ABSTRACT
Cell migration, a key feature of embryonic development, immunity, angiogenesis, and tumor metastasis, is based on the coordinated regulation of actin dynamics and integrin-mediated adhesion. Rho GTPases play a major role in this phenomenon by regulating the onset and maintenance of actin-based protruding structures at cell leading edges (i.e. lamellipodia and filopodia) and contractile structures (i.e., stress fibers) at their trailing edge. While spatio-temporal analysis demonstrated the tight regulation of Rho GTPases at the migration front during cell locomotion, little is known about how the main regulators of Rho GTPase activity, such as GAPs, GEFs and GDIs, play a role in this process. In this review, we focus on a major negative regulator of RhoA, p190RhoGAP-A and its close isoform p190RhoGAP-B, which are necessary for efficient cell migration. Recent studies, including our, demonstrated that p190RhoGAP-A localization and activity undergo a complex regulatory mechanism, accounting for the tight regulation of RhoA, but also other members of the Rho GTPase family, at the cell periphery.
KEYWORDS: cell migration, GTPase activating proteins, lamellipodia, p190RhoGAP, Rho GTPases
Introduction: Rho GTPase regulation
Human Rho (Ras homologous) GTPases are a family of signaling molecules belonging to the Ras superfamily and comprising 20 members that can be classified in 8 subfamilies: Rho, Rac, Cdc42, RhoBTB, Rnd, RhoD/RhoF, RhoH and RhoV/RhoU.1,2 Rho GTPases are major modulators of the actin cytoskeleton during cell migration, but they also play a fundamental role in microtubule dynamics, membrane trafficking, proliferation, gene expression and cell differenciation.3-5 Due to these pleiotropic functions, Rho GTPases are involved in many pathological situations, such as cancers. Indeed, their transcriptional and post-transcriptional regulation as well as their GTPase activity are often disturbed in tumors, and direct mutations on Rho GTPases coding genes have been recently enlightened in various cancers.6
Most Rho GTPases are molecular switches that oscillate between an inactive GDP- and an active GTP-bound state, except atypical Rho GTPases (i.e., Rnds and RhoH) which are always bound to GTP. Activation of Rho GTPases is triggered by extra- or intracellular stimuli and GTPases are recruited to membranes in their GTP-bound conformation, where they bind downstream effectors (i.e., kinases, lipases, oxidases and scaffolding proteins).7 The most studied members of the Rho GTPase family, RhoA, Rac1 and Cdc42 control cell locomotion by generating dynamic actin-rich structures such as stress fibers, lamellipodia and filopodia, respectively.8 To achieve efficient cell migration, these Rho GTPases are sequentially activated/inactivated to generate cycles of membrane protrusion (i.e., lamellipodia and filopodia) and assembly of adhesion sites (i.e., stress fibers). Notably, FRET-based experiments using biosensors highlighted the spatio-temporal regulation of Rho proteins at leading edges.9 Consequently, failure in both adhesion sites generation and disassembly inhibits cell migration,10 demonstrating the requirement of an intermittent and localized activation of Rho GTPases.
To this end, their GTP hydrolysis activity is controlled by a wide range of post-translational modifications (i.e., prenylation of their C-terminal CAAX box) and by 3 types of regulators: guanine nucleotide exchange factors (GEFs), which facilitate GTP loading, GTPase-activating proteins (GAPs), which promote GTPase inactivation by enhancing their low GTP hydrolysis activity and GDIs which sequester Rho GTPases in the cytoplasm and inhibit their activity. Interestingly, there are >3-fold more RhoGAPs (over 70) than Rho GTPases (20), which fine-tune Rho GTPase activity in a space-, time-, physiologic- and cell type-dependent manner. While RhoGAPs have initially been considered as signal terminators compared with GEFs, recent studies unveiled their pivotal role in Rho GTPase signaling during neuronal morphogenesis, cell differentiation, endocytosis but also as tumor suppressors or oncogenes.11
The RhoGAP family
The RhoGAP family is characterized by a RhoGAP domain of 150 amino acids, sufficient for GTP-loaded Rho GTPase binding and stimulation.5,11 This RhoGAP domain is composed of 9 α helixes and a highly conserved arginine residue localized in a loop (arginine finger) that interacts with the GTP binding core (Switch I/II regions and P-loop) of the Rho GTPase. This interaction places the catalytic arginine residue in proximity of the glutamine in the active site of the GTPase, stabilizing charges of the transition state and decreasing the energetic barrier for GTP hydrolysis, which is very slow in absence of a RhoGAP.5 Substrate specificity of the RhoGAP domain can be highly restrictive (i.e., Myr5 inactivates only RhoA)11 or moderate (i.e., CdGAP and p190RhoGAP inactivate Rac1/Cdc4211 and RhoA12,13 /RhoC14/Rac1,15 respectively). Moreover, while Rho GTPases are ubiquitous, RhoGAPs expression is mainly tissue-specific,16 accounting for their diversity compared with the Rho.
Besides their RhoGAP module, most of the RhoGAP proteins contain multiple domains, conferring plasma membrane binding (i.e., pleckstrin homology), protein interactions (i.e., SH2, SH3) or enzymatic function (i.e., kinase) which modulate their GAP activity as well as providing them with additional functions.5 For example, the interaction of the central domain of CdGAP (a GAP for Cdc42 and Rac1) with the endocytosis regulator intersectin leads to a conformational change inhibiting its activity toward Rac1.17 Furthermore, these additional domains are also responsible for the subcellular localization of the RhoGAPs, thus restricting their catalytic activity only at specific subcellular locations. Notably, interaction of the PDZ (Postsynaptic density/Disc large/ZO-1) ligand motif of the RhoGAP deleted in liver cancer-3 (DLC3) with the PDZ domain of the polarity protein scribble targets DLC3 to adherens junctions.18 Likewise, the motor domain of myosinIX-A is necessary to restrict its RhoGAP activity to nascent cell-cell contacts during collective cell migration.19 Thus, the RhoGAPs diversity can be explained by their tissue-specific expression, their substrate specificity and the resulting specific signaling pathways, which are further complexified by their additional domains.
Two p190RhoGAP isoforms
p190RhoGAP is a major negative regulator of Rho GTPases and is expressed in the cells as 2 isoforms: p190RhoGAP-A (or p190A, also known as ARHGAP35 or GRLF1) and p190RhoGAP-B (or p190B, ARHGAP5) which probably comes from a gene duplication.20 The 2 isoforms of p190RhoGAP share a common domain organization (Fig. 1): i) a N-terminal GTP-binding domain (GBD) containing GTPase-consensus sites shared by other members of the Ras superfamily (70% homology between A and B)13,21; ii) 4 FF domains that are rare 50 amino acids motifs mostly found in nuclear RNA regulating proteins and containing 2 strictly conserved phenylalanines; this domain sequesters the transcription factor TFII-I into the cytoplasm in a phosphorylation-dependent manner22 and is implicated in p190A localization23; iii) a large middle domain (MD), which in p190A is similar to the transcriptional repressor of the glucocorticoid receptor GRF-1;21 this domain is the most divergent between the 2 isoforms (∼45% homology)24 but has a similar function in mediating protein-protein interactions implicated in the regulation of p190RhoGAP localization and function (see below);23,25 iv) p190A contains a p120RasGAP interacting domain (p120BD), reported to be necessary for its function and localization;26 and finally, v) a C-terminal GAP domain (∼70% homology between A and B isoforms) similar to that of BCR and n-chimaerin.21
Figure 1.
