54 in knockdown cells. For an unknown reason, vector controls showed more activity than WT. (B) tRNA Ψ54 synthase activity (dark bars—based on analyses in Supplemental Fig. S2C; light bars—another independent replicate) presented as in A using WT, scrambled control (SC), and Pus10-knockdown (KD1) strains of HEK293T. (C) tRNA Ψ54 synthase activity (dark bars—based on analyses in Supplemental Fig. S2E; light bars—another independent replicate) as in A using HEK293T-KD1 cells transiently expressing codon-changed Pus10 and its D344A mutant. Transient transfection by empty vector (EV) was used to produce control cells.
We transiently expressed a plasmid-borne codon-changed human Pus10 gene and its catalytic mutant (D344A) in a Pus10-knockdown strain of HEK293T cells. Some codons in the gene were replaced by synonymous codons to avoid shRNA-mediated knockdown of plasmid-borne gene-product. Although, both Pus10 proteins were expressed in the cells (Supplemental Fig. S2D), only the normal Pus10, not the mutant, rescued the tRNA Ψ54 synthase activity of the extracts (Fig. 4C; Supplemental Fig. S2E). Even though the native Pus10 protein was not detected by the antiPus10 antibody in the cytoplasm (Fig. 3C), this overexpressed tagged Pus10 protein was detected both in cytoplasm and nucleus by the antibody against its His-tag (Supplemental Fig. S2F). Overall these data suggest that human Pus10 is the catalytic protein for tRNA Ψ54 synthesis.
Certain structural requirements are critical for tRNA Ψ54 synthase activity of human Pus10
As mentioned previously, most tRNAs contain a consensus GUUCRANYC sequence at position 53–61. A predominant subset of this sequence in Ψ54-containing animal tRNAs appears to be GUUCAAAUC. We changed U55 to an A or A58 to a G in H. volcanii tRNATrp and A57, A59, and U60 of human tRNATrp to the other three bases and treated the transcripts with HeLa cell extracts (Fig. 5A). The U55A and A58G changes abolished Ψ54 formation. The changes at positions 57, 59, and 60 considerably reduced Ψ54 formation relative to unaltered tRNA. A triple change of these three residues showed similar reduction. These results suggest that the tRNA Ψ54 synthase activity prefers the 53-GUUCAAAUC-61 sequence and that U55 and A58 are absolutely necessary for this activity.
FIGURE 5.
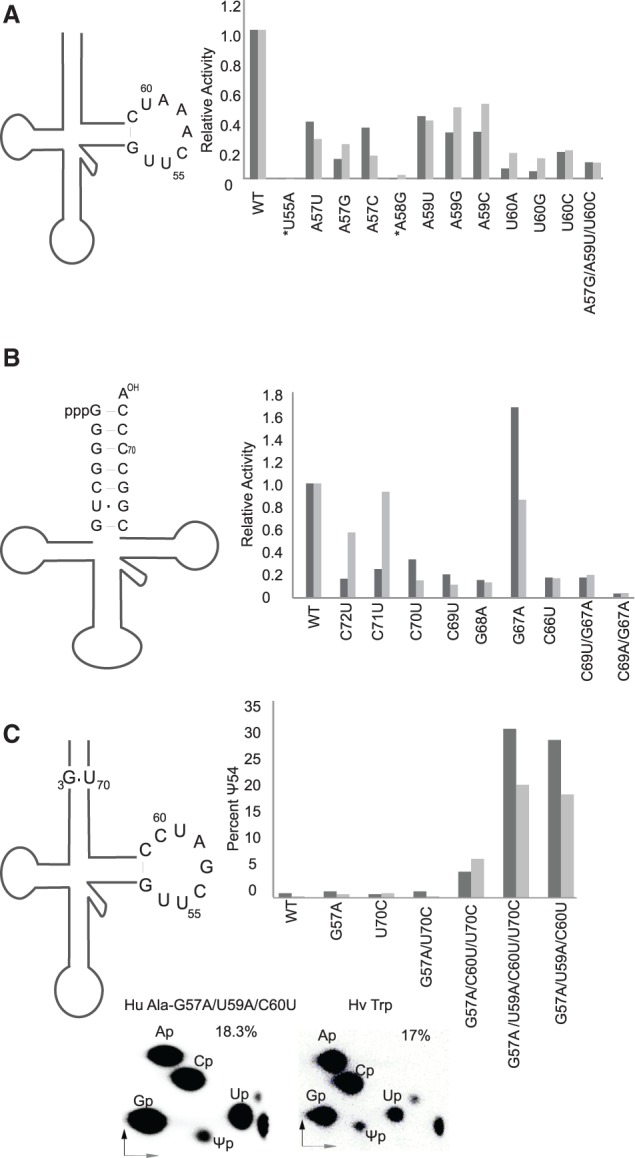
Importance of TΨC loop sequence and stable structure of acceptor stem for tRNA Ψ54 synthase activity of Pus10. (A) TΨC loop mutants were assessed for Ψ54 formation. RNase T2 digests of [α-32P]UTP-labeled transcripts of different human and H. volcanii (marked by *) tRNATrp mutants treated with HeLa cell extracts were separated by 2D-TLC, as in Figure 2A. U55A transcripts were [α-32P]ATP-labeled. The amount of Ψ54 produced in each case was determined and is presented here in bar graphs as the relative activity (Ψ54 in mutant/Ψ54 in WT using the same extract). Dark and light bars are individual results of two independent experiments. Mutants of human and H. volcanii tRNAs are compared with their own WT. The original (WT) TΨC loop sequence of the transcript is shown on the left. (B) Acceptor stem mutants of [α-32P]UTP-labeled H. volcanii tRNATrp transcripts were treated and the data are presented as in A. The original (WT) acceptor stem sequence of the transcript is shown on the left. (C) Mutants of [α-32P]UTP-labeled human tRNAAla transcripts were treated as in A and the data are presented as percent of U54 converted to Ψ54, not as relative activity, because the WT activity is negligible. The relevant sequence of the original (WT) transcript is shown on the left. Representative figures of 2D-TLC analyses of the [α-32P]UTP-labeled G57A/U59A/C60U mutant of tRNAAla and H. volcanii tRNATrp treated with the same HeLa extract are presented to show that the activity of the two transcripts are about the same.
The crystal structure of human Pus10 predicts that it binds the acceptor stem of the tRNA (McCleverty et al. 2007). We changed several base pairs in the acceptor stem of H. volcanii tRNATrp and determined their effects on Ψ54 formation (Fig. 5B). There is a U6•G67 wobble pair in the original transcript. Changing it to U6–A67, making the stem fully Watson–Crick paired, slightly increased the amount of Ψ54 formed. Two non-Watson–Crick pairs in the stem, i.e., one at U6•G67 and the other at any other place in the stem, drastically reduce Ψ54 formation. Indeed, even if U6•G67 had been converted to Watson–Crick U6-A67, changing G4-C69 to either G4•U69 or G4•A69, showed that even a single non-Watson–Crick pair in the middle of the stem was sufficient to reduce the activity. In fact, the activity was nearly abolished when the pair was G4•A69. These results suggest disruptions in the paired structure of the acceptor stem of a tRNA impede Pus10-mediated Ψ54 synthesis.
To confirm the above-described requirements for Ψ54 synthesis, we modified human tRNAAla to generate a Ψ54 synthase consensus similar to tRNATrp and determined Ψ54 formation (Fig. 5C). tRNAAla in all domains of life contains a G3•U70 pair. The sequence of human tRNAAla at position 53–61 is GUUCGAUCC, which differs from the Ψ54 synthase consensus sequence in having G57, U59, and C60, instead of A57, A59, and U60 (Supplemental Fig. S1A). Changing these three residues to the consensus, along with or without changing G3•U70 to G3-C70, brought the activity to about the same level as WT tRNATrp (Fig. 5C). Changing only two of these, i.e., G57A and C60U, along with U70C in the acceptor stem also gave some activity. G57A and U70C, either independently or in combination did not show activity.
