Abstract
Background
Treatment options of glioblastoma, the most aggressive primary brain tumor with frequent relapses and high mortality, are still very limited, urgently calling for novel therapeutic targets. Expression of the glycoprotein podoplanin correlates with poor prognosis in various cancer entities, including glioblastoma. Furthermore, podoplanin has been associated with tumor cell migration and proliferation in vitro; however, experimental data on its function in gliomagenesis in vivo are still missing. Hence, we have functionally investigated the impact of podoplanin on glioblastoma in a preclinical mouse model to evaluate its potential as a therapeutic target.
Methods
Fluorescence activated cell sorting, genome-wide expression analysis, and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)–mediated deletion of podoplanin in patient-derived human glioblastoma cells were combined with organotypic brain slice cultures and intracranial injections into mice.
Results
We defined a malignant gene signature in tumor cells with high podoplanin expression. The increase and/or maintenance of high podoplanin expression in serial transplantations and in podoplaninlow-sorted glioblastoma cells during outgrowth indicated the association of high podoplanin expression and poor outcome. Unexpectedly, similar rates of proliferation, apoptosis, angiogenesis, and invasion were observed in control and podoplanin-deleted tumors. Accordingly, neither tumor growth nor survival was affected upon podoplanin loss.
Conclusion
We report that tumor progression occurs independently of podoplanin. Thus, in contrast to previous suggestions, blocking of podoplanin does not represent a promising therapeutic approach. However, as podoplanin is associated with tumor aggressiveness and progression, we propose the cell surface protein as a biomarker for poor prognosis.
Keywords: brain tumor, intracranial injection, patient-derived xenografts, therapy
Key Points
High podoplanin expression in glioma cells is part of a malignant gene signature.
Glioma cells acquire high podoplanin expression during in vivo tumor growth.
CRISPR/Cas9-mediated knockout of podoplanin in glioma cells does not affect malignancy.
Importance of the Study
Previously, podoplanin was reported to regulate glioblastoma proliferation and migration in vitro. In combination with studies that correlated high podoplanin expression with poor prognosis, podoplanin was hypothesized as a major driver for glioblastoma and considered as a potential therapeutic target. Unexpectedly, we found that deletion of podoplanin in primary glioblastoma cells and glioblastoma cell lines does not affect tumor progression in a mouse xenograft model, suggesting either that the potential malignant function of podoplanin is compensated by another yet unknown protein or that podoplanin is associated with, but not functionally implicated in, malignant features. Thus, podoplanin inactivation does not represent a promising option for glioblastoma therapy. However, the evident upregulation and the association with high aggressiveness in glioblastoma rather prompt the clinical usage of tumor-specific podoplanin as a marker for tumor progression and a cell surface molecule for targeted cytotoxic therapy.
Glioblastoma is the most frequent and most malignant primary brain tumor in humans, with a median survival of 12–15 months after diagnosis.1 Despite gross resection of the tumor mass, harsh radiation, and chemotherapy, the prognosis for glioblastoma patients remains very dismal, and current therapeutic means add mere months to patient survival.1 Thus, there is a great need for the identification of novel targets and development of more effective treatments to combat therapy resistance and diffuse infiltration of tumor cells into adjacent healthy tissue. Podoplanin (PDPN) has recently been implicated in progression and invasion of various cancer entities, including glioblastoma.2–4 PDPN is a cell surface protein expressed in various tissues throughout the body.5 During embryonic development, PDPN in the neuroepithelium mediates the maturation and integrity of the developing vasculature in the murine brain in interaction with C-type lectin-like receptor 2 on platelets.6 In the adult brain, PDPN expression is restricted to neural stem cells,7 ependymal cells, choroid plexus,8 and reactive astrocytes.9 However, the physiological function of PDPN in the adult brain has not been clarified yet. Previously, we and others have shown strong expression of PDPN in high-grade gliomas and furthermore presented PDPN as a marker for malignant progression and poor prognosis in glioma patients.2,10,11 High levels of podoplanin have also been observed in many other cancer entities, such as squamous cell carcinoma.3 In vitro overexpression studies in Michigan Cancer Foundation 7 breast cancer cells,4 Madin–Darby canine kidney cells,12 and U373MG and U87MG glioma cells13 resulted in a significant increase in migration. Consistently, we found that a short hairpin (sh)–mediated knockdown of PDPN in glioma cells resulted in decreased proliferation, 2D migration, and invasion into a collagen matrix.10,11 However, only a few publications dealing with other cancer entities have included functional studies in vivo. PDPN overexpression has been linked with tumor cell invasion in pancreatic adenocarcinoma and lung metastasis.4,14 Conformably, one recent study has associated downmodulation of PDPN expression with decreased tumor growth and invasion of xenotransplanted epidermoid carcinoma cells.15 In summary, there are many indications that PDPN might be a driver for tumor cell invasion and malignant progression; however, whether this applies for high-grade glioma in a preclinical patient-derived xenograft model remains to be examined.
Here we have functionally investigated the effect of PDPN on glioblastoma progression in vivo. Analysis of PDPN high and low expressing subpopulations of primary glioblastoma has experimentally confirmed the finding that PDPN is associated with poor prognosis in glioblastoma. However, loss-of-function experiments demonstrated that PDPN deletion in human glioma cell lines as well as in primary glioblastoma cells affected neither tumor growth nor vascularization, apoptosis, or tumor cell invasion, contradicting the hypothesis of a driving function for PDPN in glioblastoma. In summary, we show that PDPN does not represent a major driver for glioblastoma but may constitute both a valuable biomarker for progressive disease and, due to its exposition on the cell surface, a possible entry point for antibody- or chimeric antigen receptor (CAR) T-cell–mediated cytotoxic therapy.