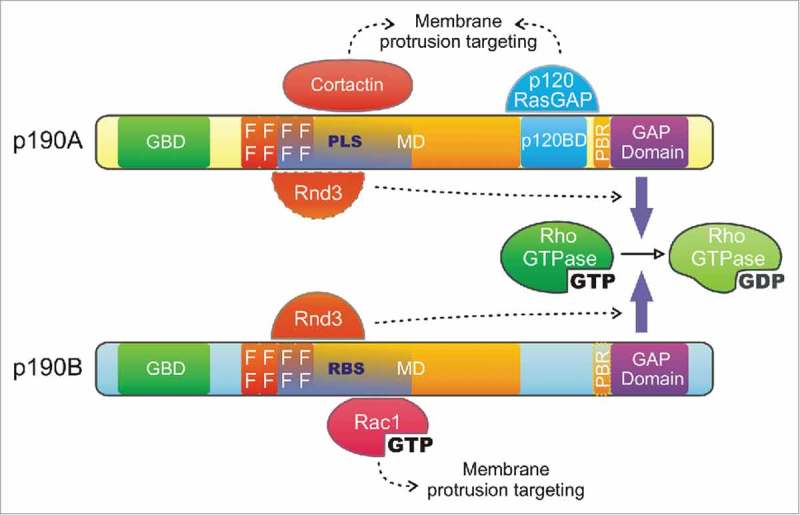
p190RhoGAP domain organization - The 2 p190RhoGAP isoforms share a common domain organization. From the N- to the C-terminus, these proteins contain: a GTP-binding domain (GBD), 4 FF motifs (FF), a large middle domain (MD) and a GAP domain (GAP), responsible for their catalytic activity. Binding of cortactin and p120RasGAP to the protrusion localization sequence (PLS) and p120RasGAP binding domain (p120BD) of p190A, respectively, as well as Rac1-GTP to the RBS of p190B targets p190RhoGAP isoforms to membrane protrusions. Rnd3 interaction through the Rac1-GTP binding sequence (RBS) of p190B and presumably the PLS of p190A stimulates their GAP activity toward RhoA. Interaction of the polybasic region (PBR, amino acids 1213–1236) with acidic phospholipids switches p190RhoGAP substrate preference from RhoA to Rac1.
The 2 isoforms are expressed in most of the tissues, with a high expression in the brain leading to perinatal mortality due to neural development defects when the genes coding p190A (ARHGAP35) and p190B (ARHGAP5) are knocked out in mouse models. Specifically, depletion of p190A leads to cell adhesion defects, i.e., abnormal eye development, hemispheres fusion and neural tube closure27 together with axon guidance and fasciculation defects.28 Mice lacking p190B also present neurogenesis and differentiation defects as well as a reduced size due to dysregulation of the insulin and IGF-1 pathways, affecting adipogenesis.29,30
To date, only few functional differences have been reported between p190RhoGAP isoforms. Like in central nervous system, both isoforms have similar but distinct roles in mammary gland development (i.e., ductal outgrowth or differentiation).31 Furthermore, p190A has also been implicated in lineage-type specification in the cardiac tissue,32 epithelial differentiation and polarity,33 cell-cell junctions34 and cell cycle.35 P190B has been shown to play a specific role in insulin/IGF-1 signaling,29,30 in RhoA inhibition upon tensional homeostasis at cell-cell contacts36 and in MCT-1-dependent neoplastic multinucleation that only stimulates the Src/p190B pathway.37 Finally, we have demonstrated in endothelial cells that p190A and p190B colocalize but have distinct functions in podosome assembly and extracellular matrix remodelling.38
Dysregulation of p190RhoGAP in tumors
Many data support that p190A is a tumor suppressor. First, at the cellular level, p190A inhibition results in transformation of NIH/3T3 fibroblasts whereas overexpression of the full-length protein or its GAP domain leads to dendritic-like phenotype, multi-nucleation, apoptosis and inhibition of Ras-dependent transformation, respectively.39,40 Moreover, its chromosomal locus (19q13.3) is often rearranged in solid tumors (pancreas, ovarian, thyroid tumors and gliomas)24 and p190A is under-expressed or deleted in various cancers (i.e., hepatocellular carcinoma or oligodendriogliomas).41,42 Recently, a large-scale study highlighted that mutations in p190A encoding gene, ARHGAP35 are found in 2% of global cancers and in 15% of endometrial tumors.43 In this study, the numerous nonsense mutations support the tumor suppressor function of p190A. However, p190A may also be considered as oncogenic, since high expression of p190A mRNA is associated with advanced states of lung carcinoma and colorectal cancer, and its expression in lung adenocarcinoma and breast carcinoma correlates with cell proliferation, migration, and invasion.44-46 Interestingly, we have recently identified cancer-occurring p190A mutations altering its function in an opposite way, i.e., S866F and Δ865–870 enhance while R997* decreases p190A RhoGAP activity.23 Hence, mutations in ARHGAP35 may have opposite consequences on p190A catalytic activity, which probably explains its role as both an oncogene and a tumor suppressor and underlines the requirement of a balanced Rho GTPase activity to preserve cells from tumorigenesis.
The implication of p190B has also been described in cancers and argues for an oncogenic role. ARHGAP5, the gene coding for p190B, is located in a locus (14q12) often rearranged in hepatocellular carcinoma (HCC) and p190B is overexpressed in Huh7, a HCC cell line.47 Furthermore, p190B haploinsufficiency inhibits breast tumorigenesis48 and it is critical for HCC cell migration in response to CD147.49 More recently, ARHGAP5 was found to be targeted by several miRNAs such as miR-744 and miR-486–5p to exert its pro-tumorigenic function in nasopharyngeal carcinoma and lung cancer, respectively.50,51
p190RhoGAP functions in cells
Both p190A and B have mainly a catalytic activity toward RhoA and Rac1 in cells,12,13,15 where p190A represents 60% of the RhoGAP activity in cultured fibroblasts.52
Negative regulation of RhoA
P190A was first described as a tyrosine phosphorylated protein in v-Src–transformed cells21,53 that inactivates RhoA upon integrin signaling in fibroblasts during cell migration/invasion.26,54-56 Overexpression of p190A in rat-1 fibroblasts inhibits premature stress fibers assembly during cell spreading and enhances protrusion formation and cell migration.56 Conversely, failure of RhoA inactivation by overexpression of the p190A GAP deficient mutant R1283A or p190A knockout increases stress fibers and inhibits protrusion formation and cell migration,56,57 demonstrating the importance of p190A in this process.
P190B is also recruited to the plasma membrane upon integrin cross-linking.13 Furthermore, p190B knockdown increases RhoA activity in Huh7 and inhibits cell spreading and migration.47 Likewise, p190B+/+, but not p190B−/− 3T3 fibroblasts elicited RhoA inactivation, stress fibers collapse and cell rounding upon interaction with Rnd1/3.58
Not only RhoA
Although an extensive amount of data demonstrated the effects of p190A on RhoA, its cellular functions may involve more GTPases. Indeed, p190A has been shown to control the onset and functionality of actin-rich structures, such as lamellipodia and invadopodia, by regulating RhoC activity in a local antagonism with p190RhoGEF.14,59 Moreover, the phenotype caused by overexpression of p190A in cells could be overcome by constitutively active mutants of either RhoA or Rac1.60 Indeed, both p190RhoGAP isoforms can modulate their substrate specificity by switching their GAP activity from RhoA to Rac1 upon direct interaction with plasma membrane acidic phospholipids (PPL).15,61,62 For p190A, interaction with PPL occurs through a patch of negatively charged amino acids (polybasic region) in its middle domain (amino acids 1191–1252) (Fig. 1).62 Phosphorylation of 2 serine residues by PKCα within this polybasic region dissociates p190A from PPL and inhibits its activity toward Rac1 while enhancing its activity toward RhoA. This process controls its cellular effects, i.e., cell spreading, formation of stress fibers and multinucleation.15,62 Accordingly, loss-of-function mutation L1396Q in the GAP domain of p190A has been shown to affect its catalytic activity toward both RhoA and Rac1 in the kidney cells primary cilium.63 Interestingly, while Rac1 is a substrate of p190A, it is a positive regulator of p190B activity by targeting and activating p190B at the plasma membrane (Fig. 1).25 Thus, to precisely control Rho GTPase activity, p190RhoGAP subcellular localization and function have to be tightly regulated themselves.
