ABSTRACT
Enzymes of the cytosine-5 RNA methyltransferase Trm4/NSun2 family methylate tRNAs at C48 and C49 in multiple tRNAs, as well as C34 and C40 in selected tRNAs. In contrast to most other organisms, fission yeast Schizosaccharomyces pombe carries two Trm4/NSun2 homologs, Trm4a (SPAC17D4.04) and Trm4b (SPAC23C4.17). Here, we have employed tRNA methylome analysis to determine the dependence of cytosine-5 methylation (m5C) tRNA methylation in vivo on the two enzymes. Remarkably, Trm4a is responsible for all C48 methylation, which lies in the tRNA variable loop, as well as for C34 in tRNALeuCAA and tRNAProCGG, which are at the anticodon wobble position. Conversely, Trm4b methylates C49 and C50, which both lie in the TΨC-stem. Thus, S. pombe show an unusual separation of activities of the NSun2/Trm4 enzymes that are united in a single enzyme in other eukaryotes like humans, mice and Saccharomyces cerevisiae. Furthermore, in vitro activity assays showed that Trm4a displays intron-dependent methylation of C34, whereas Trm4b activity is independent of the intron. The absence of Trm4a, but not Trm4b, causes a mild resistance of S. pombe to calcium chloride.
KEYWORDS: RNA methylation, tRNA, Trm4, NSun2, cytosine methylation, m5C
Introduction
The Trm4 family of methyltransferases catalyses the methylation of position 5 on cytosine to create 5-methyl-cytosine (m5C) in several tRNAs. Trm4 (also termed Ncl1) from S. cerevisiae generates m5C at four different positions in yeast tRNA [1]: C34, C40, C48 and C49. Position 48 lies at the junction between the variable loop and the TΨC-stem of tRNAs, and C49 is at the first position of the TΨC-stem. Either C48 or C49 alone, but not both simultaneously, are frequently methylated (m5C48 in 13 of 15 cases, m5C49 in 12 of 13 cases in S. cerevisiae) [2]. Furthermore, tRNAPheGAA carries Trm4-dependent m5C40 in S. cerevisiae, which lies in the anticodon stem-loop [1]. Finally, tRNALeuCAA is methylated at the wobble position C34 by Trm4. Interestingly, both respective tRNAPhe and tRNALeu genes carry an intron, and Trm4 requires the presence of the intron to carry out C34 and C40 methylation, whereas its activity on C48 and C49 is independent of the intron [1].
Functionally, m5C34 wobble base modification affects the efficiency of tRNALeu to act as an amber suppressor tRNA in vivo [3], whereas m5C40 is important for the spatial organization of the anticodon stem-loop and for formation of the Mg2+ binding pocket (reviewed in [4]). The absence of Trm4 in S. cerevisiae causes sensitivity towards the translational inhibitor paromomycin and synthetic growth defects in combination with the absence of other tRNA modifications, which is due to the fact that undermodified tRNAs are subject to rapid tRNA decay [5–7]. Also, the level of m5C34 methylation in tRNALeu is increased when S. cerevisiae cells are under oxidative stress, and this results in a codon-biased translation of proteins required for the response to the stress condition [8].
The Trm4 homolog from higher eukaryotes, NSun2 (NOP2/Sun domain family, member 2; MYC-induced SUN domain-containing protein, Misu) also is a tRNA methyltransferase, but NSun2 has also been reported to methylate non-tRNA substrates, including rRNA and other non-coding and coding RNAs [9,10]. For instance, NSun2 has been shown to methylate vault ncRNAs, which affects their processing into small regulatory RNAs [10,11]. The absence of NSun2 furthermore increases cleavage of tRNAs by angiogenin and causes higher accumulation of 5ʹ tRNA fragments [9], which affects translation and cellular stress response. These effects of NSun2 on RNA processing are hypothesized to contribute to disease states in humans, where homozygous mutation of the NSUN2 gene leads to the Dubowitz-like syndrome [12].
Intriguingly, widespread m5C in mRNAs has been reported in human [13,14] and plant cells [15,16], where hundreds of mRNA methylation sites were reported, and their methylation in Arabidopsis has been shown to depend on the TRM4B methyltransferase. However, this view of pervasive mRNA methylation has been challenged, since many methylation sites as determined by high-throughput bisulfite sequencing do not withstand stringent statistical filtering [17].
Most organisms, including human, mouse, Drosophila and S. cerevisiae, contain a single Trm4/NSun2 homolog. In contrast, the fission yeast Schizosaccharomyces pombe [18] as well as Arabidopsis thaliana and some other plants carry two homologs that are termed Trm4a and Trm4b [15] (Suppl. Figure 1). This raises the question how the two homologs affect (t)RNA methylation, and whether they have additional substrates. In an earlier study of tRNA methylation in S. pombe, we observed that m5C34 is present not only on tRNALeu, but also on a novel site, tRNAPro (CGG) [19], though the dependence of Trm4a or Trm4b remained to be determined.
In this study, we have used tRNA methylome analysis to determine the specificity of Trm4a and Trm4b from S. pombe. Interestingly, we found a clear division of labour in that Trm4a conducted all C48 methylation as well as C34 methylation on both tRNALeu and tRNAProCGG, whereas Trm4b methylated all C49 sites on tRNAs. In vitro, Trm4a was only able to methylate C34 of tRNAProCGG in the presence of the intron of the tRNA. Trm4b methylated C49 of tRNAProCGG in vitro independently of the intron, and it showed in vitro activity on C34. The absence of Trm4a causes S. pombe cells to be mildly resistant to calcium chloride, an indicator of mitochondrial function, whereas no obvious defects were observed in the absence of Trm4b. Altogether, our results highlight a strict distribution of tRNA substrates between the two NSUN2 homologs, which suggests distinct functions for methylation at two adjacent tRNA sites, positions C48 and C49.
