Abstract
Efforts are being directed to systematically analyse the non-coding genome for cancer-driving mutations1–6. Cis-regulatory elements (CREs) represent a highly enriched subset of the non-coding genome in which to search for such mutations. We use capture Hi-C for 19,023 promoter fragments to catalogue the regulatory landscape of colorectal cancer (CRC) in cell lines, mapping CREs and integrating these with TCGA whole genome sequence and expression data7,8. We identify a recurrently mutated CRE interacting with the ETV1 promoter affecting gene expression. ETV1 expression influences cell viability and is associated with patient survival. We further refine our understanding of the regulatory effects of copy-number variations (CNVs), showing RASL11A to be targeted by a previously identified enhancer amplification1. This study reveals new insights into the complex genetic alterations driving tumour development, providing a paradigm for employing chromosome conformation capture to decipher non-coding CREs relevant to cancer biology.
The identification of driver mutations as distinguished from passenger mutations is fundamental to understanding cancer and its response to therapy. With the large number of exome-sequenced tumours, all genes with coding changes that contribute substantially to tumourigenesis are likely to be catalogued shortly. Motivated by the identification of recurrent mutations in regulatory elements in genes associated with oncogenesis, such as the TERT promoter9,10, and TAL111 and PAX512 enhancers, efforts are now being directed to systematically analyse non-coding regions of cancer genomes for driver mutations1–6.
Although regional excess of somatic mutations is suggestive of positive selection in tumours, the size of the non-coding genome places a high burden on robustly establishing statistical significance. Coding regions provide obvious, discrete intervals in which to search for mutations, and it would be highly propitious to define similar functional elements for non-coding regions. CREs modulating gene expression represent a highly-enriched subset of the non-coding genome in which to search for driver mutations. These CREs however, can be highly tissue-specific, are often dispersed over long ranges, and only a small fraction of distal enhancers target the nearest transcript8,13. The recent development of high-throughput chromosome conformation capture techniques (Hi-C) has allowed researchers to map such regulatory regions, and importantly, link these to their respective target genes8,14–16.
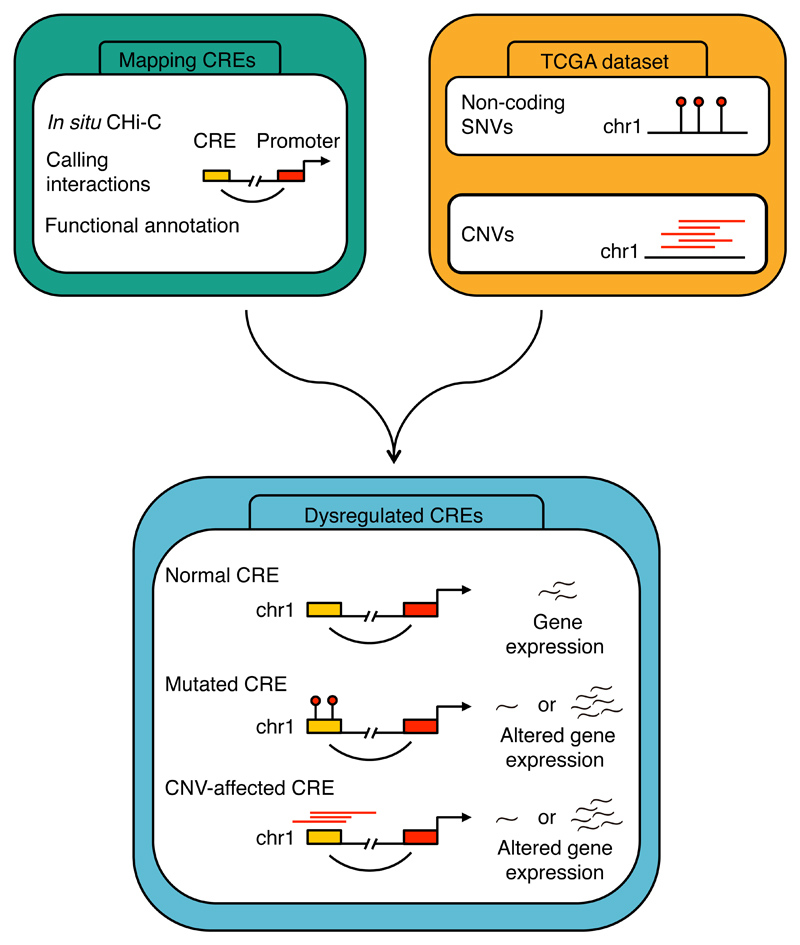
Here we have used capture Hi-C (CHi-C) for 19,023 promoter fragments to catalogue the CRE landscape of CRC, identifying putative enhancers8,16. Using these data in conjunction with TCGA whole genome sequencing (WGS), RNA sequencing (RNAseq) and CNV data7, we report the identification of novel non-coding driver mutations for CRC (Fig. 1, Supplementary Fig. 1, Supplementary Note).
Figure 1. Workflow for the identification of mutated cis-regulatory elements in colorectal cancer.
CRE, cis-regulatory element; SNV, single nucleotide polymorphism; CNV, copy-number variation.
We prepared in situ HindIII-digested Hi-C libraries from CRC HT29 and LoVo cell lines, which represent the two major molecular subtypes of CRC – microsatellite stable (MSS) and microsatellite instable (MSI), respectively. To examine the interactions underlying CREs in CRC, we generated a biotinylated RNA bait library, specifically targeting 19,023 promoter-encompassing HindIII fragments representing 2.3% of all HindIII fragments. We hybridised in situ Hi-C libraries to the RNA baits to capture promoter-associated di-tags and sequenced the resulting libraries, from which we identified 96,458 and 118,758 significant contacts in LoVo and HT29 respectively (Supplementary Tables 1-3). In both cell lines, the majority of interactions were within topologically associated domains (TADs) (74% and 83% in HT29 and LoVo, respectively; Supplementary Tables 4 and 5, Supplementary Note). Across 127 cell lines and tissues17 the LoVo and HT29 CREs showed strong enrichment of histone marks identified in colonic tissue (Supplementary Table 6).
