ABSTRACT
Long non-coding RNAs (lncRNAs) are a new arm of gene regulatory mechanism as discovered by sequencing techniques and follow-up functional studies. There are only few studies on lncRNAs as related to gene expression regulation and anti-viral activity during influenza virus infection. We sought to identify and characterize lncRNAs involved in influenza virus replication. Using RNA sequencing analysis, we found that 1,912 lncRNAs were significantly changed in human lung epithelial A549 cells infected with influenza A/Puerto Rico/8/34. Gene ontology analysis on neighboring genes of these lncRNAs revealed that the genes involved in type I interferon signaling and cellular response were highly enriched. Seven selected up-regulated lncRNAs (AC015849.2, RP-1-7H24.1, PSMB8-AS1, CTD-2639E6.9, PSOR1C3, AC007283.5 and RP11-670E13.5) were verified by real-time PCR. These lncRNAs were also induced by other two influenza H1N1 virus strains (A/WSN/1933 and A/Oklahoma/3052/09) and interferon β1. Repression of PSMB8 antisense RNA 1 (PSMB8-AS1) using CRISPR interference reduced viral mRNA and protein levels as well as the release of progeny influenza virus particles. Our study suggests that lncRNA PSMB8-AS1 could be a new host factor target for developing antiviral therapy against influenza virus infection.
KEYWORDS: Long non-coding RNAs (lncRNAs), anti-viral activity, PSMB8-AS1
Introduction
Only approximately 2% of human genome is used for protein-coding genes [1]. Recent advances in sequencing technologies enable discovering vast portion of non-coding transcripts. Long non-coding RNAs (lncRNAs) are non-coding transcripts that have a length of > 200 nucleotides and do not encode any proteins.
Although very few lncRNAs have been studied, lncRNAs play roles in development and diseases. LncRNA expression shows greater cell- and tissue-specificity than protein-coding genes [2,3]. The number of lncRNAs increases with developmental complexity [4], leading to the idea that lncRNAs play an important role in giving rise to the diversity of cell differentiation programs underlying development in multicellular organisms. Dysregulation of lncRNAs has been observed under many pathological conditions including respiratory diseases [5], cancers [6,7] and heart diseases [8], indicating that abnormal expression of lncRNAs contributes to the development of pathophysiological conditions.
The functions of lncRNAs are diverse including chromatin remodeling, transcription and post transcription regulation, decoying, scaffolding, and microRNA sponging. Examples of epigenetic gene regulation by lncRNAs are X-inactive specific transcript (Xist) and HOX transcript antisense RNA (HOTAIR). They define epigenetic changes by interacting with various chromatin modifiers to alter their structure and in turn govern the accessibility of DNA to transcription factors and polymerase [6,9]. One example where lncRNA can directly interfere with polymerase II (Pol II) activity is the inhibition of the major coding transcript of dihydrofolate reductase (DHFR) [10]. In dormant cells, an upstream minor promoter of DHFR produces a lncRNA that apparently interrupts formation of the transcription preinitiation complex at the major promoter. Several lncRNAs act as RNA decoys. These lncRNAs titrate transcription factors away from their DNA targets by directly binding to them and thus suppress the transcription. LncRNA PANDA is one such example, which sequester nuclear transcription factor Y, alpha (NF-YA) away from its pro-apoptotic gene [11]. At the post-transcriptional level, lncRNAs can function as microRNA target site decoys, titrating microRNA effector complexes away from their mRNA targets. For example, the tumor suppressor pseudogene PTENP1 sequesters miR-19b and miR-20a to regulate the target gene expression of these microRNAs [12]. LncRNAs also act as a scaffold by binding specific combinations of regulatory proteins, enforcing a transcription silent state or contributing to the assembly of DNA–RNA–protein interactions at specific transcribed locations. Two lncRNAs, Mistral and HOTTIP, have been implicated in recruiting MLL, an H3K4 trimethylase to chromatin [13,14].
The innate and adaptive immune responses provide immunity against a variety of pathogens. Innate immunity presents the first line of defense against pathogens. LncRNAs such as THRIL [15] lincRNA-Cox2, Lethe [16] and PACER [17] have been shown to regulate gene expression in innate immune cells. LincRNA-Cox2 regulates the expression of different sets of inflammatory genes in unstimulated and TLR2 ligand-stimulated macrophage cell line. The silencing of lincRNA-Cox2 up-regulates certain chemokines (CCL5, CX3CL1), chemokine receptors (CCR1), and interferon-stimulated genes (ISGs) (IRF7, OAS1A, OAS1L, OAS2, IFI204 and ISG15) in the unstimulated cells, but down-regulates TLR1, IL6 and IL23 in the stimulated cells. This negative regulation of the genes is dependent on the interactions of lincRNA-Cox2 with heterogeneous nuclear ribonucleoprotein A/B and A2/B1 [18]. Adaptive response is orchestrated through T and B lymphocytes. LncRNAs play a role in lineage-specific differentiation and activation of these cells. Several studies highlight the role of lncRNA in adaptive immune response including NRON [19], NeST [20], Gas5 [21,22] and LncR-Ccr2-5ʹAS [23]. Knockdown of lincRNA LincR-Ccr2-5′AS results in impaired recruitment of TH2 cells to the lungs [23]. LincR-Ccr2-5′AS is expressed specifically on TH2 subset of helper T cells and is regulated by GATA-3 transcription factor.
The roles of lncRNAs in viral infections have been documented [24–29]. Negative regulator of antiviral response (NRAV) is a lncRNA that is downregulated by various viruses including influenza virus, sendai virus, muscovy duck reovirus, and herpes simplex virus [27]. Overexpression of NRAV increases virus replication whereas knockdown of NRAV has an opposite effect. The down-regulation of NRAV by influenza virus infection activates the marks of transcription (H3K4m3) and a decrease in repression signal of transcription (H3K27m3) at mxA and ifitm3 transcription start sites and thus regulates histone modification of these genes. Nuclear enriched abundant transcript 1 (NEAT1) upregulates interleukin-8 (IL8) transcription during influenza virus infection by recruiting splicing factor proline/glutamine-rich (SFPQ/PSF) to nuclear paraspeckle bodies [30]. Gene regulation by lncRNA can also take place in a locus-specific manner. BST2 Interferon Stimulated Positive Regulator (BISPR) acts as a positive regulator of the flanking antiviral gene, bone marrow stromal cell antigen 2 (BST2) and dictates the potency of the antiviral interferon (IFN) response [31]. Virus inducible lncRNA (VIN) is highly induced by influenza A virus and vesicular stomatitis virus, but not by interferons and is required for influenza virus replication [29]. These studies suggest that lncRNAs play a crucial role in influenza virus pathogenesis.
In the current study, we identified dys-regulated lncRNAs in influenza virus-infected human lung epithelial A549 cells using RNA sequencing (RNA_seq). We showed that the up-regulated lncRNA, PSMB8 antisense RNA 1 (PSMB8-AS1) was induced by various strains of influenza virus and type I IFN. We further demonstrated that repression of PSMB8-AS1 resulted in the reduction of influenza virus infection and synthesis. Our results suggest that PSMB8-AS1 is a viral lncRNA that promotes influenza virus replication.
Materials and methods
Cell culture
Human lung epithelial A549, human lung adenocarcinoma epithelial H441 cells, Madin-Darby canine kidney epithelial (MDCK), and human embryonic kidney (HEK) epithelial 293T cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). A549 and H441 cells were maintained in F12-K and RPMI-1640 medium with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (PS), respectively. HEK 293T and MDCK cells were cultured in DMEM plus 10% FBS and 1% PS at 37°C in a 5% CO2 incubator.
Influenza virus stocks
Stocks of H1N1 strains of influenza virus, A/Puerto Rico/8/34 (PR/8), A/WSN/1933 (WSN) and A/Oklahoma/3052/09 (pdmH1N1) (pdm/OK) were propagated in the allantoic cavity of 10-day specific-pathogen-free embryonated chicken eggs (Charles River Laboratories, Wilmington, MA, USA) at 35°C. The allantoic fluid was harvested, centrifuged at 2,000 g for 10 min, and stored at −80°C. Virus titer was determined by a Tissue Culture Infective Dose (TCID50) assay. Briefly, MDCK cells were seeded in 96-well plates at a density of 25,000 cells per well. The next day cells were washed with serum-free medium twice. A series of ten-fold dilutions of virus stock ranging from 10−1 to 10−8 were prepared in serum-free medium with 2 µg/ml L-1-Tosylamide-2-phenylethyl chloromethyl ketone treated trypsin (TPCK-trypsin). Cells were infected with each diluted virus stock in triplicate. After a 72 h culture, cells were analyzed for cytopathic effect (CPE) and TCID50 was calculated using the Reed-Muench method [32].
