ABSTRACT
Ciliates represent a morphologically and genetically distinct group of single-celled eukaryotes that segregate germline and somatic functions into two types of nuclei and exhibit complex cytogenetic events during the sexual process of conjugation, which is under the control of the so-called “mating type systems”. Studying conjugation in ciliates may provide insight into our understanding of the origins and evolution of sex and fertilization. In the present work, we studied in detail the sexual process of conjugation using the model species Euplotes vannus, and compared these nuclear events with those occurring in other ciliates. Our results indicate that in E. vannus: 1) conjugation requires about 75 hours to complete: the longest step is the development of the new macronucleus (ca. 64h), followed by the nuclear division of meiosis I (5h); the mitotic divisions usually take only 2h; 2) there are three prezygotic divisions (mitosis and meiosis I and II), and two of the eight resulting nuclei become pronuclei; 3) after the exchange and fusion of the pronuclei, two postzygotic divisions occur; two of the four products differentiate into the new micronucleus and macronucleus, respectively, and the parental macronucleus degenerates completely; 4) comparison of the nuclear events during conjugation in different ciliates reveals that there are generally three prezygotic divisions while the number of postzygotic divisions is highly variable. These results can serve as reference to investigate the mating type system operating in this species and to analyze genes involved in the different steps of the sexual process.
KEYWORDS: Ciliate, Euplotes vannus, conjugation, mating type, life cycle
Introduction
Ciliates represent a monophyletic group of eukaryotic microorganisms characterized by the presence of two types of nuclei within the same cytoplasm: a small diploid germline micronucleus (MIC) which is transcriptionally silent in cell’s vegetative life, and a large polyploid somatic macronucleus (MAC) which is transcriptionally active and regulates the cell phenotype [1–6]. In cell reproduction, MIC undergoes mitosis while MAC divides by “amitosis”, a division which does not involve spindle formation and chromosome condensation but is simply a split of the DNA content into two equal halves [7–12].
The sexual process of ciliates is typically represented by conjugation, which involves a temporary union of two mating partners, although in some groups such as the peritrichs and chonotrichs the mating partners merge permanently [13,14]. Once mating partners are united in pairs, MIC undergoes meiosis to generate two gametic pronuclei, one resident and one migratory. The migratory pronuclei are exchanged between the two mating partners through a cytoplasmic bridge, and after mutual fertilization between the resident and migratory gametic pronuclei, the two cells separate. The new nuclear apparatus eventually develops in each exconjugant cell from the mitotic products of the synkaryon, while the old MAC fragments and degrades [15]. The genome of the MIC remains organized in large chromosomes, while the genome of the new developing MAC is subjected to extensive rearrangements which includes chromosome fragmentation, DNA elimination and gene amplification to generate unconventional linear nano-chromosomes [7,16–20].
Mating is genetically controlled through a mechanism known as the “mating type system”. Some species have only two mating types, such as Paramecium aurelia [21]: mating type E depends on expression of the transmembrane protein mtA, while type O is determined during MAC development by scnRNA-dependent excision of the mtA promoter [22]. Others have more than two types. Tetrahymena thermophila has seven mating types, each one determined by a specific mating type gene pair that is stochastically assembled in the MAC during somatic differentiation by homologous recombination of incomplete mating type gene pairs that are linearly arranged in the MIC [23]. Euplotes Ehrenberg, 1830, one of the most highly diverse and cosmopolitan ciliate genera, has evolved high-multiple mating type systems [4,24–29]. In E. patella, E. octocarinatus, E. raikovi, and E. crassus, each mating type is determined by the allele combination at the “mating type locus” of the germinal MIC [30–32]. These alleles are converted into gene-size DNA molecules in the MAC, where they are co-dominantly expressed to control the synthesis of type-specific chemical markers usually referred as pheromones [13,32–35].
Euplotes vannus is a cosmopolitan marine ciliate that has been used as model organism in a wide range of disciplines [27,36,37]. It has more than ten mating types; however, the molecular mechanism of its mating type determination is still largely unknown. Detailed knowledge of the different steps taking place during conjugation in this species thus appears to be crucial for analyzing genes controlling the sexual process. In this paper, we describe the time-course analysis of the nuclear events occurring during and after conjugation in E. vannus strains collected along the Yellow Sea coast of Qingdao, China, and compare them with those of other species.
Results
The conjugation process step-by-step
Conjugation was induced by mixing cells of different mating types (Figure 1(e–g)), and the initial formation of mating pairs was taken as time 0 of the process (Figure 1(l)). A detailed description of the different steps of nuclear events is as follows.
Figure 1.