Subcellular localizations
In eukaryotic cells, p190A localizes homogeneously throughout the cytoplasm and accumulates in actin-rich structures where a regulation of RhoA is necessary, such as lamellipodia/ruffles,51 podosomes/invadosomes,43 new adhesion sites,50 intercellular junctions29 and cleavage furrow.35 Upon integrin signaling, p190A phosphorylation by Src family kinases (i.e., Src, Arg, Fyn), PKC27 or FAK64 promotes its association with p120RasGAP through its p120BD and its recruitment to the plasma membrane65-68 where p120RasGAP binds paxillin,69 allowing p190A to turn off RhoA26,54,55,70 (Fig. 2). The same mechanism has been shown to target p190A to adhesion sites upon semaphorin binding to plexin A1 and B1 for focal adhesion disassembly71 to cell-cell contacts through interaction between p190A and the p120-catenin/cadherin complex72 and to invadopodia.55 Moreover, at the rear of the lamellipodia, recruitment of active ERK by RACK1 to mature focal adhesions induces C-terminal inhibitory phosphorylation of p190A, allowing RhoA to promote the formation of stress fibers (Fig. 2).73 p190B is also targeted to adhesion sites upon integrin engagement13 and positively regulates cell migration.49 Interestingly, whereas p190B shares most of p190A binding partners (i.e., TFII-I or Rnd3),22,58 it is less tyrosine-phosphorylated and seems to not interact with p120RasGAP.74 This raises the possibility that p190B may be recruited to adhesions sites through a distinct mechanism. Accordingly, Bustos and collaborators found that p190B, but not p190A, interacts with Rac1 and is targeted to the plasma membrane in a Rac1-GTP-dependent manner (Fig. 1).25
Figure 2.
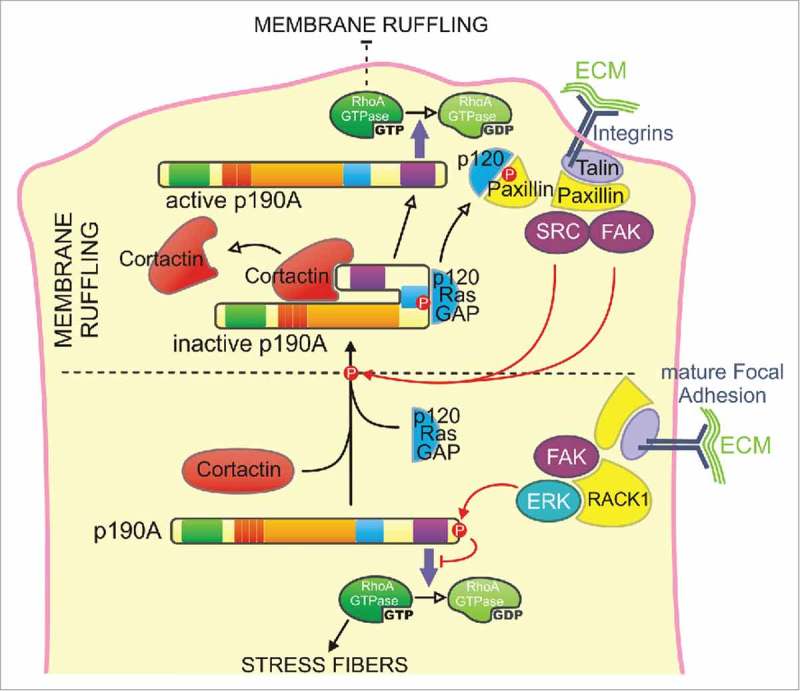
Model for the regulation of p190A targeting and activity at the cell leading edge - Two cooperative mechanisms may control p190A targeting to membrane protrusions. Cortactin binding to the PLS domain constitutively targets p190A in a closed/inactive conformation from the cytoplasm to the leading edge. Cortactin dissociation will result in the opening of the molecule, which locally turns off RhoA, thus allowing the maintenance of the newly expended membrane protrusion. Upon adhesion of the protrusion to the substratum, integrin signaling leads to the phosphorylation of p190A by Src or FAK on its p120BD and subsequent interaction with p120RasGAP (p120) that locally enhances p190A recruitment to the leading edge. p120RasGAP-paxillin interaction thus releases p190A that inactivates RhoA to limit premature adhesion maturation by stress fibers assembly. At the rear of the lamellipodia, recruitment of active ERK by RACK1 to mature focal adhesions induces C-terminal inhibitory phosphorylation of p190A, allowing RhoA to promote the formation of stress fibers.
Regulation
To ensure adapted Rho GTPase regulation, p190RhoGAPs catalytic activity is fine-tuned by post-translational modifications, such as phosphorylation, ubiquitylation/proteasome-dependent degradation and nitration as well as interaction with proteins and lipids. Phosphorylation, the most studied regulatory process of p190RhoGAPs activity toward RhoA, is achieved by various kinases which either stimulate, i.e., Src or FAK75; or inhibit their GAP activity, i.e., GSK3β57 ERK,76 PKSδ77 or the feedback loop p190A/RhoA/ROCK.78 For all these kinases, except PKCδ, phosphorylation data were obtained both on isolated protein and in the cellular context. This post-translational regulation of p190RhoGAP activity is different between isoforms. For instance, p190B is not subjected to C-terminal phosphorylation by GSK3β, which is necessary for p190A function in cell migration.57 These fast phosphorylation-induced activation/inhibition events are counterbalanced by phosphatases, i.e., low molecular weight (LMW) phosphatase,79,80 PTP2081 and Ptprz82 which control its inhibitory effect on RhoA activity and actin cytoskeleton remodeling.79,80
Coordinated implication of p190RhoGAP in Rho GTPases interplay at the leading edge
Analysis of the sequence of activation of Rho GTPases during cell migration using Rho GTPases biosensors demonstrated that RhoA plays a role in the onset of the protrusion, whereas Rac1 and Cdc42 are involved in the reinforcement and stabilization of the newly expanded protrusion.9 An explanation for the reciprocal balance between these Rho GTPases could be a local antagonistic crosstalk between them. Indeed, Cdc42 has been initially proposed to activate Rac1, which in turn activates or inactivates RhoA.8,83
Many data support that p190RhoGAP is a key molecule in Rho GTPases interplay at cell protrusions.25,84 First, the inhibition of RhoA by Rac1 has been shown to follow the “Bar-Sagi pathway,” involving consecutively the formation of reactive oxygen species (ROS), which inactivate the LMW phosphatases and activate p190RhoGAP. This mechanism involves other Rho GTPases, i.e., RhoG and Rnd3 that activate Dock180 (a GEF for Rac1) and p190A, respectively.85 Second, p190RhoGAP is the convergence point of integrin α5β1 and syndecan-4 in Rac1 and RhoA antagonism during cell attachment. Indeed, syndecan-4 activation of PKCα activates Rac1 and inhibits RhoA through phosphorylation and activation of p190A, which reinforces integrin-mediated activation of Rac1 and inactivation of RhoA via p190A,70,86 a phenomenon that may be tightly controlled by the PKCα-mediated inhibition of p190A RacGAP activity.62 Finally, caveolin-1 regulates cell migration by both activating Rac1 and inhibiting RhoA through the Src-dependent phosphorylation of p190A.87 Thus, many migratory signaling pathways concomitantly activate Rac1 and inactivate RhoA via p190RhoGAP, raising the possibility that these proteins physically interact during their targeting to and residency at actin-rich structures.