The CREs identified by CHi-C were evolutionally conserved (P<1.0×10-3) and enriched for transcription factor (TF) binding as compared to a random set of genomic fragments (3-fold and 2-fold enrichment for HT29 and LoVo respectively, P<1.0×10-3; Supplementary Fig. 2). Based on the level of gene expression using RNAseq for respective target genes we classified interactions as either active or inactive. As previously documented8, a higher frequency of interactions and shorter contact distances typified promoters of active genes (Supplementary Fig. 3). For genes that were actively transcribed, both promoters and their respective CREs had a higher number of bound TFs (P<2.2×10-16; Supplementary Fig. 4). Moreover, a relationship between elevated gene expression and higher proportion of TFs bound to CREs was shown, consistent with the functional role of identified CREs (Supplementary Fig. 5).
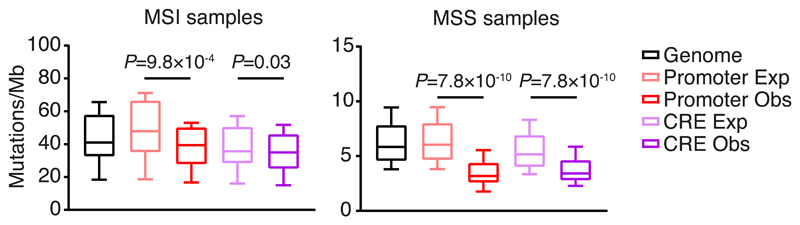
To investigate the frequency of single nucleotide variants (SNVs) in CREs delineated by CHi-C we analysed colon and rectal adenocarcinoma WGS data from TCGA7. After applying stringent quality control and filtering, we retained data on 50 MSS and 12 MSI cancers. The frequencies of SNV base changes were consistent with those previously documented for CRC18,19 (Supplementary Fig. 6, Supplementary Table 7). Both promoters and CREs showed a significantly lower rate of mutation compared to the genome rate in MSS and MSI cancers (Fig. 2). This property was also seen after taking into account mutational-signature-derived substitution probabilities (Fig. 2). This is consistent with previous reports that have shown that functional regulatory regions are less likely to be mutated than non-functional regions of the genome, either as a consequence of selective evolutionary pressure or chromatin accessibility9,20. Since MSI cancers exhibit a significantly higher mutational rate to MSS cancers (median mutations 92,968 and 14,290, respectively) reflective of mismatch repair deficiency19 (Supplementary Table 8), we analysed the two CRC subtypes separately.
Figure 2. Non-coding mutations in cis-regulatory elements.
Mutation rates in promoters, CREs and genome-wide in MSI (n=12) and MSS (n=50) cancers. Shown are both the mutation rates observed in promoters and CREs, and the mutation rates expected considering the sample-specific occurrence of mutations of each of the 96 substitution types, and the trinucleotide composition of the fragment classes. Box-plots denote quartiles. Whiskers correspond to the 10th and 90th percentiles. Difference assessed using a two-sided paired Wilcoxon test.
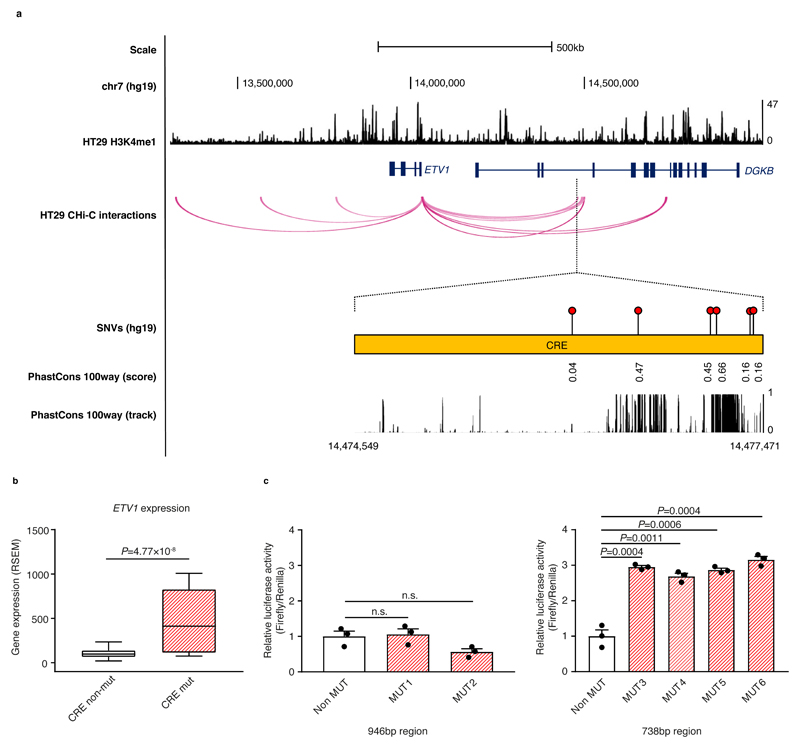
To identify non-coding driver mutations in CREs, we integrated results from three driver discovery methods (Supplementary Fig. 7a). First, we assessed the transcriptional affects of CRE mutations by comparing the expression levels of respective target genes in mutated and non-mutated cancers9,10,21. To avoid confounding, cancers in which either the CRE or target gene were subject to CNVs were excluded from the analysis. Second, we tested for regional excess of mutations to provide evidence of positive selection6,9,21,22. Third, we assessed the clustering of CRE mutations, as this can be suggestive of events in specific TF binding sites21. In MSS cancers this integrated analysis yielded a CRE interacting with the ETV1 promoter, associated with differential target gene expression and an excess of clustered mutations (Fig. 3a-b, Supplementary Tables 9 and 10). Conversely, no CREs were identified in MSI cancers, and we therefore restricted further SNV analysis to MSS cancers.
Figure 3. Cis-regulatory element mutation affects ETV1 expression.
(a) Chromatin looping interactions between the ETV1 promoter and CREs in HT29. Also detailed are the relative positions of SNVs for the significantly mutated CRE and the evolutionary conservation of the region, as measured using PhastCons 100-way smoothed scores and track. (b) Relationship between mutation status and ETV1 expression in MSS cancers. One sample containing a CNV overlapping ETV1 was excluded. Box-plots denote quartiles. Whiskers correspond to the 10th and 90th percentiles. Difference between samples in which the CRE is mutated (mut, n=5) and not mutated (non-mut, n=41) assessed by negative binomial test. (c) A 946bp and a 738bp putative regulatory regions containing the six mutations were cloned upstream of the SV40 promoter in the pGL3-promoter vector. The resultant reporter constructs were transiently transfected into HT29 for 24 hours and the relative luciferase activity was measured for each reporter gene construct. The luminescence ratio of the experimental vector to the Renilla internal control, pRL-TK, was normalised to the backbone pGL3-SV40 promoter vector. Data shown are mean ± SEM relative to the non-mutated (Non MUT) samples from three independent experiments and assessed by two-tailed t-test, n.s., non-significant.
