ABSTRACT
Genome-wide mapping of lamin-B1-genome interactions has shown that gene-poor and transcriptionally inactive genomic regions are associated with the nuclear lamina. Numerous studies have suggested that lamins, the major structural components of the nuclear lamina, play a role in global chromatin organization and gene expression. How lamins could influence the 3D genome organization and transcription from the nuclear periphery has, however, remained unclear. Our recent studies showed that lamins differentially regulate distinct lamina-associated chromatin domains (LADs) at the nuclear periphery, which can in turn influence global 3D genome organization and gene expression. In this Extra View, we discuss how by using various genomics tools, it has become possible to reveal the functions of lamins in orchestrating 3D genome organization and gene expression.
Abbreviations: 3D: three dimensional; LAD: lamina-associated chromatin domain; 3C: Chromosome Conformation Capture; TAD: topologically associated domain; HiLands: Histone and lamina landscape; NL: nuclear lamina; mESC: mouse embryonic stem cell; DamID: DNA adenine methyltransferase identification;
KEYWORDS: 3D genome, Hi-C, HiLands, LADs, TADs, lamin, nuclear lamina
Introduction
In mammalian cells, the linear genomic DNA stretches for more than 2 meters, which is approximately 200,000-fold the diameter of the cell nucleus. Thus, genomic DNA is extensively packaged into the nucleus. Recently, chromosome conformation capture (3C)-based techniques, including 3C, 4C, 5C, Hi-C and ChIA-PET, have visualized 3D genome organization at an unprecedented high resolution. Data from these 3C-based techniques showed that, within interphase nuclei, genomic DNA forms domain-like structures highly organized in a hierarchical manner, which can play a critical role in orchestrating cell type-specific gene expression during development and homeostasis [1]. The most fundamental structural unit of the genome is the chromosome. Each chromosome occupies a specific space within interphase nuclei, referred to as a chromosome territory (CT) [2]. Genomic regions with similar transcriptional activities tend to co-localize in the nuclear space, forming active A compartments and inactive B compartments that reflect euchromatin and heterochromatin, respectively [3]. Studies of 3D chromatin organization in human and mouse cell lines have revealed that the genome is further organized into high-order structural domains known as topologically associating domains (TADs), which range from several hundred kilobases (kb) to several megabases (Mb) in size [4,5]. To date, the densest Hi-C map to probe 3D genome organization in human lymphoblastoid cells contains 4.9 × 109 contacts, achieving 1 kb resolution [6]. This high-resolution Hi-C map revealed that the genome is additionally partitioned into sub-TAD scale domains with a median length of 185 kb. Finally, the genome contains approximately 10,000 long-range interactions referred to as loops, which frequently link promoters and enhancers within a TAD and correlate with gene activation. Increasing evidence suggests that various combinations of chromatin architectural proteins such as CTCF, cohesins, and mediators are responsible for the assembly of chromatin loops of different lengths and transcriptional activities, which cooperate to establish sub-TADs and TADs [7–10]. Genome-wide studies using Hi-C and extensive analyses of local chromatin interactions using 5C have suggested that extrusion of the chromatin fiber by cohesin contributes to the establishment of chromatin loops and stochastic aspects of the transcription process [11]. However, many of these studies have focused on the 3D organization of actively transcribing euchromatin regions, whereas the folding principle of heterochromatin regions, especially the heterochromatin found at the nuclear periphery, and their impact on gene expression are not as well understood.
Classic microscopic observations have shown that the nuclear lamina (NL) is associated with highly condensed heterochromatin in the nucleus. The NL is a protein meshwork that forms beneath the inner nuclear membrane. Lamins, the intermediate filament proteins, are the major constituents of the NL. Mutations in lamins are associated with many different human diseases referred to as laminopathies. Genetic analyses of mice or mouse embryonic stem cells (mESCs) lacking all three lamins (lamin triple knockout, TKO) or just lamin-B1 and -B2 double knockout (lamin-B DKO) suggested that lamins are not essential for cell survival and proliferation, but are required for proper organ building and maintenance [12–14]. High-resolution maps of lamin-B1-genome interactions via DNA adenine methyltransferase identification (DamID) revealed that gene-poor and transcriptionally inactive genomic regions preferentially interact with the NL, leading to the hypothesis that the NL provides an environment that silences their interacting genes [15]. However, tethering genes to the NL does not necessarily lead to gene silencing [16–18]. Similarly, removing NL proteins does not always lead to de-repression of NL-associated genes [12]. Therefore, the influence of the NL on chromatin organization and gene expression is not explained by the simple NL-chromatin tethering model.