The PLS: a novel functional domain that allows p190A targeting to leading edges
While p190A targeting to actin-rich structures has been extensively studied and relies both on protein interactions (i.e., via anillin at cleavage furrow88) and phosphorylation (i.e., Src at focal adhesions54) little is known about the mechanisms ensuring its targeting to the leading edge of migrating cells. By a structure/function analysis of p190A domains, we characterized the region necessary and sufficient for p190A targeting to actin-rich cell edges.23 This domain that we called PLS for “protrusion localization sequence,” corresponds to the region encompassing amino acids 380–971 and containing the last 2 FF motifs and part of the MD but not the GAP domain nor the p120BD (Fig. 1). These data demonstrate that p190A is not targeted to the cell leading edge by its substrate, RhoA, but by cortactin that interacts with the PLS (Fig. 2). The PLS-dependent targeting of p190A further demonstrates that p120RasGAP interaction is not a prerequisite for its lamellipodial recruitment but rather improves its targeting upon specific stimuli leading to p190A phosphorylation (i.e., integrin engagement or growth factors). In this report, we also showed that cancer-occurring point mutation (S866F) and deletion (delta 865–870) phenocopy PLS deletion in abrogating p190A targeting to cells leading edge.23
Strikingly, a sequence similar to the PLS (amino acids 382–1007) has been found to mediate p190B interaction with Rnd3 and Rac1 and subsequent Rac1-GTP-dependent plasma membrane recruitment and activation (Fig. 1).25 While Rnd family members (Rnd1/2/3) interact and stimulate both p190A and B activity58 and Rnd1/3 are thought to recruit and activate p190A at plasma membrane lipid rafts,89 p190A, contrary to p190B, is targeted to lamellipodia in a Rnd3- and Rac1-independent manner, but cortactin-dependent manner.23,25 Interestingly, cortactin is itself targeted to the cell periphery by Rac1-GTP,90 which also recruits p190B.25 Thus, cortactin may play a scaffolding role for p190RhoGAPs recruitment to the cell periphery. Furthermore, cortactin targeting to lamellipodia depends on another RhoGAP, BPGAP191 and cortactin interacts with other Rho GTPases regulators, i.e., Fgd1 and Dock4, GEFs of Cdc42 and Rac1, respectively.92,93 Collectively, these data suggest that Rho GTPase spatio-temporal activation is likely to occur through the recruitment of multi-molecular protein complexes involving actin-binding proteins (i.e., cortactin, myosins), Rho GTPases and their specific GEFs and GAPs. Consistently, it has been shown that p190RhoGAP belongs to a protein complex containing the polarity protein Dvl and Tiam1, a GEF for Rac1.94 Whether p190B targeting to lamellipodia is also dependent on cortactin remains to be tested. However, such a different mechanism would explain the cellular difference in both p190RhoGAP functions, in addition to their distinct post-translational modifications and specific partners.
The PLS: a novel functional domain that regulates p190A GAP activity
A fast regulation of p190RhoGAP activity at a precise subcellular location by protein interactions and phosphorylation raised the hypothesis of a model based on a conformational switch.25,58 Our experiments unveiled that PLS domain deletion or mutations (S866F and delta 865–870) dramatically enhance p190A RhoGAP activity toward RhoA, assigning a double-role for the PLS as a regulator of p190A localization and function (Figs. 2 and 3).23 Likewise, p190B RhoGAP activity is increased upon deletion of its Rac1-GTP binding sequence (RBS) (Fig. 1).25 This suggests that p190A and B are regulated by a similar middle domain with distinct targeting properties (i.e., p120RasGAP/Cortactin and Rac1-GTP/Rnd3, respectively) but with a similar catalytic regulation. In this model, p190RhoGAPs are in a closed/less active conformation in the cytoplasm and are open/fully active only where a local regulation of RhoA is needed (Fig. 2). This mechanism would avoid an uncontrolled cellular RhoA inactivation, which would have deleterious effects, such as cell rounding and formation of long extensions (dendritic-like phenotype)60 that are intermediates of cell apopotosis.39 We further found that cancer-associated mutations in the PLS of p190A (S866F or delta 865–870) lead to a constitutively active form of the protein, as PLS domain removal, and favor loss of focal adhesion and random migration of tumor cells (Fig. 3).23 Auto-inhibition of the GAP activity by protein folding has been proposed for several GAPs such as p50RhoGAP, oligophrenin, Abr, chimerins5 and p190B25 and our data support this hypothesis for p190A.23 Whereas Rac1-GTP has been proposed to relieve p190B auto-inhibition, p190A is targeted to the leading edge through a Rac1- and Rnd3-independent pathway,23,25 and the mechanism for full activation of p190A at cell leading edges remains to be established. Indeed, we show that cortactin interaction is necessary for p190A targeting to cell edges but seems to maintain p190A in its closed state.23 Rnd3 activates p190A58 and limiting their interaction by competition with Plexin B2 inhibits the GAP activity of p190A on RhoA.95 This suggests that p190A and B are targeted to cell leading edge through a distinct mechanism but opening of the molecule may be ensured by a common mechanism, involving Rnd3. Finally, we showed that serine 866, not found in p190B, is a key amino acid for p190A localization and function,23 and may reveal a new phosphorylation-dependent regulation of p190A localization, consistent with its Abl/Arg phosphorylation-dependent recruitment to the plasma membrane shown previously.96 Considering that kinase localization is sometimes restricted to cell leading edge (i.e., aPKC97), it is tempting to hypothesize that phosphorylation or dephosphorylation of this serine may be responsible for the local auto-inhibitory regulation of p190A, dependent on migration cues.
Figure 3.
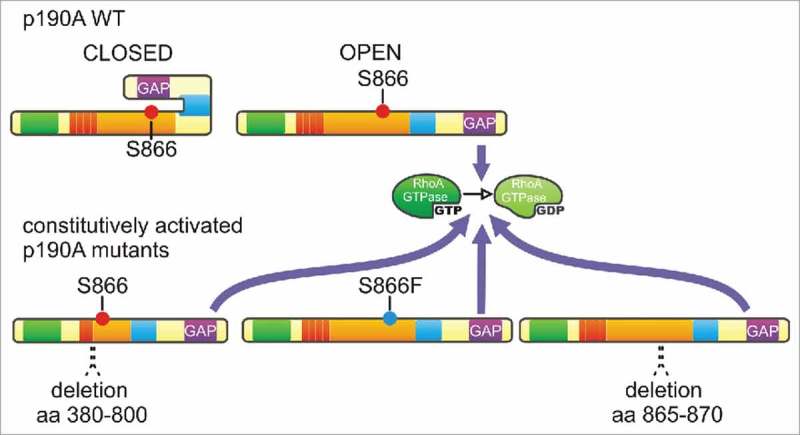
Auto-inhibition of the GAP activity is lost in p190A mutants - In the model of p190RhoGAP auto-inhibition, wild-type p190A (p190A WT) oscillates between a closed/inactive and an open/active form to ensure intermittent RhoA activity at the leading edge. PLS removal (deletion 380–800) and cancer-occurring point mutation (S866F) and deletion (deletion 865–870) maintain p190A in a constitutively active conformation that leads to enhanced and sustainable RhoA inactivation.