Results
tRNA methylome analysis shows specialization in S. pombe of Trm4a for C34 and C48 methylation and Trm4b for C49
Trm4/NSun2-dependent tRNA methylation is known on C34, C40, C48 and C49 [1]. Since S. pombe carries two homologs, Trm4a and Trm4b, we asked which m5C sites on tRNAs depended on which of the two enzymes. For this purpose, we determined the complete tRNA methylome of S. pombe wt, trm4a∆, trm4b∆ and trm4a∆ trm4b∆ strains by RNA bisulfite treatment and next-generation sequencing of tRNAs. The resulting reads were mapped to the known S. pombe tRNAs [20] and analyzed for cytosine methylation. Comparison of tRNA methylation patterns in the individual strains allowed us to assign methylation sites to Trm4a and Trm4b (Figure 1(a), Suppl. Figure 1).
Figure 1.
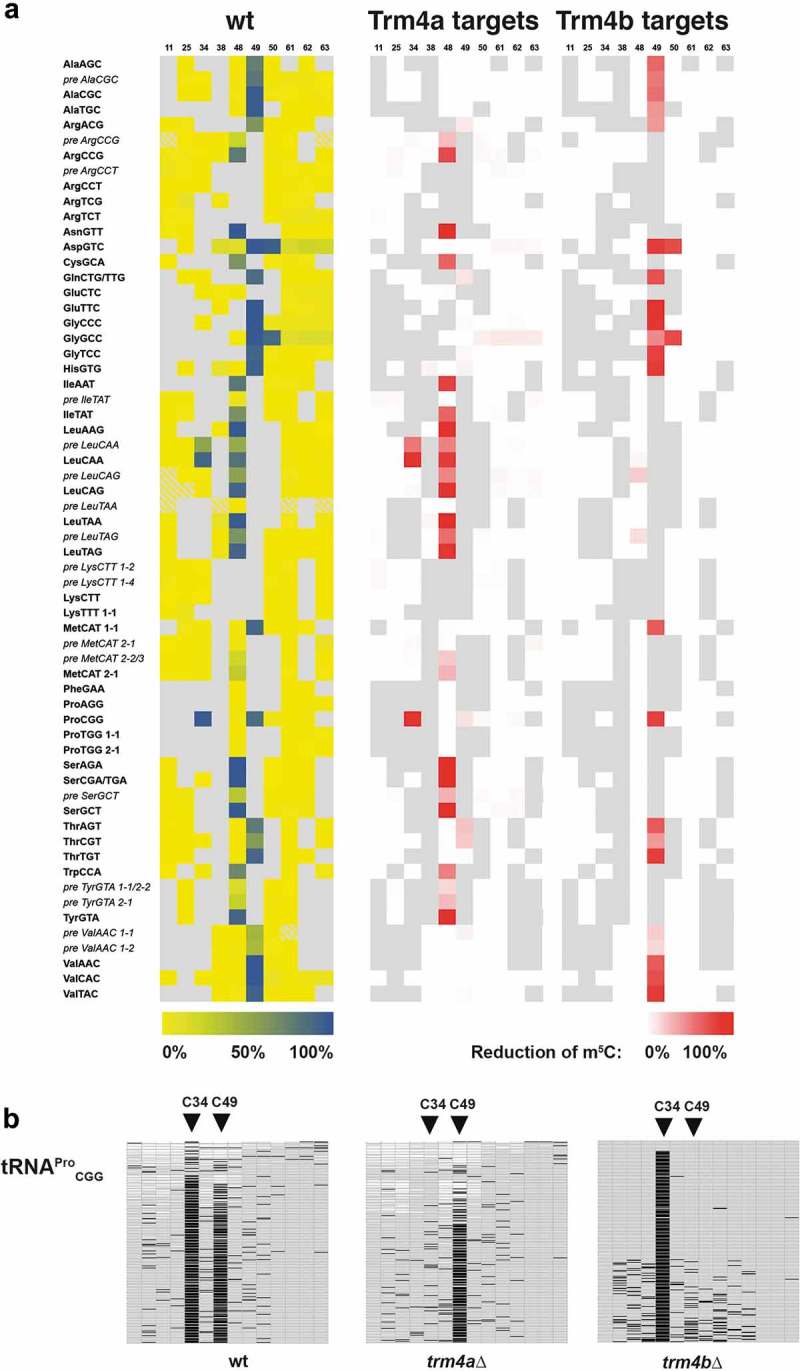
Genome-wide high-throughput tRNA bisulfite sequencing in Schizosaccharomyces pombe reveals selectivity of Trm4a for C34 and C48 methylation and Trm4b for C49. (a) Left, cytosine methylation levels are shown for individual tRNA species in wt cells. The top row indicates the position of the cytosine using standard tRNA numbering. tRNAs are sorted alphabetically by the respective amino acids they encode. The prefix ‘pre’ designates the unspliced version of selected tRNAs. Blue, strong methylation; yellow, no methylation. Grey fields indicate the absence of a C at that position. Yellow/grey cross-hatched fields indicate positions that constitute a C, but were not covered or had too low coverage in the high-throughput sequencing data. Middle, difference in tRNA methylation between wt and trm4a∆ cells. Red colours indicate a strong loss of methylation in trm4aΔ compared to wt. Right, difference in tRNA methylation between wt and trm4b∆ cells. Representation as in the middle panel. (b) Methylation of tRNAProCGG at C34 depended on Trm4a, and C49 methylation on Trm4b. Methylation levels were determined by targeted sequencing of bisulfite-treated tRNAProCGG.