To better understand how the NL influences genome organization and gene expression, it is essential to examine the relationship of the NL-chromatin interaction along with other genomic and epigenetic features. Indeed, combinatorial analyses of lamin-B1 DamID, histone modifications, and core and linker histone occupancy in mESCs revealed that formerly defined uniform lamina-associated chromatin domains (LADs) could be separated into two distinct chromatin domains referred to as Histone and Lamina landscape (HiLands)-B and -P [19]. Comparison of LADs from different cell lineages derived from mESCs also showed that LADs could be separated into facultative LADs (fLADs) and constitutive LADs (cLADs), which are correlated with HiLands-B and -P, respectively [20]. Studies of lamin-B1 DamID in different cell lineages showed that fLADs were detached from the NL in a cell type-specific manner, while cLADs were fairly consistently associated with the NL in all tested cell types, suggesting that cLADs and fLADs behave differently depending on their underlying genomic features. Another study showed that lamin-A/C and lamin-B receptor (LBR, an inner nuclear envelope transmembrane protein) were required to tether heterochromatin to the nuclear periphery, and the absence of these proteins led to large-scale relocation of heterochromatin to the nuclear center in cells such as the rod photoreceptors in nocturnal mammals [21]. These studies suggest that NL proteins play a role in organizing the 3D chromatin structure at the nuclear periphery. However, the precise mechanism by which NL proteins regulate 3D genome organization is poorly understood. By combining various experimental approaches, including Hi-C, 4C analysis, fluorescence in situ hybridization (FISH) with DamID and HiLand, epigenome and transcriptome analyses of wild-type (WT) and lamin null (Lmnb1, Lmnb2, and Lmna TKO) mESCs [13], we recently reported that lamins orchestrated 3D genome organization from the nuclear periphery by differentially regulating different classes of LADs, which in turn influenced chromatin interactions and gene expression in neighboring non-LAD genomic regions [22]. In the following sections, we discuss how our studies using various genomics tools have begun to reveal the functions of lamins in orchestrating 3D genome organization and gene expression.
Absence of lamins leads to changes in inter-TAD chromatin interactions without affecting overall TAD structure
To explore how lamins influence 3D genome organization, we mapped genome-wide chromatin interactions in lamin null mESCs using an in situ ligation-based Hi-C method [6,23]. After filtering and mapping the raw reads, we obtained approximately 3.04 × 108 and 3.74 × 108 validated read pairs for WT and lamin null mESCs, respectively. To assess the consistency between our Hi-C datasets, as well as between our data and other datasets, we normalized the filtered and mapped Hi-C data using the iterative correction and eigenvector decomposition method [24]. We found that our Hi-C datasets from WT and lamin null mESCs were consistent between biological replicates, and our WT mESC Hi-C datasets showed consistency with published WT E14 mESC Hi-C datasets.
Hi-C analyses of genomes from organisms such as mammals and fly have shown that TADs, which are self-interacting and functional chromatin domains, are demarcated with sharp TAD boundaries [4,5,25]. Through insulation score calculation [26], a TAD calling method, we obtained 3,268 and 3,206 TAD boundaries for WT and lamin null mESCs, respectively. We found that more than 90% of TAD boundaries overlapped between WT and lamin null mESCs, indicating that the overall TAD structure is maintained in the absence of all lamins (Figure 1(a,b)). However, a closer analysis of WT and lamin null datasets revealed that the interactions between TADs were evidently changed in lamin null cells (Figure 1(c)). EdgeR analysis of Hi-C datasets from our WT and lamin null mESCs showed that 4,352 TAD pairs presented altered inter-TAD interactions upon lamin loss. We obtained similar results by comparing our lamin null and published E14 WT mESC datasets. Therefore, this difference is not related to a random variation in chromatin interactions in different datasets. Taken together, our Hi-C studies demonstrated that depletion of all lamins did not disrupt the overall TAD structure, but it led to alterations in TAD-TAD interactions.
Figure 1.