Methods
Tumor Cell Cultivation
Primary human glioblastoma tumors GBMF2, GBMF3, and GBMF10 were freshly obtained from the University Hospital/Edinger Institute in Frankfurt, Germany. For cell isolation, tumor tissue was minced and digested in Leibovitz medium supplemented with 12 U/mL papain, 200 U/mL DNase, and 0.5 mM EDTA for 25 min at 37 °C. After filtration (70 µm) and lysis of erythrocytes, tumor cells were cultured as spheroids in serum-free Neurobasal medium (Life Technologies) containing B27 supplement, 20 ng/mL of each epidermal growth factor and basic fibroblast growth factor, 2 μg/mL heparin sodium salt, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin at 37°C and 5% CO2. Established human glioma cell lines LN308 and LN319 were cultured as adherent monolayers in Dulbecco’s modified Eagle’s medium 10% fetal calf serum, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin at 37°C and 5% CO2.
The cell lines LN319 and LN308 were authenticated in April 2018 using Multiplex Cell Authentication by Multiplexion (Heidelberg, Germany), as described recently.16 The single nucleotide polymorphism profiles matched known profiles.
Experiments involving human patient material were performed in accordance with the Declaration of Helsinki and were approved by the ethics committee of the University Cancer Center Frankfurt, project number SNO_01_13.
Fluorescence Activated Cell Sorting and Flow Cytometry
In line with PDPN being a substrate for the cysteine protease calpain-1,17 we found PDPN to be cleaved upon papain treatment, a common and in our hands the most efficient method of primary glioblastoma cell isolation. Using established primary in vitro cultures we detected full PDPN reconstitution on the cell surface 2 days after papain treatment. Thus, isolated primary cells were cultivated for 2 days before flow cytometry. For this purpose, single cell suspensions were stained for human PDPN (Biolegend #337008) and/or human leukocyte antigen (HLA) class I (Biolegend #311413) for 20 min at 4°C. Flow cytometry was performed using a Becton Dickinson (BD) FACSCalibur. Tumor cells were sorted for either low or high PDPN expression (PDPNlow and PDPNhigh), ablation of PDPN (PDPNKO), or HLA expression (HLA+) by a BD FACSAria I or BD FACSAria Fusion Cell Sorter device at the German Cancer Research Center (DKFZ) Flow Cytometry Service Unit. In order to obtain sufficient material for intracranial injections of PDPNhigh or PDPNlow cells, GBMF2 and GBMF3 cells were passaged once in vivo (i.c.) before fluorescence activated cell sorting (FACS).
Gene Expression Profiling
Six different human long-term tumor spheroid cultures were sorted into PDPNlow and PDPNhigh populations. RNA was extracted using the RNeasy kit (Qiagen) and sent to microarray analysis using Illumina Human-HT-12 Expression BeadChip according to the manufacturer’s instructions at the DKFZ Genomics and Proteomics Core Facility. Quality control, reverse transcription with labeling, chip hybridization, and calculation of mean averages were conducted in the core facility for each probe. R was used for quantile normalization of the raw microarray data. Differential gene expression was analyzed based on a fold change analysis. The ratios for the 6 samples were averaged and compared. Gene annotation enrichment analysis was performed using the bioinformatics resources software of DAVID (Database for Annotation, Visualization and Integrated Discovery).18 Enrichment score is reported as the minus log transformation of the geometric mean of P-values (modified Fisher’s exact test). All functional clusters with enrichment score >1.3 (corresponding to minus log of P < 0.05) were considered significant. Raw and normalized data are deposited in the Gene Expression Omnibus (GEO) database with the accession number GSE114915.
Intracranial Injections
Using a motorized stereotaxic instrument (Neurostar) 2 × 104 primary tumor cells or 1 × 105 tumor cells from established lines were injected in 2 µL phosphate buffered saline 2 mm lateral (right) and 3 mm ventral to the bregma with a speed of 0.2 µL/min. Used as recipients were severe combined immunodeficient (SCID)–beige mice 8–10 weeks old (C.B-Igh-1b/GbmsTac-Prkdcscid-Lystbg N7; Taconic). For serial xenotransplantations, tumor cells re-isolated from mice were sorted for HLA+ in order to re-inject a pure tumor cell population. With the exception of knockout studies, primary glioblastoma cells were cultivated at the most for 7 days before injection. Mice were sacrificed when exhibiting termination criteria such as loss of >20% body weight or poor general condition. Length of animal survival was measured by means of Kaplan–Meier estimate.
All animal experiments were approved by the responsible authority for animal experiments (Regierungspräsidium Karlsruhe, Germany) and performed in conformity with the German Law for Animal Protection.
Immunohistochemistry
Brains of sacrificed mice were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin. Six-micrometer histological sections were stained according to standard immunohistochemistry protocols. The antibodies used were specific for human PDPN (D2-40, Covance, #SIG-3730; 1:100); STEM121 (Cellartis Takara, #Y40410; 1:1000), which targets a to our knowledge undisclosed protein used to identify xenotransplanted human cells; laminin (Progen Biotech, #10765; 1:100); Ki67 (Abcam, #ab15580; 1:500); and Iba1 (Wako, # 019-19741), and counterstained with hematoxylin.
CRISPR/Cas9-Mediated Gene Deletion
We used lentivirus-mediated transduction for the stable transfer of the required plasmids. For virus production we transfected one 10 cm dish of HEK293T cells with 8 µg lentiCRISPRv2, 4 µg psPAX2, 2 µg pVSVg, and 42 µg polyethylenimine (Alfa Aesar). Medium was changed to Neurobasal the next day. Virus-containing medium was transferred to the target cells. In order to transduce sufficient cells of tumor GBMF3, cells were propagated in vivo (i.c.). Upon recovery from infection, recipient cells were selected for transfer plasmid integration by puromycin treatment for one week. Additionally, cells transduced with the single-guide (sg)RNA targeting PDPN were sorted for PDPN deletion by FACS.
The lentiCRISPRv2 plasmid encoded either the PDPN-targeting sgRNA (AGACTTATAGCGGTCTTCGC) or the control sgRNA against renilla luciferase (GGTATAATACACCGCGCTAC). Cloning was performed according to the depositor’s protocol. The sgRNA sequence targeting the renilla luciferase gene was provided by Kwang Lee. The lentiCRISPRv2 (Addgene #52961), psPAX2 (Addgene #12260), and pCMV-VSV-G (Addgene #8454) plasmids were gifts from Dr Feng Zhang, Dr Didier Trono, and Dr Robert Weinberg, respectively.