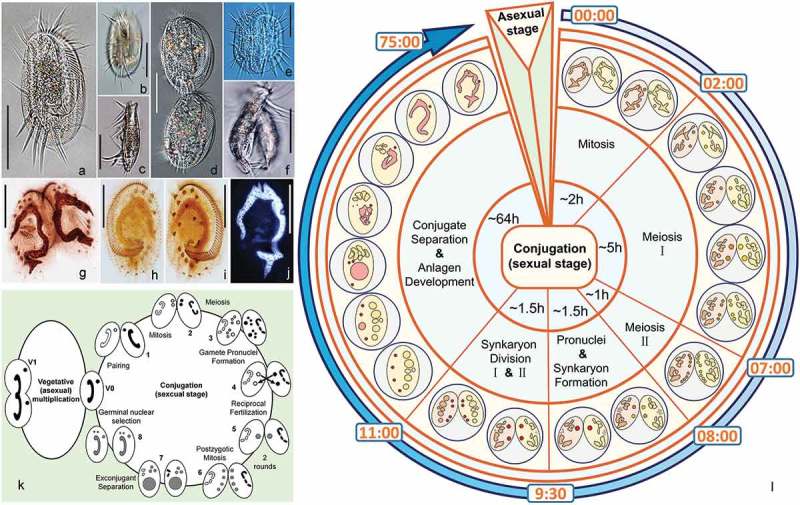
Morphology and life cycle of Euplotes vannus. (a–b) Ventral side of a representative vegetative cell; (c) Lateral view of a vegetative cell; (d) Ventral side of the divider at late divisional stage; (e–f) Ventral and lateral views of a conjugating pair. (g) Ventral view of a conjugating pair to show the infraciliature after protargol staining; (h–i) Dorsal and ventral views of the same specimen at vegetative stage to show the infraciliature; (j) A vegetative cell after Hoechst 33342 staining to show the micronucleus and macronucleus; (k) Life cycle of E. vannus. Left: vegetative (asexual) reproduction phase. V0: vegetative cell. V1: cell undergoing binary fission. Micronucleus and macronucleus divide mitotically and amitotically, respectively. Right: conjugation, the sexual stage of the life cycle. (l) Timing of nuclear events during conjugation. The initial formation of mating pairs was taken as time 0 of the process. Scale bars = 50 μm.
Step 1: mitosis of the MIC (first prezygotic division).
Soon after cell-cell union, MIC migrates out of the concavity of the “C”-shaped macronucleus and the concaved surface of MAC flattens (Figure 2(a)). At this point, MIC divides by mitosis, forming two nuclear products (Figure 2(b)). This step lasts about 2 h (Figure 1(l)).
Figure 2.
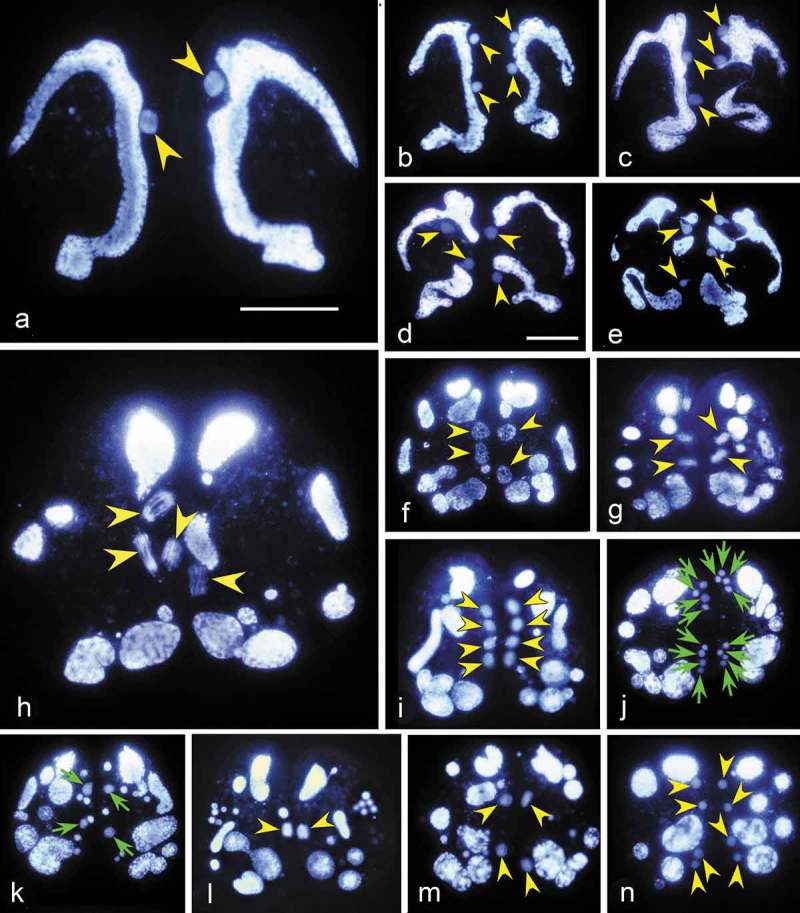
Hoechst 33342 stained conjugating pairs of Euplotes vannus to show the nuclear events before the pair separation. Yellow arrowhead: nucleus that has the same ploidy with the micronucleus. Green arrow: gamete nucleus. (a) Micronucleus migrates out of the concavity of macronucleus after the pair formation; (b) A conjugating pair after mitosis of the MIC; (c-d) Mitosis products become inflated. Meanwhile, the macronucleus begins to degenerate; (e) Zygotene stage of the first meiosis showing that chromatin polymerizes into a typical “bouquet” shape; (f–i) Various stages of the first meiosis division; (j) The second meiotic division resulting in eight pronuclei; (k) Gamete nuclei (indicated by green arrows) inflate while other division products degenerate; (l) Synkaryon formation after migration and fusion of pronuclei; (m) The first synkaryon division; (n) The second synkaryon division. Scale bars = 30 μm.
Step 2: meiosis (second and third prezygotic divisions)
The two products of the first MIC division undergo a classical two-step meiosis. The first meiotic division (Figure 2(c–i)) (second prezygotic division) takes about 5h (Figure 1(l)), during which chromatin polymerizes assuming the “bouquet-like” shape, a typical zygotene stage when chromosomes approximately line up with each other into homologous chromosome pairs [38,39] (Figure 2(e–g)). The four products enter the second meiotic division (third prezygotic division) which lasts about 1 hour (Figure 1(l)), with the final formation of eight pronuclei (Figure 2(j)).