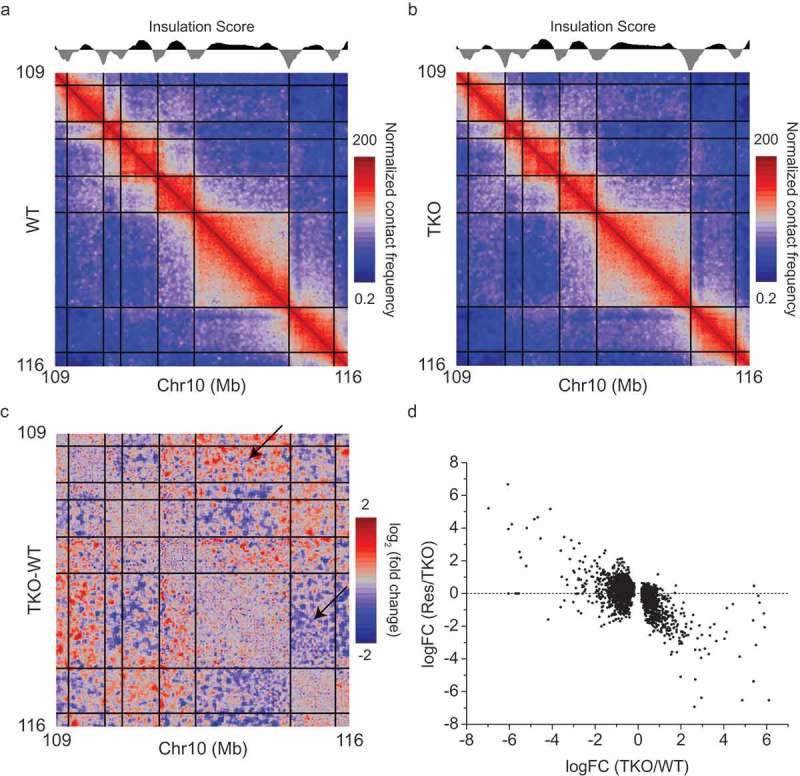
Changes of inter-TAD interactions upon lamin loss and a rescue by expressing lamin-B1. (a,b) Heat map delineates normalized chromatin interaction frequency in a selected region of chromosome 10 from wild-type (WT, a) and lamin null (TKO, b) mESCs. (c) The log2 fold changes of inter TAD interactions between WT and lamin null mESCs. Arrows represents increased or decreased inter-TAD interactions upon lamin loss. Black lines in (a) – (c) demarcate the TAD boundaries. (d) Comparison of log2 fold changes (FC) of inter-TAD interactions between rescued (Res, lamin null mESCs expressing lamin-B1) and lamin null mESCs as a function of those between lamin null and WT mESCs. Figures are from Zheng et al. 2018 [22], courtesy of the authors.
We next analyzed NL associations with genomic regions exhibiting altered inter-TAD interactions by comparing lamin-B1 DamID values in each region. We found that most TAD pairs exhibiting altered inter-TAD interactions were associated with the NL in at least one TAD of each TAD pair. Interestingly TAD pairs exhibiting increased inter-TAD interactions showed strong lamin-B1 associations in both TADs of the TAD pairs, whereas TAD pairs exhibiting decreased inter-TAD interactions showed strong lamin-B1 associations in one TAD and weak or no lamin-B1 associations in the other TAD of each TAD pair. We also found that restoring lamin-B1 expression in lamin null mESCs significantly reversed the alterations in inter-TAD interactions in both types of TAD pairs exhibiting increased and decrease interactions (Figure 1(d)).
Lamin loss causes decondensation of HiLands-P
To understand how lamins regulate the two different types of TAD pairs exhibiting changed inter-TAD interactions, it is essential to analyze their underlying genomic features along with their NL-association. Our Hidden Markov Model of core histone modifications (H3K4me1, H3K27me3, and H3K9me3), core and linker histone occupancy (H3, H1c, and H1d), and chromatin interactions with lamin-B1 revealed that genomic regions can be separated into 6 distinct chromatin states referred to as HiLands-R (red), -O (orange), -Y (yellow), -G (green), -B (blue), and -P (purple) (Figure 2(a,b)). HiLands-B and -P represent distinct LADs, while HiLands-R, -O, -Y, and -G delineate interior chromatin domains [19]. This new classification of chromatin states not only provides an additional tool for analyzing chromatin domains but also establishes a foundation for analyzing distinct LADs when studying the functions of NL proteins in genome organization at the nuclear periphery and their influence on interior chromatin domains.
Figure 2.