Virus infection
Cells were cultured in 6- or 12-well plates overnight and washed with serum-free medium twice. The cells were infected with influenza virus at various multiplicity of infection (MOIs) as indicated in serum-free medium containing TPCK-Trypsin (1 µg/ml) at 37°C and 5% CO2 incubator for 1 h for all of the strains used. The cells were then washed with phosphate buffered saline (PBS) once and serum-free medium containing TPCK-Trypsin (0.1 µg/ml) was added. At indicted times post infection, RNAs were extracted for lncRNA expression by real-time PCR.
RNA sequencing and data analysis
A549 cells were infected with PR/8 at a MOI of 2 for 24 h. Total RNAs from 3 control and 3 infected cells were extracted. The RNA samples were assessed for their quality using the RNA 6000 pico kit (Agilent, catalog# 5067–1513) and Agilent Bioanalyzer following manufacture’s recommendations. High quality RNA were used to prepare sequencing library with the Illumina TruSeq Stranded mRNA Library Preparation kit (Illumina, catalog# RS-122–2101). Briefly, poly-adenylated mRNAs were selected using oligo-dT beads and fragmented to 300–500 bp. Each fragment underwent cDNA synthesis using random hexamer. Indexed adapters were ligated to the resulting cDNA. The indexed libraries were pooled in equal molar ratio and sequenced on the Illumina NextSeq 500 system using 2 × 75 bp paired-end sequencing. Each sample was sequenced to a minimum of 20 million reads. Paired-end reads were directionally mapped to the genomic loci of mRNA and lncRNA (GRCh37/hg19) by TopHat2. CuffDiff analysis was then run to identify the dysregulated RNAs. RNAs with a fold change of ≥ 2 and a FDR of < 0.05 were considered to be differentially expressed. Gene ontology (GO) functional annotation of mRNA expression profile was conducted by STIRNG analysis (http://string-db.org/). KEGG pathway enrichment in the altered mRNAs was also performed by STIRNG analysis. STRING is a web-based tool to investigate protein-protein interactions, KEGG pathway, and GO annotation.
Interferon treatment and lncRNA induction
Cells were seeded in 12-well plates and cultured overnight. Cells were then treated with 1,000 U/ml human IFNβ1a (#11,415–1, PBL assay science, Piscataway Township, NJ, USA) in medium with 10% FBS and 1% PS for different times as indicated. At each time point, RNA was extracted for real-time PCR analysis.
CRISPR interference
LentiGuide-Puro vector for expressing hU6-driven gRNA was a gift from Feng Zhang (Addgene plasmid # 52,963) and pHAGE EF1α dCas9-KRAB vector was a gift from Rene Maehr & Scot Wolfe (Addgene plasmid # 50,919). gRNA sequences targeting PSMB8-AS1 and its promoter were designed using a web-based tool (http://crispr.mit.edu/). We selected gRNAs with a high rank and G at the first position, which is usually the transcription start site of the U6 promoter to avoid the addition of extra G. The sequences of gRNAs are as follows: gRNA1, GAAGCCGGTCCCCAGTGTGA; gRNA2, GCGGACAGATCTCTGGGTGC; gRNA3, GATCTGTCCGCTCTCGGAGG. gRNA1 targets the transcript and gRNA2 and gRNA3 target the promoter. The gRNA control sequence is GGTGGTAGAATAACGTATTAC as described in the published paper [33]. Each pair of oligos were annealed and ligated into the lentiGuide-Puro vector using BsmBI sites.
To produce lentivirus, HEK293T cells were co-transfected with a lentiviral vector and packaging plasmid psPAX2 (Addgene, 12,260), and the envelope plasmid pMD2.G (Addgene, 12,259) by polyethylenimine (PEI). Virus-containing medium was harvested after 48 h of transfection. The titer was determined by colony formation assay.
A549 cells were cultured in 12-well plates (1 x 104/well) for 24 h and infected with a lentivirus stably expressing dCas9-KRAB at a MOI of 100 in the presence of polybrene (4 μg/ml) for 24 h. The virus was removed. Fresh medium was added and cells were cultured for another 24 h. Cells were then cultured in medium containing 1 µg/ml of puromycin for 7 days with change of medium every 2–3 days. The cells stably expressing dCas9-KRAB were then stored in liquid N2.
To repress PSMB8-AS1 expression, the cells stably expressing dCas9-KRAB were cultured in 24-well plates (1 x 105/well) in complete medium containing 1 µl/ml of puromycin for 24 h. Cell were then infected with a lentivirus expressing PSMB8-AS1 gRNA at a MOI of 200 in serum-free medium containing 4 μg/ml polybrene and 0.1 µg/ml of puromycin for 48 h.
Cells were washed with serum-free media and infected with A/PR/8/34 PR8 (MOI, 0.01) for 1 h in serum-free medium containing 0.5 µg/ml TPCK-trypsin. Influenza virus was then removed and cells were cultured in serum-free medium containing 0.3% BSA and 0.5 µg/ml TPCK-trypsin for 48 h. Cells were used for western blotting or real-time PCR and media were used for plague assay.
Real-time PCR
Total RNAs were treated with TURBO DNA-freeTM (Ambion Thermo Fischer, Waltham, MA, USA) to eliminate any genomic DNA contaminations. One µg of RNA was reverse-transcribed into cDNA using Moloney Murine Leukemia Virus reverse transcriptase and Oligo dT and random primers. The primers used in this study are listed in Table 1. The real-time PCR thermal conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s and 65°C for 30 s. β-actin and 18S rRNA were used as endogenous reference genes. Real-time PCR was performed using SYBR Green master mix from Eurogentec (Seraing, Belgium) on an ABI 7500 fast system (Applied Biosystems, Foster City, CA, USA).
Table 1.
Primers used for real-time PCR.