To further confirm our results, we tested whether Ψ54 can be produced in human tRNACys. The TΨC loop sequence of human tRNACys (based on its gene) matches the consensus sequence for Ψ54 synthesis (Supplemental Fig. S1A). However, the sequence of any native animal tRNACys is not available. Therefore, the modification status of no animal tRNACys is known. As we expected, Ψ54 was produced in human tRNACys, which was confirmed by its conversion to m1Ψ by TrmY (Fig. 2A). We believe the leftover Ψ in this case (third column, Hu Cys in Fig. 2A) is derived from Ψ27 produced by Pus1 present in HeLa cell extracts. Although the TΨC loop sequence of human tRNALys3 differs from the Ψ54 synthase consensus sequence in having G59 instead of A59 (Supplemental Fig. S1A) and there is no report of the presence of Ψ54 in tRNALys3, our HeLa cell extracts could produce some Ψ54 in its transcript (Fig. 2A). This is consistent with the results shown in Figure 5A, where A59G mutation reduces, but does not abolish Ψ54 synthesis.
All these results suggest that the 53-GUUCAAAUC-61 sequence of the TΨC loop and the stable structure of the acceptor stem of tRNA are the major determinants for Ψ54 synthesis.
m1A58 modification in tRNA is very important for tRNA Ψ54 synthesis by cell extracts
Our cell extracts produced a mixture of Ψ54 and T54 in tRNA transcripts (Figs. 2A, 6), although in vivo, these tRNAs contain only Ψ54. This T54 is produced by Trmt2 methyltransferases normally present in the extracts. The amounts of T54 varied due to varying amounts of S-adenosyl-methionine (SAM) present in the extracts. Addition of SAM in the reactions increased the amount of T54 and reduced Ψ54 (A58 + SAM in Fig. 6). Therefore, to suppress T54 production we added S-adenosyl-homocysteine (SAH), an inhibitor of SAM-dependent methyltransferases, to our reactions. As expected, it abolished T54, but surprisingly it also drastically reduced Ψ54 (A58 + SAH in Fig. 6). We speculated that SAH is inhibiting m1A58 formation, which may be a necessary precondition for in vivo Ψ54 synthesis. Indeed, our cell extracts do produce m1A58 (Supplemental Fig. S3A). We checked the effects of SAM and SAH on Ψ54 production by cell extracts in m1A58-containing transcripts. There was not much difference in Ψ54 production between A58- and m1A58-containing transcripts in the presence or absence of SAM (first four sets of bars in Fig. 6). However, Ψ54 production was substantially increased in m1A58-containing transcripts in the presence of SAH, which inhibited both T54 and the m1A58 methyltransferase activity of the extracts (compare the last two sets of bars in Fig. 6). All these data suggest that the tRNA m1A58 modification substantially increases Ψ54 synthesis by cell extracts.
FIGURE 6.
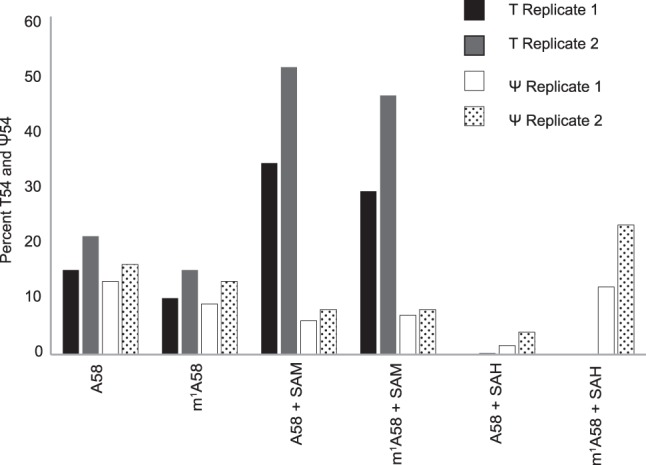
m1A58 in tRNA is needed for tRNA Ψ54 formation by cell extracts. [α-32P]UTP-labeled H. volcanii tRNATrp transcripts were first treated with TrmI to produce m1A58 and then treated with cell extracts in the presence of SAM or SAH. TrmI does produce m1A58 in this transcript (Supplemental Fig. S3B). RNase T2 digests of the products were separated by 2D-TLC as in Figure 2A. The percentage of U54 converted to Ψ54 and T54 were determined from the same 2D-TLC separation and are presented here in bar graphs. The presence of A (TrmI-untreated) or m1A (TrmI-treated) at position 58 and the presence of SAM or SAH are indicated. Replicate 1 and 2 refer to two independent experiments.
Recombinant human Pus10 can form Ψ54 in select tRNAs and tRNA fragments
Recombinant human Pus10 protein derived from E. coli cells did not show any tRNA Ψ synthase activity (Supplemental Fig. S4). Ψ54 or Ψ55 was not produced even in the presence of m1A58. However, recombinant human Pus10 from SF9 cells (Supplemental Fig. S5A) did show some Ψ54 synthesis with the H. volcanii tRNATrp transcript, which was substantially enhanced when the transcript contained m1A58 (Fig. 7). This protein did not produce Ψ54 in tRNAAla, even when it contained m1A58 (Fig. 7). However, it did produce Ψ54 in a mutant of tRNAAla, whose TΨC loop was made similar to tRNATrp (Fig. 7). Treatment of a human tRNALys3 transcript that has G59 instead of A59 in the Ψ54 consensus (Supplemental Fig. S1A) with the recombinant Pus10 produced Ψ54 in the transcript (Fig. 7) as seen before with cell extracts (Fig. 2A). This was enhanced when the transcript contained m1A58. The recombinant protein also produced some Ψ54 in human tRNAArg that has G57 and C59 instead of the A57 and A59 of consensus (Fig. 7). These results suggest that the SF9 cells-derived recombinant protein has the same specificity as the tRNA Ψ54 synthase activity of the cell extracts and human Pus10 does not require any associated factor for its tRNA Ψ54 synthase activity.
FIGURE 7.