We demonstrated the benefit of using CHi-C data to discover non-coding driver mutations by comparing the number of CREs identified by the integrated driver discovery analysis using real and randomised CHi-C data. Specifically, we randomised the CHi-C data by changing the HindIII fragments contacting each gene, sampling from non-interacting fragments within 1Mb of the gene, whilst maintaining the number of fragment contacts (Supplementary Fig. 7b), and applied the integrated driver discovery analysis in full to each randomised CHi-C data set. Through this procedure we estimated the empirical false discovery rate (FDR) for the ETV1 CRE to be 0.023 (Supplementary Table 9, Supplementary Fig. 8).
ETV1, an oncogene encoding an ETS TF, is altered in several cancers by translocations in Ewing's sarcoma23 and prostate cancer24,25, amplification in melanoma26, and oncogenic dysregulation in gastrointestinal stromal tumours27,28. The ETV1 interaction was confirmed in a panel of MSS CRC cell lines (Supplementary Fig. 9). Considering only the five cancers without a structural variation (SV) within 1 Mb from ETV1 (Supplementary Table 11), CRE mutations were associated with a 4-fold increased expression of ETV1 compared to non-mutated samples (Fig. 3b). Additionally, increased ETV1 expression was also seen in two cancer samples without CRE mutations but with ETV1 amplifications (P=4.1×10-3; Supplementary Fig. 10a). Four of the six mutations map proximally to an evolutionary conserved region (Fig. 3a), and in HT29 each was associated with a 3-fold increase in luciferase activity, consistent with regulatory impact (Fig. 3c, Supplementary Figure 10b). The ETV1 CRE, which is proximal to an enhancer H3K4me1 chromatin mark, uniquely interacts with the ETV1 promoter, and the contact is not present in LoVo or 17 blood-specific CHi-C data (Fig. 3a, Supplementary Fig. 11). While the putative enhancer maps within the intron of DGKB, there was no relationship between CRE mutation and DGKB expression (Supplementary Fig. 12). Although no TF was bound to the CRE on the basis of HT29 cell line ChIPseq data, in silico analysis identified numerous potential disrupted TF binding sites29–31 including those for BCL6, HNF1A, HNF1B and MAFK, all of which are expressed in colonic tissue32 (Supplementary Table 12 and 13, Supplementary Fig. 13, Supplementary Note).
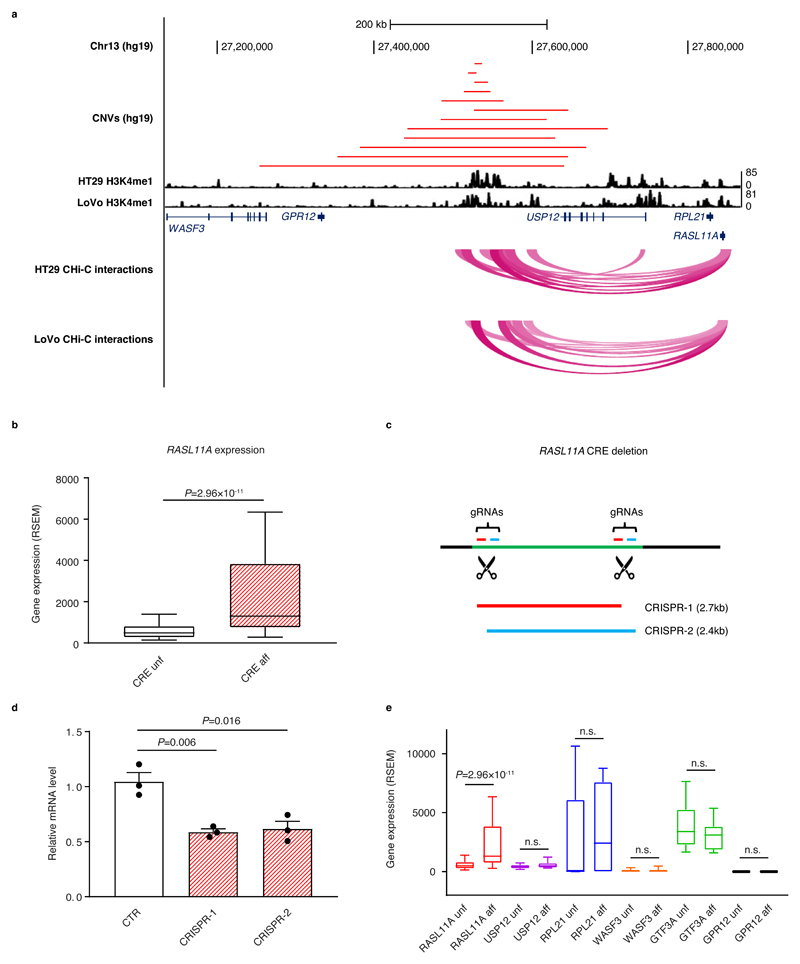
We made use of Affymetrix SNP Array 6.0 data from 615 TCGA CRC samples to identify CREs subject to somatic CNVs. CNV-positive CREs were assessed for correlation with expression of their interacting gene, where it was not encompassed by the same CNV, using matched RNAseq data. The RASL11A promoter showed interactions with a putative enhancer characterised by H3K4me1 marks in both HT29 and LoVo cell lines (Fig. 4a, Supplementary Fig. 14). While a role for RASL11A in tumourigenesis has yet to be defined, it is reported to be a GTPase chromatin-associated modulator of pre-ribosomal RNA synthesis, acting to facilitate initiation of transcription by RNA polymerase 133. The RASL11A interaction was confirmed in a panel of MSS CRC cell lines (Supplementary Fig. 9). This CRE was amplified in 12 cancer samples and these had significantly higher RASL11A expression (P=2.96×10-11; Fig. 4b, Supplementary Table 14). Using CRISPR-mediated genome editing, disruption of the interacting CRE was shown to reduce RASL11A expression (Fig. 4c and 4d, Supplementary Fig. 15). USP12 has previously been implicated as a target of this specific CRE1. However, after exclusion of samples in which CNVs overlay the gene itself, amplification of the CRE was not associated with differential expression of USP12 or any other gene in the proximity of the CNV segments (Fig. 4e, Supplementary Table 15).
Figure 4. Amplification of cis-regulatory element upregulates RASL11A expression.