| Gene Name | Forward primer | Reverse primer |
|---|---|---|
| AC015849.2 | AACTGTCAAGCTCAATTTCCCTCT | GTGGAAAGGTTCGCTGGGAC |
| PSMB8-AS1 (common) | GGAAAGACATCGGACCGTCA | TGGGAAACGTTGGTGTCCTT |
| PSMB8-AS1-1 | TGGCGATGGGTTACAGTAAGAG | TCGACAGTTGCTGGGTAGATG |
| PSMB8-AS1-2 | GCTTCTCTGCTCTCCCGTTA | GTGTGGCCAGCACTCTGAA |
| PSMB8-AS1-3 | TGGGTTACAGTAAGAGGTTTCC | GCTGCCTCCAGGATGAGTTA |
| PSMB8-AS1-4 | ACAGTAAGAGGAAGTCTGCATT | GAGAACCACAGCTGCAGAGT |
| RP1-71H24.1 | TTCCAGCTGTCTCCTAATTTCC | CTTTGTCCTGGTTGTCTTCCT |
| CTD-2639E6.9 | AAGTCTGACTCCAGTCCCCG | GTTTGCGCTGCGAGATAAGG |
| PSORS1C3 | CATCATGGCACACAACAACC | CCGGTCTAGGAAACCACTTATT |
| AC007283.5 | GTACTTTGGGAGGCTGAGTG | CTGAAGTGCAGTGGTGTGA |
| RP11-670E13.5 | AAATAGCATTTTGTACCCGCACT | GCCCGATTCCTCTTAGAAGGTT |
| β-actin | GCCGGGACCTGACTGACTAC | TTCTCCTTAATGTCACGCACGAT |
| 18S | CGTTGATTAAGTCCCTGCCCTT | TCAAGTTCGACCGTCTTCTCAG |
| UGGT2 | CCTTCGCAATCTTGGGATCAA | GCCGGATCAATAAACAGAACCA |
| UBE2G2 | ATCTACCCTGATGGGAGAGTCT | CTCCACTTTCGTCATTGGGC |
| GANAB | TGGGGATTACCCTTGCTGTG | CCGTATGCTTCTCTGTCGCT |
| BAG2 | ATCAACGCTAAAGCCAACGAG | CGTCACTGATCTGCCTCATGT |
| SIL1 | CTGCCTTCATCTAGGATGGCT | GGGTTGGTCAGGGCAAACTC |
| EIF2AK1 | ACCCCGAATATGACGAATCTGA | CAAGTGCTCCAGCAAAGAAAC |
| UBE4B | CTACCTCCCCAATAGGTGCAT | GGCGAGCTGCTGAGAGAAC |
| snU2RNA | CATCGCTTCTCGGCCTTTTG | TGGAGGTACTGCAATACCAGG |
| NS1 | CCGACATGACTCTTGAGGAAAT | CGCCTGGTCCATTCTGATAC |
| NP | TGTGTATGGACCTGCCGTAGC | CCATCCACACCAGTTGACTCTTG |
Western blot analysis
Cells were lysed in M-PER™ Mammalian Protein Extraction Reagent (Thermo Fischer, Waltham, MA, USA). Proteins (30 µg/lane) were separated by 12% of SDS-PAGE and transferred onto nitrocellulose membranes using Trans-Blot® Turbo™ Transfer System (Bio-Rad, Hercules, CA, USA). Membranes were blocked with 5% non-fat milk and then probed with different primary antibodies prepared in 5% non-fat milk overnight. Membranes were washed with TRIS-buffered saline for 5 min × 3 times and incubated with species-appropriate horseradish peroxidase-labeled secondary antibody for 1 h at room temperature. Membranes were again washed with TRIS-buffered saline for 5 min × 3 times. Finally, the membranes were developed using SuperSignal™ West Pico Chemiluminescent Substrate (Thermo Fischer, Waltham, MA, USA). Protein bands were detected with Amersham Western blot detection system (GE healthcare system, Pittsburgh, PA, USA). Protein levels were normalized to β-actin and quantitation was determined using Image Quant software from GE healthcare system. Mouser anti-β-actin antibodies (Catalog No, MA5-15,739-1MG, lot#QK229411) were purchased from Sigma-Aldrich (St Louis, MO, USA). Influenza viral protein antibodies, mouse anti-NP (Catalog No, HB-65) and mouse anti-NS1 (cat# SC-130,568, lot#D0517) were purchased from ATCC and Santa-Cruz biotechnology (Paso Robles, CA, USA), respectively. Anti-H3 (cat# ab1791), anti-H3K27m3 (cat# ab6002), and anti-H3K27Ac (cat# ab4729) antibodies were purchased from Abcom (Cambridge, MA, USA). Antibody dilutions were 1:1,000 for β-actin, 1:40 for NP, 1:1,000 for NS1, H3K27m3 and H3K27Ac, 1:2,000 for H3. HRP-conjugated secondary antibodies against mouse (cat#115–0.5–003, lot#89,918) were purchased from Jackson Immuno Research (West Grove, PA, USA) and used at a dilution of 1:2,000.
Isolation of cytoplasmic and nuclear RNAs
Cytoplasmic and nuclear fractions were prepared from A549 cells using a kit (#21,000, Norgen, Canada). cDNA was prepared using 1 µg RNA and real-time PCR was performed to analyze both cellular fractions using primers for β-actin, GAPDH, PSMB8-AS1 and U2snRNA. The expression of mRNA or lncRNA in nucleus and cytoplasm was calculated with the equation 2−Ct. The percentage of each RNA in the nucleus and cytoplasm were calculated.
Results
LncRNAs are differentially expressed in influenza virus-infected A549 cells
To identify lncRNAs that are dysregulated during influenza virus infection, we infected lung epithelial A549 cells with PR/8 at a MOI of 2 for 24 h and performed RNA_seq analysis. Tophat was used for read mapping and Cufflinks/Cuffdiff was used for gene expression quantification. Fig. 1A depicts the design of the experiment. Using a p value of < 0.05, we found that 8,983 mRNAs were significantly changed by PR/8 infection (Fig. 1B). Of them, 3,649 mRNAs were upregulated and 5,334 mRNAs were downregulated (Fig. 1C). 1,625 up-regulated and 2,784 down-regulated mRNAs were changed 2-fold or more (Table 2). The numbers of mRNAs with various fold changes are also listed in Table 2. Up-regulation of ISGs was observed (Table 3), indicative of successful influenza virus infection.
Figure 1.
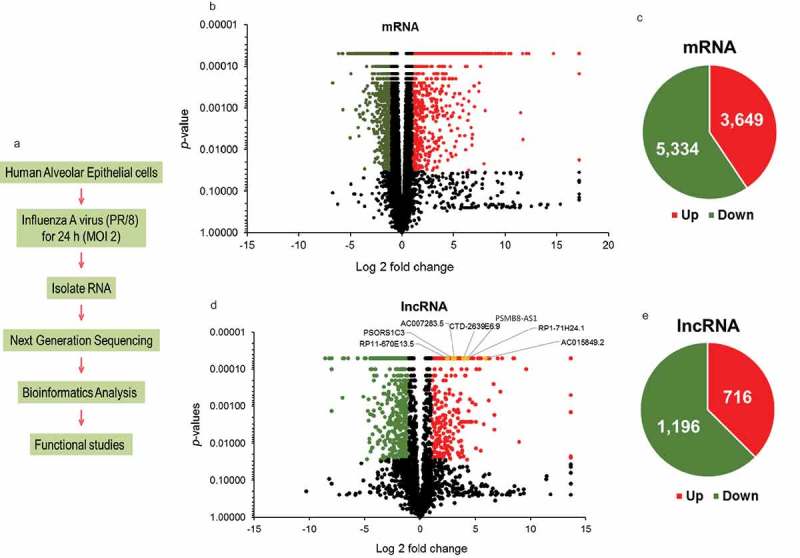
lncRNAs are differentially regulated during influenza virus infection. (a) Flowchart of RNA-seq experiment design. (b, d) Volcano plot of differentially expressed mRNAs and lncRNAs in PR/8-infected A549 cells at MOI of 2 for 24 h. Downregulated genes are denoted in green color and upregulated genes in red color based on a p-value of ≤ 0.05 and a fold change of ≥ 2. Genes with a p-value of > 0.05 or a fold change of < 2 are marked with black color. The yellow colored lncRNAs are the selected lncRNAs for further analysis (see the text). (c, e) Pie charts of significantly changed mRNAs and lncRNAs with a p-value of ≤ 0.05. Green and red colors denote downregulated and upregulated genes, respectively.
Table 2.
Summary of RNA_seq data sets.
| lncRNA |
mRNA |
|||
|---|---|---|---|---|
| Fold change | Up-regulated | Down-regulated | Up-regulated | Down-regulated |
| > 100 | 68 | 11 | 80 | 0 |
| 50–100 | 5 | 1 | 45 | 2 |
| 10–50 | 71 | 59 | 238 | 79 |
| 2–10 | 274 | 612 | 1,262 | 2,703 |
Table 3.
Interferon-stimulated antiviral genes.
| Anti-viral genes | Expression level (FPKM) |
Fold change | |
|---|---|---|---|
| Control | Influenza virus | ||
| OAS1 (2ʹ-5ʹ-Oligoadenylate Synthetase 1) | 21 | 421 | 20 |
| OAS2 (2ʹ-5ʹ-Oligoadenylate Synthetase 2) | 0.12 | 173 | 1,468 |
| OAS3 (2ʹ-5ʹ-Oligoadenylate Synthetase 3) | 19 | 181 | 10 |
| OASL (2ʹ-5ʹ-Oligoadenylate Synthetase-Like) | 0.46 | 1,539 | 3,327 |
| MX1(MX Dynamin-Like GTPase 1) | 0.42 | 222 | 526 |
| ISG15 (ISG15 Ubiquitin-Like Modifier) | 3 | 974 | 367 |
| ISG20 (Interferon Stimulated Exonuclease Gene) | 6 | 251 | 41 |
| IRF1 (Interferon Regulatory Factor 1) | 9 | 237 | 27 |
| IRF2 (Interferon Regulatory Factor 2) | 9 | 27 | 3 |
| IRF7 (Interferon Regulatory Factor 7) | 2 | 163 | 74 |
| IRF9 (Interferon Regulatory Factor 9) | 39 | 164 | 4 |
| TRIM25 (Tripartite Motif Containing 25) | 27 | 199 | 7 |
Similarly, we identified 1,912 lncRNAs that were significantly changed in PR/8-infected A549 cells (Fig. 1D). Of them, 716 lncRNAs were up-regulated and 1,196 lncRNAs were down-regulated (Fig. 1E). There were 418 up-regulated and 683 down-regulated lncRNAs based on a fold change of 2 or more. Interestingly there were only 11 down-regulated lncRNAs compared to 68 up-regulated lncRNAs based on a fold change of > 100 (Table 2).