TUNEL Staining
Apoptosis was evaluated in tumor sections by staining by terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL). Tissue was permeabilized by 15 min treatment with 20 µg/mL proteinase K at 37°C. TUNEL labeling solution (Sigma #11767291910) and terminal transferase (New England Biolabs #M0315S) were incubated for 60 min at 37°C. Five pictures each from the tumor margin and tumor core were taken and number of apoptotic cells counted.
Ex Vivo Invasion Assay
The ex vivo invasion assay based on organotypic brain slice cultures was conducted as previously reported.19 Briefly, 350-µm-thick brain slices were prepared from 6-week-old C57Bl/6 mice using a vibratome (Leica VT1200 S) and cultured on 0.4-µm pore size filters (Millipore, #PICM03050) in 6-well plates. Added in the lower compartment was brain slice medium (Minimum Essential Medium, 25% heat-inactivated horse serum, 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid), 1 mM L-glutamine, 5 mg/mL glucose, 100 U/mL penicillin/streptomycin). Primary glioblastoma spheroids labeled with DiD (DiIC18(5); 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt) (5 µg/mL; Biotium, #60014) were manually implanted into the cortical tissue using a blunt Hamilton syringe (701N; 10 μL; 26s/51/3). Brain slices were fixed 2 days later in 4% PFA at 4°C o/n and spheroids imaged by confocal microscopy. Z-stack images were transformed to a maximum projection image by using ImageJ,20 and image quality was optimized by adjustment of brightness, contrast, and gamma. Migration was quantified by measuring the average cumulative sprout length per spheroid.
RNA Interference–Mediated Knockdown of PDPN
Viral transduction of target cells was performed as described above. MISSION pLKO.1-puro (Sigma-Aldrich) encoding a nontarget (HC002V, Sigma-Aldrich) or anti-PDPN (TRC-61926, Sigma-Aldrich) shRNA served as target vector.
Quantitative Real-Time PCR
For quantitative gene expression analysis, 40 cycles of real-time PCR was performed on the StepOnePlus real-time detection system (Applied Biosystems). Every PCR reaction was carried out in duplicates with 2.5 ng of cDNA in a final volume of 12.5 μL Power SYBR Green PCR Master Mix (Applied Biosystem). StepOne Software v2.2 was used for data analysis. Importin-8 and TATA-box binding protein were used as housekeeping genes to normalize target gene expression. Primer sequences are given in Supplementary Table 1.
Cell Viability in the Presence of Chemotherapeutic Agents
LN308 and LN319 PDPNKO and control cells were seeded in 96-well plates (5000 cells/well) one day prior to addition of the temozolomide (in a half-logarithmic concentration range from 1 mM to 30 nM) or etoposide (in a half-logarithmic concentration range from 100 µM to 3 nM) with each condition analyzed in duplicate. Dimethyl sulfoxide (1%) was used as vehicle control. Read-out for cell viability was metabolic activity (ATP content) assessed 72 hours after compound addition using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s protocol. Curve fitting and determination of half-maximal inhibitory concentration values and area under the curve were performed using the GraphPad Prism nonlinear regression tool.
Results
PDPNhigh Glioma Subpopulation Shows a Malignant Gene Signature
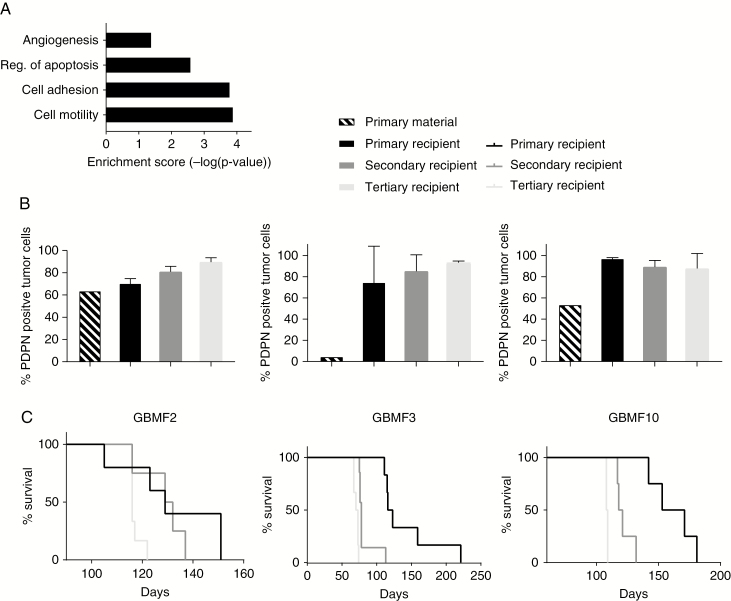
In order to assess whether PDPNhigh glioma cells are enriched in tumor-promoting properties, we sorted PDPNhigh and PDPNlow cells from 6 long-term patient-derived glioblastoma cultures. The percentage of these populations differed among individual cultures; however, most cultures exhibited higher amounts of PDPNhigh than PDPNlow populations (Supplementary Figure 1A). We then isolated RNA and performed a genome-wide expression analysis. The microarray data were validated by quantitative real-time PCR analysis of selected genes which were either up- or downregulated in PDPNhigh cells (Supplementary Figure 1B). Gene annotation enrichment analysis revealed a significantly higher expression of genes functionally associated with cell adhesion and motility, negative regulation of apoptosis, and angiogenesis in PDPNhigh glioma cells (Fig. 1A). As these gene ontologies are associated with tumor development and progression, we concluded that high PDPN expression is part of the malignant gene signature in glioma.
Fig. 1.
PDPN expression is associated with malignancy. (A) Microarray data show overrepresentation of genes associated with given biological functions in the PDPNhigh glioma subpopulations of 6 human glioblastoma cultures. Enrichment score corresponds to the minus log transformation of the geometric mean of P-values (modified Fisher’s exact test). (B) Decreased survival of the recipients (n = 6 each) with successive transplantation rounds. (C) Flow cytometry shows gradual increase in PDPN levels during serial transplantations of GBMF2 (n = 2, n = 4, n = 4) and GBMF3 (n = 3, n = 5, n = 3), and strong increase with subsequent maintenance of high PDPN levels in GBMF10 (n = 3, n = 4, n = 3).