(a) The amplification of the CRE interacting with the RASL11A promoter; upper track, CNVs overlapping the CRE in 12 samples; middle track, H3K4me1 mark in HT29 and LoVo; lower track, CHi-C interactions in HT29 and LoVo. (b) RASL11A expression in CRC stratified by CRE amplification status (samples with CNV overlapping RASL11A excluded). Box-plots denote quartiles. Whiskers correspond to the 10th and 90th percentiles. Difference between samples in which the CRE is affected (CRE aff, n=12) and unaffected (CRE unf, n=522) by a CNV amplification assessed by negative binomial test. (c) Schematic representation of the CRISPR/Cas9-mediated deletion using two plasmids expressing different sets of gRNAs. (d) Barplots showing RASL11A mRNA levels relative to GAPDH in control (CTR) and CRISPR-edited cells (CRISPR-1 and CRISPR-2). Differences assessed by two-tailed t-test from three independent experiments, mean ± SEM. (e) Expression levels of six genes in the proximity of the CRE in the samples where the CRE is affected (aff) or unaffected (unf) by CNV amplification (RASL11A unf n=522, RASL11A aff n=12, USP12 unf n=524, USP12 aff n=7, RPL21 unf n=522, RPL12 aff n=12, WASF3 unf n=523, WASF3 aff n=14, GTF3A unf n=521, GTF3A aff n=12, GPR12 unf n=523, GPR12 aff n=13). Samples containing CNVs overlapping the respective gene are excluded from each comparison. Box-plots denote quartiles. Whiskers correspond to the 10th and 90th percentiles. Differences assessed by negative binomial test, n.s., non-significant.
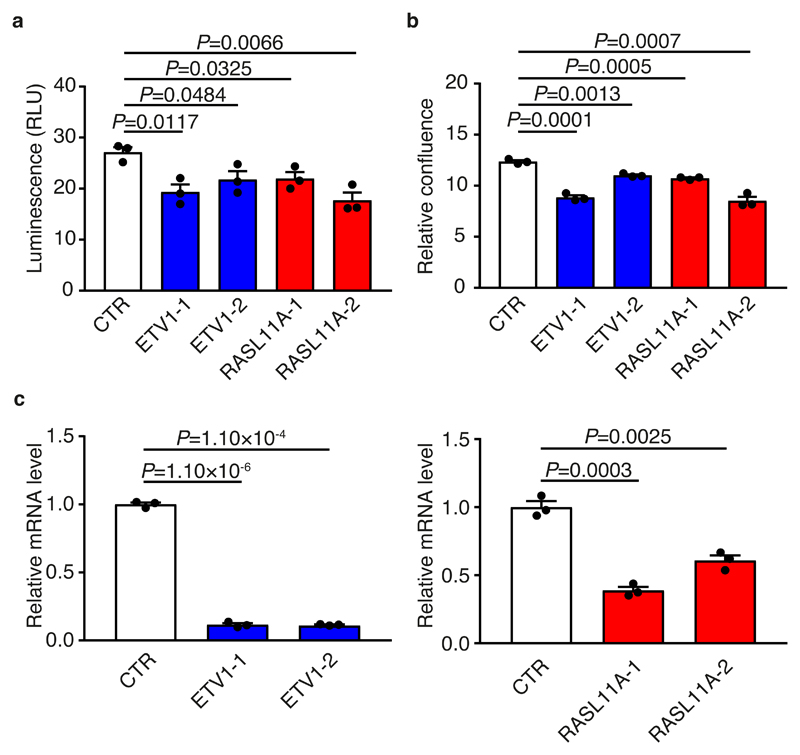
In HT29, reduction of endogenous ETV1 and RASL11A levels using siRNA was associated with decreased cell viability and cell proliferation (Fig. 5, Supplementary Fig. 16, Supplementary Note). Using patient outcome data from three independent series totalling 1,282 CRC cases7,34,35, high levels of ETV1 expression were associated with worse relapse-free and overall survival in a multivariate analysis. Respective meta-analysis hazard ratios associated with elevated expression were 1.32 (95% confidence interval [CI]: 1.06-1.64, P=1.5×10-2) and 1.14 (95% CI: 1.03-1.27, P=9.9×10-3) (Supplementary Fig. 17, Supplementary Table 16, Supplementary Note). No significant association was shown between RASL11A expression and patient outcome, although data were not available for all series (Supplementary Table 16).
Figure 5. ETV1 and RASL11A levels are associated with differential cell growth.
(a) Relative luminescence of ETV1, RASL11A and control (CTR) knockdowns. Differences assessed 72 hours post-transfection by two-tailed t-test using three independent experiments, mean ± SEM. (b) Relative confluence of ETV1, RASL11A and control treated cells. Values normalised to the 0 hour time-point confluence. Differences assessed 96 hours post-transfection by two-tailed t-test using three independent experiments, mean ± SEM. (c) Barplots showing ETV1 and RASL11A mRNA levels, normalised to GAPDH, after siRNA treatment. Differences assessed by two-tailed t-test using three independent experiments, mean ± SEM.
It is arguable that despite high-profile, genuine successes, the number of new driver genes from exome sequencing projects has been disappointingly small compared with expectations. Many cancers have no observable driver mutation, and the full complement of molecular lesions that are individually necessary, and together sufficient, to cause malignancy are still unknown. The Encyclopedia of DNA Elements (ENCODE) project has proposed that around 80% of the genome contains elements linked to biochemical functionality36, however others have estimated that the percentage of the human genome that is functional may actually be 10-fold lower37. Hence, rather than exploiting the epigenomic landscape we identified physical contacts between promoters and the interacting CREs, thereby reducing the genomic space in which to search for non-coding driver mutations.
In our analysis, we identified a CRE whose mutation was associated with altered expression in CRC. This CRE, interacting with the ETV1 promoter, was mutated in 12% of MSS cancers with mutation being associated with significant upregulation of ETV1 expression. While not previously assessed as a determinant of CRC outcome38, upregulation of ETV1 was seen to be associated with poor patient prognosis in CRC (Supplementary Fig. 17, Supplementary Table 16). The fact that upregulation of ETV1 has previously been linked to reduced survival in other cancers, including gastric cancer, is therefore entirely consistent with increased ETV1 expression inducing aberrant activation of transcriptional programs that generically governs multiple facets of tumourigenesis. In addition to SNVs having a role in dysregulation of gene expression, we have provided evidence that implicates RASL11A, through amplification of an interacting CRE, in CRC oncogenesis.