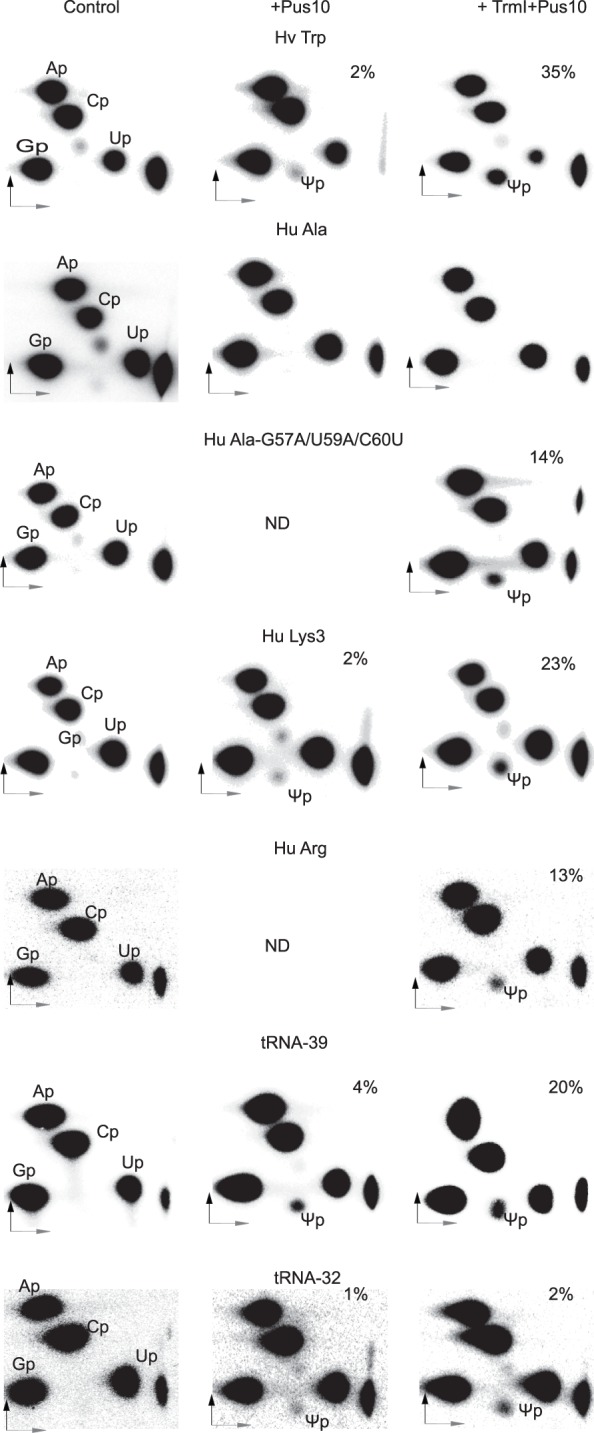
Recombinant human Pus10 produced in SF9 cells can synthesize tRNA Ψ54 in select tRNAs and tRNA fragments. Different [α-32P]UTP-labeled transcripts (indicated above panels) after TrmI treatment (to produce m1A58) or without TrmI treatment were incubated with SF9 cells-derived recombinant human Pus10, and RNase T2 digests of the products were separated by 2D-TLC as in Figure 2A. We confirmed TrmI-mediated m1A58 production in several of these transcripts (Supplemental Fig. S3B). Hu Ala-G57A/U59A/C60U is the same mutant shown in Figure 5C. tRNA-32 is a fragment of H. volcanii tRNATrp containing only the TΨC and anticodon arms, and tRNA-39 is the tRNA-32 with a seven-base bulge between the two arms (see sequences in Supplemental Fig. S1B). The percent Ψ54 produced in each case is indicated. ND, not determined.
SF9 cells-derived recombinant Pus10 also produced some Ψ54 equivalent in tRNA fragments that contained an acceptor stem and a TΨC stem either contiguous or having a bulge between the two (tRNA-32 and tRNA-39 in Supplemental Fig. S1B and Fig. 7). The amount significantly increased in tRNA-39 that contained the m1A58-equivalent modification. Therefore, it appears that the acceptor stem, but not the other parts of a tRNA, makes some contribution in Ψ54 synthesis. We did not study Ψ54 production in tRNA fragments by cell extracts because these RNAs were degraded by the extracts.
These results with different tRNAs and tRNA fragments also suggest that the Ψ54 catalytic activity of the recombinant Pus10 does not absolutely require m1A58 modification, but is substantially increased by it.
Recombinant human Pus10 can also form Ψ55 in tRNAs and tRNA fragments
Recombinant human Pus10 from SF9 cells also showed robust Ψ55 synthesis in H. volcanii tRNATrp and human tRNAAla transcripts, which was not significantly enhanced when the transcript contained m1A58 (Fig. 8A). This was determined by nearest-neighbor analyses of [α-32P]CTP-labeled transcripts. The absence of any Ψ production in nuclease P1 digests of the [α-32P]UTP-labeled U55A mutant of H. volcanii tRNATrp (Supplemental Fig. S6) confirmed that recombinant Pus10 has both Ψ54 and Ψ55 synthase activity. Ψ55 is not produced here because of the absence of its substrate U55, and Ψ54 is not produced, because as observed with the cell extracts (U55A in Fig. 5A), recombinant Pus10 also requires the presence of U55 to produce Ψ54. Recombinant protein can also produce some Ψ55 equivalent in tRNA-32 and tRNA-39, the tRNA fragments containing acceptor and TΨC stems (Fig. 8B). Overall these results suggest that the recombinant Pus10 can produce Ψ55 even in those tRNAs where it cannot produce Ψ54, e.g., tRNAAla, and so, the specific Ψ54 recognition sequence is not required for Ψ55 formation.
FIGURE 8.
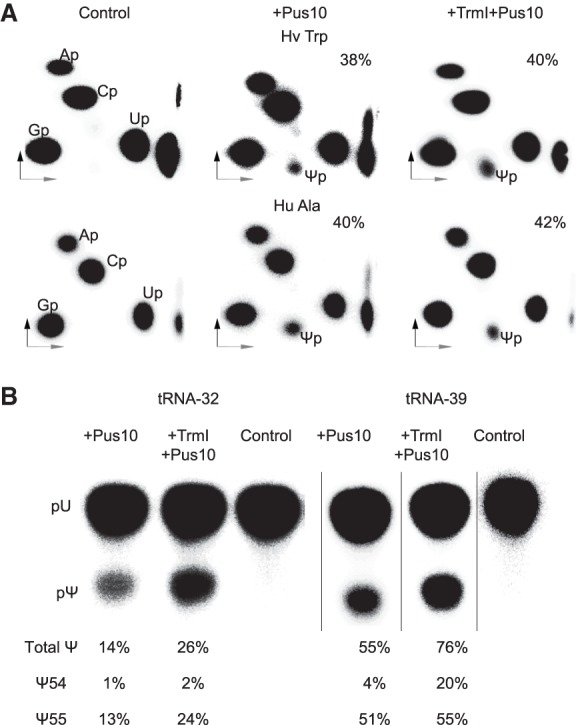
SF9 cells-derived recombinant human Pus10 has tRNA Ψ55 synthase activity. (A) [α-32P]CTP-labeled H. volcanii tRNATrp and human tRNAAla transcripts (indicated above panels) after TrmI treatment or without TrmI treatment were incubated with SF9 cells-derived recombinant human Pus10 and RNase T2 digests of the products were separated by 2D-TLC as in Figure 2A. Ψ here is derived from position 55 of the tRNAs, because the only common UC sequence in the two tRNAs is at position 55–56 (Supplemental Fig. S1A). The percent Ψ55 produced in each case is indicated. (B) Reaction products of [α-32P]UTP-labeled tRNA-32 and tRNA-39, in addition to the RNase T2 digestion shown in Figure 7, were also digested by nuclease P1, separated by 1D-TLC, and the total amount of Ψ produced was determined. The amount of Ψ55 produced was calculated by subtracting the amount of Ψ54 determined from the TLC separations represented in Figure 7.
Cellular Pus10 and active recombinant Pus10 are larger than their predicted sizes
Although Ψ synthesizing Pus10 protein cannot be detected in the cytoplasm by immunoblotting (Fig. 3C), both nonfunctional E. coli-derived and functional SF9 cells-derived human Pus10 can be detected by anti-Pus10 antibody (Supplemental Fig. S5B). The predicted sizes of the two recombinant proteins are about the same, but the SF9 cells-derived protein appears to be larger in immunoblots than the E. coli-derived protein (Supplemental Fig. S5B). The native cellular protein (presumably nuclear Pus10, see Fig. 3C) is also substantially larger than the size predicted by its open reading frame (Supplemental Fig. S5B). Furthermore, cytoplasmic Pus10 is expected to be of the same size as the nuclear Pus10, because His-tagged proteins that are observed in both nucleus and cytoplasm after transient transfection are of about the same size (Supplemental Fig. S2F).