Interestingly, C34 methylation depended on Trm4a in both tRNALeuCAA and tRNAProCGG (Figure 1(a), middle), since methylation levels at this site were at background levels in the trm4a∆ strain (Suppl. Figure 2). To further confirm this for tRNAProCGG, we performed high-throughput tRNA bisulfite sequencing on tRNAProCGG (Figure 1(b)). m5C34 and m5C49 levels were determined to be 89 and 83%, respectively, in a wild-type strain. In trm4a∆, the C34 methylation level dropped to background levels (8%), whereas C49 methylation remained at 82%. Conversely, m5C34 was at 94% in trm4b∆, and m5C49 dropped to 9%, thus confirming that Trm4a is responsible for m5C34 and Trm4b for m5C49 on tRNAProCGG in vivo.
For the methylation of C48 and C49, we observed a surprising selectivity of the Trm4 enzymes. Namely, Trm4a was responsible for all methylation of C48 (Figure 1(a), middle), and conversely, methylation of all C49 sites depended only on Trm4b (Figure 1(a), right). This was also apparent when the tRNA methylome data was not sorted alphabetically by the respective amino acid they carry (Figure 1(a)), but by the degree of methylation at C48 and C49 (Suppl. Figure 3). Also, as has been noted earlier [1], C48 and C49 are never methylated in the same tRNA. Thus, a picture emerged where C48 is carried out exclusively by Trm4a and C49 methylation by Trm4b. tRNAPro(CGG) is the only tRNA that is methylated both by Trm4a (C34) and Trm4b (C49).
In vitro activity of Trm4b on C49 and Trm4a on C34 of intron-containing tRNAPro
To verify the specificity of Trm4a and Trm4b, we investigated their in vitro methylation activity on in vitro-transcribed tRNAs (Figure 2(a)). Both proteins were heterologously expressed and purified from E. coli and used in in vitro tRNA methylation assays using tRNAProCGG as a substrate, which is methylated in vivo on C34 by Trm4a and on C49 by Trm4b. We also generated mutant versions of the tRNA in which either C34 or C49 was mutated to adenine (C34A, C49A).
Figure 3.
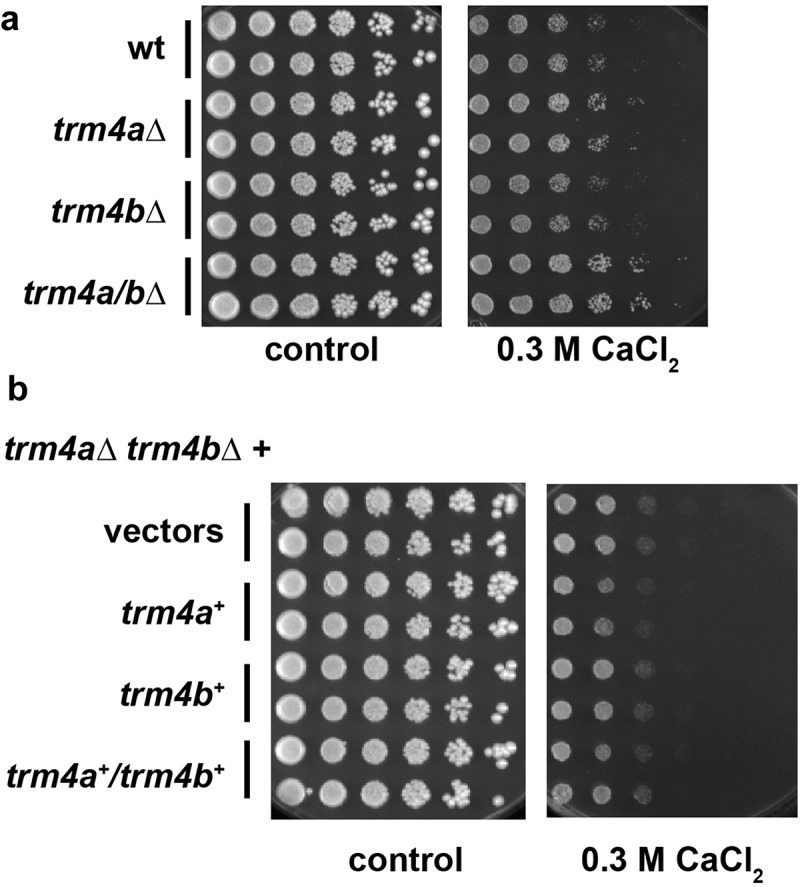
trm4a∆ causes mild resistance to calcium chloride. (a) The indicated strains (AEP1, AEP499, AEP391, AEP501) were serially diluted and spotted on full medium with or without 0.3M CaCl2. Plates were incubated for four days at 30ºC. (b) The CaCl2 resistance of trm4a∆ trm4b∆ (AEP501) was complemented by plasmid-borne trm4a+, but not trm4b+.
Significantly, Trm4b showed robust in vitro activity on wild-type tRNAProCGG. However, it was inactive on the C49A mutant version under the reaction conditions used here while maintaining activity on the C34A mutant version (Figure 2(b)). To further evaluate this, a time course of methylation was conducted with Trm4b and wild-type tRNAProCGG (wt) or the C34A and C49A versions. This showed that Trm4b was active on the wt tRNA; its activity was slightly enhanced on C34A and completely abrogated on C49A (Figure 2(c)), indicating that Trm4b has in vitro activity on tRNAProCGG and is specific for C49, which was in agreement with in vivo methylation data.