Because the functions of most lncRNAs are unknown and lncRNAs often regulate their neighboring genes [34], we selected the genes within 10,000 kb of significantly changed lncRNAs during influenza virus infection for GO analysis including cellular components, molecular pathways and biological processes. Prediction terms with a p-value of less than 0.05 were selected and ranked. Enrichment scores (−log10(p-value) were plotted on x-axis. The most enriched cellular components were related to endosome (Fig. 2A). The genes involved in the α, β, and γ IFN and immune signaling or cellular responses are enriched in the molecular and biological pathways (Fig. 2B, C). The GO analysis revealed the neighboring genes of the lncRNAs changed by influenza virus were enriched in the pathways and processes that are known to be involved in influenza virus infection.
Figure 2.
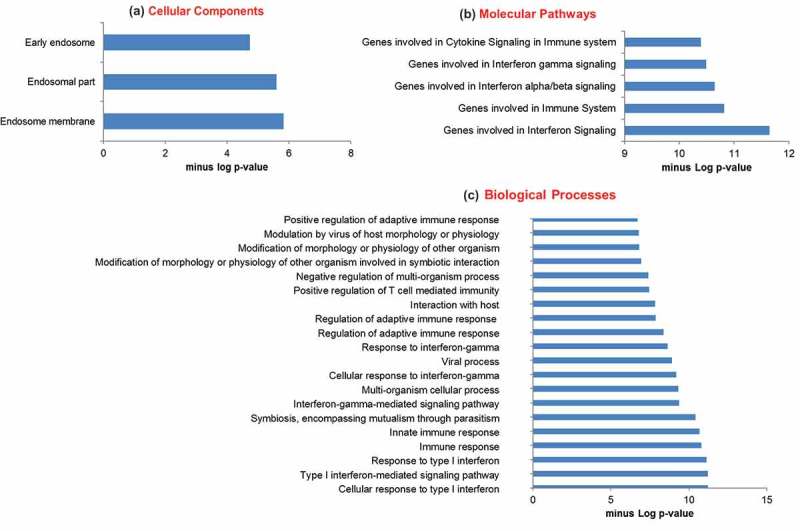
GO pathway analysis of lncRNA co-expressed mRNAs. Protein-coding genes within 10,000 kb distance of the significantly dysregulated lncRNAs during influenza virus infection were selected for GO analysis. GO analysis includes 3 annotations- cellular components (a), molecular pathways (b) and biological process (c). GO id with a p-value of < 0.05 were selected.
Given a greater number of lncRNAs that are up-regulated compared to the down-regulated lncRNAs during influenza virus infection based on a fold change of > 100 (Table 2), we decided to focus on the up-regulated lncRNAs for further studies. To narrow down our selection, we used the following criteria: (a) a fold change of >2 for up-regulated lncRNAs, (b) significant changes in the expression of neighboring genes (up or down) within 10,000 kb of lncRNAs, (c) neighboring genes involved in > 5 pathways based on GO analysis, and (d) >500 base pair long with no ORF (open reading frame). Seven lncRNAs met these criteria and are listed in Table 4 and marked in yellow color in the scatter plot (Fig. 1B). These lncRNAs were composed of different types of transcripts (Table 4). Four of them were anti-sense lncRNAs that overlap the genomic span of a protein-coding locus on the opposite strand. AC015849.2 has two neighboring genes, Chemokine (C-C Motif) Ligand 5 (CCL5) and TATA Box Binding Protein (TBP)-Associated Factor (TAF15). CCL5 was highly upregulated (147,688-fold), which is consistent with a previous report [35] whereas TAF15 was downregulated (5-fold). The other two anti-sense lncRNAs, RP-1-7H24.1 and RP11-670E13.5 have well-known antiviral genes, OAS2, OAS3 and TRIM25 as their neighbors [36]. They were also highly upregulated. The neighboring genes of PSMB8-AS1, PSMB8 and TAP1 were also upregulated. CTD-2639E6.9 is an intergenic lncRNA (lincRNA) and neighboring genes of CTD-2639E6.9 were either downregulated (FTL) or not detected (BAX) by RNA_seq. AC007283.5 is 3 prime overlapping lncRNA that overlaps the 3ʹ-UTR of a protein-coding locus on the same strand and its neighboring genes, CASP10 and CFLAR were upregulated. PSOR1C3 is a sense intronic lncRNA that lies within introns and do not overlap with exons. Its neighboring genes, POU5F1 and HLA-C, were highly upregulated.
Table 4.
Selected lncRNAs and their properties.
| FPKM |
FPKM |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| lncRNA | Con | Flu | Fold change | Chromosome locus | bp | Type | Neighboring genes | Con | Flu | Fold change |
| AC015849.2 | 8 | 491 | 60 | chr17:34,195,970–34,212,867 | 662 | anti-sense | CCL5 | 0.02 | 2,988 | 147,688 |
| TAF15 | 219 | 44 | —5 | |||||||
| PSMB8-AS1 | 11 | 211 | 19 | chr6:32,811,862–32,814,272 | 1,267 | anti-sense | PSMB8 | 9 | 51 | 5 |
| TAP1 | 8 | 253 | 33 | |||||||
| RP1-71H24.1 | 14 | 257 | 18 | chr12:113,345,432–113,455,556 | 575 | anti-sense | OAS2 | 0.12 | 173 | 1,470 |
| OAS3 | 19 | 181 | 10 | |||||||
| CTD-2639E6.9 | 10 | 159 | 15 | chr19:49,467,231–49,468,415 | 819 | lincRNA | FTL | 8,488 | 3,245 | —3 |
| BAX | ND | ND | ND | |||||||
| AC007283.5 | 142 | 1,261 | 9 | chr2:202,031,687–202,032,269 | 445 | 3 prime overlapping | CASP10 | 2 | 14 | 6 |
| CFLAR | 27 | 137 | 5 | |||||||
| PSORS1C3 | 31 | 245 | 8 | chr6:31,141,511–31,145,676 | 593 | Sense intronic | POU5F1 | 2 | 22 | 13 |
| HLA-C | 24 | 412 | 17 | |||||||
| RP11-670E13.5 | 337 | 1,880 | 6 | chr17:54,966,240–54,969,202 | 538 | anti-sense | TRIM25 | 27 | 199 | 7 |
| DGKE | ND | ND | ND | |||||||
Fold changes: from RNA-seq data
bp: length in terms of base pairs
ND: Not detected by RNA-seq
- number: Downregulation
FPKM: Fragments Per Kilo base of transcript per Million mapped reads
CCL5: Chemokine (C-C motif) ligand 5
TAF15: TATA Box Binding Protein (TBP)-Associated Factor
PSMB8: Proteasome Subunit Beta 8
TAP1: Transporter 1, ATP-Binding Cassette
OAS2: 2ʹ-5ʹ-Oligoadenylate Synthetase 2
OAS3: 2ʹ-5ʹ-Oligoadenylate Synthetase 3
FTL: Ferritin, Light Polypeptide
BAX: BCL2-Associated X Protein
CASP10: Caspase 10, Apoptosis-Related Cysteine Peptidase
CFLAR: CASP8 and FADD-Like Apoptosis Regulator
POU5F1: POU Class 5 Homeobox 1
HLA-C: Major Histocompatibility Complex, Class I, C
TRIM25: Tripartite Motif Containing 25
DGKE: Diacylglycerol Kinase, Epsilon
We then utilized real-time PCR to validate the results from RNA_seq analysis using the same RNAs from the RNA_seq analysis. Real-time PCR confirmed that all of the 7 lncRNAs were induced by PR/8 (Fig. 3A) although the absolute fold changes varied between real-time PCR and RNA_seq for some lncRNAs. For the mRNA real-time PCR validation, we selected the genes that are implicated in endoplasmic reticulum (ER) stress since ER stress is involved in viral replication and vice-versa [37,38]. The genes that were included for this analysis were UDP-Glucose Glycoprotein Glucosyltransferase 2 (UGGT2), Ubiquitin-Conjugating Enzyme E2G 2 (UBE2G2), Glucosidase, Alpha; Neutral AB (GANAB), BCL2-Associated Athanogene 2 (BAG2), SIL1 Nucleotide Exchange Factor (SIL1), Eukaryotic translation initiation factor 2-alpha kinase 1 (ELF2AK1) and Ubiquitination Factor E4B (UBE4B). All of the genes were down-regulated and similar changes were observed using real-time PCR and RNA_seq (Fig. 3B).