Concluding remarks
Compelling data suggest that p190RhoGAP is fundamental in the coordinated, localized and intermittent Rho family GTPase activity at the leading edge of migrating cells. Rac1-GTP and cortactin-mediated translocation of p190RhoGAPs to the leading edge is responsible for RhoA inhibition and is counterbalanced by RhoA GEFs that may belong to the same complex. Consistently, it has been shown that lamellipodia maintenance relies on p190RhoGAP activity toward RhoC that is restricted to the far leading edge, while RhoC activity is activated behind the leading edge by p190RhoGEF.14 The model of fast local release of p190RhoGAPs auto-inhibition could provide the time resolution compatible with the fast turnover of Rho GTPase activity at the leading edge (protrusion/retraction periodicity of ∼100s).9 Furthermore, p190A may have the intrinsic capacity to regulate Rho GTPases crosstalk at actin-rich protrusions by dynamically interacting with PPL at the leading edge, a phenomenon that switches its substrate specificity between RhoA and Rac1.15
The discovery of the complex, and sometimes opposite role played by the abundant Rho GTPase regulators depending on the physiologic context, highlights the complexity of Rho GTPases function in cell migration and proliferation and their pivotal role as oncogenes or tumor suppressors. Targeting Rho GTPases may consequently be inoperative as a cancer cure and understanding the various roles played by RhoGAPs, identifying their partners and post-translational modifications may help to specifically target their effect on tumor proliferation, epithelial-to-mesenchymal transition and metastasis.
Abbreviations
- GAP
GTPase activating protein
- GDI
guanine nucleotide dissociation inhibitor
- GEF
Guanine nucleotide exchange factor
- PLS
protrusion localization sequence
- PPL
phospholipids
- Rho
Ras homologous.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
ABM was supported by a PhD scholarship from the French Ministry of Research and an end-of-thesis short-term fellowship from the Fondation pour la Recherche Médicale, comité Aquitaine. FB was supported by an Idex Postdoctoral fellowship from the Université de Bordeaux. This work was supported by grants from Institut National du Cancer PLBIO-2014, La Ligue Contre le Cancer “Equipe labellisée 2016” and Comité de la Charente Maritime, and Fondation ARC pour la Recherche sur le Cancer.
References
- [1].Grise F, Bidaud A, Moreau V. Rho GTPases in hepatocellular carcinoma. Biochim Biophys Acta 2009; 1795:137-51; PMID: 19162129; http://dx.doi.org/ 10.1016/j.bbcan.2008.12.003 [DOI] [PubMed] [Google Scholar]
- [2].Haga RB, Ridley AJ. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016; 7:207-21; PMID:27628050; http://dx.doi.org/ 10.1080/21541248.2016.1232583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature 2002; 420:629-35; PMID:12478284; http://dx.doi.org/ 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- [4].Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21:247-69; PMID:16212495; http://dx.doi.org/ 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- [5].Ligeti E, Welti S, Scheffzek K. Inhibition and termination of physiological responses by GTPase activating proteins. Physiol Rev 2012; 92:237-72; PMID:22298657; http://dx.doi.org/ 10.1152/physrev.00045.2010 [DOI] [PubMed] [Google Scholar]
- [6].Porter AP, Papaioannou A, Malliri A. Deregulation of Rho GTPases in cancer. Small GTPases 2016; 7:123-8; PMID:27104658; http://dx.doi.org/ 10.1080/21541248.2016.1173767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays 2007; 29:356-70; PMID:17373658; http://dx.doi.org/ 10.1002/bies.20558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995; 81:53-62; PMID: 7536630; http://dx.doi.org/ 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- [9].Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature 2009; 461:99-103; PMID: 19693013; http://dx.doi.org/ 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells – over and over and over again. Nat Cell Biol 2002; 4:E97-100; PMID:11944043; http://dx.doi.org/ 10.1038/ncb0402-e97 [DOI] [PubMed] [Google Scholar]
- [11].Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell 2007; 99:67-86; PMID:17222083; http://dx.doi.org/ 10.1042/BC20060086 [DOI] [PubMed] [Google Scholar]
- [12].Ridley AJ, Self AJ, Kasmi F, Paterson HF, Hall A, Marshall CJ, Ellis C. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J 1993; 12:5151-60 PMID: 8262058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Burbelo PD, Miyamoto S, Utani A, Brill S, Yamada KM, Hall A, Yamada Y. p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J Biol Chem 1995; 270:30919-26; PMID: 8537347; http://dx.doi.org/ 10.1074/jbc.270.52.30919 [DOI] [PubMed] [Google Scholar]
- [14].Bravo-Cordero JJ, Sharma VP, Roh-Johnson M, Chen X, Eddy R, Condeelis J, Hodgson L. Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J Cell Sci 2013; 126:3356-69; PMID:23704350; http://dx.doi.org/ 10.1242/jcs.123547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Levay M, Bartos B, Ligeti E. p190RhoGAP has cellular RacGAP activity regulated by a polybasic region. Cell Signal 2013; 25:1388-94; PMID:23499677; http://dx.doi.org/ 10.1016/j.cellsig.2013.03.004 [DOI] [PubMed] [Google Scholar]
- [16].Csepanyi-Komi R, Safar D, Grosz V, Tarjan ZL, Ligeti E. In silico tissue-distribution of human Rho family GTPase activating proteins. Small GTPases 2013; 4:90-101; PMID:23518456; http://dx.doi.org/ 10.4161/sgtp.23708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jenna S, Hussain NK, Danek EI, Triki I, Wasiak S, McPherson PS, Lamarche-Vane N. The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J Biol Chem 2002; 277:6366-73; PMID:11744688; http://dx.doi.org/ 10.1074/jbc.M105516200 [DOI] [PubMed] [Google Scholar]
- [18].Hendrick J, Franz-Wachtel M, Moeller Y, Schmid S, Macek B, Olayioye MA. The polarity protein Scribble positions DLC3 at adherens junctions to regulate Rho signaling. J Cell Sci 2016; 129:3583-3596; PMID:27505894; http://dx.doi.org/ 10.1242/jcs.190074 [DOI] [PubMed] [Google Scholar]
- [19].Omelchenko T, Hall A. Myosin-IXA regulates collective epithelial cell migration by targeting RhoGAP activity to cell-cell junctions. Curr Biol 2012; 22:278-88; PMID:22305756; http://dx.doi.org/ 10.1016/j.cub.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burbelo PD, Finegold AA, Kozak CA, Yamada Y, Takami H. Cloning, genomic organization and chromosomal assignment of the mouse p190-B gene. Biochim Biophys Acta 1998; 1443:203-10; PMID: 9838117; http://dx.doi.org/ 10.1016/S0167-4781(98)00207-3 [DOI] [PubMed] [Google Scholar]
- [21].Settleman J, Narasimhan V, Foster LC, Weinberg RA. Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell 1992; 69:539-49; PMID: 1581965; http://dx.doi.org/ 10.1016/0092-8674(92)90454-K [DOI] [PubMed] [Google Scholar]