We acknowledge that the present analysis has limitations. Firstly, we have used a cellular model to map the CREs, which is unlikely to fully recapitulate the spectrum of pathogenic SNVs and CNVs seen in CRC. Secondly, the low-resolution of the defined CNVs has not permitted the study of smaller structural changes, potentially affecting the discovery of deleted or amplified CREs. Thirdly, inevitably as our WGS dataset is modest, this has restricted the study power to identify non-coding drivers. Fourthly, our strategy may not identify genes that are up or down regulated in most cancers, either when driven by the same or alternative mechanisms.
Accepting these caveats, the generation of a genome-wide promoter-CRE map of CRC using in situ CHi-C has allowed us to perform a highly focused search for functional non-coding mutations. As sequencing costs are significantly reduced, this will afford the opportunity to perform CHi-C directly on respective WGS samples, thereby providing the opportunities to define individualised networks.
In conclusion, our work supports the existence of non-coding drivers for CRC, and more broadly provides a paradigm for employing chromosome conformation capture to decode disease-specific regulatory elements. Such discoveries facilitate the identification of novel therapeutic and chemoprevention agents, and classification of patients into molecular subgroups to personalise therapy. For example, it allows for the repositioning of ETV1 inhibitors, one of which has been employed in a clinical trial for gastrointestinal stromal tumours28,39.
Online Methods
Cell culture
HT115 and SW948 were obtained from ECACC, and all other cell lines were obtained from ATCC. All cell lines were cultured at 37°C; LoVo was cultured in Ham's F-12 Nutrient Mix, HT29 was cultured in McCoy's 5A (Modified) medium, SW480 and SW1116 were both cultured in DMEM, SW948 were cultured in Leibovitz’s L-15, all supplemented with 10% FBS. Caco2 was cultured in MEM supplemented with 20% FBS, HT115 was cultured in DMEM supplemented with 15% FBS. Cell line identity was confirmed by STR-profiling. Cells were regularly tested for mycoplasma contamination (PromoCell, PK-CA91).
Hi-C analysis
In situ Hi-C library preparation
In situ Hi-C libraries were prepared as previously described14,16. Briefly, 25 million cells were fixed in 1% formaldehyde for 10 min. Cross-linked DNA was digested by restriction enzyme HindIII (NEB, R0104). Digested chromatin ends were filled and marked with biotin-14-dATP (ThermoFisher, 19524-016). The resulting blunted ended fragments were ligated at 16°C in the nucleus with T4 DNA ligase (NEB, M0202) to minimise random ligation. DNA purified after crosslinking was reversed by proteinase K (Ambion, AM2546) treatment. DNA was sheared by sonication (Covaris, M220) and 200-650bp fragments selected. Biotin tag DNA was pulled down with streptavidin beads and ligated with Illumina paired end adapters (Illumina). Six cycles of PCR were performed to amplify libraries before capture.
Promoter capture Hi-C library
Promoter capture was based on 32,313 biotinylated 120-mer RNA baits (Agilent Technologies) targeting both ends of HindIII restriction fragments that overlap Ensembl promoters of protein-coding, non-coding, antisense, snRNA, miRNA and snoRNA transcripts8 (Supplementary Table 17). After library enrichment, a post-capture PCR amplification step was carried out using 5 amplification cycles. Hi-C and CHi-C libraries were sequenced using HiSeq 2000 technology16 (Illumina).
Interaction calling
Reads were aligned to the GRCh37 build using Bowtie2 v2.2.640 and identification of valid di-tags was performed using HiCUP v0.5.941. To declare significant contacts, HiCUP output was processed using CHiCAGO v1.1.842. For each cell line, data from three independent biological replicates were combined to obtain a definitive set of contacts. As previously advocated, interactions with a score ≥5.0 were considered to be statistically significant42.
ChIPseq analysis
ChIPseq was performed on H3K4me1, H3K9me3, H3K27me3 and H3K36me3 for LoVo, and H3K4me1 and H3K9me3 for HT29. Description of the procedures performed can be found in the Supplementary Note.
Gene expression in LoVo and HT29
Analysis of RNAseq data on LoVo and HT29 cell lines was performed as previously described7. Briefly, RNAseq BAM files were downloaded from the Broad Institute Cancer Genomics Hub and analysed using Cufflinks43. Gene-level FPKM read counts were derived using GENCODE v7 annotated mRNA transcripts. Genes with FPKM >0 were divided into quartiles based on their expression levels, with genes with either 0 FPKM or in Q1 considered to be inactive, and genes in Q2-Q4 considered to be active8,44. For promoter fragments associated with multiple genes, we excluded those with discordant expression.
Annotation of cis-regulatory elements in LoVo and HT29
Reads that did not align uniquely were removed by HiCUP as they are liable to result in false positive contacts. The detection of duplicate regions is dictated by genomic build. Therefore, an additional filtering step was performed to remove contacts between fragments mapping to regions that did not map to unique locations in hg38. Briefly, hg19/GRCh37 was split into windows of 100bp before being aligned to hg38 using BWA. A base was considered to be poorly mapped if the majority of reads containing it could be mapped elsewhere in the genome with at most one mismatch or gap, as described in SNPable (see URLs). A contact region was kept if 95% of its constituent bases were not poorly mapped imposing this metric. Conservation was measured using PhastCons 100way, considering the average conservation score in 100bp windows centred on each mutation45. The proportion of CREs, not overlapping any coding regions, containing runs of at least 8 conserved sites (defined by PhastCons score >0.5) was compared for the set of LoVo and HT29 CREs to sets of randomly generated fragments. For enrichment calculations, random non-coding regions of the genome were selected matching the identified CREs in fragment size and number, restricting start positions to HindIII sites. Random fragments not overlapping coding regions were resampled 1,000 times. Significance was determined by permutation. We annotated each CHi-C contact identified in LoVo and HT29 cells with chromatin features and TF binding information. HindIII interacting fragments were overlaid with histone marks from 127 tissues and cell lines from the ROADMAP Epigenomics project17 and ChIPseq experiments46 (198 and 29 experiments in LoVo and HT29 respectively). Enrichment calculations were performed as described above. The activity of each promoter on its corresponding gene was defined as active or inactive based on the analysis of the matching RNAseq data. To assess whether there was a relationship between TF binding and expression, we selected only the fragments that interact with promoters of genes belonging to the same expression quartile and allocated TF counts to each CRE as previously described8. For promoter fragments associated with multiple genes, all genes’ expression was required to be in the same expression quartile. For each expression class, we calculated the proportion of CREs bound by the TF divided by the proportion of all the CREs bound by the same TF. Corresponding values were log2 transformed8,44.