Figure 3.
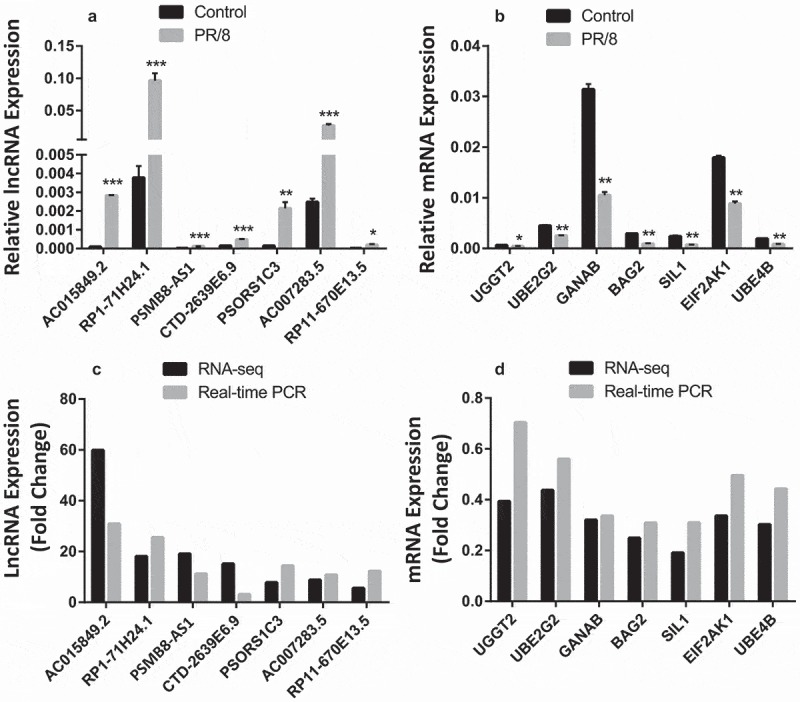
Validation of RNA-seq results with real-time PCR. (a, b) Relative expression of selected lncRNAs and mRNAs performed on the same samples as for RNA-seq using real-time PCR. Data was normalized to β-actin and expressed as means ± SE. n = 3 independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. control (paired student’s t-test). (c, d) Fold changes as determined by RNA-seq and real-time PCR.
Effects of influenza virus strains on lncRNA expression
We used a single MOI for our RNA_seq analysis. We further examined whether the induction of the upregulated lncRNAs identified above by PR/8 is dose-dependent. We found that lncRNA induction by PR/8 was increased as a MOI of PR/8 increased (Fig. 4). RP1-7H24.1, PSMB8-AS1, and RP11-670E13.5 expression reached maximum at a MOI of 0.2 while AC015849.2, CTD-2639E6.9, AC007283.5, and PSORS1C3 had a highest expression at a MOI of 2.
Figure 4.
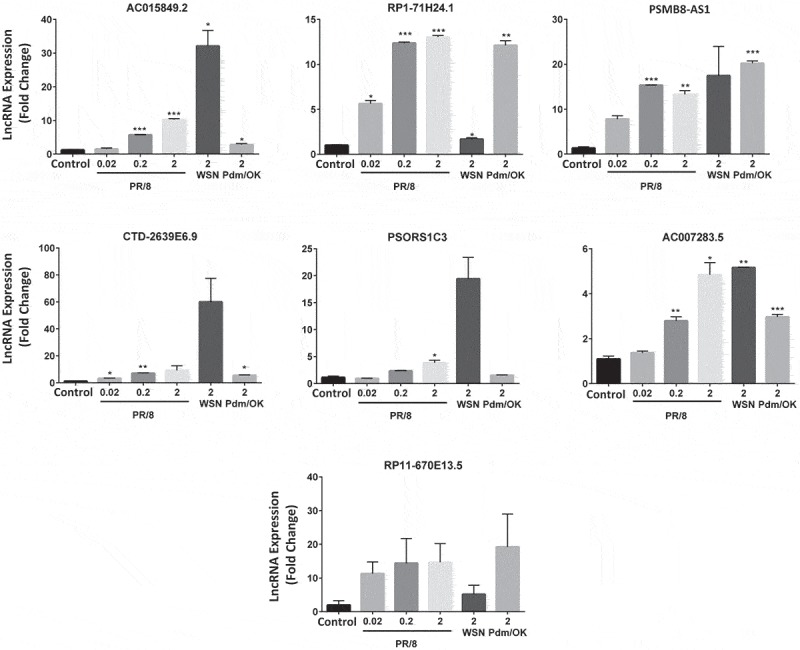
lncRNA induction by different influenza viruses. A549 cells were infected with influenza A viruses, PR/8 (MOI: 0.02, 0.2 and 2), WSN (MOI 2) and Pdm/OK (MOI 2) for 24 h. lncRNA expression levels were determined by real-time PCR and normalized to β-actin to determine ΔΔct. Results are represented as means ± SE from three independent experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. mock control (One-way ANOVA post hoc Dunnett).
Influenza A virus strains exhibit different virulence for a particular host [39]. We thus examined if the up-regulated lncRNAs by PR/8 are also induced by other strains of influenza viruses. Since a maximal induction was observed by PR/8 at a MOI of 2 in the MOIs tested for all of the lncRNAs, we compared the effects of 3 influenza A virus strains on the lncRNA expression using a MOI of 2 in A549 cells: PR/8, WSN and Pdm/OK. WSN and Pdm/Ok are another commonly used laboratory strain and a clinical isolate of 2009 Oklahoma pandemic influenza H1N1 virus of swine origin, respectively. Six of the 7 lncRNAs were induced by all the strains except that Pdm/OK had no effects on PSORS1C3 expression and that PR/8-, WSN- and Pdm/Ok-induced RP11-670E13.5 expressions did not reach a significant level due to the variation between the experiments (Fig. 4). However, the magnitude of induction varied among strains and lncRNAs. Similar inductions were observed for PSMB8-AS1 by 3 strains. WSN induced much higher expression of AC015849.2, CTD-2639E6.9 and PSORS1C3 compared to PR/8 and Pdm/OK while PR/8 and Pdm/OK increased RP1-7H24.1 expression more than WSN. PR/8 and WSN were more effective in the induction of AC007283.5 than Pdm/OK.
Effect of IFNβ1 on lncRNA expression
Influenza viruses are known to induce IFN response. Hence, we examined whether type I IFN induce the expression of lncRNAs identified above. We treated A549 cells with 1,000 U/ml IFNβ1 for different times (0, 3, 9 and 24 h) and determined the expression levels of lncRNAs by real-time PCR. IFNβ1 treatment markedly increased the expression of OAS1, a known ISG gene [40] (Fig. 5). We found that 6 of the 7 lncRNAs were significantly induced by IFNβ1. PSORS1C3 level was also increased by IFNβ1, but did not reach a significant level. The IFNβ1-induced expression of lncRNAs occurred as early as 3 h post treatment and reached a maximum at 3 h for AC007283.5, RP11-670E13.5 and PSMB8-AS1, 9 h for RP1-71H24.1 and 24 h for AC015849.2.
Figure 5.

lncRNAs are induced by IFNβ1. A549 cells were treated with IFNβ1a (1,000 U/ml) for different times. Interferon inducible gene, OAS1 and lncRNA levels were determined by real-time PCR and normalized to18S rRNA. Fold change was calculated based on 0 h and results are represented as means ± SE from three independent experiments. *p < 0.05, and **p < 0.01 vs. 0 h (Student’s t-test).