- [22].Jiang W, Sordella R, Chen GC, Hakre S, Roy AL, Settleman J. An FF domain-dependent protein interaction mediates a signaling pathway for growth factor-induced gene expression. Mol Cell 2005; 17:23-35; PMID: 15629714; http://dx.doi.org/ 10.1016/j.molcel.2004.11.024 [DOI] [PubMed] [Google Scholar]
- [23].Biname F, Bidaud-Meynard A, Magnan L, Piquet L, Montibus B, Chabadel A, Saltel F, Lagrée V, Moreau V. Cancer-associated mutations in the protrusion-targeting region of p190RhoGAP impact tumor cell migration. J Cell Biol 2016; 214:859-73; PMID:27646271; http://dx.doi.org/ 10.1083/jcb.201601063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tikoo A, Czekay S, Viars C, White S, Heath JK, Arden K, Maruta H. p190-A, a human tumor suppressor gene, maps to the chromosomal region 19q13.3 that is reportedly deleted in some gliomas. Gene 2000; 257:23-31; PMID: 11054565; http://dx.doi.org/ 10.1016/S0378-1119(00)00387-5 [DOI] [PubMed] [Google Scholar]
- [25].Bustos RI, Forget MA, Settleman JE, Hansen SH. Coordination of Rho and Rac GTPase function via p190B RhoGAP. Curr Biol 2008; 18:1606-11; PMID: 18948007; http://dx.doi.org/ 10.1016/j.cub.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bradley WD, Hernandez SE, Settleman J, Koleske AJ. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol Biol Cell 2006; 17:4827-36; PMID: 16971514; http://dx.doi.org/ 10.1091/mbc.E06-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, Silver J, Bronson RT, Settleman J. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development 2000; 127:4891-903; PMID: 11044403 [DOI] [PubMed] [Google Scholar]
- [28].Brouns MR, Matheson SF, Settleman J. p190 RhoGAP is the principal Src substrate in brain and regulates axon outgrowth, guidance and fasciculation. Nat Cell Biol 2001; 3:361-7; PMID:11283609; http://dx.doi.org/ 10.1038/35070042 [DOI] [PubMed] [Google Scholar]
- [29].Sordella R, Classon M, Hu KQ, Matheson SF, Brouns MR, Fine B, Zhang L, Takami H, Yamada Y, Settleman J. Modulation of CREB activity by the Rho GTPase regulates cell and organism size during mouse embryonic development. Dev Cell 2002; 2:553-65; PMID: 12015964; http://dx.doi.org/ 10.1016/S1534-5807(02)00162-4 [DOI] [PubMed] [Google Scholar]
- [30].Sordella R, Jiang W, Chen GC, Curto M, Settleman J. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell 2003; 113:147-58; PMID: 12705864; http://dx.doi.org/ 10.1016/S0092-8674(03)00271-X [DOI] [PubMed] [Google Scholar]
- [31].Heckman-Stoddard BM, Vargo-Gogola T, Herrick MP, Visbal AP, Lewis MT, Settleman J, Rosen JM. P190A RhoGAP is required for mammary gland development. Dev Biol 2011; 360:1-10; PMID:21945077; http://dx.doi.org/ 10.1016/j.ydbio.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kshitiz AJ, Kim DH, Levchenko A. Concise review: Mechanotransduction via p190RhoGAP regulates a switch between cardiomyogenic and endothelial lineages in adult cardiac progenitors. Stem Cells 2014; 32:1999-2007; PMID:24710857; http://dx.doi.org/ 10.1002/stem.1700 [DOI] [PubMed] [Google Scholar]
- [33].Bryant DM, Roignot J, Datta A, Overeem AW, Kim M, Yu W, Peng X, Eastburn DJ, Ewald AJ, Werb Z, et al.. A molecular switch for the orientation of epithelial cell polarization. Dev Cell 2014; 31:171-87; PMID:25307480; http://dx.doi.org/ 10.1016/j.devcel.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell 2006; 127:1027-39; PMID:17129786; http://dx.doi.org/ 10.1016/j.cell.2006.09.046 [DOI] [PubMed] [Google Scholar]
- [35].Su L, Pertz O, Mikawa M, Hahn K, Parsons SJ. p190RhoGAP negatively regulates Rho activity at the cleavage furrow of mitotic cells. Exp Cell Res 2009; 315:1347-59; PMID:19254711; http://dx.doi.org/ 10.1016/j.yexcr.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ponik SM, Trier SM, Wozniak MA, Eliceiri KW, Keely PJ. RhoA is down-regulated at cell-cell contacts via p190RhoGAP-B in response to tensional homeostasis. Mol Biol Cell 2013; 24:1688-99, S1681-1683; http://dx.doi.org/ 10.1091/mbc.E12-05-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu MH, Chen YA, Chen HH, Chang KW, Chang IS, Wang LH, Hsu HL. MCT-1 expression and PTEN deficiency synergistically promote neoplastic multinucleation through the Src/p190B signaling activation. Oncogene 2014; 33:5109-20; PMID:24858043; http://dx.doi.org/ 10.1038/onc.2014.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Guegan F, Tatin F, Leste-Lasserre T, Drutel G, Genot E, Moreau V. p190B RhoGAP regulates endothelial-cell-associated proteolysis through MT1-MMP and MMP2. J Cell Sci 2008; 121:2054-61; PMID: 18505793; http://dx.doi.org/ 10.1242/jcs.025817 [DOI] [PubMed] [Google Scholar]
- [39].Ludwig K, Parsons SJ. The tumor suppressor, p190RhoGAP, differentially initiates apoptosis and confers docetaxel sensitivity to breast cancer cells. Genes Cancer 2011; 2:20-30; PMID:21779478; http://dx.doi.org/ 10.1177/1947601911402680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang DZ, Nur EKMS, Tikoo A, Montague W, Maruta H. The GTPase and Rho GAP domains of p190, a tumor suppressor protein that binds the M(r) 120,000 Ras GAP, independently function as anti-Ras tumor suppressors. Cancer Res 1997; 57:2478-84 PMID: 9192829 [PubMed] [Google Scholar]
- [41].Kurokawa Y, Matoba R, Takemasa I, Nakamori S, Tsujie M, Nagano H, Dono K, Umeshita K, Sakon M, Ueno N, et al.. Molecular features of non-B, non-C hepatocellular carcinoma: a PCR-array gene expression profiling study. J Hepatol 2003; 39:1004-12; PMID: 14642619; http://dx.doi.org/ 10.1016/S0168-8278(03)00473-2 [DOI] [PubMed] [Google Scholar]
- [42].Smith JS, Jenkins RB. Genetic alterations in adult diffuse glioma: occurrence, significance, and prognostic implications. Front Biosci 2000; 5:D213-31; PMID: 10702383; http://dx.doi.org/ 10.2741/Smith [DOI] [PubMed] [Google Scholar]
- [43].Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505:495-501; PMID:24390350; http://dx.doi.org/ 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shen CH, Chen HY, Lin MS, Li FY, Chang CC, Kuo ML, Settleman J, Chen RH. Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res 2008; 68:7779-87; PMID:18829532; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0997 [DOI] [PubMed] [Google Scholar]
- [45].Notsuda H, Sakurada A, Endo C, Okada Y, Horii A, Shima H, Kondo T. p190A RhoGAP is involved in EGFR pathways and promotes proliferation, invasion and migration in lung adenocarcinoma cells. Int J Oncol 2013; 43:1569-77; PMID: 24043274; http://dx.doi.org/ 10.3892/ijo.2013.2096 [DOI] [PubMed] [Google Scholar]
- [46].Li L, Li YM, Zhou P, Wang XS, Wang GY, Zhao XH, Cui BB, Ren YL, Dong XS, Chen ZQ. Abnormal expression of p190RhoGAP in colorectal cancer patients with poor survival. Am J Transl Res 2016; 8:4405-14;PMID: 27830024 [PMC free article] [PubMed] [Google Scholar]
- [47].Gen Y, Yasui K, Zen K, Nakajima T, Tsuji K, Endo M, Mitsuyoshi H, Minami M, Itoh Y, Tanaka S, et al.. A novel amplification target, ARHGAP5, promotes cell spreading and migration by negatively regulating RhoA in Huh-7 hepatocellular carcinoma cells. Cancer Lett 2009; 275:27-34; PMID:18996642; http://dx.doi.org/ 10.1016/j.canlet.2008.09.036 [DOI] [PubMed] [Google Scholar]