DISCUSSION
Nuclear and cytoplasmic versions of human Pus10 have different activities
Previously, we showed by immunofluorescence and immunoblotting that human Pus10 is mainly present in the nucleus, and that it translocates to the mitochondria during TRAIL-induced apoptosis (Jana et al. 2017). Here again, by immunoblotting, we detect Pus10 only in the nucleus (Fig. 3C). However, we observed Ψ54 synthase activity only in the cytoplasmic extracts and not in the nuclear extracts. Knockdown and subsequent rescue experiments indicate this activity is due to Pus10. Although anti-Pus10 antibody did not detect native Pus10 in the cytoplasm (Fig. 3C), anti-His antibody could detect His-tagged Pus10 both in the nucleus and in the cytoplasm when Pus10 was transiently overexpressed in a knockdown cell line (Supplemental Fig. S2F). These observations about the locations of the Pus10 suggest that there are at least two different versions of Pus10 and that the anti-Pus10 antibody used here detects only the nuclear version. Nuclear and cytoplasmic Pus10 are distinct because the addition of nuclear extract to cytoplasmic extract does not alter Ψ54 synthase activity of the latter. The two versions of human Pus10 appear to harbor two distinct but apparently unrelated activities, pseudouridylation by cytoplasmic Pus10 and involvement of nuclear Pus10 in apoptosis.
Nuclear and cytoplasmic versions of the Pus10 protein are not the products of paralogous genes. We reported previously (Fitzek et al. 2018) that only a single pus10 gene per genome is observed both in archaeal and eukaryotic genomes. This is despite the occurrence of many genome duplications that have occurred during evolution of several eukaryotic lines. We believe that both versions of the protein are post-translationally modified and the modifications of the two versions are not identical. Our reasoning is based on the following. The predicted size of the E. coli-derived inactive Pus10 (Supplemental Fig. S5B) is 63.76 kDa (including purification tags). The predicted size of the cellular Pus10 (presumably nuclear) is 60.24 kDa, but it appears larger than E. coli-derived Pus10 (Supplemental Fig. S5B), suggesting that it has post-translational modifications. Similarly, SF9 cells-derived Pus10 is expected to be 64.46 kDa (including purification tags), but it also appears to be larger than its expected size (Supplemental Fig. S5B), again suggesting that it has post-translational modifications. Since (presumably) unmodified E. coli-derived protein is inactive and SF9 cells-derived protein can produce Ψ, we believe that cytoplasmic Ψ synthesizing Pus10 protein would also have post-translational modifications. However, the modifications of the SF9 cells-derived and human cytoplasmic proteins appear to be somewhat different, such that the anti-Pus10 antibody can recognize the former but not the latter.
In principle, nuclear and cytoplasmic Pus10 could be splice variants of the same gene product. However, based on the available information (https://www.proteinatlas.org/ENSG00000162927-PUS10/cell#human and http://useast.ensembl.org/Homo_sapiens/Transcript/ProteinSummary?db=core;g=ENSG00000162927;r=2:60940222-61018259;t=ENST00000421319), this is less likely. Human Pus10 is reported to have six splice variants. Of these, only four are protein coding, two of which give rise to annotated full length Pus10 (529 aa), and the other two are expected to produce proteins of 62 aa and 120 aa. Alignment of the shorter versions with the 529 amino acid version suggest that they match with only the N terminus of human Pus10, which would not contain the catalytic domain of the protein (McCleverty et al. 2007). The epitopes of the antibodies used in our study would not detect these smaller proteins. Human Pus10 splice variants that code for the 529 aa protein contain 18 exons.
Mammalian tRNAs containing the sequence 57-AAAU-60 have Ψ at position 54
All sequenced mammalian tRNAs that have 57-AAAU-60 (part of 53-GUUCAAAUC-61) contain Ψ54 instead of T54 (Jühling et al. 2009). Our transcripts containing this sequence also produced maximum levels of Ψ54 (Fig. 5A). However, our extracts produced some T54 in all tRNA transcripts, even where Ψ54 was also produced. The T54 and Ψ54 modifications in a tRNA are mutually exclusive and cannot be directly interconverted. Therefore, cells in vivo must have some mechanism that does not allow U54 to T54 conversion in 57-AAAU-60-containing tRNAs.
Requirement of the 57-AAAU-60 sequence for tRNA Ψ54 synthesis is probably relaxed in dividing and virus-infected cells
We could not produce Ψ54 in tRNALys1,2 by either cell extracts or recombinant Pus10 protein. This tRNA has 57-GAGC-60 (Supplemental Fig. S1A). However, as mentioned before, the amount of tRNALys2 increases in SV40-transformed mouse fibroblast cells and other dividing and proliferating tissues (Raba et al. 1979). This increased tRNA contains some Ψ54 (Raba et al. 1979), suggesting Pus10 may modify this tRNA under certain conditions. Similarly, one mouse leukemia tRNAArg has been shown to contain Ψ54 (Harada and Nishimura 1980). We could produce Ψ54 in the same tRNAArg isoacceptor by recombinant protein (Fig. 7). This tRNA has 57-GACU-60 (Supplemental Fig. S1A). Using a primer corresponding to this tRNAArg, we could also detect Ψ54 in mouse liver tRNAs (Fig. 1). These reports suggest that certain tRNAs increase in dividing and virus-infected cells and at least a fraction of these tRNAs may contain Ψ54, even if they do not contain the 57-AAAU-60 sequence. It is possible that a similar situation may exist in metabolically active tissues, e.g., liver cells. Probably certain factors or conditions in these cells relax the 57-AAAU-60 requirement of Pus10-mediated Ψ54 synthesis. This may be possible because some tRNA genes for most amino acids in animal genomes contain sequences that differ from 57-AAAU-60 only at one position (Jühling et al. 2009), and we showed (Fig. 5A) that variation at one position (except A58) from this 57-AAAU-60 reduces, but does not abolish Ψ54 synthesis.
tRNA Ψ54 and m1A58 modifications may be associated with retroviral DNA synthesis
Ψ54-containing tRNAs for Trp, Pro, and Gln of the host cell have been identified as primers for replication of retroviruses (Harada et al. 1975, 1979; Hu and Dahlberg 1983; Colicelli and Goff 1986; Marquet et al. 1995; Mak and Kleiman 1997). In addition, tRNAs for Lys are also suggested as primers for certain retroviruses—tRNALys1,2 for Mason-Pfizer monkey virus and human foamy virus, and tRNALys3 for mouse mammary tumor virus and lentiviruses, including HIV (Marquet et al. 1995; Mak and Kleiman 1997). These mammalian Lys tRNAs are reported to contain Tm54 (Raba et al. 1979; Jühling et al. 2009). However, as mentioned before, tRNALys2 can have Ψ54 under certain conditions (Raba et al. 1979) and we could produce Ψ54 in tRNALys3 both by using cell extract and recombinant protein (Figs. 2A, 7), suggesting that Ψ54 can be produced even in Lys tRNAs. To initiate DNA synthesis, 18 residues at the 3′ end of the primer tRNA pair with the primer binding site (PBS) of the viral genome and m1A58 of the tRNA terminates plus-strand strong-stop synthesis (Renda et al. 2001). The m1A58 also appears to be necessary for Ψ54 synthesis in vivo, because suppression of m1A58 formation by SAH, inhibits Ψ54 activity of cell extracts (Fig. 6). In HIV, several tRNAs are packaged in the virus but only tRNALys3 functions as a primer (Jiang et al. 1993; Eckwahl et al. 2016). HIV is known to modulate the tRNA pool to improve the translation efficiency of its late proteins (van Weringh et al. 2011). Therefore, theoretically HIV might also increase the population of Ψ54-containing tRNALys3, if present in low amounts, as is the case for tRNALys2 in some cells (Raba et al. 1979). tRNALys3 tightly associates with the HIV genome (Jiang et al. 1993). So far, the identity of the residue at position 54 of this tightly associated population of tRNALys3 has not been determined. It could be Tm or Ψ.