Figure 2.
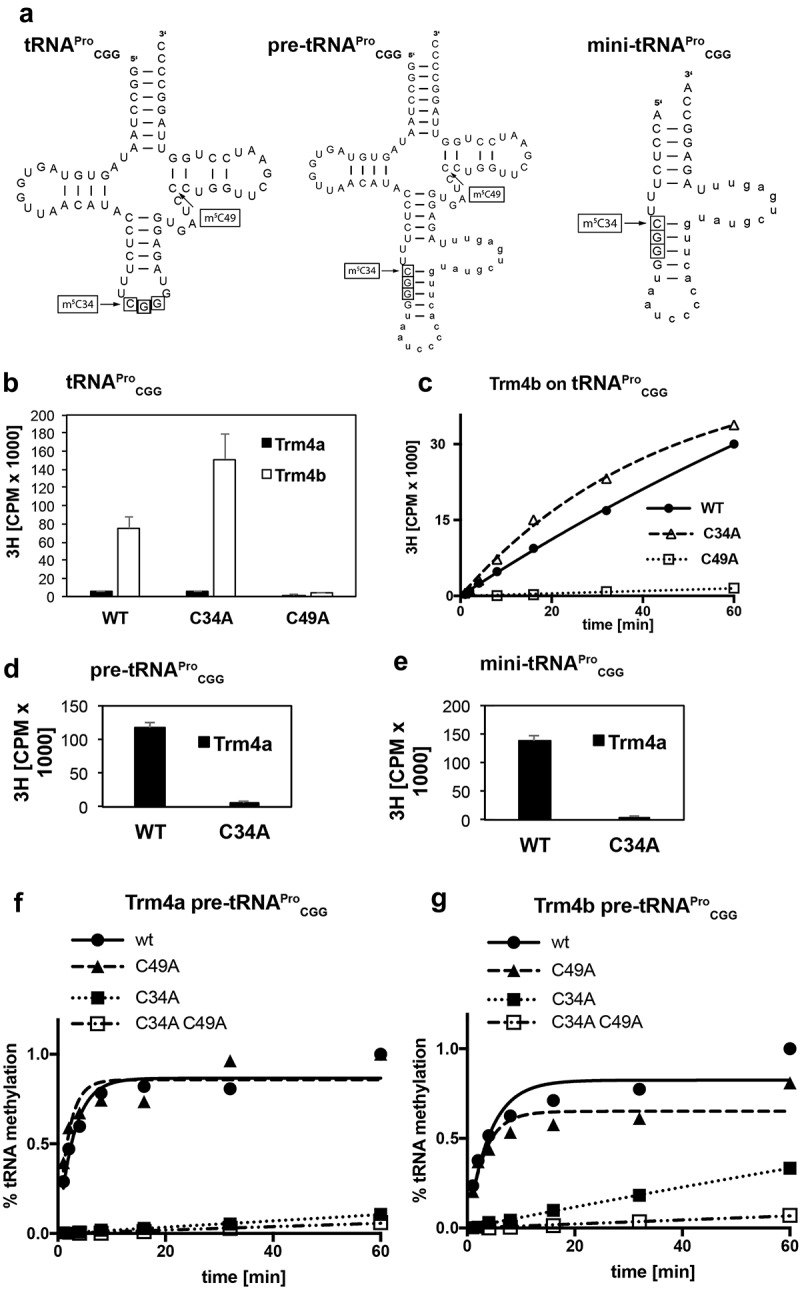
In vitro methylation activity of Trm4a and Trm4b shows that Trm4a catalyses intron-dependent C34 methylation in pre-tRNAProCGG. (a) tRNA sequences of tRNAProCGG (left), the intron-containing tRNAProCGG (middle, intron sequence in lower case) and a mini-tRNAProCGG version comprising the anticodon stem-loop and the intron. Anticodon nucleotides are boxed, and the position of C34 and C49 are indicated. (b) In vitro methylation of tRNAProCGG by Trm4a and Trm4b as determined by incorporation of 3H-labeled methyl groups into in vitro-transcribed tRNA substrates. The activity of Trm4b is abrogated by mutation of C49 to adenine (C49A), and Trm4a is inactive on tRNAProCGG. Error bars show the standard deviation (n = 3; reaction time 60 minutes). (c) Time course of tRNAProCGG methylation by Trm4b. Trm4b is inactive on the tRNA when C49 is mutated. (d, e) In vitro activity of Trm4a on pre-tRNAProCGG (d) and mini tRNAProCGG (e) is abrogated by mutation of C34 to adenine (C34A). The experiment was performed as in (b). (f) Time course of methylation of the intron-containing pre-tRNAProCGG by Trm4a. Trm4a activity is abrogated on pre-tRNAProCGG – C34A and – C34A C49A. (g) Time course of methylation of the intron-containing pre-tRNAProCGG by Trm4b. Trm4b activity was reduced on pre-tRNAProCGG-C34A and abrogated on – C34A C49A.
In contrast to Trm4b, Trm4a was inactive on tRNAPro and its mutant versions (Figure 2(b)). Since C34 methylation in tRNALeu by Trm4 from S. cerevisiae has previously been shown to depend on the presence of an intron in this tRNA [1], we asked whether Trm4a from S. pombe also depended on intronic sequences to be active. Therefore, we tested the activity of Trm4a on the intron-containing tRNAProCGG (pre-tRNAPro, Figure 2(a)) and a mini-tRNAPro, which consists of the anticodon loop that is extended by the intron (Figure 2(a)). Significantly, Trm4a activity was readily detected on the wild-type pre-tRNAProCGG and mini-tRNAProCGG substrates, but not in the C34A mutant versions (Figure 2(d,e)), indicating that Trm4a is a C34 methyltransferase for the intron-containing tRNAProCGG precursor.