TCGA colorectal cancer whole genome sequencing data
Whole-genome sequencing (WGS) data on 50 MSS (36 colon adenocarcinoma [COAD] and 14 rectal adenocarcinoma [READ]) and 12 MSI were obtained from TCGA. Description of these data and mutation calling can be found in the Supplementary Note.
Mapping non-coding mutations to chromatin-looping interactions
We mapped non-coding mutations to CREs defined by promoter CHi-C generated on CRC cell lines, LoVo and HT29. We annotated only cis-interactions involving a promoter and CRE, excluding trans-interactions and promoter-promoter contacts.
Genome-wide analysis of non-coding mutations
Mutations were allocated to promoters or CREs, and the mutation rates for each fragment class were calculated. Mutation rates were determined as the number of mutations in each fragment class, divided by the size of all the fragments in that class, minus all regions overlapping ORFs, 3’ UTRs, 5’ UTRs and regions with poor mappability (Supplementary Note). Mutational signatures have however been shown to be critical for estimating purifying selection pressures in cancer somatic mutation data47. Taking this into account the expected mutation rates were estimated considering the sample-specific occurrence of mutations of each of the 96 substitution types defined by Alexandrov et al.18, and the trinucleotide composition of the fragment class.
where i is each of the possible mutation substitutions, ui is the genome-wide number of mutations of type i, vi is the genome-wide number of positions at which mutations of type i can occur, wi is the number of positions in the considered fragment class at which mutations of type i can occur, and y is the size of all fragments in the class. Excluded regions as defined above were not considered.
Integrated analysis of non-coding mutations in CREs
To identify potential non-coding driver mutations we integrated three driver discovery methods (Supplementary Fig. 7a). In this integrated analysis, we (i) assessed the transcriptional effects of non-coding mutations in CREs, and (ii) tested for an excess of non-coding mutations in CREs. P-values computed in these two analyses were adjusted for multiple testing using the Benjamini-Hochberg procedure and CREs excluded if Q≥0.05 in either analysis. Finally we (iii) assessed the clustering of non-coding mutations in the remaining CREs. P-values computed in this clustering analysis were adjusted for multiple testing using the Benjamini-Hochberg procedure and CREs excluded if Q≥0.05. These three driver discovery methods are outlined below.
Testing the transcriptional effects of non-coding mutations in CREs
Gene expression profiles were based on RNA sequencing of 12 MSI and 50 MSS CRC cases obtained from TCGA that had matched WGS data (normalised gene-level values, accessed 20 January 2017). Differences between samples analysed on Genome Analyzer and HiSeq were batch-corrected using the ComBat method48. To reduce spurious long distance contacts, interactions were filtered such that the distance between the promoter and CRE was <1Mb49. For each CRE, samples were divided between non-mutated and mutated based on the presence of non-coding mutations in the corresponding CRE. We tested for differential gene expression between mutated and non-mutated groups using a negative binomial model9, implemented in edgeR50. Samples with copy number alterations at either the gene or the related CRE were excluded9. CREs were not tested if gene expression data were not available for the interacting gene, or if the CRE was mutated in fewer than two samples, after the removal of samples with overlapping copy number alterations. CREs observed to interact with the promoters of multiple genes were tested multiple times. Only CREs interacting with protein-coding genes were tested.
Testing for an excess of non-coding mutations in CREs
We tested regions for an excess of non-coding mutations using a global approach, as per Weinhold et al9. This assumes that the observed number of tumour samples mutated in any specific region follows the binomial distribution, under the null hypothesis that mutations in the region are not under positive selection. As replication timing affects the nucleotide mutation rate, we estimated the mutation rate using data from HeLa, K562, HEPG2, MCF7 and SKNSH cell lines51. Again as per Weinhold et al.9, we subsetted the genome into 100kb bins and derived mean replication times for each cell line across each bin. For each interacting region, we then computed a single replication time for each cell line by taking the replication time of the 100kb bin in which the region is located, or the average replication time if the region spanned multiple 100kb bins. For each interacting region, we then identified the top 5% of interacting regions with the most similar replication times across cell lines, measured using the Euclidian distance between the vectors of times. The background nucleotide mutation rate qi could then be estimated by dividing the number of mutations in this 5% of interacting regions across all samples by the total number of samples with mutation data and the total effective length of the regions. These results are robust to the percentage of interacting regions considered (Supplementary Table 18). We excluded the areas of the interacting regions overlapping ORFs, 3’ UTRs, 5’ UTRs and areas with poor mappability (Supplementary Note). The removal of these areas is incorporated into the effective lengths of the regions. The estimated sample mutation rate si is dependent on the estimated nucleotide mutation rate qi of the region under the null hypothesis, and the effective size of the region Li:
Mutational excess P-values follow discreet distributions and we therefore computed randomised P-values for each region i using the right tail masses, as per Imielinski et al22:
where n is the total number of cancers, k the number of cancers with ≥ 1 mutation in region i and pi is the corresponding randomised P-value. Inflation factor estimation was conducted with the regression method implemented in the GenABEL R-package52. Using all P-values indicated that the statistics generated under this model are weakly inflated (λ100%=1.08; Supplementary Fig. 18).
Testing for clustering of non-coding mutations in CREs
We evaluated whether CRE mutations cluster using the weighted average proximity (WAP) method21:
where d is the linear genomic distance between mutations i and j, and t is a weighting constant. As per Rheinbay et al.21, we used t=6, as this reflects the typical size of the core of TF binding motifs. Statistical significance of each WAP score was determined by permuting mutation positions 10,000 times, whilst maintaining the size of the CRE and excluding regions overlapping ORFs, 3’ UTRs, 5’ UTRs and regions with poor mappability (Supplementary Note). Empirical P-values were calculated as the proportion of mutation permutations with WAP scores at least as great as the WAP score computed using the observed mutation positions.
Empirical FDR estimation
We estimate an empirical FDR for the identified CRE by comparing the number of results yielded by the integrated driver discovery analysis when using real and randomised CHi-C data (Supplementary Fig. 7b). CHi-C data were randomised by changing the HindIII fragments contacting each gene, by sampling from non-interacting fragments (CHiCAGO score <1) within 1Mb of the gene, whilst maintaining the number of fragment contacts. The integrated driver discovery analysis was then applied in full to each randomised CHi-C data set. The expected number of false discoveries was estimated as the mean number of CREs yielded by the integrated driver discovery analysis across 1,000 randomised CHi-C data sets. The empirical FDR was estimated as the ratio of false discoveries to the total number of CREs detected when the real CHi-C data were used53.