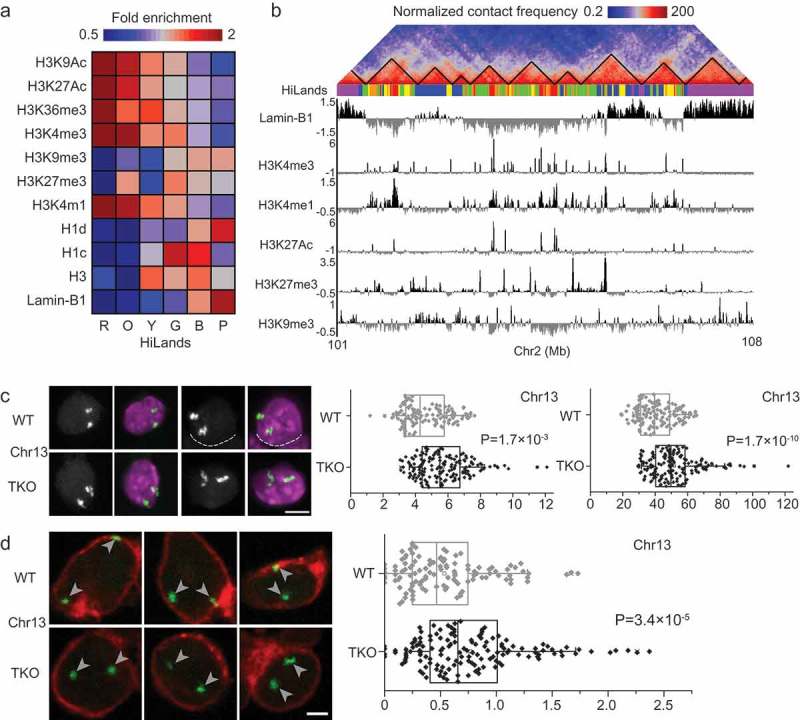
Differential changes in distinct LADs upon lamin loss. (a) Combinatory patterns of different histone modifications, histone H1 and H3, and lamin-B1 DamID reads defining the six different HiLands in mESCs. Heatmap represents the genome-wide fold enrichment of each chromatin state in mESCs. (b) A co-alignment of Hi-C heatmap, color-coded HiLands, lamin-B1 DamID and different histone modifications in a selected region of chromosome 2. Black lines in the Hi-C heatmap delineate TADs. (c) Two representative 3D-FISH images for a selected HiLands-P LAD region (left). Fluorescence images represents DAPI staining for DNA (purple) and FISH signal (white). The white dashed lines delineate the boundaries of neighboring nuclei. Scale bar, 5 μm. Quantifications of the volumes and surface areas of the HiLands-P LAD region (right); p values, Wilcoxon rank-sum test. (d) Three representative 3D-FISH images for a selected HiLands-B LAD region. Immunofluorescence staining of emerin (red) demarcates the boundaries of nuclei and the white arrow heads indicate FISH signals (green). Scale bar, 2 μm. Quantifications of the distances of FISH signals for the HiLands-B LAD region to the nuclear envelope marked by emerin (right); p values, Wilcoxon rank-sum test. Figures are from Zheng et al. 2018, courtesy of the authors.
When we mapped chromatin interactions from the Hi-C dataset of WT mESCs to the HiLands domains, we found strong intra-chromosomal interactions within all HiLands and between HiLands pairs R-O, Y-G, and B-P, whereas the HiLands-B or -P (the two distinct LADs) showed no strong interactions with interior HiLands domains. Depletion of lamins led to a strong increase in intra-chromosomal interactions within HiLands-P, whereas interactions between HiLands-P and other HiLands were decreased. When the inter-TAD interactions were overlaid on the HiLands map, most increased inter-TAD interactions in lamin null mESCs were mapped to HiLand pair P-P, while most decreased inter-TAD interactions in lamin null mESCs were mapped to HiLands-P and interior HiLands. To further analyze LADs characterized by HiLands-P in lamin null mESCs, we plotted changes in the intra-chromosomal interactions within HiLands-P in lamin null cells against the linear distance between two interacting loci. We found that upon lamin loss, interactions between loci separated by less than 1 Mb were mildly decreased, whereas interactions between distant loci located more than 1 Mb apart increased with the most significant increases being between loci separated by 10–20 Mb. Because the average size of TAD is approximately 1 Mb, it is likely that most increased interactions within HiLands-P in lamin null mESCs occur between different TADs. Indeed, overlaying all altered interactions within HiLands-P on a TAD map revealed increased inter-TAD interactions and decreased intra-TAD interactions in the absence of lamins. To visually inspect the inter-TAD interaction changes in HiLands-P regions, we employed FISH based on the Oligopaint method [27]. We generated pools of FISH probes for 4 HiLands-P regions in chromosomes 1, 4, 13, and 14 by two rounds of PCR against one common and one specific target sequences in each region. Measurement of FISH signals using the ‘Spots object’ function of Imaris revealed that all tested HiLands-P regions significantly increased their volumes and surface areas in lamin null mESCs compared to in WT mESCs (Figure 2(c)). Because FISH against the entire chromosome 1 or 13 revealed no significant changes in the volume or surface area of lamin null mESCs, the observed expansion of volume and surface area in the selected HiLands-P regions upon lamin loss did not result from whole chromosomal decondensation. Thus, local decondensation of HiLands-P regions may cause increased inter-TAD interactions in these regions upon lamin loss.