Step 3: pronuclei and synkaryon formation
At the end of meiosis, four pronuclei gather together in the anterior part and four in the posterior part of the cell. For each anterior and posterior group of the pronuclei, one out of four swells while the other three become pyknotic and degenerate (Figure 2(k)). Of the two swollen nuclei, one is the migratory pronucleus and the other is the stationary pronucleus. After the reciprocal exchange of the migratory pronuclei, the fusion of pronuclei generates the synkaryon in each mating cell (Figure 2(l)). The degenerating meiotic products accumulated on the edge of the cell are still visible at this stage. This step takes about 1.5 hours (Figure 1(l)).
Step 4: first and second postzygotic divisions
The synkaryon is subjected to two successive mitotic divisions generating four postzygotic nuclei (Figure 2(m,n)). After approximately 11 hours, mating cells complete the second synkaryon division and start to separate. Mating pairs usually separate completely within 1 hour after the second synkaryon division; at this point, fragments of the old MAC and the four products of the synkaryon division are visible inside each mating cell (Figure 3(a)).
Figure 3.
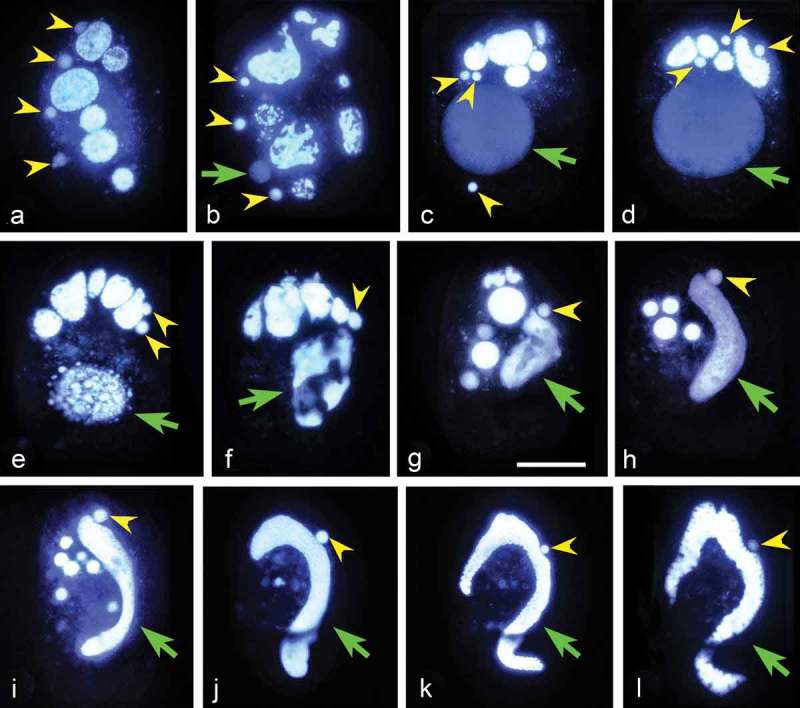
Hoechst 33342 stained exconjugants of Euplotes vannus to show the nuclear events. Yellow arrowhead: synkaryon division product or micronucleus. Green arrow: macronucleus anlagen or macronucleus. (a) There are four products of synkaryon division in each exconjugant (as indicated by the yellow arrowheads); (b–d) One out of four products (usually the third one from anterior to posterior) of the synkaryon divisions swells and differentiates into the macronucleus anlagen (indicated by the green arrow); (e–g) Complex DNA rearrangements during the development of macronucleus anlagen. (h–k) Development of macronucleus anlagen and micronucleus selection, while the old macronucleus and the residual nuclei gradually degrade; (l) Well-developed new macronucleus and micronucleus. Scale bars = 30 μm.
Step 5: new MIC and MAC development
Soon after mate separation, one out of four products (usually the third one from anterior to posterior) of the synkaryon divisions swells and differentiates into the macronuclear anlagen (Figure 3(b)). In the early stage, the macronuclear anlagen is eccentrically placed but moves to occupy the mid-region of the posterior half of the cell and enlarges rapidly, increasing in size to 40 μm within the next 24 hours, to reach about 1/3 of the cell length (Figure 3(c,d)). The time of anlagen development appears to be directly correlated with the uptake of food by the cell: in the absence of food, the anlagen will remain in the cell for more than 40 hours, whereas this time is reduced by at least a half when food is present. The parental macronuclear fragments degrade completely and the newly developing macronucleus elongates to acquire the typical “C” shape (Figure 3(e–l)).
The new MIC derives from one of the three residual products of synkaryon divisions. The remaining two eventually degenerate, gathering together with fragments of the old macronucleus in the front part of the cell (Figure 3(d-f)). On 200 exconjugant cells analyzed at the stage of maximum expansion of macronuclear anlagen, 66 cells had 3 residual nuclei, 51 cells had 2, and 83 cells had only 1. Residual nuclei could be observed in some exconjugant cells even after the new macronucleus is completely formed.
Fate of the parental macronucleus
From the beginning of conjugation to the end of the first prezygotic division, the parental MAC is “C”-shaped without obvious morphological changes. From the second prezygotic division (Figure 2(c)), the macronucleus begins to break into several oval fragments, which then cluster in the anterior part of the cell. With the development of the macronuclear anlagen, these fragments become irregular and move toward the center of the cell and they reduce in size (Figure 3(a–i)). Finally they disappear when a new macronucleus is completely developed (Figure 3(j-l)).