- [48].Heckman-Stoddard BM, Vargo-Gogola T, McHenry PR, Jiang V, Herrick MP, Hilsenbeck SG, Settleman J, Rosen JM. Haploinsufficiency for p190B RhoGAP inhibits MMTV-Neu tumor progression. Breast Cancer Res 2009; 11:R61; PMID:19703301; http://dx.doi.org/ 10.1186/bcr2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen R, Wang SJ, Zhang Y, Hou R, Jiang JL, Cui HY. CD147 promotes cell motility via upregulation of p190-B RhoGAP in hepatocellular carcinoma. Cancer Cell Int 2016; 16:69; PMID:27601938; http://dx.doi.org/ 10.1186/s12935-016-0344-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fang Y, Zhu X, Wang J, Li N, Li D, Sakib N, Sha Z, Song W. MiR-744 functions as a proto-oncogene in nasopharyngeal carcinoma progression and metastasis via transcriptional control of ARH.GAP5. Oncotarget 2015; 6:13164-75; PMID:25961434; http://dx.doi.org/ 10.18632/oncotarget.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang J, Tian X, Han R, Zhang X, Wang X, Shen H, Xue L, Liu Y, Yan X, Shen J, et al.. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene 2014; 33:1181-9; PMID:23474761; http://dx.doi.org/ 10.1038/onc.2013.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vincent S, Settleman J. Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur J Cell Biol 1999; 78:539-48; PMID:10494860; http://dx.doi.org/ 10.1016/S0171-9335(99)80019-3 [DOI] [PubMed] [Google Scholar]
- [53].Ellis C, Moran M, McCormick F, Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature 1990; 343:377-81; PMID: 1689011; http://dx.doi.org/ 10.1038/343377a0 [DOI] [PubMed] [Google Scholar]
- [54].Arthur WT, Petch LA, Burridge K. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr Biol 2000; 10:719-22; PMID: 10873807; http://dx.doi.org/ 10.1016/S0960-9822(00)00537-6 [DOI] [PubMed] [Google Scholar]
- [55].Nakahara H, Mueller SC, Nomizu M, Yamada Y, Yeh Y, Chen WT. Activation of beta1 integrin signaling stimulates tyrosine phosphorylation of p190RhoGAP and membrane-protrusive activities at invadopodia. J Biol Chem 1998; 273:9-12; PMID: 9417037; http://dx.doi.org/ 10.1074/jbc.273.1.9 [DOI] [PubMed] [Google Scholar]
- [56].Arthur WT, Burridge K. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol Biol Cell 2001; 12:2711-20; PMID: 11553710; http://dx.doi.org/ 10.1091/mbc.12.9.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jiang W, Betson M, Mulloy R, Foster R, Lévay M, Ligeti E, Settleman J. p190A RhoGAP is a glycogen synthase kinase-3-beta substrate required for polarized cell migration. J Biol Chem 2008; 283:20978-88; PMID: 18502760; http://dx.doi.org/ 10.1074/jbc.M802588200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, Settleman J, Der CJ, Hansen SH. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr Biol 2003; 13:1106-15; PMID: 12842009; http://dx.doi.org/ 10.1016/S0960-9822(03)00418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bravo-Cordero JJ, Oser M, Chen X, Eddy R, Hodgson L, Condeelis J. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr Biol 2011; 21:635-44; PMID:21474314; http://dx.doi.org/ 10.1016/j.cub.2011.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tatsis N, Lannigan DA, Macara IG. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J Biol Chem 1998; 273:34631-8; PMID: 9852136; http://dx.doi.org/ 10.1074/jbc.273.51.34631 [DOI] [PubMed] [Google Scholar]
- [61].Ligeti E, Dagher MC, Hernandez SE, Koleske AJ, Settleman J. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J Biol Chem 2004; 279:5055-8; PMID: 14699145; http://dx.doi.org/ 10.1074/jbc.C300547200 [DOI] [PubMed] [Google Scholar]
- [62].Levay M, Settleman J, Ligeti E. Regulation of the substrate preference of p190RhoGAP by protein kinase C-mediated phosphorylation of a phospholipid binding site. Biochemistry 2009; 48:8615-23; PMID: 19673492; http://dx.doi.org/ 10.1021/bi900667y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Stewart K, Gaitan Y, Shafer ME, Aoudjit L, Hu D, Sharma R, Tremblay M, Ishii H, Marcotte M, Stanga D, et al.. A point mutation in p190A RhoGAP affects ciliogenesis and leads to glomerulocystic kidney defects. PLoS Genet 2016; 12:e1005785; PMID:26859289; http://dx.doi.org/ 10.1371/journal.pgen.1005785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tomar A, Lim ST, Lim Y, Schlaepfer DD. A FAK-p120RasGAP-p190RhoGAP complex regulates polarity in migrating cells. J Cell Sci 2009; 122:1852-62; PMID:19435801; http://dx.doi.org/ 10.1242/jcs.046870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].McGlade J, Brunkhorst B, Anderson D, Mbamalu G, Settleman J, Dedhar S, Rozakis-Adcock M, Chen LB, Pawson T. The N-terminal region of GAP regulates cytoskeletal structure and cell adhesion. EMBO J 1993; 12:3073-81 PMID: 8344248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bryant SS, Briggs S, Smithgall TE, Martin GA, McCormick F, Chang JH, Parsons SJ, Jove R. Two SH2 domains of p120 Ras GTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein. J Biol Chem 1995; 270:17947-52; PMID: 7629101; http://dx.doi.org/ 10.1074/jbc.270.30.17947 [DOI] [PubMed] [Google Scholar]
- [67].Hu KQ, Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J 1997; 16:473-83; PMID: 9034330; http://dx.doi.org/ 10.1093/emboj/16.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Roof RW, Haskell MD, Dukes BD, Sherman N, Kinter M, Parsons SJ. Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of p190 RhoGAP-p120 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol Cell Biol 1998; 18:7052-63; PMID: 9819392; http://dx.doi.org/ 10.1128/MCB.18.12.7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tsubouchi A, Sakakura J, Yagi R, Mazaki Y, Schaefer E, Yano H, Sabe H. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J Cell Biol 2002; 159:673-83; PMID: 12446743; http://dx.doi.org/ 10.1083/jcb.200202117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bass MD, Morgan MR, Roach KA, Settleman J, Goryachev AB, Humphries MJ. p190RhoGAP is the convergence point of adhesion signals from alpha 5 beta 1 integrin and syndecan-4. J Cell Biol 2008; 181:1013-26; PMID: 18541700; http://dx.doi.org/ 10.1083/jcb.200711129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Barberis D, Casazza A, Sordella R, Corso S, Artigiani S, Settleman J, Comoglio PM, Tamagnone L. p190 Rho-GTPase activating protein associates with plexins and it is required for semaphorin signalling. J Cell Sci 2005; 118:4689-700; PMID:16188938; http://dx.doi.org/ 10.1242/jcs.02590 [DOI] [PubMed] [Google Scholar]
- [72].Noren NK, Arthur WT, Burridge K. Cadherin engagement inhibits RhoA via p190RhoGAP. J Biol Chem 2003; 278:13615-8; PMID: 12606561; http://dx.doi.org/ 10.1074/jbc.C200657200 [DOI] [PubMed] [Google Scholar]
- [73].Klimova Z, Bráborec V, Maninová M, Čáslavský J, Weber MJ, Vomastek T. Symmetry breaking in spreading RAT2 fibroblasts requires the MAPK/ERK pathway scaffold RACK1 that integrates FAK, p190A-RhoGAP and ERK2 signaling. Biochim Biophys Acta 2016; 1863:2189-200; PMID:27212270; http://dx.doi.org/ 10.1016/j.bbamcr.2016.05.013 [DOI] [PubMed] [Google Scholar]