Further characterization of PSMB8-AS1
We chose PSMB8-AS1 for further characterization and functional studies because all of the influenza virus strains tested were able to induce PSMB8-AS1 expression with a high fold change. To determine whether PSMB8-AS1 is also induced in other cells, we repeated the experiments above in two additional cell lines, human embryonic kidney HEK293 cells and human lung epithelial H441 cells. The cells were infected with 0.2 MOI of PR/8, WSN, or Pdm/OK or treated with IFNβ1 for 24 h. PSMB8-AS1 were induced by all of the treatments except Pdm/OK. The induction was more potent in HEK293 cells than H441 cells (Fig. 6).
Figure 6.

PSMB8-AS1 induction in HEK293 and H441 cells by influenza virus and IFNβ1. HEK293 and H441 cells were infected with PR/8, WSN and Pdm/OK at a MOI of 0.2 for 24 h or treated with IFNβ1a (1,000 U/ml) for 24 h. The results are represented as means ± SE from three independent experiments. *P < 0.05 vs. Control, **P < 0.01 vs. Control, ***P < 0.0001 vs. Control. (One-way ANOVA post hoc Tukey’s Test).
There are 4 transcripts for PSMB8-AS1 (https://genome.ucsc.edu). The studies on the expression of PSMB8-AS1 were performed using the primer pair that amplifies all of the transcripts 4 (Fig. 7A). To determine whether other transcripts are induced by influenza A virus, we designed additional 4 real-time PCR primers to specifically amplify different transcripts of PSMB8-AS1. All of the transcripts were induced by PR/8 in A549 cells and HEK293 cells although the transcript 4 appears to be the major transcript (Fig. 7B, C). PR/8 also induced the expression transcript 2 and 4, but not the transcripts 1 and 3 in H441 cells (Fig. 7D). To further confirm the identities of the PSMB8-AS1 transcripts, we performed end-point PCR with the samples from PR/8-infected A549 cells and cloned the PCR products into the T-vector. DNA sequencing confirmed all of the 4 transcripts.
Figure 7.

Induction of PSMB8-AS1 transcripts by PR/8. (A) The PSMB8-AS1 transcripts and real-PCR primers and gRNA locations. (B-C) Human alveolar epithelial (A549) cells (B), human embryonic kidney cells (HEK 293) (C) and human lung adenocarcinoma cell line (H441) (D) were infected with PR/8 at a MOI of 0.2 for 24 h and the relative expression of the four PSMB8-AS1 transcripts were determined by real-time PCR. Data was normalized to β-actin and expressed as means ± SE. n = 3 independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 (Student’s t-test).
Repression of PSMB8-AS1 reduces influenza virus replication
To determine the functional role of PSMB8-AS1 in influenza virus replication, we repressed PSMB8-AS1 using CRISPR interference. We designed 3 gRNAs, two target the promoter region and one targets the transcript. A549 cells stably expressing dCas9-KRAB were infected with a lentiviral PSMB8-AS1 gRNA or its control and selected with puromycin. The expression level of PSMB8-AS1 was effectively reduced by each of 3 gRNAs (Fig. 8A). The reduction of PSMB8-AS1 decreased mRNA expression of viral genes, NP and NS1 and protein expression of NP, NS1 and PB1. The percentage of NP-positive cells and the virus titer in the culture medium were also reduced by gRNAs (Fig. 8E-G). gRNAs had no effects on cell viability (Fig. 8H). These results indicate that PSMB8-AS1 repression inhibits influenza virus replication.
Figure 8.
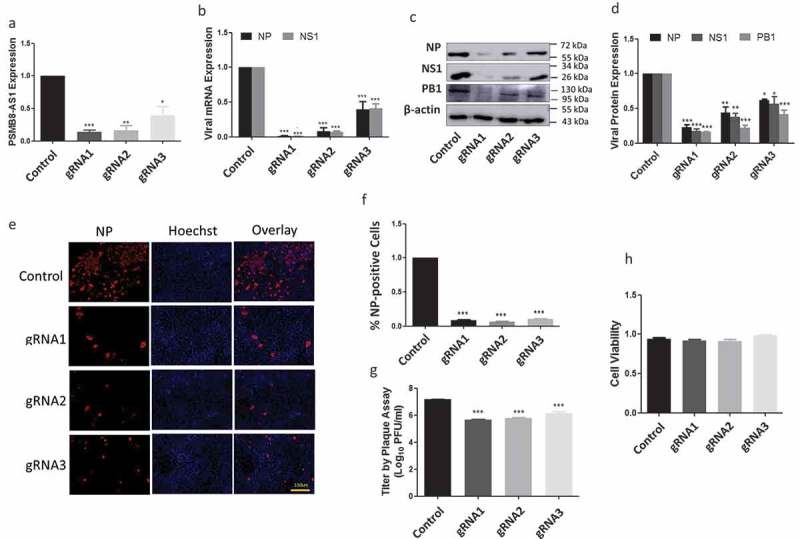
Repression of PSMB8-AS1 reduces influenza virus replication. A549 cells stably expressed dCas9-KRAB were infected with lentiviral gRNAs at a MOI of 200 in the presence of 0.1 µg/ml of puromycin for 48 h and infected with PR/8 at a MOI of 0.01 for 48 h. (A) PSMB8-AS1 expression level and (B) NP and NS1 mRNA levels as determined by real-time PCR. (C, D) NP, NS1 and PB1 protein levels as determined by western blotting. The protein bands were quantitated using Image Quant software and normalized to β-actin. (E, F) % NP-positive cells as revealed by immunostaining. (G)Virus titer in medium as determined by a plaque assay. (H) Cell viability as determined by a Cell-Titer Blue assay (Promega G8080). *P < 0.05, **p < 0.01, ***p < 0.001 (n = 3, one-way ANOVA, Tukey’s).
Effects of PSMB8-AS1 on epigenetic markers
As the first step in elucidating mechanisms of PSMB8-AS1-mediated influenza virus replication, we determined the location of PSMB8-AS1 in cells. We isolated cytoplasmic and nuclear fractions from A549 cells and determined PSMB8-AS1 levels in both fractions using real-time PCR. As shown in Fig. 9A, PSMB8-AS1 was enriched in the nucleus similar to nuclear U2 snRNA. Cytoplasmic β-actin and GAPDH mRNAs were primarily located in the cytoplasm.
Figure 9.
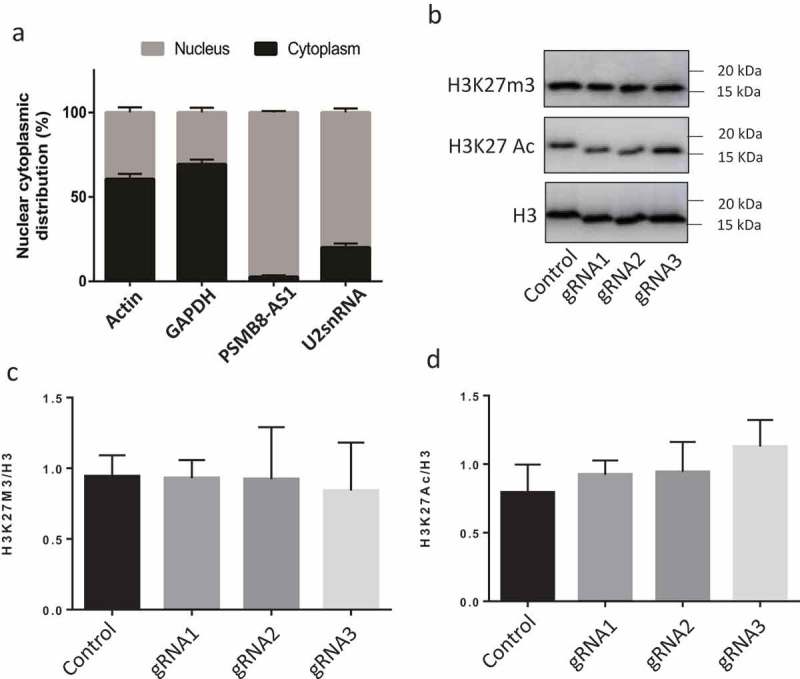
Localization of PSMB8-AS1 in cells and effects of PSMB8-AS1 on epigenetic markers. (A) The levels of PSMB8-AS1, GAPDH and β-actin mRNAs (cytoplasmic RNA positive controls), and U2snRNA (nuclear RNA positive control) in cytoplasmic and nuclear fractions of A549 cells were determined by real-time PCR. Results are represented for each gene as means ± SE from three independent experiments. (B-D) A549 cells stably expressed dCas9-KRAB were infected with lentiviral gRNAs at a MOI of 200 in the presence of 0.1 µg/ml of puromycin for 48 h and infected with PR/8 at a MOI of 0.01 for 48 h. H3K27m3 and H3K27Ac levels in the nuclei were determined by western blotting and normalized to H3. Representative blots and quantitation were presented in B and C, D, respectively. Results are represented as means ± SE from three independent experiments. No significance was detected by one-way ANOVA.