To further characterize the activities of Trm4a as well as Trm4b on pre-tRNAPro, time courses of methylation on the wt as well as the mutant versions were conducted. Trm4a was active on wt pre-tRNAPro and pre-tRNAPro with C49A, but was inactive when C34 or C34 and C49 were mutated (Figure 2(f)). This supported the notion that Trm4a is only active on the intron-containing pre-tRNAPro to methylate C34.
The activity of Trm4b on pre-tRNAPro was more complex (Figure 2(g)). It showed robust activity on wt pre-tRNAPro. Surprisingly, Trm4b retained activity on C49-mutated pre-tRNAPro, which was in disagreement with our observation that all m5C49 in vivo depends on Trm4b and suggested that Trm4b methylated another cytosine in vitro on pre-tRNAPro. Furthermore, Trm4b showed reduced activity on the pre-tRNAPro version with C34A, which may indicate that full Trm4b activity requires methylation at C34. In vitro activity of Trm4b was only abrogated when both C34 and C49 were mutated, which is consistent with the notion that Trm4b methylates both C34 and C49 in vitro, even though it does not methylate C34 in vivo.
Altogether, these data suggest a scenario in which Trm4a first methylates C34 on the intron-containing tRNA, which subsequently is spliced. Trm4b then methylates C49, and its activity is stimulated by prior Trm4a-mediated methylation on C34. Alternatively, Trm4b activity could be stimulated by other modifications on mature tRNAPro.
Absence of Trm4, but not Trm4b, causes mild resistance to CaCl2
We next investigated whether the absence of Trm4a or Trm4b had phenotypic consequences in S. pombe. To this end, we determined how their absence affected the growth characteristics of S. pombe cells as well as their drug sensitivity. None of the strains showed a phenotype under a variety of conditions (Suppl. Table 1). In particular, the strains did not display sensitivity to paromomycin or oxidative stress, though the absence of TRM4 in S. cerevisiae has been shown to cause sensitivity to these agents [8,21]. Thus, the Trm4 homologs in S. pombe have a different function than their equivalent in S. cerevisiae.
Table 1.
S. pombe strains used in this study.
| Designation | Genotype | Source |
|---|---|---|
| AEP1 | h− leu1-32 ura4-D18 his3-D3 | YGRC |
| AEP102 | h+ leu1-32 ade6-216 ura4-D18 trm4a∆::kanMX | Bioneer |
| AEP162 | h+ leu1-32 ade6-216 ura4-D18 trm4a∆::kanMX trm4b::NatMX | [18] |
| AEP391 | h− leu1-32 ura4-D18 his3-D3 trm4b∆::NatMX | This study |
| AEP499 | h− leu1-32 ura4-D18 his3-D3 trm4a∆::NatMX | This study |
| AEP501 | h− leu1-32 ura4-D18 his3-D3 trm4a∆::kanMX trm4b∆::NatMX | This study |
Among the growth conditions we tested was cellular survival on increased levels of CaCl2, which is an indicator of mitochondrial function in S. cerevisiae [22]. Indeed, we observed that trm4a∆ showed a slightly increased resistance to CaCl2 (Figure 3(a)), and no effect was detected for trm4b∆. Accordingly, trm4a∆ trm4b∆ cells showed a similar CaCl2 resistance as trm4a∆ alone.
Since the CaCl2 resistance of trm4a∆ was rather mild, we sought to verify whether it could be complemented with trm4a+. For this purpose, the trm4a∆ trm4b∆ strain was transformed with a trm4a+, a trm4b+, or with both plasmids and tested for growth on CaCl2. The strain grew less well on CaCl2 when it carried the trm4a+ plasmid compared to the vector control, and this was irrespective of the presence of the trm4b+ plasmid (Figure 3(b)). This confirmed that the CaCl2 resistance was due to absence of Trm4a.
The CaCl2 resistance phenotype indicates that Trm4a may affect mitochondrial function in S. pombe, though how this occurs is unclear. The S. pombe mitochondrial genome encodes 25 tRNAs [23], necessitating the import of tRNAs into the mitochondria that are modified by Trm4a and Trm4b. However, both enzymes are localized to the nucleus (S. pombe Postgenome Database, http://www.riken.jp/SPD), and the coordination of methylation, splicing and subcellular localization of the tRNAs remains to be determined. An alternative possibility is that tRNA methylation by Trm4a affects translation of a protein involved in CaCl2 uptake or metabolism.
Discussion
Unlike most model organisms, S. pombe carries two paralogs of the Trm4/NSun2 family of RNA methyltransferases, which are termed Trm4a and Trm4b. Here, we report that the two enzymes have distinct specificities in that Trm4a is responsible for methylation of all C48 sites, whereas Trm4b methylates all C49 sites in tRNA. Furthermore, Trm4a, but not Trm4b, carries out C34 methylation in vivo. C34 methylation is found in tRNALeu, and we have reported a second C34 methylation site in tRNAProCGG in S. pombe whose modification also depends on Trm4a. In vitro, recombinant Trm4a showed activity on C34, though it was only active on an intron-containing version of the tRNA. Thus, with respect to its methylation activity, Trm4a most closely reflects the characteristics of Trm4/NSun2 homologs from other organisms that also depend on the presence of the intron for activity [1], whereas Trm4b methylates its tRNA substrates at C49 regardless of the presence or absence of the intron. Surprisingly, Trm4b in vitro was active on C34, although m5C34 in vivo was independent of Trm4b. At the cellular level, the absence of C34 and C48 in trm4a∆, but not C49 methylation in trm4b∆, causes a mild increase in resistance to CaCl2, which may indicate an effect of C34 and C48 methylation in mitochondria and/or calcium homeostasis.