Discussion
Conjugation is unique to ciliates and has been extensively studied since its discovery [40]. Cell mating and restoration of the vegetative state (Figure1(k)) have been described in many different ciliates, but mostly intensively in Tetrahymena, Paramecium, Oxytricha, Chilodonella, and Euplotes, and primarily focused on nuclear events [41–43] (Figure 5). In E. vannus, we determined the time needed to complete the prezygotic and postzygotic divisions by using a fluorescent dye to visualize the nuclear division products. Our results indicate that the prezygotic nuclear division of meiosis I, which lasts about five hours, is much longer than mitosis, which requires about two hours. The three prezygotic divisions are carried out in about eight hours. After approximately 11 hours from mating pair formation, most mating partners complete the second synkaryon division and start to separate. An additional ca. 64 hours are needed to complete the development of the new nuclear apparatus in each exconjugant cell (Figure 1(l)).
Figure 5.
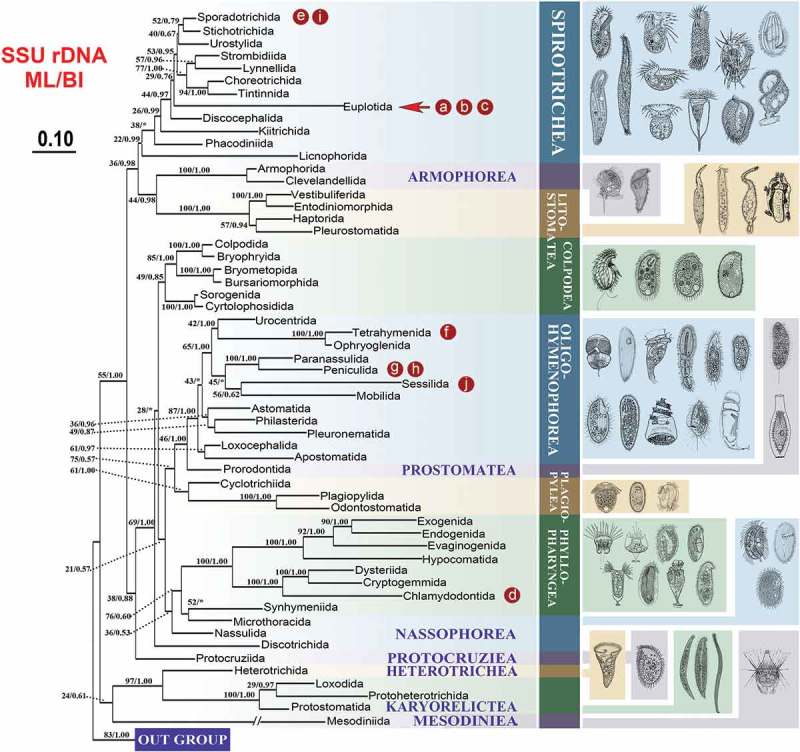
Maximum likelihood (ML) trees of the phylum Ciliophora based on the SSU rDNA to show the position of the ciliates that were compared in the present study. Numbers at nodes represent the bootstrap values of maximum likelihood (ML) out of 1000 replicates and the posterior probability of Bayesian analysis (BI). Asterisk (*) indicates disagreement between ML and BI analyzes. The scale bar corresponds to 10 substitutions per 100 nucleotide positions. The letters marked in the tree are corresponding to the taxa shown in Figure 4. (a) Euplotes vannus; (b) Euplotes woodruffi; (c) Euplotes cristatus; (d) Chilodonella uncinata; (e) Halteria grandinella; (f) Tetrahymena thermophila; (g) Paramecium aurelia; (h) Paramecium caudatum; (i) Oxytricha trifallax; (j) Carchesium polypinum. The illustrations on the right side are according to the previous study [44].
Our observations confirmed that, as previously reported for the E. crassus-vannus-minuta group [45], MIC undergoes only three prezygotic divisions (mitosis, meiosis I and meiosis II) in E. vannus, whreas in other Euplotes species, such as E. woodruffi, E. charon, E. raikovi, E. octocarinatus and E. cristatus, MIC divides four times (a preliminary mitotic division, meiotic I and II, and a second mitotic division) (Figure 4) [38,46–48]. Therefore, the stationary and migratory pronuclei in E. vannus derive directly from two of the eight meiotic products and, as such, may be genetically different.
Figure 4.
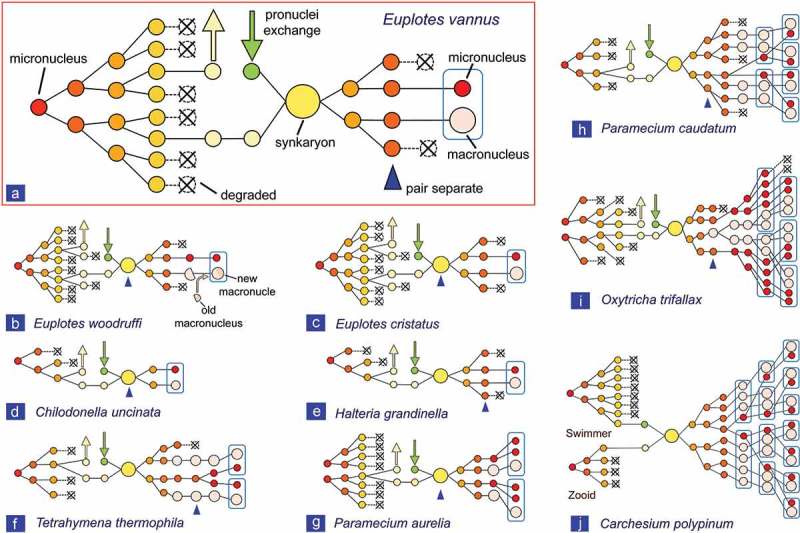
Comparison of the nuclear events during conjugation in Euplotes vannus with those in other ciliate species. (a) Euplotes vannus from present study; (b) Euplotes woodruffi [38]; (c) Euplotes cristatus [46]; (d) Chilodonella uncinata [43]; (e) Halteria grandinella [50]; (f) Tetrahymena thermophila [15]; (g) Paramecium aurelia [21,53]; (h) Paramecium caudatum [49]; (i) Oxytricha trifallax [42,51]; (j) Carchesium polypinum [52].