Because of the nuclear location of PSMB8-AS1, we suspected that PSMB8-AS1 might play a role in gene expression in response to influenza viral infection. We thus examined whether PSMB8-AS1 affects epigenetic markers. The control and PSMB8-AS1-repressed A549 cells were infected with PR/8 for 48 h and nuclei were isolated and examined for H3K27m3 and H3K27Ac by western blotting. As shown in Fig. 8B-D, H3K27m3 and H3K27Ac levels were not changed in all of the 3 gRNAs-treated cells.
Discussion
Influenza virus requires host cellular factors to function as revealed by several high throughput and genome wide studies [41–43]. Since virus infection leads to global translation inhibition [44], a translation-independent system is needed to prepare cells for antiviral responses. lncRNAs represent a such potential class of host factors and are new alternatives for development of host-centric antiviral strategies. In this study we identified a lncRNA PSMB8-AS1 whose repression reduced influenza virus replication. PSMB8-AS1 was induced by different influenza virus strains and type I IFN.
Our present studies showed that a large number of human lncRNAs (3,158), along with protein-coding genes (8,638), were differentially expressed after influenza virus infection. A recent microarray analysis reported a similar number of non-coding transcripts (3,415 and 3,401) altered by influenza virus infection using two commercial microarrays, NCode™ and Sureprint™ G3 [29]. However, RNA_seq technique yields more comprehensive datasets of both lncRNA and mRNA along with less false-positive hits [45].
It is well known that influenza virus hijacks cellular protein synthesis machinery to make more viral proteins [33]. It is conceivable that many of the cellular mRNAs are downregulated [46]. Our RNA_seq datasets revealed a higher number of lncRNAs and mRNAs were up-regulated than these that were down-regulated. Previous studies have shown that lncRNAs have similar functions as their neighboring genes [34]. GO analysis using upstream and downstream protein coding genes within 10,000 kb distance of the dysregulated lncRNAs indicated that many of the lncRNAs were associated with pathogen-related immune signaling pathways and responses based on molecular pathway and biological process analysis and endosomes based on cellular component enrichment. This is consistent with the fact that influenza virus enters cells via endocytosis [47]. Thus there is a correlation between influenza virus infection and the pathways and processes that are highly enriched for immune signaling and responses.
Among 1,298 up-regulated lncRNAs in PR/8-infected cells, 7 selected lncRNAs in our studies were also induced by other H1N1 strains, WSN and Pdm/OK, but the magnitude of induction varied, indicating that there are differences in virus strains in induction of lncRNA expression. lncRNA expression can be induced by not only influenza viruses but also other RNA viruses including human immunodeficiency virus (HIV) [24] and hepatitis C virus (HCV) [28] and DNA viruses such as herpesviruses [25]. Efforts have been made to understand if lncRNA induction is due to a direct viral effect or changes in host cellular signaling. One study has demonstrated that live influenza virus is necessary for induction of lncRNA VIN [29]. Although it is induced by various strains of influenza virus such as H1N1, H3N2 and H7N7, VIN is not inducible by IFN or viral RNA mimics. On other hand, host IFN signaling also contributes to the induction of lncRNA expression as demonstrated by using IFNs [31,48,49] or NS1-mutant virus which is unable to counteract IFN response from host [29]. The seven lncRNAs used in our studies were also induced by type I IFN.
Functional analysis of one of the highly upregulated lncRNA PSMB8-AS1 indicated that it acts as pro-viral for influenza virus replication. In addition to this PSMB8-AS1 is also induced by IFN treatment which suggests that role of PSMB8-AS1 may not be only limited to viral pathogenesis but may be involved in modulation of IFN response. Similar observations were also recorded that lncRNAs may play role in different viral infection models [27] and reviewed as bio-entities having important role in virus infections [50]. LncRNAs NRAV [27] and VIN [29] have been shown to reduce virus replication. VIN lncRNA is required for influenza virus replication and its deletion reduces virus yield and protein synthesis however the exact mechanism is yet to be clarified. NRAV has been shown to be involved in downregulation of Mx1 and interferon induced transmembrane protein 3 (IFITM3) through histone modification of these genes which are crucial part of anti-viral immune response during influenza virus infection.
Localization of lncRNAs in cells may provide significant information about how their functions are achieved. lncRNA in the cytoplasm such as DANCR can compete for microRNA binding sites [51]. lncRNAs in the nucleus can regulate gene transcription through chromatin modification [52]. NEAT1 and MALAT1 are present in paraspeckles and nuclear speckles inside the nucleus, respectively [30,53]. NEAT1 is responsible for maintaining structure of paraspeckles [54] as well as regulation of transcription of IL-8 genes [30]. MALAT1 is known to regulate genes associated with lung cancer metastasis [55]. Another nuclear lncRNA, NRAV functions as a histone modification factor of anti-viral genes, MxA and IFITM3 [27]. PSMB8-AS1 is predominantly located in the nucleus of A549 cells which suggest that it may play a role in transcription, chromatin remodeling or post-transcriptional processing [56]. However, our results showed that PSMB8-AS1 had no effects on the epigenetic markers (H3K27m3 and H3K27Ac). Exact role and location inside the nucleus of PSMB8-AS1 during influenza virus infection remains to be determined. Taken together, our results indicate, for the first time, a role of lncRNA PSMB8-AS1 in influenza virus replication.
Funding Statement
This work was supported by the National Institutes of Health [HL116876]; National Institutes of Health [AI121591]; National Institutes of Health [GM103648]; National Institutes of Health [HL135152]; the Lundberg-Kienlen endowment fund; the Oklahoma Center for Adult Stem Cell Research; the Oklahoma Center for the Advancement of Science & Technology.