Shortened Survival Is Paralleled by Increased PDPN Expression in Serial Xenotransplantations
We have previously reported that PDPN expression is correlated with high grade and shorter overall survival in human glioma patients.10 Glioblastoma tumors are characterized by high intratumoral heterogeneity.21 During the course of the disease the diverse population of cancer cells is subject to selective pressure of limited supply of nutrients, oxygen, or therapeutic means. Selection results in competition among subclones and prevalence of the most aggressive ones. Serial transplantations of tumor material resulting in increased tumor growth rate and enhanced invasion are frequently used to model this evolution of aggressive tumor cells.22,23 Thus, this experimental approach was used conducting 3 serial transplantations of the 3 human primary glioblastoma tumors GBMF2, GBMF3, and GBMF10 into immunocompromised mice. Indeed, with every stage of transplantation we observed a reduced survival of the recipients, indicative for increased aggressiveness of the tumor (Figure 1B). To gain insight into the expression pattern of PDPN during the disease progression, its expression level was monitored by flow cytometry in the acute primary patient material before in vivo transplantation and after every isolation step. To prevent the contamination of re-isolated tumor material with murine cells, we performed FACS for HLA class I positive cells before re-injections. In both GBMF2 and GBMF3 glioblastoma tumors we observed a steady increase in the percentage of PDPN-expressing cells with every stage of transplantation, with GBMF2 and GBMF3 reaching almost 90% PDPN-positive tumor cells after the third in vivo transplantation (Fig. 1C). The GBMF10 tumor showed PDPN expression in close to 100% of all tumor cells from the first transplantation round and maintained this percentage in the subsequent 2 rounds (Fig. 1B). In summary, we observed in all analyzed tumors a strong increase in PDPN expression and/or maintenance of high PDPN levels after their transplantations. This may be explicable by an adaptation reaction of the tumor cells in response to their changed microenvironment. On the other hand, the elevated PDPN levels may be associated with the observed increased tumor aggressiveness (Fig. 1B).
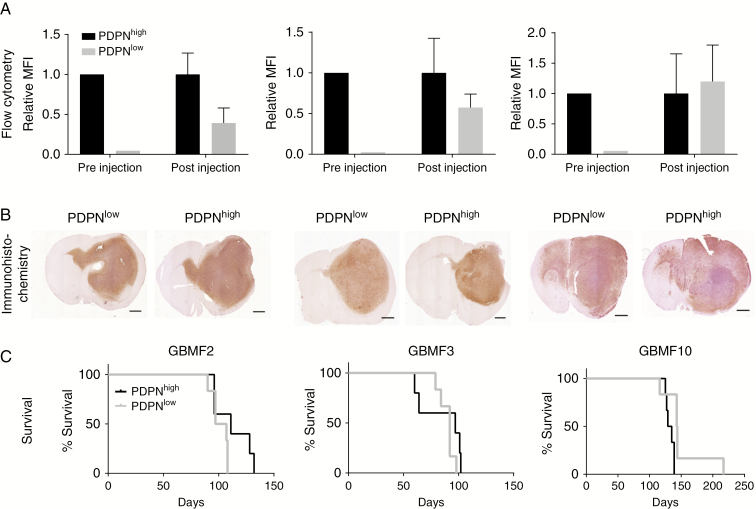
PDPNlow Sorted Glioma Cells Regain PDPN Expression In Vivo
In order to test whether the increased number of PDPN-positive cells is based on a higher proliferative capacity compared with PDPN-negative or PDPNlow cells or whether the latter cells gain PDPN expression, we sorted PDPNhigh and PDPNlow tumor cells from 3 human primary glioblastoma tumor cell isolates (GBMF2, GBMF3, and GBMF10) (Fig. 2A pre-injection and Supplementary Figure 1C). PDPNhigh and PDPNlow cells were intracranially injected into 6 immunodeficient mice each. When animals had to be sacrificed, brains were either fixed and embedded in paraffin for histological examination or dissociated for flow cytometry analysis. Although the FACS of PDPNhigh and PDPNlow cells prior to intracranial transplantation was efficient (Fig. 2A pre-injection and Supplementary Figure 1C), histological sections and flow cytometry of re-isolated tumors revealed a strong increase in PDPN expression of initially PDPNlow cells (Fig. 2A, B and Supplementary Figure 1D). This assimilation of PDPN levels probably accounts for the similar survival times of both groups (Fig. 2C). The upregulation of PDPN expression in all tumors that developed from PDPNlow cells argues for a positive selection of high PDPN expression during tumor outgrowth, either due to adaptation to a new environment in response to xenotransplantation or due to the proposed malignant function of PDPN.
Fig. 2.
PDPNlow sorted glioma cells regain high PDPN expression in vivo. (A) PDPN levels indicated by mean fluorescence intensity (MFI) before orthotopic injection (n = 1 per primary tumor) and 2 days after isolation of GBMF2 (n = 2 recipients), GBMF3 (n = 3), and GBMF10 (n = 3) tumor cells. MFI was normalized to PDPNhigh values. (B) Immunohistochemical staining for PDPN, scale bar 1 mm. (C) Survival of mice injected with PDPNlow (n = 6) and PDPNhigh (n = 6) sorted glioma cells.