Ψ54 also appears to be involved in the interaction of tRNA with the retroviral reverse transcriptase, because interaction of avian and mammalian tRNATrp with avian myeloblastosis virus reverse transcriptase either requires or is stabilized by Ψ at positions 54 and 55 (Hu and Dahlberg 1983). All retroviral RNA genomes contain a motif called the primer activation signal (PAS), which pairs with an 8-nt segment (anti-PAS) of tRNA at positions 48–55, and this interaction is important for both primer selection and efficient reverse transcription (Beerens and Berkhout 2002). This anti-PAS element contains Ψ54 and Ψ55 in several primer tRNAs, e.g., tRNATrp, tRNAPro, and tRNAGln (Jühling et al. 2009).
TruB and Pus10 in mammalian cells seem to have redundant Ψ55 synthase activities
The tRNA Ψ55 synthase activities of Pus10 and TruB appear to be redundant, because they can both produce Ψ55 in all tRNAs and recognize the same TΨC arm sequence. Mammals have two paralogs of TruB, TruB1, and TruB2 (Zucchini et al. 2003; Rintala-Dempsey and Kothe 2017). TruB2 is present in the mitochondria (Antonicka et al. 2017) and probably produces Ψ55 in all four (out of 22 total) human mitochondrial tRNAs that contain this modification (Suzuki and Suzuki 2014). We cannot distinguish Ψ55 synthase activity of Pus10 from that of TruB1 in our extracts because active TruB1 is reported to be present in both nucleus and cytoplasm (Safra et al. 2017a).
Pus10 may produce Ψ in certain mRNAs
The orthologs, Pus4 and TruB1 have been shown to produce Ψ in mRNAs of yeast and mammals, respectively (Carlile et al. 2014; Schwartz et al. 2014; Safra et al. 2017a), in a consensus sequence GUUCNANNC (underlined U is converted to Ψ). Both TruB and Pus10 produce Ψ55 at the corresponding position in the TΨC arm consensus sequence GUUCRANYC, which is a subset of the mRNA consensus sequence. It is suggested that TruB1-mediated Ψ in mRNAs is predominantly produced in the nucleus (Safra et al. 2017a). In this case, these Ψ-containing mRNAs, when transported to the cytoplasm, would retain Ψ till they are degraded, because Ψ modification in RNA cannot be reversed. Therefore, it is possible that cytoplasmic versions of both TruB1 and Pus10 could produce Ψ in mRNAs under some inducible conditions that might have a regulatory role.
We observed that m1A58 in tRNA is required for Ψ54 synthesis. Both these modifications occur in tRNA because the Ψ54 consensus sequence GUUCAAAUC is a subset of the m1A consensus sequence GUUCNANNC observed in mRNAs in a 17-nt TΨC arm-like stem–loop structure (Safra et al. 2017b). This association suggests that Pus10 may also convert the first U of GUUCAAAUC in cytoplasmic mRNAs to Ψ (equivalent to Ψ54 of tRNAs) under inducible conditions, especially when m1A is present in the mRNA four residues 3′ to the Ψ site. There is evidence of the presence of two adjacent Ψ's in human mRNAs like OCIAD1, AK2, AKT1S1, and PMPCB (GEO accession number GSE60047) in a GUUCNANNC consensus motif (Schwartz et al. 2014). Possibly the first of the two Ψ’s in these cases are produced by Pus10.
Thus, to conclude, we have shown here that Pus10 is responsible for Ψ54 production in certain mammalian tRNAs. This activity is enhanced by another tRNA modification (m1A58). While this activity is generally dependent on a specific tRNA structural requirement, there exist certain exceptional conditions under which this requirement might be slightly relaxed. In addition to its tRNA Ψ54 activity, we show evidence that mammalian Pus10 can also form tRNA Ψ55. Based on several observations, we speculate that Pus10 may even produce Ψ in certain mRNAs. Finally, there is compelling evidence supporting the possible association between tRNA Ψ54 modification, and retroviral DNA synthesis. Our study demonstrating the function of Pus10 in tRNA modifications, opens up new questions for future studies, such as, whether the Ψ55 activity of Pus10 is just redundant with that of TruB1 or has a distinct role, whether Pus10-mediated tRNA modification is essential for DNA synthesis of HIV and other retroviruses, whether Pus10 is involved in the regulation of protein expression by modifying mRNAs in association with m1A modification, and whether the two activities (RNA modification and a role in apoptosis) of Pus10 indicate a role of RNA modifications in apoptosis.
MATERIALS AND METHODS
Standard molecular biology procedures (Green and Sambrook 2012) were used unless specifically described. Oligonucleotides used in this work are listed in Supplemental Table S1.
Determination of the presence of Ψ at specific positions in tRNAs
Total RNA from HeLa cells and C57BL/6 mouse livers was isolated using TRI Reagent according to the manufacturer's protocol. tRNAs were separated from other RNAs by PAGE on 6% denaturing gel and eluted. The presence of Ψ in the tRNA was determined by primer extension following CMCT treatment, as described previously (Majumder et al. 2016). Briefly, 20 µg tRNA enriched total RNA (6 µg tRNA and 14 µg total RNA) was treated with CMCT for 20 min at 37°C, while an untreated sample was simply incubated at 37°C for 20 min as control. After precipitation of the RNA, CMCT-treated samples were treated with 50 mM Na2CO3, pH 10.4, for 3 h at 37°C. Again, after precipitation, the RNAs were used for primer extension using [5′-32P]-labeled primers (listed in Supplemental Table S1) that hybridized close to and on the 3′ side of the position to be mapped for Ψ. M-MLV reverse transcriptase (Promega) was used for primer extension according to the manufacturer's instructions. Reaction products were separated by 12% sequencing gels. CMCT forms adducts with Ψ, U, and G. Alkali removes all CMCT groups except those attached to N3 of Ψ. The extension stops at one residue before the CMCT modified Ψ. A dark band in CMCT-treated lane indicates the presence of Ψ at that position. The position of the band in the gel that corresponds to Ψ is determined by its distance from the end of the primer and correlation with the known sequence of the RNA. This method is not suitable for quantitation of Ψ at a particular position in the RNA, because partial reaction conditions are used here for the CMCT reactions. Furthermore, reverse transcriptase reactions used for primer extensions show varying amounts of sensitivity to secondary structures and different modified residues present in the RNA, which can produce dark bands in both untreated and CMCT-treated lanes. We were not successful in conducting CMCT-primer extension analyses of tRNAAla and tRNACys. We believe this was due to the presence of four and five G's in the primers for these tRNAs that were needed to hybridize to four and five C's in the 3′ regions of these tRNAs (see tRNAAla and tRNACys sequences in Supplemental Fig. S1A).
DNA template construction and in vitro transcription
All human (Hu) tRNA genes were cloned in the pBluescript KS+ (Stratagene) vector between the EcoRI and HindIII sites. PCR products for cloning were prepared by using HeLa genomic DNA as template. tRNA PheU27A mutant was prepared by site directed mutagenesis. The previously described pVT9P11Δi plasmid (Gurha et al. 2007) was used for H. volcanii (Hv) tRNATrp. PCR-amplified DNAs using plasmids of tRNA gene clones (for Hu Trp, Hv Trp, Hu Phe, Hu PheT27A, Hu Lys1, Hu Lys3, Hu Gln, Hu Ala, Hu Asp, and Hu Cys) and HeLa genomic DNA (for Hu Pro, Hu Arg, and Hu Lys1,2) as template were used for in vitro transcription to obtain tRNA transcripts. Forward primers for PCR contained T7 RNA polymerase promoter sequences. The same forward primers were used to prepare PCR products for variants of Hu Trp, Hv Trp, and Hu Ala (see Fig. 5) transcripts, but the reverse primer contained the desired mutations. PCR products of T7P oligonucleotide hybridized to HVTRP39-R and HVTRP32-R were used to generate the transcripts for tRNA-32 and tRNA-39, respectively. In vitro transcriptions to prepare 32P-labeled transcripts were carried out as described before (Gurha et al. 2007; Gurha and Gupta 2008).