The specificity of Trm4b on C49/C50 is in apparent conflict with our earlier work, where we reported that tRNAAsp methylation at C49, C50 and C61 – C63 depended on both Trm4a and Trm4b [18]. However, re-analysis of the trm4b∆ strain employed in that study revealed that it does not contain the trm4b+ deletion.
This apparent specialization of the two Trm4 homologs for C48 or C49 methylation raises the question of the evolutionary origin of these paralogs. Interestingly, the phylogenetic analysis (Suppl. Figure 1) shows that they derive from a relatively recent duplication event that occurred independently of the duplication in plants (e.g. Arabidopsis, Oryza sativa, Zea mays) and is not shared by other closely related fungi from the Candida or Aspergillus families. It remains to be seen what the relevance of this duplication in S. pombe is, especially since the absence of the two proteins causes only mild phenotypes.
A further question is what distinguishes the tRNA groups that are methylated at C48 or C49 from each other. Notably, those tRNAs that carried a methylated C48 at the edge of the variable loop did not carry a cytosine at position C49. Thus, it can be hypothesized that Trm4a is specific for the cytosine that is within this variable loop, adjacent to the TΨC-stem. If a tRNA carries both C48 and C49 (e.g. tRNAThr, tRNAVal, tRNAGln), then this tRNA is not a substrate for C48 methylation by Trm4a, but is methylated at C49 by Trm4b (Figure 1(a)).
In contrast to C48, C49 lies within the TΨC-stem and forms the first base-pair of the stem loop. We hypothesize that methylation by Trm4b is specific for this structure, since it methylates only C49 even in those tRNAs that also possess C48 (e.g. tRNAThr, tRNAVal, tRNAGln). For those tRNAs with C49 only, but no C48 (e.g. tRNAArgACG, tRNAGluTTC, tRNAGlyTCC), this position invariably is methylated by Trm4b.
Interestingly, while most tRNAs are methylated at either C48 or C49, but not both, we observed two tRNAs, tRNAAsp and tRNAGlyGCC, that are methylated at two adjacent cytosines, the positions C49 and C50. These two sites lie within the TΨC-stem. There exist other tRNAs with C49 and C50 that are only methylated at C49, but not C50 (e.g. tRNAAlaAGC, tRNAArgACG). Why some, but not other such C50-carrying tRNAs are targets for Trm4b methylation remains to be determined.
There also are some tRNAs that possess a C48, but are not subject to C48 methylation by Trm4a (e.g. tRNAGluCTC, tRNAMetCAT2-1, 2–2, 2–3; tRNAProTGG). Again, it is unclear what distinguished these tRNAs from the Trm4a targets. The 3ʹ base seems not to play a role, since both targets and non-targets can carry G or A at the position following C48.
Of note, Trm4 has previously been reported to methylate C40 in tRNAPhe. tRNAPhe in S. pombe possesses a cytosine at position 40, which our data indicate to be unmethylated, though the coverage of this site was very low in our tRNA methylome data, such that this has to be interpreted with caution (data not shown).
Next to determining the effects of both Trm4a and Trm4b on the full S. pombe tRNA methylome, we also investigated the phenotypic consequences of the absence of the two enzymes. Surprisingly, neither trm4a∆ nor trm4b∆ or both deletions combined caused strong phenotypes, even though trm4∆ in S. cerevisiae is known to cause sensitivity to the translation inhibitor paromomycin [21] as well as to oxidative stress [8]. Also, several genome-wide studies have reported phenotypes for trm4∆ in S. cerevisiae, including sensitivity to benomyl [24], hydroxyurea [25], caffeine [26] and cycloheximide [25]. S. cerevisiae trm4∆ furthermore shows a temperature-sensitive growth defect in combination with trm8∆ [5]. TRM8 encodes the methyltransferase for m7G46, a position that lies close to C48 and C49 in the variable loop of the tRNA. The temperature sensitivity arises due to the rapid tRNA decay (RTD) of tRNAValAAC [5]. Also, trm4∆ in S. cerevisiae is temperature-sensitive in the absence of Trm1, which generates N2, N2-dimethylguanosine at G26. Both trm4∆ trm1∆ and trm4∆ trm8∆ are suppressed by overexpression of the translation elongation factor EF-1A, which thus counteracts tRNA decay by the RTD pathway [7]. Whether trm4a∆ and trm4b∆ of S. pombe show similar synthetic genetic interactions with deletions in genes encoding other tRNA-modifying enzymes remains to be determined. In mice, the deletion of NSun2 is synthetically lethal with the deletion of Dnmt2, the m5C methyltransferase for C38 of tRNAAsp [27]. However, S. pombe cells lacking both Trm4 homologs as well as the Dnmt2 homolog Pmt1 are viable and have no obvious phenotype [18].
It will also be interesting to see whether Trm4a and Trm4b methylate mRNAs or other small RNAs in S. pombe, as has been reported for NSun2/TRM4B in mouse and plants [9–11,15,16], especially since the presence of m5C in mRNA has been widely discussed, with some reports arguing for wide-spread mRNA methylation [13], while stringent statistical filtering points towards only few m5C-modified mRNAs [17].