Similar to E. vannus, Chilodonella uncinata, Halteria grandinella, Tetrahymena thermophila and Paramecium caudatum have only one MIC and three prezygotic divisions occur in these species (Figure 4(d–g)). In P. caudatum [49] and T. thermophila [15], however, MIC first enters two meiotic divisions (meiosis I and II) and generates four haploid nuclei, only one of which divides mitotically to form gametic nuclei. In H. grandinella [50] and C. uncinata [43], one product is selected at the end of each division to complete the process (Figure 4(d–e)). Consequently, in these four species stationary and migratory gametic pronuclei are genetically identical.
In Paramecium aurelia [21] and Oxytricha trifallax [51], which have two MICs, there are three prezygotic divisions (meiosis I and II, and mitosis). Conjugation in peritrichs such as Carchesium polypinum [52] differs from most other ciliates because the two mating cells are anisometric comprising a stationary macrogamete and a motile microgamete. The MIC divides three times also in the microgamete. Therefore, it appears that three prezygotic divisions (mitosis, meiosis I and meiosis II) represent a general rule, although in each species different products are selected for generating gametic pronuclei. Euplotes vannus conforms to this rule, whereas the other Euplotes species with four prezygotic divisions represent exceptions.
The number of postzygotic divisions is more variable in different ciliates (Figure 4). In C. uncinata the synkaryon divides only once to produce the new MIC and MAC [43]; in H. grandinella and all Euplotes spp. it divides twice [38,46,50]; in T. thermophila and P. aurelia it divides twice followed by one cell division [15,21,53]; in O. trifallax it divides twice followed by two cell divisions [42,51]; in P. caudatum it divides three times followed by two cell divisions [49]; in C. polypinum it divides three times followed by three cell divisions [52]. Also the selection of nuclei for MIC and MAC differentiation appears to be species-specific.
In conclusion, conjugation in E. vannus requires about 75 hours to complete all the different steps, from mating pair formation to the development of new MIC and MAC in exconjugant cells. Pre- and postzygotic nuclear divisions occur rapidly and, soon after the second postzygotic division, two of the four nuclei are selected to generate the new MIC and the MAC anlagen, while the remaining two nuclei degenerate. Compared to the pre- and postzygotic nuclear divisions, a longer time is needed to complete the development of a new MAC. Different species of Euplotes, however, vary with regard to the fate of the old macronucleus. In E. woodruffi and E. patella, the remnants of the old MAC reorganize and fuse with the new MAC anlagen [38,54]. In other species of Euplotes, including E. vannus, this phenomenon has never been observed, and fragments of the old MAC degenerate completely.
Conjugation in ciliates is equivalent to, but more complicated than sexual reproduction in higher eukaryotes. First, due to the nuclear dimorphism, the daughter nuclei of the last postzygotic division have to differentiate into both germline and somatic nuclei within a single cell. Although recent studies indicate that nucleus differentiation in ciliates requires biased assembly of the nuclear pore complex [55], the entire process has not yet been fully clarified. The most complicated nucleus differentiation was observed in the species Blepharisma japonicum in which macronucleus could differentiate through both sexual and asexual paths proceeded synchronously in each cell with one path eventually dominated the other [56]. Second, conjugation is performed under the control of the so-called “mating type systems”, which exhibit significant variety in terms of mating type number, mating type determinants, and mating type inheritance [25]. The mechanisms of mating type determination in Paramecium [57], Tetrahymena [41,58], and Euplotes [24,35,59,60] are strikingly different at the molecular level. Therefore, this study may provide basic information to investigate the evolution of mating type systems, sexual phenomena and differentiation of dimorphic nuclei in ciliates and, more in general, may shed new light on the origins and evolution of sex and fertilization in eukaryotes.
Materials and methods
Species identification and mating type determination
Strains used in this study were collected in July 2015 from the Silver Sand Beach of Qingdao, China (35°55ʹN, 120°12ʹE), and have been assigned to the species E. vannus on the basis of morphological characters observed both in vivo and after protargol staining (Figure 1(a–d,h,i)) as previously described [61–64]. Single cells of the exconjugants were separated to obtain 17 monocultures. Six mating types were identified using pairwise mixtures of the 17 monocultures.
Cell culture and conjugation induction
Cells were maintained at room temperature (ca. 25°C) in sterilized seawater, using Escherichia coli as food source. To obtain highly reactive cells, cells were centrifuged (300 g, 3 min), re-suspended in fresh seawater at a concentration of 4000 cell/ml, and starved for two days. Conjugation was induced by mixing reactive cells of different mating types. The rate of conjugation was usually more than 80%.
Staining and observation
When the cells started to form pairs, the paired cells were picked out and were considered as synchronized (time 0). Cells were sampled every 30 min or 1 hour after initial mating pair formation (Table (S1)) and stained with Hoechst 33342 (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China) [49]. Hoechst stock solutions (10x, 100 µg/ml) were prepared in sterilized distilled water and kept at 4°C until further processing. Aliquots of 2.5 µl Hoechst stock solution were added to 200 µl cell suspensions (final concentration: 1.1 μg/ml), and incubated at room temperature for 15 min. Cells were then transferred to a glass microscope slide, covered by a coverslip, and observed under a “ZEISS AXIO Imager. D2” fluorescence microscope, equipped with an Axiocom 506 camera for photographic documentation. For each time point, about 20–50 pairs were recorded.