Pseudouridylation by cell extracts and thin layer chromatography
To prepare total cell extracts, the cells were grown to 80%–90% confluency in 100 mm plates. The cells were trypsinized and collected by centrifugation at 500g for 5 min. Cells were washed with PBS (137 mM NaCl, 27 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4). The cells were then suspended in buffer containing 10 mM Tris-Cl, pH 7.5, 150 mM KCl, 3 mM MgCl2, 0.3% NP-40, 10% glycerol and protease and phosphatase inhibitor (Pierce A32961, 1 mini tablet/10 mL buffer), and incubated on ice for 10 min. Cells were sonicated three times at 20× power for 15 sec each with 2 min incubation on ice after each sonication. This was followed by centrifugation at 18,000g for 15 min. The supernatant was aliquoted and flash frozen in liquid nitrogen and stored at −80°C for enzymatic assays. Nuclear and cytoplasmic extracts were prepared as described previously (Jana et al. 2017) and supplemented with 10% glycerol before storage at −80°C. Protein concentrations of the extracts were determined by the Coomassie Protein Assay reagent (Thermo Scientific 1856209).
Activities of extracts were determined by treating 4 pmole of in vitro transcribed tRNAs with 200 µg (protein) of extracts in a 50 µL reaction containing 20 mM Tris-Cl, pH 7.5, 2 mM DTT, 120 mM KCl, and 1 unit of Ribolock (ThermoScientific EO0382). Assays were done at 37°C for 30 min. The reactions were supplemented with 0.1 mM SAM (New England Biolabs B9003S) or 0.1 mM SAH (Sigma A9384) as needed. The reactions were stopped by adding 150 µL stop solution (0.2 mM EDTA, 0.1% SDS) followed by phenol–chloroform extraction, ethanol precipitation and digestion with RNase T2 or Nuclease P1. All assays were repeated twice. The data for each of the two independent experiments are presented in appropriate figures. Sometimes one control assay was used for multiple different assays when the same preparation of the labeled transcript was used for all those assays. The digests were resolved by two-dimensional thin-layer chromatography on cellulose plates (EMD Millipore) except for those in Supplemental Figure S2E, where PEI-cellulose plates (EMD Millipore) were used. The solvents used were Solvent I (isobutyric acid/0.5 N NH4OH, 5:3, v/v) for the first dimension and Solvent II (isopropanol/HCl/H2O, 70:15:15, v/v/v) or Solvent III [0.1 M sodium phosphate, pH 6.8/(NH4)2SO4/n-propanol, 100:60:2, v/w/v] for the second dimension (Gupta 1984; Gurha et al. 2007; Gurha and Gupta 2008). Radioactivity in the plates was revealed and quantified by phosphorimaging using Optiquant software. The percent Ψ or T produced in RNase T2 digests was determined by the formula: (radioactivity in Ψp or Tp spot x the number of labeled nucleotides per transcript × 100)/(sum of the radioactivity in all Np spots). The percent Ψ produced in nuclease P1 digests was determined by the formula: (radioactivity in pΨ spot × the number of U's in the RNA × 100)/(sum of the radioactivity in pU and pΨ spots).
Preparation of HEK293T Pus10-knockdown strain and expression of Pus10 in the strain
HeLa, MCF7 and PC3 cells were cultured as described previously (Jana et al. 2017). HEK293T cells were grown in DMEM media with 10% FBS and 100 U/mL penicillin and 100 µg/mL streptomycin. Pus10 knockdown strains KD1 and KD2 of PC3 cells have been described before (Jana et al. 2017). These were prepared using shRNAs TRCN0000134366 and TRCN0000133677, respectively, and pLKO.1 (RHS4080) as control vector (all from ThermoFisher). Pus10 knockdown strain KD1 of HEK293T cells was prepared just as described before for PC3 cells (Jana et al. 2017) by using shRNA TRCN0000134366, and the scrambled control was prepared by using nontargeting shRNA RHS6848 (ThermoFisher). Human Pus10 (HuP10) cDNA clone (8069212) was purchased from Open Bio Systems Dharmacon, Lafayette, CO, USA, and used as a template for amplification of the HuP10 gene. It was cloned in pEF6/V5-His-TOPO (Invitrogen) between the BamHI and XbaI sites to have V5 and six-His tags at the C terminus of the protein. The Kozak sequence was added to the forward primer for expression in mammalian cells and the protein also has a TEV protease cleavage site. This clone was codon changed by site directed mutagenesis. A catalytic Asp to Ala mutant clone was generated by a second site directed mutagenesis. HuP10 clones were transiently transfected in HEK293T knockdown cells using Turbofect (ThermoFisher) and extracts were prepared after 48 h incubation.
Cloning, purification and assays of recombinant Pus10
Pus10 was PCR amplified from the above-described HuP10 clone in the pEF/V5-His-TOPO vector to include the V5 and 6-His tags, and cloned in pFastBac (Invitrogen) between the BamHI and SstI sites. The PCR product generated was cut with PmeI and BamHI because HuP10 contains a restriction site for SstI. PmeI and SstI generate compatible overhangs for cloning. The Bac-to-Bac baculovirus expression system was used to overexpress the Pus10 protein. A bacmid was generated using the manufacturer's protocol (Invitrogen). SF9 cells were grown and transfected with the bacmid as described in the protocol (Invitrogen). Six hundred milliliters of SFX media was seeded with 1 × 106 cells/mL, and 60 mL of the viral P2 stock was added to it and grown for 72 h at 27°C at 80 rpm. The cells were pelleted and suspended in lysis buffer (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 20% glycerol) supplemented with EDTA free protease inhibitor (Pierce Protease Inhibitor, 88666, 1 mini tablet/10 mL buffer). The suspension was sonicated three times at 20× power and the lysate was centrifuged at 14,000 rpm for 20 min. The clear supernatant was bound to a pre-equilibrated Ni-NTA column with lysis buffer containing 3 mM imidazole. Binding was done by recirculation of the supernatant through the column for 1 h. The column was washed with five column volumes of wash buffer (20 mM Tris-Cl, pH 8.0, 300 mM NaCl, 20% glycerol, and 40 mM imidazole). The protein was eluted in fractions in elution buffer (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 20% glycerol, and 250 mM imidazole). The eluted fractions were run on an SDS-PAGE gel and the fractions containing the protein were pooled together and dialyzed against dialysis buffer (20 mM Tris-Cl, pH 8.0, 100 mM NaCl, 3 mM MgCl2, 20% glycerol, 30 pM ZnCl2, and 2 mM DTT). Concentrations of all recombinant proteins were determined by the Coomassie Protein Assay reagent.
The Ψ synthase activities of recombinant protein were determined by treating 4 pmoles of in vitro transcribed RNA with 20 µg of Pus10 in a 50 µL reaction consisting of 100 mM Tris-Cl, pH 8.0, 10 mM MgSO4, 0.1 mM EDTA, 100 mM NH4OAc, 0.5 mM DTT at 37°C for 1 h. The reaction was stopped by adding 150 µL stop solution (0.2 mM EDTA, 0.1% SDS) followed by phenol–chloroform extraction and ethanol precipitation. Further RNase digestion and TLC were done as described for cell extract assays. E. coli-derived recombinant human protein, which did not show any Ψ synthase activity, was prepared as described before (Jana et al. 2017) and used under the same conditions as SF9 cells-derived protein.