Acknowledgments
L.L. is supported by NIH grants AI121591, HL116876, HL135152 and GM103648, the Oklahoma Center for the Advancement of Science & Technology, the Oklahoma Center for Adult Stem Cell Research, the Lundberg-Kienlen endowment fund and seed grants from the Center of Veterinary Health Sciences, Oklahoma State University.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Heward JA, Lindsay MA.. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014. September;35(9):408–419. PubMed PMID: 25113636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009. July 14;106(28):11667–11672. PubMed PMID: 19571010; PubMed Central PMCID: PMCPMC2704857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012. September 06;489(7414):101–108. PubMed PMID: 22955620; PubMed Central PMCID: PMCPMC3684276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004. April;5(4):316–323. PubMed PMID: 15131654. [DOI] [PubMed] [Google Scholar]
- [5].Liu Y, Zhang R, Ying K. Long noncoding RNAs: novel links in respiratory diseases (review). Mol Med Rep. 2015. June;11(6):4025–4031. PubMed PMID: 25647639. [DOI] [PubMed] [Google Scholar]
- [6].Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010. April 15;464(7291):1071–1076. PubMed PMID: 20393566; PubMed Central PMCID: PMCPMC3049919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011. October;1(5):391–407. PubMed PMID: 22096659; PubMed Central PMCID: PMCPMC3215093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015. February 13;116(4):737–750. PubMed PMID: 25677520. [DOI] [PubMed] [Google Scholar]
- [9].Plath K, Mlynarczyk-Evans S, Nusinow DA, et al. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. PubMed PMID: 12429693. [DOI] [PubMed] [Google Scholar]
- [10].Martianov I, Ramadass A, Serra Barros A, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007. February 08;445(7128):666–670. PubMed PMID: 17237763. [DOI] [PubMed] [Google Scholar]
- [11].Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011. June 05;43(7):621–629. PubMed PMID: 21642992; PubMed Central PMCID: PMCPMC3652667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010. June 24;465(7301):1033–1038. PubMed PMID: 20577206; PubMed Central PMCID: PMCPMC3206313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bertani S, Sauer S, Bolotin E, et al. The noncoding RNA mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011. September 16;43(6):1040–1046. PubMed PMID: 21925392; PubMed Central PMCID: PMCPMC3176448. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011. April 07;472(7341):120–124. PubMed PMID: 21423168; PubMed Central PMCID: PMCPMC3670758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014. January 21;111(3):1002–1007. PubMed PMID: 24371310; PubMed Central PMCID: PMCPMC3903238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rapicavoli NA, Qu K, Zhang J, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013. July 23;2:e00762 PubMed PMID: 23898399; PubMed Central PMCID: PMCPMC3721235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. Elife. 2014. April 29;3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013. August 16;341(6147):789–792. PubMed PMID: 23907535; PubMed Central PMCID: PMCPMC4376668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sharma S, Findlay GM, Bandukwala HS, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A. 2011. July 12;108(28):11381–11386. PubMed PMID: 21709260; PubMed Central PMCID: PMCPMC3136327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gomez JA, Wapinski OL, Yang YW, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013. February 14;152(4):743–754. PubMed PMID: 23415224; PubMed Central PMCID: PMCPMC3577098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu AY, Torchia BS, Migeon BR, et al. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics. 1997. January 15;39(2):171–184. PubMed PMID: 9027504. [DOI] [PubMed] [Google Scholar]
- [22].Mourtada-Maarabouni M, Hedge VL, Kirkham L, et al. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci. 2008. April 01;121(Pt 7):939–946. PubMed PMID: 18354083. [DOI] [PubMed] [Google Scholar]
- [23].Hu G, Tang Q, Sharma S, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013. November;14(11):1190–1198. PubMed PMID: 24056746; PubMed Central PMCID: PMCPMC3805781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imam H, Bano AS, Patel P, et al. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Sci Rep. 2015. March 02;5:8639 PubMed PMID: 25728138; PubMed Central PMCID: PMCPMC4345339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Massimelli MJ, Majerciak V, Kruhlak M, et al. Interplay between polyadenylate-binding protein 1 and Kaposi’s sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J Virol. 2013. January;87(1):243–256. PubMed PMID: 23077296; PubMed Central PMCID: PMCPMC3536381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Saayman S, Ackley A, Turner AW, et al. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther. 2014. June;22(6):1164–1175. PubMed PMID: 24576854; PubMed Central PMCID: PMCPMC4048891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ouyang J, Zhu X, Chen Y, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014. November 12;16(5):616–626. PubMed PMID: 25525793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xiong Y, Jia M, Yuan J, et al. STAT3regulated long noncoding RNAs lnc7SK and lncIGF2AS promote hepatitis C virus replication. Mol Med Rep. 2015. November;12(5):6738–6744. PubMed PMID: 26328522; PubMed Central PMCID: PMCPMC4626162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Winterling C, Koch M, Koeppel M, et al. Evidence for a crucial role of a host non-coding RNA in influenza a virus replication. RNA Biol. 2014;11(1):66–75. PubMed PMID: 24440876; PubMed Central PMCID: PMCPMC3929426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014. February 06;53(3):393–406. PubMed PMID: 24507715. [DOI] [PubMed] [Google Scholar]
- [31].Kambara H, Gunawardane L, Zebrowski E, et al. Regulation of interferon-stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front Immunol. 2014;5:676 PubMed PMID: 25688240; PubMed Central PMCID: PMCPMC4311693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Reed L, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27:493–497. [Google Scholar]
- [33].Inglis SC. Inhibition of host protein synthesis and degradation of cellular mRNAs during infection by influenza and herpes simplex virus. Mol Cell Biol. 1982. December;2(12):1644–1648. PubMed PMID: 14582206; PubMed Central PMCID: PMCPMC369973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015. February 03;16(2):3251–3266. PubMed PMID: 25654223; PubMed Central PMCID: PMCPMC4346893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Buggele WA, Johnson KE, Horvath CM. Influenza a virus infection of human respiratory cells induces primary microRNA expression. J Biol Chem. 2012. September 07;287(37):31027–31040. PubMed PMID: 22822053; PubMed Central PMCID: PMCPMC3438935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008. July;8(7):559–568. PubMed PMID: 18575461; PubMed Central PMCID: PMCPMC2522268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hassan IH, Zhang MS, Powers LS, et al. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem. 2012. February 10;287(7):4679–4689. PubMed PMID: 22194594; PubMed Central PMCID: PMCPMC3281634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Roberson EC, Tully JE, Guala AS, et al. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-beta release in lung epithelial cells. Am J Respir Cell Mol Biol. 2012. May;46(5):573–581. PubMed PMID: 21799120; PubMed Central PMCID: PMCPMC3359902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dittmann J, Stertz S, Grimm D, et al. Influenza a virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J Virol. 2008. April;82(7):3624–3631. PubMed PMID: 18199636; PubMed Central PMCID: PMCPMC2268464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Melchjorsen J, Kristiansen H, Christiansen R, et al. Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res. 2009. April;29(4):199–207. PubMed PMID: 19203244. [DOI] [PubMed] [Google Scholar]
- [41].Benitez AA, Panis M, Xue J, et al. In Vivo RNAi Screening Identifies MDA5 as a significant contributor to the cellular defense against influenza a virus. Cell Rep. 2015. June 23;11(11):1714–1726. PubMed PMID: 26074083; PubMed Central PMCID: PMCPMC4586153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cheng H, Koning K, O’Hearn A, et al. A parallel genome-wide RNAi screening strategy to identify host proteins important for entry of Marburg virus and H5N1 influenza virus. Virol J. 2015. November 24;12:194 PubMed PMID: 26596270; PubMed Central PMCID: PMCPMC4657351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wilk E, Pandey AK, Leist SR, et al. RNAseq expression analysis of resistant and susceptible mice after influenza a virus infection identifies novel genes associated with virus replication and important for host resistance to infection. BMC Genomics. 2015. September 02;16:655 PubMed PMID: 26329040; PubMed Central PMCID: PMCPMC4557482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Garfinkel MS, Katze MG. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5ʹ-untranslated region. J Biol Chem. 1993. October 25;268(30):22223–22226. PubMed PMID: 8226725. [PubMed] [Google Scholar]
- [45].Li J, Hou R, Niu X, et al. Comparison of microarray and RNA-Seq analysis of mRNA expression in dermal mesenchymal stem cells. Biotechnol Lett. 2016. January;38(1):33–41. PubMed PMID: 26463369. [DOI] [PubMed] [Google Scholar]
- [46].Beloso A, Martinez C, Valcarcel J, et al. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J Gen Virol. 1992. March;73(Pt 3):575–581. PubMed PMID: 1545220. [DOI] [PubMed] [Google Scholar]
- [47].Lakadamyali M, Rust MJ, Zhuang X. Endocytosis of influenza viruses. Microbes Infect. 2004. August;6(10):929–936. PubMed PMID: 15310470; PubMed Central PMCID: PMCPMC2715838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Josset L, Tchitchek N, Gralinski LE, et al. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014;11(7):875–890. PubMed PMID: 24922324; PubMed Central PMCID: PMCPMC4179962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42(16):10668–10680. PubMed PMID: 25122750; PubMed Central PMCID: PMCPMC4176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang Q, Jeang KT. Long non-coding RNAs (lncRNAs) and viral infections. Biomed Pharmacother. 2013. March 01;3(1):34–42. PubMed PMID: 23645970; PubMed Central PMCID: PMCPMC3641704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016. February;63(2):499–511. PubMed PMID: 25964079. [DOI] [PubMed] [Google Scholar]
- [52].Nakagawa S, Kageyama Y. Nuclear lncRNAs as epigenetic regulators-beyond skepticism. Biochim Biophys Acta. 2014. March;1839(3):215–222. PubMed PMID: 24200874. [DOI] [PubMed] [Google Scholar]
- [53].Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. Rna. 2012. August;18(8):1487–1499. PubMed PMID: 22718948; PubMed Central PMCID: PMCPMC3404370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009. March 27;33(6):717–726. PubMed PMID: 19217333; PubMed Central PMCID: PMCPMC2696186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013. February 01;73(3):1180–1189. PubMed PMID: 23243023; PubMed Central PMCID: PMCPMC3589741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang K, Shi Z-M, Chang Y-N, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014. August 15;547(1):1–9. PubMed PMID: 24967943. [DOI] [PubMed] [Google Scholar]