Relationship between gene expression and CNV at CREs
To identify CNVs overlapping CREs, we utilised Affymetrix Genome-Wide Human SNP Array 6.0 copy number data from the TCGA COAD study of 450 cancers and the READ study of 165 cancers. Focal deletions and amplifications were defined as abs(log2ratio) ≥0.354 and size <3Mb55. We analysed matched RSEM RNAseq and CNV data from the TCGA COAD and READ studies (n=606). To identify cancers with deleted or amplified CREs correlated with expression of an interacting gene, we applied the following filters to each CRE-promoter interaction: (i) we identified cancers with a focal amplification or deletion of the CRE; (ii) we excluded cancers with evidence of a CNV at the interacting gene (a GISTIC2 score ≠0); (iii) we restricted our analysis to CREs affected in ≥7 samples (representing 1% of samples). We compared gene expression between affected and unaffected samples using edgeR50 with default parameters. Samples were considered unaffected if there was no evidence of a CNV at the CRE (a GISTIC2 score =0). Genes that showed differential expression between affected and unaffected samples and an absolute correlation coefficient ≥0.4 between GISTIC2 score and gene expression value were taken forward for further analysis.
Relationship between CRE mutation and translocations and inversions
Since translocations and inversions can dysregulate gene expression we examined for translocations and inversions in the proximity of mutated CRE target genes. Translocation and inversion breakpoints, called using dRanger56, were downloaded from the International Cancer Genomic Consortium Data Portal (accessed 30 July 2017).
Genomic DNA extraction
Cells were collected, washed twice in PBS and genomic DNA was extracted using QIAamp DNA Blood Mini Kit (Qiagen). DNA concentration was measured using Qubit dsDNA BR Assay (ThermoFisher Scientific).
3C-PCR validation
3C was used to validate ETV1 and RASL11A interactions in a panel of colon derived MSS CRC cell lines. Two cell culture replicates of in situ 3C libraries were prepared using HT29, SW480, SW1116, Caco2, HT115 and SW948 cells. Cell pellets were cross-linked, digested with HindIII, and ligated using the same conditions described in the Hi-C section, excluding the biotin step. Digestion and ligation efficiency were assessed on agarose gel before proceeding to phenol-chloroform purification. Ligation primer pairs were designed to amplify ligation junctions between the promoter and interacting HindIII fragment (promoter-CRE) (Supplementary Table 19). Genomic DNA was used as control for the possibility that amplification across ligation junctions could be the result of structural variations. To prove fidelity of genomic DNA as template, primer pairs were also designed to amplify the genomic region around each of the ligation primers (Supplementary Table 19). All primers were designed using Primer3. Regions were amplified using Multiplex PCR Kit (Qiagen); 100ng template DNA amplified using the following procedure: initial 15 min denaturation at 95°C followed by 38 cycles of 95°C for 30 seconds, 60°C for 90 seconds, 72°C for 45 seconds. 5μl of each PCR reaction was visualised on 2% agarose gels stained with ethidium bromide. Identity of fragments visualised on agarose gels was confirmed by Sanger sequencing (Supplementary Fig. 9).
Plasmid construction and luciferase assays
The length of the full region spanning the six mutations is over 1.2kb and therefore a 1.6kb fragment would have been required to ensure that all mutations mapped centrally within the fragment. In view of this we decided to clone the two fragments separately, so as to have cloned inserts <1kb. A 946bp and a 738bp genomic region within the ETV1 CRE were amplified from human genomic DNA using primers detailed in Supplementary Table 19. Gel-purified PCR products (Qiagen) were A-tailed using 2U Thermoprime DNA polymerase (ThermoFisher Scientific) and 200μM dATP for 30 min at 70°C. The products were cloned into the PCR8/GW/TOPO vector and single bacterial colonies containing the vector were cultured and purified (Qiagen Mini-prep Kit). The six somatic mutations were generated with site-directed mutagenesis (SDM) (Agilent Quick Change XL kit) using primers detailed in Supplementary Table 19. SDM changes were confirmed by Sanger-sequencing. Regulatory regions with both non-mutated and mutated sequences were cloned into pGL3 luc2-promoter vector (Promega) using Gateway LR Clonase II technology (ThermoFisher Scientific). The reporter constructs were transfected into HT29 using Lipofectamine 2000 (ThermoFisher Scientific). Briefly, 7.5×105 cells were seeded and transfected the following day with 3μg reporter constructs and 150ng of internal control plasmid (pRL-TK). Transiently transfected cells were cultured for 24 hours, following which the luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega) as per manufacturer's recommendations. Firefly and Renilla luciferase luminescence were measured in triplicate on a Fluoroskan Ascent FL plate reader (ThermoFisher Scientific). The ratio of luminescence from the experimental reporter to the luminescence from the control reporter was calculated for each sample, defined as the relative luciferase activity.
CRISPR-Cas9-mediated enhancer disruption
Two set of guide RNAs (gRNAs) were designed to target the CRE interacting with the RASL11A promoter using E-CRISP57. To check that the required PAM sequence was not altered in the HT29 cell line, the region where the gRNAs were mapped was Sanger-sequenced using genomic DNA. Each set of gRNAs was designed to target upstream and downstream of the enhancer region, and cloned in one plasmid expressing the Cas9 gene and the RFP-marker. A control plasmid was generated containing all the elements of the targeting plasmids other than the gRNAs. Custom plasmids were obtained from ATUM. gRNA sequences are reported in Supplementary Table 19. HT29 were seeded at 7.5x105 density in 6-well plates and transfected the following day with 10μl of Lipofectamine 2000 (ThermoFisher Scientific) and 2μg of the respective plasmids. 48 hours post-transfection cells expressing RFP were selected and seeded into using FACSAriaIIμ cell sorter (BD Biosciences) in a 24-well plate. After 2-3 weeks cells were harvested to extract either genomic DNA to confirm the enhancer deletion, or total RNA to assess gene expression. Primers used to assess the deletion are listed in Supplementary Table 19.