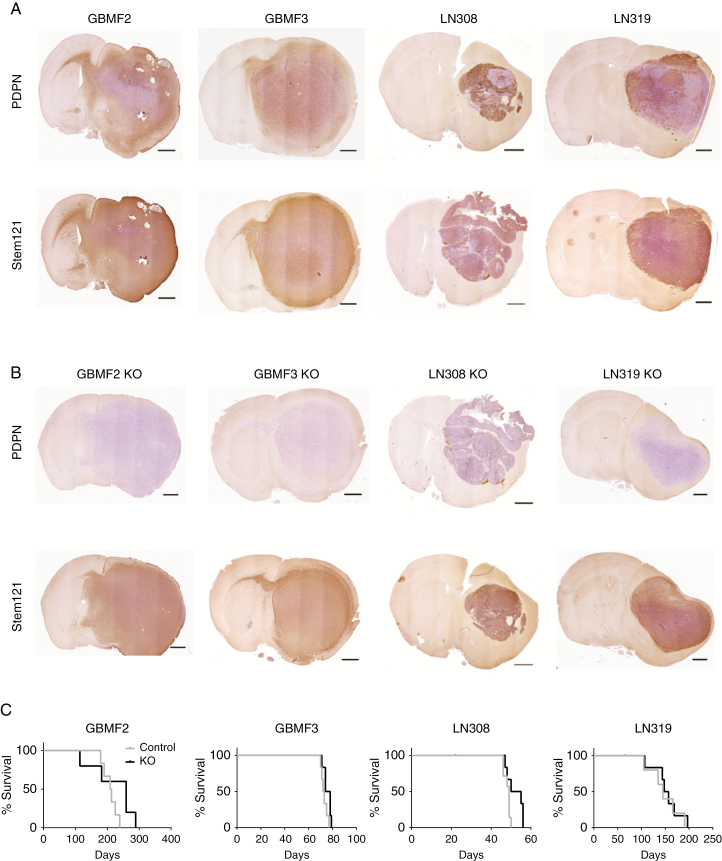
CRISPR/Cas9-Mediated Knockout of PDPN in Glioma Cells Does Not Affect Tumor Growth In Vivo
In order to clarify whether dynamic, high PDPN levels are causative for glioma development and progression or whether its high expression is a concomitant phenomenon, we proceeded with a loss-of-function approach. First, primary glioblastoma cultures were transduced with shRNA constructs targeting PDPN. However, we observed a gradual escape of the PDPN knockdown (Supplementary Figure 2A), which prevented further investigations. Thus, we deleted PDPN using the CRISPR/Cas9 system in 2 primary human glioblastoma cultures (GBMF2, GBMF3) as well as in 2 human glioma cell lines (LN308, LN319). An sgRNA targeting renilla luciferase was used as a control. The PDPNKO population was purified by FACS, avoiding the generation of single cell clones and related clonal artifacts and to furthermore retain the heterogeneity of the primary glioblastoma culture. PDPN deletion of the sorted bulk was validated by western blot (Supplementary Figure 2B). Control and PDPNKO cells were orthotopically injected into immunodeficient SCID-beige mice. Immunohistochemistry confirmed the deletion of PDPN in PDPNKO tumors and showed intermediate to strong PDPN expression in control tumors (Figure 3A, B). Of note, we also observed areas devoid of PDPN in LN308 and LN319 control tumors. The deletion of PDPN in PDPNKO tumors altered neither histology nor tumor extension or invasion to the contralateral hemisphere, as indicated by STEM121 staining, a marker for human and, thus in this case, tumor cells. Importantly, we did not record an altered survival of the recipients (Fig. 3C). In contrast to previously obtained data, this result demonstrates that PDPN is dispensable for glioma growth in vivo.
Fig. 3.
PDPN deletion does not affect tumor growth in vivo. Immunohistochemical staining of (A) control and (B) PDPNKO tumors for PDPN and STEM121; scale bar 1 mm. (C) Survival of mice injected with PDPNKO (n = 6) or control cells (n = 6). The survival time was not significantly altered between the 2 groups; p(GBMF2) = 0.16; p(GBMF3) = 0.07; p(LN308) = 0.06; p(LN319) = 0.59; log-rank test.
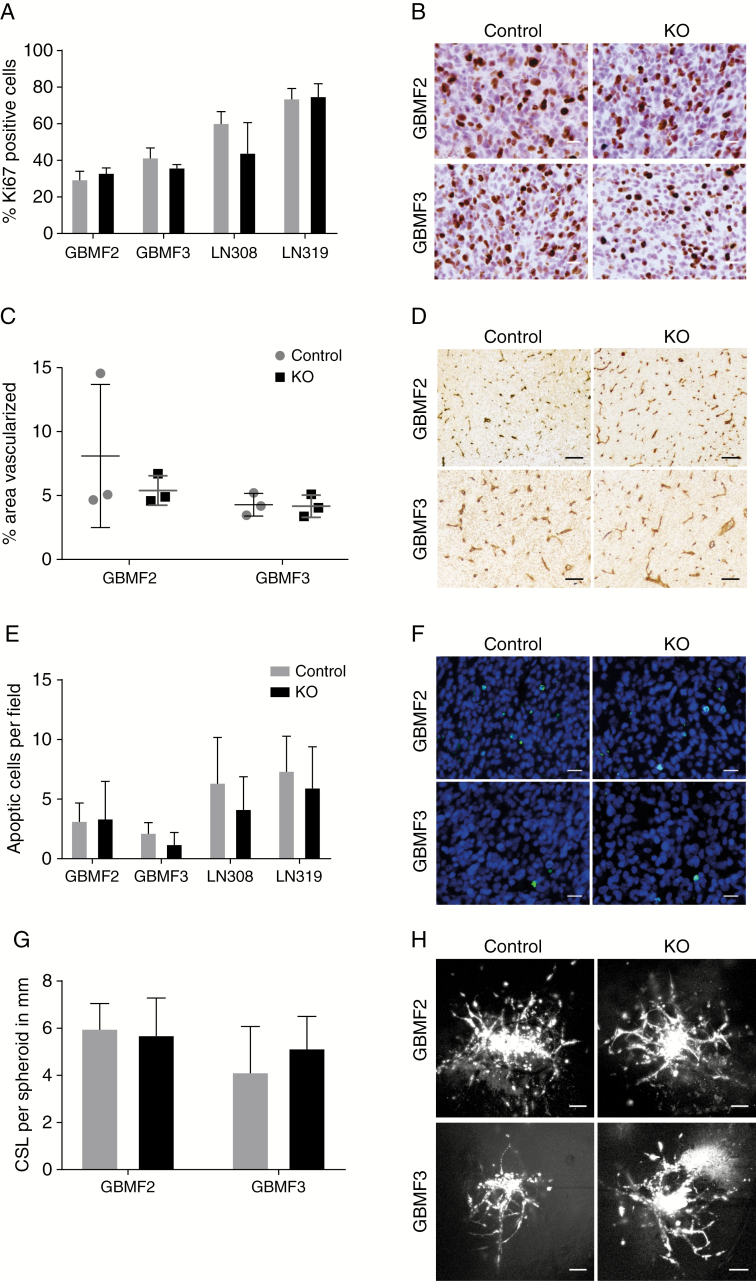
Deletion of PDPN Affects Neither Tumor Cell Proliferation nor Apoptotic Events, Tumor Vascularization, or Invasion
In order to determine whether tumor cell proliferation is altered by the absence of PDPN, we performed a Ki67 staining but did not observe any difference between PDPNKO and control tumors (Fig. 4A, B), consistent with the identical in vitro doubling times of PDPNKO and control cells (Supplementary Figure 2C). We next investigated whether PDPN affects in vitro cell proliferation or viability in LN308 and LN319 PDPNKO and control cells under stress conditions such as chemotherapy. However, we did not observe a difference in the response between the 2 groups to temozolomide or to etoposide (Supplementary Figure 3). Thus, we conclude that PDPN does not affect the therapeutic response to chemotherapy in vitro.