Lamin loss leads to compartment shift of HiLands-B
In a previous study, we demonstrated that HiLands-B domains were detached from the nuclear periphery in the absence of lamins as determined by the decreased emerin DamID values, while emerin DamID values at the HiLands-P domains in lamin null cells were increased compared to in WT mESCs [19]. These data clearly show that lamins differentially regulate distinct LADs. To further analyze how lamins regulate HiLands-B regions, we conducted A/B compartment analysis of the Hi-C datasets from WT and lamin null mESCs. Comparison of A/B compartments with HiLands domains showed that LADs consisting of HiLands-B and -P correspond to inactive B compartments, while interior domains consisting of HiLands-R, -O, -Y, and -G constitute active A compartments. Because emerin DamID analysis indicated that HiLands-B regions are detached from the nuclear periphery upon lamin loss, it is reasonable to examine whether HiLands-B regions undergo a compartment shift from the B to the A compartment in lamin null mESCs. Because the original compartment analysis based on the eigenvector method [3] could not quantitatively compare Hi-C datasets from different samples, we used our new compartment analysis tool known as Cscoretool [28] to compare Hi-C datasets of WT and lamin null mESCs. In WT mESCs, compartments defined by the original method and Cscoretool were highly correlated with positive C score values corresponding to B compartments, while negative C score values corresponded to A compartments. C score values were also highly consistent between biological repeats of our WT Hi-C datasets and between our WT and published E14 Hi-C datasets. In lamin null mESCs, more than 75% of HiLands-B regions showed decreased C scores compared to WT mESCs, which is consistent with the decreased emerin DamID values in HiLands-B regions upon lamin loss. We observed minor increases in C score values in HiLands-P regions upon lamin loss, which is consistent with HiLands-P remaining in the B compartment in lamin null mESCs. The increases in emerin DamID values in the lamin null mESCs may reflect the decondensation of HiLands-P regions upon lamin loss leading to more frequent associations with the NL. Detachments of HiLands-B regions from the NL were also visualized by FISH analyses using Oligopaint. For two selected HiLands-B regions, FISH signals in lamin null mESCs were farther away from the nuclear periphery marked by emerin than those in WT mESCs (Figure 2(d)). We did not observe significant changes in volume and surface area in FISH analysis of HiLands-B regions upon lamin loss. This may be because the size of the HiLands-B regions is too small to detect changes in volume or surface area at fluorescent microscopic resolution. Alternatively, enrichment in H3K27me3 in HiLands-B but not in HiLands-P [19] may lead to PRC1-mediated chromatin condensation, preventing decondensation of HiLands-B regions upon lamin loss.