Phylogenetic analyzes
In order to show the phylogenetic positions of species used in the present study, phylogenetic analyzes based on small subunit ribosomal DNA (SSU rDNA) sequences (accession number as shown in Table (S2)) were performed as previous described [65,66]. In brief, 59 SSU rDNA sequences (including the representatives from 57 orders covering all the major lineages of ciliates and two outgroup taxa) were downloaded from the National Center for Biotechnology Information (NCBI) database and aligned using the GUIDANCE2 Server with default parameters [67]. The alignment was manually modified using BioEdit v.7.0.1 [68] to generate a matrix of 57 taxa with 1,484 nucleotide sites. Both maximum likelihood (ML) and Bayesian inference (BI) analyzes were performed in CIPRES Science Gateway (URL: http://www.phylo.org/sub_sections/portal) [69]. ML tree was constructed using RAxML-HPC2 on XSEDE v.8.2.10 [70] with GTR+I + G model and 1000 bootstrap replicates. BI analysis was performed using MrBayes on XSEDE v.3.2.6 with GTR+I + G model which was selected by MrModeltest v.2.0 [71] and PAUP [72]. Markov chain Monte Carlo (MCMC) simulations were run for 10,000,000 generations with a frequency of 100 generations and a burn-in of 10,000 trees. A majority rule consensus tree with posterior probabilities (PP) was constructed by all remaining trees. Tree topologies were visualized with MEGA v.6.06 [73].
Funding Statement
This work was supported by the Natural Science Foundation of China [31772428]; The Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) [2018SDKJ0406-1]; Fundamental Research Funds for the Central Universities [201841013]; Young Elite Scientists Sponsorship Program by CAST [2017QNRC001].
Acknowledgments
This research was funded by the Natural Science Foundation of China (No. 31772428), the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0406-1), Young Elite Scientists Sponsorship Program by CAST (2017QNRC001) and the Fundamental Research Funds for the Central Universities (201841013) to FG. We gratefully acknowledge Prof. Weibo Song, Dr. Alan Warren and Dr. Xiaotian Luo (OUC) for their help in revising this paper and species identification.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary material data for this article can be accessed here.
References
- [1].Juranek SA, Lipps HJ.. New insights into the macronuclear development in ciliates. Int Rev Cytol. 2007;262:219–251. [DOI] [PubMed] [Google Scholar]
- [2].Nanney DL. Experimental ciliatology: an introduction to genetic and developmental analysis in ciliates. New York (NY): Wiley; 1980. [Google Scholar]
- [3].Wang YR, Wang YY, Sheng Y, et al. A comparative study of genome organization and epigenetic mechanisms in model ciliates, with an emphasis on Tetrahymena, Paramecium and Oxytricha. Eur J Protistol. 2017;61:376–387. [DOI] [PubMed] [Google Scholar]
- [4].Song WB, Warren A, Hu X. Free-living ciliates in the Bohai and Yellow Seas, China. Beijing (BJ): Science Press; 2009. [Google Scholar]
- [5].Gao F, Huang JA, Zhao Y, et al. Systematic studies on ciliates (Alveolata, Ciliophora) in China: progress and achievements based on molecular information. Eur J Protistol. 2017;61:409–423. [DOI] [PubMed] [Google Scholar]
- [6].Wang YY, Chen X, Sheng Y, et al. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 2017;45:11594–11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prescott DM. The DNA of ciliated Protozoa. Microbiol Rev. 1994;58:233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yan Y, Rogers AJ, Gao F, et al. Unusual features of non-dividing somatic macronuclei in the ciliate class Karyorelictea. Eur J Protistol. 2017;61:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen XM, Lu XT, Luo XT, et al. Researches on formation of cortical patterns during morphogenesis in ciliates supported by the IRCN-BC and NSFC projects. Eur J Protistol. 2017;61:439–452. [DOI] [PubMed] [Google Scholar]
- [10].Raikov IB. The protozoan nucleus–morphology and evolution. Berlin: Springer-Verlag; 1982. [Google Scholar]
- [11].Zhao X, Wang YY, Wang YR, et al. Histone methyltransferase TXR1 is required for both H3 and H3.3 lysine 27 methylation in the well-known ciliated protist Tetrahymena thermophila. Sci China Life Sci. 2017;60:264–270. [DOI] [PubMed] [Google Scholar]
- [12].Chen X, Wang YR, Sheng Y, et al. GPS it: an automated method for evolutionary analysis of nonculturable ciliated microeukaryotes. Mol Ecol Resour. 2018;18:700–713. [DOI] [PubMed] [Google Scholar]
- [13].Luporini P, Alimenti C, Pedrini B, et al. Ciliate communication via water-borne pheromones In: Witzany G, Nowacki M, editors. Biocommunication of ciliates. Cham: Springer; 2016. p. 159–174. [Google Scholar]
- [14].Orias E. Ciliate conjugation In: Gall JG, editor. The molecular biology of ciliated protozoa. New Yok (NY): Academic Press; 1986. p. 45–84. [Google Scholar]
- [15].Orias E, Cervantes MD, Hamilton EP. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol. 2011;162:578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang TT, Wang C, Katz LA, et al. A paradox: rapid evolution rates of germline-limited sequences are associated with conserved patterns of rearrangements in cryptic species of Chilodonella uncinata (Protista, Ciliophora). Sci China Life Sci. 2018;61:1071–1078. [DOI] [PubMed] [Google Scholar]