- [74].Matheson SF, Hu KQ, Brouns MR, Sordella R, VanderHeide JD, Settleman J. Distinct but overlapping functions for the closely related p190 RhoGAPs in neural development. Dev Neurosci 2006; 28:538-50; PMID:17028431; http://dx.doi.org/ 10.1159/000095116 [DOI] [PubMed] [Google Scholar]
- [75].Holinstat M, Knezevic N, Broman M, Samarel AM, Malik AB, Mehta D. Suppression of RhoA activity by focal adhesion kinase-induced activation of p190RhoGAP: role in regulation of endothelial permeability. J Biol Chem 2006; 281:2296-305; PMID:16308318; http://dx.doi.org/ 10.1074/jbc.M511248200 [DOI] [PubMed] [Google Scholar]
- [76].Pullikuth AK, Catling AD. Extracellular signal-regulated kinase promotes Rho-dependent focal adhesion formation by suppressing p190A RhoGAP. Mol Cell Biol 2010; 30:3233-48; PMID:20439493; http://dx.doi.org/ 10.1128/MCB.01178-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Fordjour AK, Harrington EO. PKCdelta influences p190 phosphorylation and activity: events independent of PKCdelta-mediated regulation of endothelial cell stress fiber and focal adhesion formation and barrier function. Biochim Biophys Acta 2009; 1790:1179-90; PMID:19632305; http://dx.doi.org/ 10.1016/j.bbagen.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mori K, Amano M, Takefuji M, Kato K, Morita Y, Nishioka T, Matsuura Y, Murohara T, Kaibuchi K. Rho-kinase contributes to sustained RhoA activation through phosphorylation of p190A RhoGAP. J Biol Chem 2009; 284:5067-76; PMID:19103606; http://dx.doi.org/ 10.1074/jbc.M806853200 [DOI] [PubMed] [Google Scholar]
- [79].Chiarugi P, Cirri P, Taddei L, Giannoni E, Camici G, Manao G, Raugei G, Ramponi G. The low M(r) protein-tyrosine phosphatase is involved in Rho-mediated cytoskeleton rearrangement after integrin and platelet-derived growth factor stimulation. J Biol Chem 2000; 275:4640-6; PMID: 10671492; http://dx.doi.org/ 10.1074/jbc.275.7.4640 [DOI] [PubMed] [Google Scholar]
- [80].Alho I, Costa L, Bicho M, Coelho C. Low molecular weight protein tyrosine phosphatase isoforms regulate breast cancer cells migration through a RhoA dependent mechanism. PLoS One 2013; 8:e76307; PMID:24086724; http://dx.doi.org/ 10.1371/journal.pone.0076307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Shiota M, Tanihiro T, Nakagawa Y, Aoki N, Ishida N, Miyazaki K, Ullrich A, Miyazaki H. Protein tyrosine phosphatase PTP20 induces actin cytoskeleton reorganization by dephosphorylating p190 RhoGAP in rat ovarian granulosa cells stimulated with follicle-stimulating hormone. Mol Endocrinol 2003; 17:534-49; PMID:12554790; http://dx.doi.org/ 10.1210/me.2002-0187 [DOI] [PubMed] [Google Scholar]
- [82].Tamura H, Fukada M, Fujikawa A, Noda M. Protein tyrosine phosphatase receptor type Z is involved in hippocampus-dependent memory formation through dephosphorylation at Y1105 on p190 RhoGAP. Neurosci Lett 2006; 399:33-8; PMID:16513268; http://dx.doi.org/ 10.1016/j.neulet.2006.01.045 [DOI] [PubMed] [Google Scholar]
- [83].Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol 1999; 147:1009-22; PMID: 10579721; http://dx.doi.org/ 10.1083/jcb.147.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Herbrand U, Ahmadian MR. p190-RhoGAP as an integral component of the Tiam1/Rac1-induced downregulation of Rho. Biological chemistry 2006; 387:311-7; PMID: 16542153; http://dx.doi.org/ 10.1515/BC.2006.041 [DOI] [PubMed] [Google Scholar]
- [85].Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol 2003; 5:236-41; PMID: 12598902; http://dx.doi.org/ 10.1038/ncb938 [DOI] [PubMed] [Google Scholar]
- [86].Bass MD, Roach KA, Morgan MR, Mostafavi-Pour Z, Schoen T, Muramatsu T, Mayer U, Ballestrem C, Spatz JP, Humphries MJ. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J Cell Biol 2007; 177:527-38; PMID:17485492; http://dx.doi.org/ 10.1083/jcb.200610076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Grande-Garcia A, Echarri A, de Rooij J, Alderson NB, Waterman-Storer CM, Valdivielso JM, del Pozo MA. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J Cell Biol 2007; 177:683-94; PMID:17517963; http://dx.doi.org/ 10.1083/jcb.200701006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Manukyan A, Ludwig K, Sanchez-Manchinelly S, Parsons SJ, Stukenberg PT. A complex of p190RhoGAP-A and anillin modulates RhoA-GTP and the cytokinetic furrow in human cells. J Cell Sci 2015; 128:50-60; PMID:25359885; http://dx.doi.org/ 10.1242/jcs.151647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Oinuma I, Kawada K, Tsukagoshi K, Negishi M. Rnd1 and Rnd3 targeting to lipid raft is required for p190 RhoGAP activation. Mol Biol Cell 2012; 23:1593-604; PMID:22357615; http://dx.doi.org/ 10.1091/mbc.E11-11-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Weed SA, Du Y, Parsons JT. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J Cell Sci 1998; 111(Pt 16):2433-43 PMID: 9683637 [DOI] [PubMed] [Google Scholar]
- [91].Lua BL, Low BC. BPGAP1 interacts with cortactin and facilitates its translocation to cell periphery for enhanced cell migration. Mol Biol Cell 2004; 15:2873-83; PMID:15064355; http://dx.doi.org/ 10.1091/mbc.E04-02-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Ueda S, Negishi M, Katoh H. Rac GEF Dock4 interacts with cortactin to regulate dendritic spine formation. Mol Biol Cell 2013; 24:1602-13; PMID:23536706; http://dx.doi.org/ 10.1091/mbc.E12-11-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Hou P, Estrada L, Kinley AW, Parsons JT, Vojtek AB, Gorski JL. Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Hum Mol Genet 2003; 12:1981-93; PMID: 12913069; http://dx.doi.org/ 10.1093/hmg/ddg209 [DOI] [PubMed] [Google Scholar]
- [94].Gon H, Fumoto K, Ku Y, Matsumoto S, Kikuchi A. Wnt5a signaling promotes apical and basolateral polarization of single epithelial cells. Mol Biol Cell 2013; 24:3764-74; PMID:24088568; http://dx.doi.org/ 10.1091/mbc.E13-07-0357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Azzarelli R, Pacary E, Garg R, Garcez P, van den Berg D, Riou P, Ridley AJ, Friedel RH, Parsons M, Guillemot F. An antagonistic interaction between PlexinB2 and Rnd3 controls RhoA activity and cortical neuron migration. Nat Commun 2014; 5:3405; PMID:24572910; http://dx.doi.org/ 10.1038/ncomms4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Peacock JG, Couch BA, Koleske AJ. The Abl and Arg non-receptor tyrosine kinases regulate different zones of stress fiber, focal adhesion, and contractile network localization in spreading fibroblasts. Cytoskeleton (Hoboken) 2010; 67:666-75; PMID:20737438; http://dx.doi.org/ 10.1002/cm.20479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Rosse C, Linch M, Kermorgant S, Cameron AJ, Boeckeler K, Parker PJ. PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol 2010; 11:103-12; PMID:20094051; http://dx.doi.org/ 10.1038/nrm2847 [DOI] [PubMed] [Google Scholar]