Transcriptional changes upon lamin loss are correlated with alteration of 3D chromatin interactions
To understand how lamins regulate the epigenetic landscape and gene expression in mESCs, we carried out ChIP-seq analyses of epigenetic markers and transcriptome profiling. We found no significant changes in global H3K27me3 or H3K9me3 levels upon lamin loss. RNA-seq revealed 385 up- and 841 down-regulated genes in lamin null cells. However, the altered genes were not enriched in HiLands-B or -P, but instead were distributed across all 6 HiLands regions. To explore the correlation between altered gene expression and chromatin interaction changes upon lamin loss, we plotted the number of changed genes in the interior HiLands regions (-R, -O, -Y, and -G) against the linear distance from the transcription start site (TSS) to their nearest HiLands-B. We found that the closer the TSSs were to HiLands-B, the more likely their gene expression would change in lamin null mESCs. Therefore, the detached HiLands-B regions from the NL in the absence of lamins may lead to alterations in chromatin interactions between HiLands-B and neighboring interior chromatin regions, which could result in transcriptional changes in the interior chromatin regions. We also analyzed the influence of lamins on enhancers marked with H3K4me1 (all enhancer) and H3K27Ac (active enhancer). ChIP-seq of H3K4me1 revealed no significant difference between WT and lamin null mESCs. However, we found that the H3K27Ac level was significantly increased in genomic regions showing decreased interactions with HiLands-P upon lamin loss. By contrast, the H3K27Ac level was decreased in the genomic regions showing increased interactions with HiLands-B and decreased interactions with HiLands-R upon lamin loss. Plotting the number of altered H3K27Ac peaks against their linear distance to the nearest HiLands-B region reveals that the closer an enhancer is to HiLands-B, the more likely it is to change its activity upon lamin loss. We also found that altered TSSs and H3K27Ac peaks in lamin null mESCs were further away from HiLands-P than from HiLandsB. Thus, transcriptional changes and altered enhancer activities in lamin null mESCs can be better explained by the detachment of HiLands-B regions than by the decondensation of HiLands-P regions.
A model explaining how lamins can regulate global 3D genome organization from the nuclear periphery
A prevailing model suggests that the NL regulates 3D chromatin organization by tethering the lamina-associated chromatin domains (LADs) at the NL [29]. This model predicts that the loss of NL proteins would lead to the detachment of LADs from the NL. By analyzing mESCs without lamins, the major components of the NL, we show while the HiLands-P LAD regions maintain their attachment to the NL, they become decondensed. On the other hand, the LADs represented by HiLands-B are detached from the NL upon lamin loss. Our findings suggest that lamins do not simply tether LADs. To explain how lamins differentially regulate HiLands-P and -B, we propose a meshwork caging model (Figure 3). Lamins assemble into a dense intermediate filament meshwork. This dense meshwork traps and cages HiLands-P strongly and HiLands-B to a lesser degree in WT cells. Upon loss of this meshwork, the decondensation of HiLands-P LADs at the nuclear periphery may push the weakly trapped HiLands-B LADs away from the nuclear periphery. The detached HiLands-B would have an increased chance of interacting with interior chromatin domains, which could, in turn, perturb interactions within interior domains and transcription. Although purified lamins exhibited weak interactions with chromatin in in vitro studies [30], the assembled lamin meshwork in vivo would provide enough multivalent docking points to cage lamina-associated chromatin domains. The lamin meshwork may form multivalent interactions with HiLands-B and -P by recognizes their differential epigenetic features or their differentially enriched linker histones [19].
Figure 3.
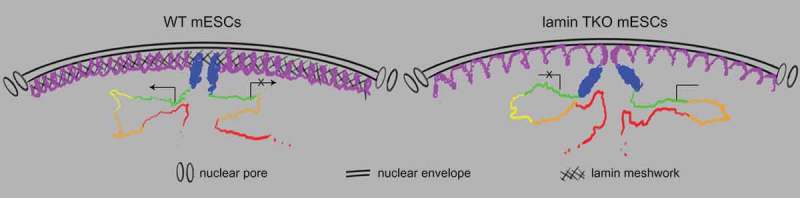
A meshwork caging model of the nuclear lamina. In WT mESCs (left), the dense meshwork of the nuclear lamina traps and cages HiLands-P LADs (purple line) strongly and HiLands-B LADs (blue line) to a lesser degree. The interior chromatin shown as HiLands-G (green), HiLands-Y (yellow), HiLands-O (orange), and HiLands-R (red) are represented by the correspondingly colored lines. Gene expression or repression are shown by arrows or arrows with crosses, respectively. In lamin null mESCs (right), HiLands-P LADs are decondensed, which, in turn, push away the weakly associated HiLands-B LADs, leading to the detachments of HiLands-B LADs from the nuclear periphery. These changes may cause the perturbations of chromatin interactions in interior chromatin regions and consequently lead to alteration of the transcriptional network. Interestingly, the linear distances between TSS in interior HiLands domins and HiLands-B are correlated with the extent of gene expression change in the TSS upon lamin loss. Figures are from Zheng et al. 2018, courtesy of the authors.