Materials and methods
S. pombe strains and growth conditions
The S. pombe strains used in this study are shown in Table 1. They were cultured in standard full medium (YES, 5 g/l yeast extract, 30 g/l glucose, 250 mg/l adenine, 250 mg/l histidine, 250 mg/l leucine, 250 mg/l uracil, 250 mg/l lysine).
To test CaCl2 sensitivity, serial dilutions of S. pombe strains were spotted on YES agar plates with or without CaCl2 (0.3 M), starting with OD600 = 2 and a dilution factor of 1:6 between consecutive spots.
Knockout of SPAC23C4.17 (trm4b+) in S. pombe was obtained by homologous recombination, and correct knockout was verified by polymerase chain reaction (PCR) analysis [28].
Plasmid constructions
For the construction of S. pombe vectors carrying trm4a+ and trm4b+, fragments with 500 bp upstream and downstream of the open reading frame were amplified from genomic DNA and cloned into PstI/SacI of pREP3x or pREP4x (Table 2).
Table 2.
Plasmids used in this study.
| Designation | Description | Source |
|---|---|---|
| pAE1688 | pJET1-tRNAAsp (S.pombe) | [18] |
| pAE2394 | pET15b-trm4a+ | This study |
| pAE2396 | pET15b-trm4b+ | This study |
| pAE2644 | pJET1-tRNAProCGG (S.pombe) | This study |
| pAE2645 | pJET1-tRNAProCGG (C34A) (S.pombe) | This study |
| pAE2646 | pJET1-tRNAProCGG (C49A) (S.pombe) | This study |
| pAE2720 | pJET1-mini tRNAProCGG (S.pombe) | This study |
| pAE2721 | pJET1-mini tRNAProCGG (C34A) (S.pombe) | This study |
| pAE2725 | pJET1-pre-tRNAProCGG | This study |
| pAE2726 | pJET1-pre-tRNAProCGG (C34A) | This study |
| pAE2931 | pREP3x- trm4a+ (LEU2) | This study |
| pAE2933 | pREP4x- trm4b+ (ura4+) | This study |
| pAE2939 | pJET1-pre-tRNAProCGG (C49A) | This study |
| pAE2949 | pJET1-pre-tRNAProCGG (C34A C49A) | This study |
tRNA methylome analysis
The methylome of S. pombe tRNAs was analyzed as described previously [19]. tRNAs from S. pombe were obtained by separating total RNA on a denaturing polyacrylamide gel and size-selection for tRNAs. Extracted RNAs were subjected to bisulfite conversion using the EZ RNA Methylation kit (Zymo). 3′ dephosphorylation and 5′ phosphorylation were performed using T4 polynucleotide kinase (TaKaRa). Library preparation for deep sequencing was done using the NEBNext Small RNA Library Prep Set for Illumina (New England Biolabs). A total of 200 ng of tRNAs per library were used as starting material and ligations done with undiluted adaptors. Adaptor-ligated cDNA was amplified with 12 cycles of PCR reaction and purified using the QIAQuick PCR Purification kit (Qiagen). Libraries were size selected with a 6% Novex tris-borate-EDTA (TBE) polyacrylamide gel (Life Technologies), extracted and ethanol precipitated according to NEBNext instruction manual and resuspended in EB buffer (Qiagen).
The libraries were multiplexed in equimolar ratios and sequenced on one lane of the Illumina HiSeq 2000 platform using paired-end 100 bp sequencing. Sequenced reads were aligned using BSMAP [29] with reference tRNA sequences downloaded from the genomic tRNA database http://gtrnadb.ucsc.edu/and from Pombase https://www.pombase.org/. Sequences were summarized so as to remove sequences that would be identical if all Cs are converted to Us using a custom bash script. Additionally, close sequences were identified using Clustal Omega https://www.ebi.ac.uk/Tools/msa/clustalo/and further pruned manually, so as to obtain a subset of sequences both representative and unique as viewed from the aligner. Aligned reads were quality-controlled, filtered for non-conversion artefacts and ambiguous alignment. Non-conversion ratio was calculated as the ratio of non-converted reads to all reads covering a specific position.
High-throughput bisulfite sequencing of individual tRNAs
Total RNA was isolated from 50 ml yeast cultures (OD600 = 1) using TriFast (PeqLab) according to the manufacturers instructions. Bisulfite sequencing of tRNAs was performed as described previously [19]. Primer sequences are listed in Suppl. Table 2. Processing included trimming of PCR primers, selection of high quality reads and sorting of the reads based on the sequence in the degenerate region of the RT-primer. Processed reads were analyzed for bisulfite conversion using BiQ Analyzer HT [30].
Purification of recombinant Trm4a and Trm4b
Total RNA extracted from wt S. pombe was used to generate cDNA with TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher). The cDNAs of trm4a+ and trm4b+ were subsequently amplified using gene specific primers and cloned into the pET15b vector using restriction sites introduced by the primers (BclI for trm4a+; XhoI and BamHI for trm4b+). His6-Trm4a and His6-Trm4b were expressed in Escherichia coli (DE3) Rosetta cells, and protein production was induced by auto-induction [31]. Purification of the recombinant protein was carried out using Profinity IMAC resin (Bio-Rad) and 200 mM imidazole (elution buffer: 30 mM potassium phosphate, 300 mM KCl, 10% glycerol, 0.1 mM DTT, 200 mM imidazole). The protein was then dialysed against dialysis buffer I (30 mM potassium phosphate pH 7, 200 mM KCl, 20% glycerol, 0.1 mM EDTA, 1 mM DTT) and dialysis buffer II (30 mM potassium phosphate pH 7, 100 mM KCl, 50% glycerol, 0.1 mM EDTA, 1 mM DTT).