Fig. 4.
The ablation of PDPN has no effect on malignant features. (A) The deletion of PDPN did not influence tumor cell proliferation in vivo as assessed by Ki67 immunohistochemistry (B), 5 fields (0.33 mm2/field) per group analyzed, p(GBMF2) = 0.15; p(GBMF3) = 0.05; p(LN308) = 0.07; p(LN319) = 0.80; Student’s t-test. (C) Tumor area covered by blood vessels (immunohistochemical staining for laminin, D) is not altered between control and PDPNKO groups, 4–5 fields (3.55 mm2/field) of 3 tumors each were analyzed, p(GBMF2) = 0.46; p(GBMF3) = 0.88; Student’s t-test. (E) Quantification of apoptotic cells per field showed no difference between control and knockout tumors, 5 fields (0.33 mm2/field) per group analyzed, p(GBMF2) = 0.90; p(GBMF3) = 0.13; p(LN308) = 0.32; p(LN319) = 0.50; Student’s t-test. (F) Representative pictures of TUNEL staining; apoptotic cells with fragmented DNA are indicated in green. (G) Invasion of PDPNKO and control cells in an ex vivo invasion assay. Quantification of cumulative sprout length (CSL) per spheroid, n(GBMF2) = 12; n(GBMF2KO) = 16; n(GBMF3) = 19; n(GBMF3KO) = 15; p(GBMF2) = 0.60; p(GBMF3) = 0.08; Student’s t-test. (H) Representative pictures of glioblastoma cell invasion. Scale bars (B, F) 20 μm; (D, H) 100 μm.
Since high PDPN expression is associated with a malignant gene signature, including genes involved in angiogenesis, negative regulation of apoptosis, and cell motility (Fig. 1A), we examined the effect of PDPN deletion on these features. Tumor vascularization assessed as coverage of PDPNKO and control tumors by blood vessels indicated by laminin staining was identical in number between the 2 groups and did not reveal any obvious structural alterations induced by the deletion of PDPN (Fig. 4C, D). Finally, TUNEL staining revealed that the rate of cell death within the tumor margin (data not shown) and tumor core was generally low, and we could not record an increase in apoptosis in the PDPNKO group (Fig. 4E, F).
As our microarray data associated high PDPN expression with genes that are involved in cell motility and migration, which is in line with previous reports describing a role of PDPN in cell migration in vitro,4,11,12 we performed an ex vivo invasion assay.19 Fluorescently labeled spheroids generated from control and PDPN-deleted primary glioblastoma cultures GBMF2 and GBMF3 were implanted into ex vivo cultured murine brain slices, and tumor cell invasion was recorded by confocal microscopy. Both PDPNKO and control cells showed a similar grade of invasion quantified by the cumulative sprout length per spheroid (Fig. 4G, H). This finding is in line with the macroscopic analysis of histological sections of PDPNKO and control tumors, where we did not observe an obvious difference in tumor cell invasion. Thus, we concluded that PDPN is dispensable for regulation of apoptosis, tumor vascularization, and tumor cell invasion in human glioblastoma.
Taken together, these functional analyses both in an in vivo setting of glioblastoma pathology and in organotypic brain slice cultures contradict previous findings of in vitro studies which had suggested a prominent role of PDPN in tumor progression.
Discussion
The identification of novel therapeutic targets to improve current treatment measures remains a major focus in glioblastoma research. Previously, we and others found PDPN overexpression in high-grade gliomas and could correlate PDPN upregulation with malignant progression and poor prognosis in glioma patients.2,10,11 In vitro studies have further supported the idea of exploiting PDPN as a therapeutic target, since the RNA interference (RNAi)–mediated knockdown10,11 or overexpression of PDPN4,12,13 in cancer cell lines of glioma and other entities indicated a function for PDPN in proliferation and migration. However, in vivo studies required to validate these in vitro findings and unequivocally establish a rate-limiting function of PDPN in glioblastoma pathology were still lacking. Thus, we investigated the functional role of PDPN in human glioblastoma using primary patient material and established cell lines in mouse xenograft models and 3D-organotypic brain slice cultures.
A microarray analysis of PDPNhigh and PDPNlow sorted human glioblastoma cells revealed a malignant gene signature of PDPNhigh cells. This, together with the fact that PDPN expression was increased in serial transplantations paralleled by a shortened survival and the strong regain of PDPN expression in tumors developed from PDPNlow sorted glioma cells strengthened the assumption of a tumor-promoting effect of PDPN. However, employing intracranial injections of PDPN-deleted primary human glioblastoma cells and established cell lines, contrary to expectations, we did not observe an effect of PDPN ablation on tumor development. No significant difference was recorded in the survival of the mice, and histology of the tumors did not differ among the 2 groups. Although gene expression profiling indicated a coexpression of PDPN and genes involved in angiogenesis, regulation of apoptosis, and cell migration, we could not find a significant difference between PDPNKO and control tumors in any of these features or in the in vitro response to 2 chemotherapeutics.