- [17].Zheng W, Wang C, Yan Y, et al. Insights into an extensively fragmented eukaryotic genome: de novo genome sequencing of the multinuclear ciliate Uroleptopsis citrina. Genome Biol Evol. 2018;10:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Swart EC, Bracht JR, Magrini V, et al. The Oxytricha trifallax macronuclear genome: A complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013;11: e1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Maurer-Alcalá XX, Knight R, Katz LA. Exploration of the germline genome of the ciliate Chilodonella uncinata through single-cell omics (transcriptomics and genomics). mBio. 2018;9:e01836–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen X, Bracht JR, Goldman AD, et al. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell. 2014;158:1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sonneborn TM. Mating types in Paramecium aurelia: diverse conditions for mating in different stocks; occurrence, number and interrelations of the types. Proc Am Philos Soc. 1938;79:411–434. [Google Scholar]
- [22].Singh DP, Saudemont B, Guglielmi G, et al. Genome-defence small RNAs exapted for epigenetic mating-type inheritance. Nature. 2014;509:447–452. [DOI] [PubMed] [Google Scholar]
- [23].Cervantes MD, Hamilton EP, Xiong J, et al. Correction: selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila. PLoS Biol. 2013;11:e1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dini F, Luporini P. Mating‐type polymorphic variation in Euplotes minuta (Ciliophora: hypotrichida) 1. J Protozool. 1985;32:111–117. [Google Scholar]
- [25].Phadke SS, Zufall RA. Rapid diversification of mating systems in ciliates. Biol J Linn Soc. 2009;98:187–197. [Google Scholar]
- [26].Nobili R, Luporini P, Dini F. Breeding systems, species relationships and evolutionary trends in some marine species of Euplotidae (Hypotrichida, Ciliata) In: Battaglia B, Beardmore JA, editors. Marine organisms: genetics, ecology and evolution.New York: Plenum Press ; 1978. p. 591–616. [Google Scholar]
- [27].Zhao Y, Yi Z, Warren A, et al. Species delimitation for the molecular taxonomy and ecology of the widely distributed microbial eukaryote genus Euplotes (Alveolata, Ciliophora). Proc R Soc B. 2018;285:20172159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lian C, Luo X, Fan X, et al. Morphological and molecular redefinition of Euplotes platystoma Dragesco & Dragesco‐Kernéis, 1986 and Aspidisca lynceus (Müller, 1773) Ehrenberg, 1859, with reconsideration of a “well‐known” Euplotes ciliate, Euplotes harpa Stein, 1859 (Ciliophora, Euplotida). J Eukaryot Microbiol. 2018;65:531–543. [DOI] [PubMed] [Google Scholar]
- [29].Yan Y, Fan Y, Luo X, et al. New contribution to the species-rich genus Euplotes: morphology, ontogeny and systematic position of two species (Ciliophora; Euplotia). Eur J Protistol. 2018;64:20–39. [DOI] [PubMed] [Google Scholar]
- [30].Vallesi A, Alimenti C, Federici S, et al. Evidence for gene duplication and allelic codominance (not hierarchical dominance) at the mating-type locus of the ciliate, Euplotes crassus. J Eukaryot Microbiol. 2014;61:620–629. [DOI] [PubMed] [Google Scholar]
- [31].Luporini P, Raffioni S, Concetti A, et al. The ciliate Euplotes raikovi heterozygous at the mat genetic locus coreleases two individual species of mating pheromone: genetic and biochemical evidence. Proc Natl Acad Sci USA. 1986;83:2889–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Luporini P, Pedrini B, Alimenti C, et al. Revisiting fifty years of research on pheromone signaling in ciliates. Eur J Protistol. 2016;55:26–38. [DOI] [PubMed] [Google Scholar]
- [33].Luporini P, Miceli C. Mating pheromones In: Gall JG, editor. The molecular biology of ciliated protozoa. New Yok (NY): Academic Press; 1986. p. 263–299. [Google Scholar]
- [34].Vallesi A, Alimenti C, Luporini P. Ciliate pheromones: primordial self-/nonself-recognition signals In: Ballarin L, Cammarata M, editors. Lessons in immunity: from single-cell organisms to mammals. Amsterdam: Academic Press ; 2016. p. 1–16. [Google Scholar]
- [35].Heckmann K, Kuhlmann HW. Mating types and mating inducing substances in Euplotes octocarinatus. J Exp Zool. 1986;237:87–96. [Google Scholar]
- [36].Zhou L, Li J, Lin X, et al. Use of RAPD to detect DNA damage induced by nitrofurazone in marine ciliate, Euplotes vannus (Protozoa, Ciliophora). Aquatic Toxicol. 2011;103:225–232. [DOI] [PubMed] [Google Scholar]
- [37].Xu H, Song W, Warren A. An investigation of the tolerance to ammonia of the marine ciliate Euplotes vannus (Protozoa, Ciliophora). Hydrobiologia. 2004;519:189–195. [Google Scholar]
- [38].Rao MVN. Nuclear behavior of Euplotes woodruffi during conjugation. J Eukaryot Microbiol. 1964;11:296–304. [Google Scholar]
- [39].Grell KG. Protozoology. Berlin: Springer-Verlag; 1973. [Google Scholar]
- [40].Dini F, Nyberg D. Sex in ciliates In: Jones JG, editor. Advances in microbial ecology. New York: Plenum Press.. 1993. p. 85–153. [Google Scholar]
- [41].Orias E, Singh DP, Meyer E. Genetics and epigenetics of mating type determination in Paramecium and Tetrahymena. Annu Rev Microbiol. 2017;71:133–156. [DOI] [PubMed] [Google Scholar]
- [42].Gregory LH. The conjugation of Oxytricha fallax. J Morphol. 2010;37:555–581. [Google Scholar]