Conclusions and perspectives
We have shown that lamin loss while not affecting the overall TAD structure, leads to alterations in inter-TAD interactions. Further analyses using DamID and FISH revealed that the two distinct LADs consisting of HiLands-B and -P respond differently to lamin loss. HiLands-P regions remain attached to the nuclear periphery but undergo expansion, which is reflected by increased inter-TAD and decreased intra-TAD interactions. HiLands-B regions, on the other hand, are detached from the nuclear periphery, which is confirmed by a shift of chromatin compartments. The detachment of HiLands-B regions perturbs chromatin interactions and correlates with gene expression changes. Our findings suggest that lamins orchestrate global 3D chromatin organization at the nuclear periphery by differentially regulating the two distinct LADs.
Because alterations in the NL are associated with defects in development, homeostasis, and aging, our findings establish a foundation for explaining the role of lamins in these processes. Previous studies suggest that interactions of fLADs with the NL are gradually changed as pluripotent stem cells differentiate into different cell lineages during development. Our findings may explain how such alterations in interactions between fLADs and the NL could establish a new 3D chromatin interactions and gene regulation program in different cell types. Furthermore, as cells differentiate, cytoskeleton activities are generally increased which is vital for cell morphological changes. The cytoskeleton-mediated morphological changes may cause alterations in the density of the NL via the linker of nucleoskeleton and cytoskeleton (LINC) complexes and nucleoporins (NPCs) in different regions of the nuclear surface, leading to reorganization of 3D chromatin interactions and gene regulation that match the needs of different cell lineages. Therefore, it would be interesting to study whether and how the cytoskeleton-mediated morphological changes in cells can specifically influence 3D chromatin interactions and gene regulation in different cell lineages, because such understanding could offer insights into how gene expression is coupled with morphogenesis of the myriads of different cell shapes during development and organogenesis.
Funding Statement
This work was supported by the National Research Foundation of Korea (grant numbers: NRF-2017R1D1A1B03035010, NRF-2016K1A4A394725), Business Belt Program of Korea (2015-DD-RD-0069), and the Soonchunhyang University Research Fund to Y.K. and National Institute of Health of US (GM110151 and GM106023) to Y.Z.
Acknowledgments
We thank Cell Press for allowing to reuse figures published in Molecular Cell.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Gorkin DU, Leung D, Ren B.. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14:762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nora EP, Lajoie BR, Schulz EG, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nora EP, Goloborodko A, Valton AL, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–44 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Phillips-Cremins JE, Sauria ME, Sanyal A, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sanborn AL, Rao SS, Huang SC, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim Y, Sharov AA, McDole K, et al. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res. 2013;23:1420–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang SH, Chang SY, Yin L, et al. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum Mol Genet. 2011;20:3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guelen L, Pagie L, Brasset E, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. [DOI] [PubMed] [Google Scholar]
- [16].Finlan LE, Sproul D, Thomson I, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reddy KL, Zullo JM, Bertolino E, et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. [DOI] [PubMed] [Google Scholar]
- [19].Zheng X, Kim Y, Zheng Y. Identification of lamin B-regulated chromatin regions based on chromatin landscapes. Mol Biol Cell. 2015;26:2685–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Peric-Hupkes D, Meuleman W, Pagie L, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Solovei I, Wang AS, Thanisch K, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. [DOI] [PubMed] [Google Scholar]
- [22].Zheng X, Hu J, Yue S, et al. Lamins organize the global three-dimensional genome from the nuclear periphery. Mol Cell. 2018;71:802–15 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imakaev M, Fudenberg G, McCord RP, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sexton T, Yaffe E, Kenigsberg E, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. [DOI] [PubMed] [Google Scholar]
- [26].Crane E, Bian Q, McCord RP, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Beliveau BJ, Joyce EF, Apostolopoulos N, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci U S A. 2012;109:21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zheng X, Zheng Y. CscoreTool: fast Hi-C compartment analysis at high resolution. Bioinformatics. 2018;34:1568–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gonzalez-Sandoval A, Towbin BD, Kalck V, et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell. 2015;163:1333–1347. [DOI] [PubMed] [Google Scholar]
- [30].Wilson KL, Foisner R. Lamin-binding proteins. Cold spring harbor perspectives in biology. 2010;2:a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]