Thus, in light of these findings we now conclude that the malignant gene signature of PDPNhigh glioma cells is most likely not directly caused by PDPN expression but rather points to malignant signaling pathways active in glioblastoma cells that induce PDPN transcription. Of note, PDPN expression is induced by multiple upstream factors that have been associated with malignancy. We have previously reported that PDPN expression in glioma cells is negatively regulated by phosphatase and tensin homolog and that the loss of this tumor suppressor and concomitant activation of the phosphatidylinositol-3 kinase‒Akt‒activator protein 1 signaling pathway results in an increased expression of PDPN.11 In addition, other oncogenic factors like transforming growth factor β and signal transducer and activator of transcription 3 activate PDPN expression.24 Thus, the malignant nature of PDPNhigh cells might be based on multiple components of the PDPN-associated gene expression profile rather than on the presence of PDPN itself—which would in this case mark but not necessarily cause malignancy. Why glioma cells do not cease but rather maintain high PDPN expression remains elusive. However, we have observed focal loss of PDPN in LN308 and LN319 control tumors (Fig. 3A), which might indicate that PDPN expression can be silenced, probably in response to selective pressure by shortage of nutrient resources or by mechanisms affecting the differentiation status of a given cell, which has also been reported to affect PDPN expression in other entities.3
In the present study, we have applied advanced cellular models including primary glioblastoma spheroid cultures, which were found to better retain the genomic and transcriptional profile of the original tumor25,26 and to display hallmarks of primary glioblastoma tumors, especially strong infiltrative growth.26 The use of these cultures might cause the discrepancies to previous in vitro studies that attributed the protein with a pro-migratory and proliferative function and that predominantly used established glioblastoma cell lines.
In general, cell-based in vitro models are very limited in their power to accurately predict the function of proteins in vivo, due to the lack of diverse multicellular interactions and environmental influences. Thus, we employed an ex vivo invasion assay, which more faithfully recapitulates the in vivo situation19 than previously applied standard methods like in vitro scratch assays or collagen gels.
However, one disadvantage of our applied in vivo model is the usage of an immunodeficient mouse strain. We used SCID-beige animals carrying profound defects in natural killer cells and the adaptive immune system27 to enable the transplantation of primary human glioblastoma cells. Although we did not observe an obvious difference between PDPNKO and control tumors in the infiltration of macrophages/microglia (based on an Iba1 staining; Supplementary Figure 2D), we cannot exclude the possibility that PDPN influences a potential interaction of tumor cells with other immune cells, which could result in different outcomes of PDPNKO and control tumors in an immunocompetent background.
In contrast to previous publications that applied RNAi, our study used the CRISPR/Cas9-based genome editing tool to functionally investigate PDPN. The key difference between the 2 techniques rests on a true loss-of-function using CRISPR/Cas9, whereas RNAi generally causes reduced protein levels. Thus, the complete ablation of PDPN by the CRISPR/Cas9-induced knockout may provoke compensatory reactions by the cell, which might not occur in cells with residual PDPN protein levels and thus result in different phenotypes. However, we did not investigate a possible compensation of PDPN, such as by other ezrin/radixin/moesin–binding proteins. Alternatively, we intended to investigate the effect of an shRNA-mediated knockdown in primary glioblastoma cells; however, only a transient knockdown was achieved, which was too short-lived for subsequent functional assays.
Our finding that deletion of PDPN does not impair glioma progression is of great importance for further preclinical studies. Previous publications that indicated a tumor-promoting role for PDPN proposed the inhibition of the protein as a therapeutic approach. However, our study suggests that the development and usage of compounds that functionally inactivate PDPN would not result in the desired tumor-suppressing effect. Instead, we suggest PDPN as a potential prognostic marker in the clinics, as we found PDPN to be part of a malignant gene signature in glioblastoma marking highly aggressive tumors with poor prognosis. Furthermore, instead of the development of PDPN-blocking therapeutic agents targeting either the extra- or intracellular domain of PDPN reviewed in 28, we propose using the surface protein as a physical target in glioblastoma therapy. Recently, cancer-specific monoclonal antibodies have been reported to detect aberrant post-translational modifications of PDPN specific for cancer cells.29,30 Hence, the membrane-bound protein could be exploited as a target to mediate apoptosis via cytotoxic anti-PDPN antibodies31 or by the targeted delivery of cytotoxic or immunogenic compounds into highly malignant glioma cells without affecting healthy PDPN positive cells. Alternatively, tumor-specific PDPN could be used as a target in CAR T-cell therapy.32 The ablation of PDPN-positive tumor cells would furthermore decrease the risk of venous thromboembolism (VTE), as high PDPN expression in primary brain tumors has recently been correlated with hypercoagulability and increased risk of VTE.33 Whether this approach will result in a lasting efficacy against glioma or provoke compensating mechanisms to escape cytotoxic therapy remains to be determined.
Funding
This work was supported by the Helmholtz Association (Helmholtz Alliance Preclinical Comprehensive Cancer to PA) and the Luxembourg National Research Fond (FNR PEARL P16/BM/11192868 grant to MM).
Supplementary Material
Acknowledgments
We kindly thank Annette Kopp-Schneider for the microarray analysis, Angelika Krischke and Sabrina Lohr for technical support, Susanne Kleber and Ana Martín-Villalba for access to stereotactic devices and primary material, the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Whole-Genome Expression Beadchips and related services, and the DKFZ flow cytometry service unit.
Conflict of interest statement. None declared.
Authorship statement. TE, PA, and HP designed the study; TE performed experiments; TE, BC, WW, MM, PA, and HP analyzed and interpreted the data; PNH and MM provided primary material and advice on isolation and cultivation; TE, PA, and HP wrote the manuscript; TE, BC, PNH, WW, MM, PA, and HP were involved in revising the manuscript; all have read and approved the final version.