- [43].Bellec L, Maureralcala XX, Katz LA. Characterization of the life cycle and heteromeric nature of the macronucleus of the ciliate Chilodonella uncinata using fluorescence microscopy. J Eukaryot Microbiol. 2014;61:313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gao F, Warren A, Zhang Q, et al. The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci Rep. 2016;6:24874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lueken WW. A marine Euplotes (Ciliophora, Hypotrichida) with reduced number of prezygotic micronuclear divisions. J Protozool. 1973;20:143–145. [Google Scholar]
- [46].Wichterman R. Mating types, breeding system, conjugation and nuclear phenomena in the marine ciliate Euplotes cristatus Kahl from the gulf of Naples. J Eukaryot Microbiol. 2010;14:49–58. [Google Scholar]
- [47].Miceli C, Luporini P, Bracchi P. Morphological description, breeding system, and nuclear-changes during conjugation of Euplotes-raikovi agamaliev from mediterranean-sea. Acta Protozool. 1981;20:215. [Google Scholar]
- [48].Valbonesi A, Ortenzi C, Luporini P. Observations on the biology of Euplotes charon (Hypotrichida, Ciliophora). Ital J Zool. 1987;54:111–118. [Google Scholar]
- [49].Yang X, Gao X, Shi X. Detection of haploid nuclear death in living Paramecium caudatum. Jpn J Protozool. 2007;40:123–130. [Google Scholar]
- [50].Agatha S, Foissner W. Conjugation in the spirotrich ciliate Halteria grandinella (Müller, 1773) Dujardin, 1841 (Protozoa, Ciliophora) and its phylogenetic implications. Eur J Protistol. 2009;45:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Adl SM, Berger JD. Timing of life cycle morphogenesis in synchronous samples of Sterkiella histriomuscorum. II. The sexual pathway. J Eukaryot Microbiol. 2000;47:443–449. [DOI] [PubMed] [Google Scholar]
- [52].Raikov I. Nuclear phenomena during conjugation and autogamy in ciliates In: Chen T-T, editor. Research in protozoology. New York: Pergmon Press; 1972. p. 147–289. [Google Scholar]
- [53].Diller WF. Nuclear reorganization processes in Paramecium aurelia, with descriptions of autogamy and “hemixis”. J Morphol. 1936;59:11–67. [Google Scholar]
- [54].Turner JP. Division and conjugation in Euplotes patella Ehrenberg with special reference to the nuclear phenomena. Univ Calif Publ Zool. 1930;33:193–258. [Google Scholar]
- [55].Iwamoto M, Koujin T, Osakada H, et al. Biased assembly of the nuclear pore complex is required for somatic and germline nuclear differentiation in Tetrahymena. J Cell Sci. 2015;128;1812–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Miyake A, Rivola V, Harumoto T. Double paths of macronucleus differentiation at conjugation in Blepharisma japonicum. Eur J Protistol. 1991;27:178–200. [DOI] [PubMed] [Google Scholar]
- [57].Siegel R, Larison L. The genic control of mating types in Paramecium bursaria. Proc Natl Acad Sci USA. 1960;46:344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nanney D, Caughey PA, Tefankjian A. The genetic control of mating type potentialities in Tetrahymena pyriformis. Genetics. 1955;40:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Akada R. Mating types and mating-inducing factors (gamones) in the ciliate Euplotes patella syngen 2. Genet Res. 1985;46:125–132. [Google Scholar]
- [60].Kimball R. The nature and inheritance of mating types in Euplotes patella. Genetics. 1942;27:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Song WB, Packroff G. Taxonomische untersuchungen an marinen ciliaten aus China mit beschreibungen von zwei neuen arten, Strombidium globosaneum nov. spec. and S. platum nov. spec. (Protozoa, Ciliophora). Arch Protistendk. 1997;147:331–360. [Google Scholar]
- [62].Yan Y, Fan Y, Chen X, et al. Taxonomy and phylogeny of three heterotrich ciliates (Protozoa, Ciliophora), with description of a new Blepharisma species. Zool J Linn Soc. 2016;177:320–334. [Google Scholar]
- [63].Pan H, Jiang J, Fan X, et al. Phylogeny and taxonomy of five poorly known species of cyrtophorian ciliates (Protozoa: Ciliophora: Phyllopharyngea) from China seas. Zool J Linn Soc. 2016;180:475–492. [Google Scholar]
- [64].Valbonesi A, Ortenzi C, Luporini P. An integrated study of the species problem in the Euplotes crassus‐minuta‐vannus Group 1. J Protozool. 1988;35:38–45. [Google Scholar]
- [65].Huang JB, Zhang TT, Zhang Q, et al. Further insights into the highly derived haptorids (Ciliophora, Litostomatea): phylogeny based on multigene data. Zool Scr. 2018;47:231–242. [Google Scholar]
- [66].Wang C, Zhang TT, Wang YR, et al. Disentangling sources of variation in SSU rDNA sequences from single cell analyses of ciliates: impact of copy number variation and experimental error. Proc R Soc B. 2017;284:20170425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sela I, Ashkenazy H, Katoh K, et al. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser; London: Information Retrieval Ltd; 1999. p. c1979–c2000. [Google Scholar]
- [69].Miller MA, Pfeiffer W, Schwartz T.. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA; IEEE; 2010. p. 1–8. [Google Scholar]
- [70].Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ronquist F, Teslenko M, Van Der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Nylander J. MrModeltest, a program to evaluate the fit of several models of evolution to a given data and unrooted tree (version 2.2). Program distributed by the author. Sweden: Evolutionary Biology Centre, Uppsala University; 2004. Available from: http://www abc se/~ nylander [Google Scholar]
- [73].Tamura K, Dudley J, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