Other protein purifications and assays
TrmI was purified according to a published protocol (Droogmans et al. 2003). Briefly, a 200 mL culture was grown in LB media containing ampicillin to an OD600 = 0.5. IPTG was added to a final concentration of 1 mM and cells were harvested after 3 h of IPTG induction. The cell pellet was suspended in 10 mL of Buffer A (50 mM Tris-Cl, pH 8.5, 500 mM KCl) and lysed by bursts of 30 sec sonication for three times with storage of samples on ice for 2 min after each sonication. The lysate was cleared by centrifugation at 14,000 rpm for 10 min at 4°C. The supernatant was bound to a nickel column by recirculation for an hour. The column was washed with Buffer A and protein was eluted using a gradient of 0–1 M imidazole. The fractions containing protein were pooled together and dialyzed against buffer A containing 200 mM imidazole. High KCl and imidazole concentrations are needed to keep the protein soluble at high concentration (Droogmans et al. 2003). Aliquots of the enzyme were flash frozen in liquid nitrogen and stored at −80°C. m1A58 containing transcripts were prepared by slight modification of a published procedure (Droogmans et al. 2003). Four picomoles of labeled RNA was treated with 5 µg of TrmI in a 100 µL reaction consisting of 50 mM Tris-Cl, pH 8.0, 10 mM MgCl2, 0.5 mM SAM for 1 h at 60°C. The reaction was stopped by phenol–chloroform extraction followed by ethanol precipitation.
AlkB was purified according to a slight modification of a published protocol (Hrabeta-Robinson et al. 2017). Briefly, E. coli BL21(DE3) pLysS was transformed with AlkB-AVA421 and grown to A600 of 0.5 and induced with 1 mM IPTG for 2 h at 37°C. After harvesting the cells the pellet was suspended in sonication buffer (20 mM Hepes, pH 7.5, 5% glycerol, 1 M NaCl, 2 mM BME) and EDTA free protease inhibitor (Pierce, 88666, 1 mini tablet/10 mL buffer), sonicated thrice for 10 sec, and centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was flash frozen and stored at −80°C. A no salt buffer (5% Glycerol, 20 mM Tris-Cl, pH 8, 2 mM BME) of equal volume was added to the frozen extract. The mixture was added to 200 µL Talon resin prewashed in 0.5 M NaCl wash buffer (5% Glycerol, 20 mM Tris-Cl, pH 8, 0.5 M NaCl, and 2 mM BME) and rotated in a nutator for 1 h. The resin bound protein was obtained by centrifugation at 4000 rpm for 5 min. The resin was washed three times with 0.5 M NaCl buffer. Nonspecific binding was removed by washing with 5 mM imidazole containing buffer followed by suspending the resin in 10 mM imidazole containing wash buffer and eluting in 300 mM imidazole containing buffer. The protein was dialyzed against 20 mM Tris-Cl, pH 8.0, 50% glycerol, 200 mM NaCl, and 2 mM DTT. Twenty to forty micrograms of total RNA was treated with equal amounts (20–40 µg) of AlkB in a 100 µL reaction volume consisting of 50 mM Hepes KOH, pH 8.0, 75 µM Ferrous ammonium sulfate, pH 5.0, 1 mM alpha-ketoglutarate, 2 mM freshly prepared sodium ascorbate and 50 µg/mL of RNase/DNase free BSA in a final concentration for 100 min at 37°C. One hundred microliters of stop solution (11 mM EDTA, 200 mM ammonium acetate) was added to stop the reaction followed by phenol–chloroform and precipitation of RNA for downstream applications.
The TrmY (Mj1640) protein was purified under denaturing conditions and used as described previously (Chatterjee et al. 2012).
Immunoblotting and qPCR
Immunoblotting and qPCR were done exactly as described previously (Jana et al. 2017). We used anti-Pus10 (HPA049582, Sigma) antibody for the immunoblots shown in Figure 3C and Supplemental Figure S5B. We also used another commercial anti-Pus10 (HPA044736, Sigma) antibody, but it showed too many nonspecific bands in both nuclear and cytoplasmic extracts and was not useful. Anti-HisTag antibody (2365S, Cell Signaling) was used to check for overexpressed Pus10.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Phizicky (University of Rochester) and Louis Droogmans (Université Libre de Bruxelles) for clones for recombinant AlkB and TrmI, respectively, Judy Davie (Southern Illinois University) for providing mouse livers, pEF6/V5-His-TOPO vector, HEK293T cells, and allowing us to work in her facilities; Farid Kadyrov and Lyudmila Kadyrova (Southern Illinois University) for providing pFastBac1 vector, HeLa S3 and SF9 cells, and DH10BAC strain of E. coli; and David Clark (Southern Illinois University) for critical reading of the manuscript. This work was supported by the National Institute of General Medical Sciences, National Institutes of Health grant GM055945 to R.G.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.068114.118.
REFERENCES
- Antonicka H, Choquet K, Lin ZY, Gingras AC, Kleinman CL, Shoubridge EA. 2017. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep 18: 28–38. 10.15252/embr.201643391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. 2003. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 12: 627–637. 10.1016/S1097-2765(03)00348-4 [DOI] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Planta RJ, Grosjean H. 1997. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 25: 4493–4499. 10.1093/nar/25.22.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Berkhout B. 2002. Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA 8: 357–369. 10.1017/S1355838202028194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crécy-Lagard V, Gupta R. 2011. Pseudouridine formation in archaeal RNAs: the case of Haloferax volcanii. RNA 17: 1367–1380. 10.1261/rna.2712811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee K, Blaby IK, Thiaville PC, Majumder M, Grosjean H, Yuan YA, Gupta R, de Crécy-Lagard V. 2012. The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 18: 421–433. 10.1261/rna.030841.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J, Goff SP. 1986. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol 57: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. 2003. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res 31: 2148–2156. 10.1093/nar/gkg314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckwahl MJ, Arnion H, Kharytonchyk S, Zang T, Bieniasz PD, Telesnitsky A, Wolin SL. 2016. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA 22: 1228–1238. 10.1261/rna.057299.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzek E, Joardar A, Gupta R, Geisler M. 2018. Evolution of eukaryal and archaeal pseudouridine synthase Pus10. J Mol Evol 86: 77–89. 10.1007/s00239-018-9827-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M, Labouesse J, Dirheimer G, Fix C, Keith G. 1978. Primary structure of bovine liver tRNATrp. Biochim Biophys Acta 521: 198–208. 10.1016/0005-2787(78)90262-9 [DOI] [PubMed] [Google Scholar]
- Fujikane R, Behm-Ansmant I, Tillault AS, Loegler C, Igel-Bourguignon V, Marguet E, Forterre P, Branlant C, Motorin Y, Charpentier B. 2018. Contribution of protein Gar1 to the RNA-guided and RNA-independent rRNA: Ψ-synthase activities of the archaeal Cbf5 protein. Scientific Rep 8: 13815 10.1038/s41598-018-32164-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Grosjean H, Edqvist J, Stråby KB, Giegé R. 1996. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J Mol Biol 255: 67–85. 10.1006/jmbi.1996.0007 [DOI] [PubMed] [Google Scholar]
- Gupta R. 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem 259: 9461–9471. [PubMed] [Google Scholar]
- Gupta R. 1985. Transfer ribonucleic acids of archaebacteria. In The archaebacteria; the bacteria: a treatise on structure and function (ed. Woese CR, Wolfe RS), Vol. 8, pp. 311–343. Academic Press, New York. [Google Scholar]
- Gupta R. 1986. Transfer RNAs of Halobacterium volcanii: sequences of five leucine and three serine tRNAs. Syst Appl Microbiol 7: 102–105. 10.1016/S0723-2020(86)80131-X [DOI] [Google Scholar]