Present address: Translational Program, Hopp Children’s Cancer Center at NCT Heidelberg, University Hospital and DKFZ Heidelberg, Heidelberg, Germany
References
- 1. Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. [DOI] [PubMed] [Google Scholar]
- 2. Mishima K, Kato Y, Kaneko MK, Nishikawa R, Hirose T, Matsutani M. Increased expression of podoplanin in malignant astrocytic tumors as a novel molecular marker of malignant progression. Acta Neuropathol. 2006;111(5):483–488. [DOI] [PubMed] [Google Scholar]
- 3. Schacht V, Dadras SS, Johnson LA, Jackson DG, Hong YK, Detmar M. Up-regulation of the lymphatic marker podoplanin, a mucin-type transmembrane glycoprotein, in human squamous cell carcinomas and germ cell tumors. Am J Pathol. 2005;166(3):913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9(4):261–272. [DOI] [PubMed] [Google Scholar]
- 5. Ugorski M, Dziegiel P, Suchanski J. Podoplanin—a small glycoprotein with many faces. Am J Cancer Res. 2016;6(2):370–386. [PMC free article] [PubMed] [Google Scholar]
- 6. Lowe KL, Finney BA, Deppermann C, et al. Podoplanin and CLEC-2 drive cerebrovascular patterning and integrity during development. Blood. 2015;125(24):3769–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kotani M, Osanai T, Tajima Y, et al. Identification of neuronal cell lineage-specific molecules in the neuronal differentiation of P19 EC cells and mouse central nervous system. J Neurosci Res. 2002;67(5):595–606. [DOI] [PubMed] [Google Scholar]
- 8. Tomooka M, Kaji C, Kojima H, Sawa Y. Distribution of podoplanin-expressing cells in the mouse nervous systems. Acta Histochem Cytochem. 2013;46(6):171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolar K, Freitas-Andrade M, Bechberger JF, et al. Podoplanin: a marker for reactive gliosis in gliomas and brain injury. J Neuropathol Exp Neurol. 2015;74(1):64–74. [DOI] [PubMed] [Google Scholar]
- 10. Ernst A, Hofmann S, Ahmadi R, et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res. 2009;15(21):6541–6550. [DOI] [PubMed] [Google Scholar]
- 11. Peterziel H, Müller J, Danner A, et al. Expression of podoplanin in human astrocytic brain tumors is controlled by the PI3K-AKT-AP-1 signaling pathway and promoter methylation. Neuro Oncol. 2012;14(4):426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martín-Villar E, Megías D, Castel S, Yurrita MM, Vilaró S, Quintanilla M. Podoplanin binds ERM proteins to activate RhoA and promote epithelial-mesenchymal transition. J Cell Sci. 2006;119(Pt 21):4541–4553. [DOI] [PubMed] [Google Scholar]
- 13. Grau SJ, Trillsch F, Tonn JC, et al. Podoplanin increases migration and angiogenesis in malignant glioma. Int J Clin Exp Pathol. 2015;8(7):8663–8670. [PMC free article] [PubMed] [Google Scholar]
- 14. Kunita A, Kashima TG, Morishita Y, et al. The platelet aggregation-inducing factor aggrus/podoplanin promotes pulmonary metastasis. Am J Pathol. 2007;170(4):1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunita A, Baeriswyl V, Meda C, et al. Inflammatory cytokines induce podoplanin expression at the tumor invasive front. Am J Pathol. 2018;188(5):1276–1288. [DOI] [PubMed] [Google Scholar]
- 16. Castro F, Dirks WG, Fähnrich S, Hotz-Wagenblatt A, Pawlita M, Schmitt M. High-throughput SNP-based authentication of human cell lines. Int J Cancer. 2013;132(2):308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martín-Villar E, Yurrita MM, Fernández-Muñoz B, Quintanilla M, Renart J. Regulation of podoplanin/PA2.26 antigen expression in tumour cells. Involvement of calpain-mediated proteolysis. Int J Biochem Cell Biol. 2009;41(6):1421–1429. [DOI] [PubMed] [Google Scholar]
- 18. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 19. Eisemann T, Costa B, Strelau J, Mittelbronn M, Angel P, Peterziel H. An advanced glioma cell invasion assay based on organotypic brain slice cultures. BMC Cancer. 2018;18(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. [DOI] [PubMed] [Google Scholar]
- 23. Yano S, Takehara K, Kishimoto H, et al. In vivo selection of intermediately- and highly-malignant variants of triple-negative breast cancer in orthotopic nude mouse models. Anticancer Res. 2016;36(12):6273–6277. [DOI] [PubMed] [Google Scholar]
- 24. Honma M, Minami-Hori M, Takahashi H, Iizuka H. Podoplanin expression in wound and hyperproliferative psoriatic epidermis: regulation by TGF-β and STAT-3 activating cytokines, IFN-γ, IL-6, and IL-22. J Dermatol Sci. 2012;65(2):134–140. [DOI] [PubMed] [Google Scholar]
- 25. Hamer PCDW, Van Tilborg AAG, Eijk PP, et al. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene. 2008;27(14):2091–2096. [DOI] [PubMed] [Google Scholar]
- 26. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 27. Shibata S, Asano T, Ogura A, et al. SCID-bg mice as xenograft recipients. Lab Anim. 1997;31(2):163–168. [DOI] [PubMed] [Google Scholar]
- 28. Krishnan H, Rayes J, Miyashita T, et al. Podoplanin: an emerging cancer biomarker and therapeutic target. Cancer Sci. 2018;109(5):1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato Y, Kaneko MK. A cancer-specific monoclonal antibody recognizes the aberrantly glycosylated podoplanin. Sci Rep. 2014;4:5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamada S, Ogasawara S, Kaneko MK, Kato Y. LpMab-23: a cancer-specific monoclonal antibody against human podoplanin. Monoclon Antib Immunodiagn Immunother. 2017;36(2):72–76. [DOI] [PubMed] [Google Scholar]
- 31. Kaneko MK, Yamada S, Nakamura T, et al. Antitumor activity of chLpMab-2, a human-mouse chimeric cancer-specific antihuman podoplanin antibody, via antibody-dependent cellular cytotoxicity. Cancer Med. 2017;6(4):768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shiina S, Ohno M, Ohka F, et al. CAR T cells targeting podoplanin reduce orthotopic glioblastomas in mouse brains. Cancer Immunol Res. 2016;4(3):259–268. [DOI] [PubMed] [Google Scholar]
- 33. Riedl J, Preusser M, Nazari PM, et al. Podoplanin expression in primary brain tumors induces platelet aggregation and increases risk of venous thromboembolism. Blood. 2017;129(13):1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.