RNA substrates for in vitro methylation
S. pombe tRNA substrates were obtained as previously described [18]. Briefly, tRNA sequences were either obtained from recursive PCR using four oligonucleotides (full-length tRNA) or from annealing of two complementary oligonucleotides (mini-tRNAs) and subsequently cloned into the pJET1 vector (Suppl. Table 2). In vitro transcription was performed using the TranscriptAid T7 High Yield Transcription Kit (ThermoFisher) according to the manufacturers instructions. 2 µg of the template vector, linearized with NcoI, was incubated for 4 hours at 37°C with nucleotides and T7 RNA polymerase in reaction buffer. After DNase I treatment, tRNAs were purified from the reaction using phenol/chloroform extraction followed by gel filtration with Sephadex G50 (GE Healthcare).
In vitro methylation assay
In vitro methylation activity of recombinant Trm4a and Trm4b was analyzed by tritium incorporation as previously described [32]. 20 µl reactions were set up by first diluting 2 µg total RNA or 250 ng tRNA in water and heating for 5 min at 65°C. After addition of methylation buffer and DTT (final concentration: 5 mM Tris–HCl pH 7.5, 5 mM NaCl, 0.5 mM MgCl2, 0.1 mM DTT) the mixture was allowed to cool to room temperature for 15 min, in order to allow for proper refolding of RNA substrates. Afterwards, 1.25 nM [methyl-3H]-AdoMet (Hartmann Analytic) and 1 µM of Trm4a or Trm4b was added to start the reaction (reaction temperature 30ºC). Assays were run for 60 min (Figure 2(b-e)) at 30°C and then spotted onto DE81 filters (Whatman). (t)RNA was precipitated by putting the filter in ice-cold 5% TCA. Filters were washed twice in 5% TCA and once in ethanol for 10 min each. Filters were dried and tritium incorporation was determined by liquid scintillation counting for 10 min per sample. Background was determined from reactions lacking an enzyme and later subtracted from obtained values. For the time courses of tRNA methylation, 500 ng of in vitro-transcribed tRNA in a 40 µl reaction was incubated with 1 µM of enzyme for the indicated times. Samples were processed as described above. Data were fitted to a single exponential reaction progress curve as described [33].
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft [SPP1784].
Acknowledgments
We wish to thank Xavi Sabaté Cadenas for work on this project and Silke Steinborn and Josta Hamann for technical support. This work was supported by the Deutsche Forschungsgemeinschaft, Priority Programme “Chemical modifications of native nucleic acid modifications” (SPP1784) to A. E. E.-M. and F. L. The authors declare no conflict of interest.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Motorin Y, Grosjean H.. Multisite-specific tRNA: m5C-methyltransferase(Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2009;38:1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Strobel MC, Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986;6:2663–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. [DOI] [PubMed] [Google Scholar]
- [5].Alexandrov A, Chernyakov I, Gu W, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. [DOI] [PubMed] [Google Scholar]
- [6].Chernyakov I, Whipple JM, Kotelawala L, et al. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5ʹ-3ʹ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dewe JM, Whipple JM, Chernyakov I, et al. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA. 2012;18:1886–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chan CT, Pang YL, Deng W, et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo J. 2014;33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hussain S, Sajini AA, Blanco S, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martinez FJ, Lee JH, Lee JE, et al. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012;49:380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].David R, Burgess A, Parker B, et al. Transcriptome-wide mapping of RNA 5-Methylcytosine in arabidopsis mRNAs and noncoding RNAs. Plant Cell. 2017;29:445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cui X, Liang Z, Shen L, et al. 5-Methylcytosine RNA methylation in arabidopsis Thaliana. Mol Plant. 2017;10:1387–1399. [DOI] [PubMed] [Google Scholar]
- [17].Legrand C, Tuorto F, Hartmann M, et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27:1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Becker M, Muller S, Nellen W, et al. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012;40:11648–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Müller M, Hartmann M, Schuster I, et al. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015;43:10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44:D184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wu P, Brockenbrough JS, Paddy MR, et al. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–117. [DOI] [PubMed] [Google Scholar]
- [22].Polevoda B, Panciera Y, Brown SP, et al. Phenotypes of yeast mutants lacking the mitochondrial protein Pet20p. Yeast. 2006;23:127–139. [DOI] [PubMed] [Google Scholar]
- [23].Bullerwell CE, Leigh J, Forget L, et al. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 2003;31:759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Parsons AB, Brost RL, Ding H, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. [DOI] [PubMed] [Google Scholar]
- [25].Kapitzky L, Beltrao P, Berens TJ, et al. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol. 2010;6:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brown JA, Sherlock G, Myers CL, et al. Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol Syst Biol. 2006;2:2006 0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tuorto F, Liebers R, Musch T, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. [DOI] [PubMed] [Google Scholar]
- [28].Janke C, Magiera MM, Rathfelder N, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. [DOI] [PubMed] [Google Scholar]
- [29].Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lutsik P, Feuerbach L, Arand J, et al. BiQ analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res. 2011;39:W551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. [DOI] [PubMed] [Google Scholar]
- [32].Tovy A, Hofmann B, Helm M, et al. In vitro tRNA methylation assay with the Entamoeba histolytica DNA and tRNA methyltransferase Dnmt2 (Ehmeth) enzyme. J vis exp. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jurkowski TP, Meusburger M, Phalke S, et al. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