- Gurha P, Gupta R. 2008. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 14: 2521–2527. 10.1261/rna.1276508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Joardar A, Chaurasia P, Gupta R. 2007. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biol 4: 101–109. 10.4161/rna.4.2.5177 [DOI] [PubMed] [Google Scholar]
- Harada F. 1989. Nucleotide sequence of threonine tRNA from mouse leukemia cells. Nucleic Acids Res 17: 7517 10.1093/nar/17.18.7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F, Nishimura S. 1980. tRNAs containing the G-Ψ-Ψ-C sequence. I. Two arginine tRNAs of mouse leukemia cells. Biochem Int 1: 539–546. [Google Scholar]
- Harada F, Sawyer RC, Dahlberg JE. 1975. A primer ribonucleic acid for initiation of in vitro Rous sarcarcoma virus deoxyribonucleic acid synthesis. J Biol Chem 250: 3487–3497. [PubMed] [Google Scholar]
- Harada F, Peters GG, Dahlberg JE. 1979. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem 254: 10979–10985. [PubMed] [Google Scholar]
- Harada F, Matsubara M, Kato N. 1989. Nucleotide sequences of two glutamine tRNAs from HeLa cells. Nucleic Acids Res 17: 8371 10.1093/nar/17.20.8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabeta-Robinson E, Marcus E, Cozen AE, Phizicky EM, Lowe TM. 2017. High-throughput small RNA sequencing enhanced by AlkB-facilitated RNA de-methylation (ARM-Seq). Methods Mol Biol 1562: 231–243. 10.1007/978-1-4939-6807-7_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, Dahlberg JE. 1983. Structural features required for the binding of tRNATrp to avian myeloblastosis virus reverse transcriptase. Nucleic Acids Res 11: 4823–4833. 10.1093/nar/11.14.4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S, Hsieh AC, Gupta R. 2017. Reciprocal amplification of caspase-3 activity by nuclear export of a putative human RNA-modifying protein, PUS10 during TRAIL-induced apoptosis. Cell Death Dis 8: e3093 10.1038/cddis.2017.476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol 67: 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joardar A, Jana S, Fitzek E, Gurha P, Majumder M, Chatterjee K, Geisler M, Gupta R. 2013. Role of forefinger and thumb loops in production of Ψ54 and Ψ55 in tRNAs by archaeal Pus10. RNA 19: 1279–1294. 10.1261/rna.039230.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalampeta R, Kothe U. 2012. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci. Rep 2: 663 10.1038/srep00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalampeta R, Keffer-Wilkes LC, Kothe U. 2013. tRNA binding, positioning, and modification by the pseudouridine synthase Pus10. J Mol Biol 425: 3863–3874. 10.1016/j.jmb.2013.05.022 [DOI] [PubMed] [Google Scholar]
- Keith G. 1984. The primary structures of two arginine tRNAs (anticodons C-C-U and mcm5a2U-C-ψ) and of glutamine tRNA (anticodon C-U-G) from bovine liver. Nucleic Acids Res 12: 2543–2547. 10.1093/nar/12.5.2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24: 2411–2415. 10.1093/nar/24.12.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Beier H, Akita N, Nishimura S. 1987. Natural UAG suppressor glutamine tRNA is elevated in mouse cells infected with Moloney murine leukemia virus. Proc Natl Acad Sci 84: 2668–2672. 10.1073/pnas.84.9.2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M, Bosmeny MS, Gupta R. 2016. Structure-function relationships of archaeal Cbf5 during in vivo RNA-guided pseudouridylation. RNA 22: 1604–1619. 10.1261/rna.057547.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J, Kleiman L. 1997. Primer tRNAs for reverse transcription. J Virol 71: 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R, Isel C, Ehresmann C, Ehresmann B. 1995. tRNAs as primer of reverse transcriptases. Biochimie 77: 113–124. 10.1016/0300-9084(96)88114-4 [DOI] [PubMed] [Google Scholar]
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. 2007. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 373: 1243–1254. 10.1016/j.jmb.2007.08.053 [DOI] [PubMed] [Google Scholar]
- Mueller EG, Ferre-D'Amare AR. 2009. Pseudouridine formation, the most common transglycosylation in RNA. In DNA and RNA modification enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 363–376. Landes Bioscience, Austin, TX. [Google Scholar]
- Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C. 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA: Ψ55-synthase and RNA-guided RNA: Ψ-synthase activities. Nucleic Acids Res 35: 5610–5624. 10.1093/nar/gkm606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Leclerc F, Behm-Ansmant I, Fourmann JB, Charpentier B, Branlant C. 2008. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res 36: 2459–2475. 10.1093/nar/gkn077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswami R, Chen J, Burnett BP, Thimmig RL, Janjic N, McHenry CS. 2002. The HIV plus-strand transfer reaction: determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence. J Mol Biol 315: 311–323. 10.1006/jmbi.2001.5205 [DOI] [PubMed] [Google Scholar]
- Nishikura K, De Robertis EM. 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol 145: 405–420. 10.1016/0022-2836(81)90212-6 [DOI] [PubMed] [Google Scholar]
- Nordlund ME, Johansson JO, von Pawel-Rammingen U, Byström AS. 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6: 844–860. 10.1017/S1355838200992422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J. 1995. Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA 1: 102–112. [PMC free article] [PubMed] [Google Scholar]
- Ny T, Björk GR. 1980. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J Bacteriol 142: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raba M, Limburg K, Burghagen M, Katze JR, Simsek M, Heckman JE, Rajbhandary UL, Gross HJ. 1979. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur J Biochem 97: 305–318. 10.1111/j.1432-1033.1979.tb13115.x [DOI] [PubMed] [Google Scholar]
- Renda MJ, Rosenblatt JD, Klimatcheva E, Demeter LM, Bambara RA, Planelles V. 2001. Mutation of the methylated tRNALys3 residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J Virol 75: 9671–9678. 10.1128/JVI.75.20.9671-9678.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala-Dempsey AC, Kothe U. 2017. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol 14: 1185–1196. 10.1080/15476286.2016.1276150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34: 4293–4301. 10.1093/nar/gkl530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Nir R, Farouq D, Slutzkin IV, Schwartz S. 2017a. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res 27: 393–406. 10.1101/gr.207613.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. 2017b. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551: 251–255. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T. 2014. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357. 10.1093/nar/gku390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H. 2008. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol 67: 323–335. 10.1111/j.1365-2958.2007.06047.x [DOI] [PubMed] [Google Scholar]
- van Weringh A, Ragonnet-Cronin M, Pranckeviciene E, Pavon-Eternod M, Kleiman L, Xia X. 2011. HIV-1 modulates the tRNA pool to improve translation efficiency. Mol Biol Evol 28: 1827–1834. 10.1093/molbev/msr005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW. 2000. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 28: 2342–2352. 10.1093/nar/28.12.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Tai LW, Agris PF, Gehrke CW, Wong TW. 1983. The nucleotide sequence of a major glutamine tRNA from rat liver. Nucleic Acids Res 11: 1991–1996. 10.1093/nar/11.7.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchini C, Strippoli P, Biolchi A, Solmi R, Lenzi L, D'Addabbo P, Carinci P, Valvassori L. 2003. The human TruB family of pseudouridine synthase genes, including the Dyskeratosis Congenita 1 gene and the novel member TRUB1. Int J Mol Med 11: 697–704. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.