Cell viability assays
In 96-well plates, cells were transfected with siRNAs and incubated with Cell Titer Glo reagent according to the manufacturer's protocol (Cell Titer Glo Luminescent Cell Viability Assay kit, Promega). For each experiment, luminescence was measured in triplicate at 0, 24, 48 and 72 hours post-transfection using a Fluoroskan Ascent FL plate reader (ThermoFisher Scientific) (Supplementary Fig. 16).
Real-time cell proliferation assays
After RNAi transfection, 96-well plates were introduced into an Incucyte Zoom imaging system (Essen BioScience) enclosed in an incubator at 37°C and 5% CO2 humidified air. Each well was imaged by phase contrast every four hours for 92 hours. Images were collected and the percentage of confluence determined using software provided by the manufacturer. Confluence was averaged over four fields of view per well. Data were normalised to the first scan to account for differences in seeding across individual samples and across replicates.
Statistical analyses
The observed and expected mutation rates in promoters and CREs were compared using two-sided paired Wilcoxon tests. CHi-C interactions involving promoters of active and inactive genes were compared using two-sided Wilcoxon tests8. For luciferase, cell viability and real-time cell proliferation assays, statistical significance was calculated using two-tailed t-tests over three independent experiments. The Benjamini-Hochberg procedure was used to adjust for multiple testing, unless otherwise specified.
Reporting summary
Further information on our experimental design is available in the Nature Research Reporting Summary document linked to this article.
Supplementary Material
Acknowledgements
This work was supported by grants from Cancer Research UK grant (C1298/A8362), the European Union Seventh Framework Programme (FP7/207–2013) under grant 258236 and FP7 collaborative project SYSCOL, all awarded to R.S.H. This publication is supported by COST Action BM1206. CIHR funded Epigenome Mapping Centre at McGill University (EP1-120608), awarded to T.P. We acknowledge the work of The Institute of Cancer Research Tumour Profiling Unit. The results published here are in part based upon data generated by The Cancer Genome Atlas established by the NCI and NHGRI. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Footnotes
URLs
SNPable, http://bit.ly/snpable; NCI Genomic Data Commons Data Portal, https://portal.gdc.cancer.gov/; UCSC Genome Browser, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwRepliSeq/; GTex portal, https://gtexportal.org/.
Data Availibility
Hi-C, CHi-C, and histone ChIPseq sequencing data have been deposited in the European Genome-phenome Archive (EGA) under the accession code EGAS00001001946. WGS, RNAseq, CNV and survival data for TCGA COAD and READ samples, and RNAseq data for HT29 and LoVo (CCLE program) were obtained from the NCI Genomic Data Commons Data Portal (see URLs). TF ChIPseq data were obtained from the Gene Expression Omnibus (GEO) (GSE49402). Survival data were obtained from GEO (GSE33113, GSE39582). Replication timing data were downloaded from the UCSC Genome Browser (see URLs). GTEx data (release v6) were obtained from the GTex portal (see URLs).
Author Contributions
G.O., P.J.L., R.S.H. conceived and designed the study; G.O. performed Hi-C and CHi-C experiments, luciferase assays, CRISPR experiments, cell viability and proliferation assays; G.O. and P.B. performed 3C validation; G.O., P.J.L., A.J.C., S.E.D., D.C. and K.L. performed bioinformatics; F.H. performed ChIPseq experiments; T.P. and J.T. contributed reagents and materials for the ChIPseq experiments; C.S.O. designed the capture baits; and G.O., P.J.L., A.J.C., S.E.D., D.C., P.B. and R.S.H wrote the manuscript with contributions from T.P., C.S.O. and J.T. All authors reviewed the final manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Zhang X, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet. 2016;48:176–82. doi: 10.1038/ng.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16:483–93. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 3.Kim K, et al. Chromatin structure-based prediction of recurrent noncoding mutations in cancer. Nat Genet. 2016;48:1321–1326. doi: 10.1038/ng.3682. [DOI] [PubMed] [Google Scholar]
- 4.Weischenfeldt J, et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet. 2017;49:65–74. doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto A, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–9. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 6.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47:710–6. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 9.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46:1160–5. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–63. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 11.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 13.Javierre BM, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384 e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jager R, et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun. 2015;6 doi: 10.1038/ncomms7178. 6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlando G, Kinnersley B, Houlston RS. Capture Hi-C Library Generation and Analysis to Detect Chromatin Interactions. Curr Protoc Hum Genet. 2018:e63. doi: 10.1002/cphg.63. [DOI] [PubMed] [Google Scholar]
- 17.Roadmap Epigenomics, C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katainen R, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–21. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 20.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041 e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rheinbay E, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imielinski M, Guo G, Meyerson M. Insertions and Deletions Target Lineage-Defining Genes in Human Cancers. Cell. 2017;168:460–472 e14. doi: 10.1016/j.cell.2016.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeon IS, et al. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–34. [PubMed] [Google Scholar]
- 24.Attard G, et al. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. 2008;99:314–20. doi: 10.1038/sj.bjc.6604472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–39. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 26.Jane-Valbuena J, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70:2075–84. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi P, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–53. doi: 10.1038/nature09409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran L, et al. Combined inhibition of MAP kinase and KIT signaling synergistically destabilizes ETV1 and suppresses GIST tumor growth. Cancer Discov. 2015;5:304–15. doi: 10.1158/2159-8290.CD-14-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulakovskiy IV, et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 2016;44:D116–25. doi: 10.1093/nar/gkv1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–4. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pistoni M, Verrecchia A, Doni M, Guccione E, Amati B. Chromatin association and regulation of rDNA transcription by the Ras-family protein RasL11a. EMBO J. 2010;29:1215–24. doi: 10.1038/emboj.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Sousa EMF, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9:476–85. doi: 10.1016/j.stem.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Marisa L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rands CM, Meader S, Ponting CP, Lunter G. 8.2% of the Human genome is constrained: variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014;10:e1004525. doi: 10.1371/journal.pgen.1004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer. 2017;17:337–351. doi: 10.1038/nrc.2017.20. [DOI] [PubMed] [Google Scholar]
- 39.Duensing A. Targeting ETV1 in gastrointestinal stromal tumors: tripping the circuit breaker in GIST? Cancer Discov. 2015;5:231–3. doi: 10.1158/2159-8290.CD-15-0116. [DOI] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingett S, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–97. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–13. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 47.Van den Eynden J, Larsson E. Mutational Signatures Are Critical for Proper Estimation of Purifying Selection Pressures in Cancer Somatic Mutation Data When Using the dN/dS Metric. Front Genet. 2017;8:74. doi: 10.3389/fgene.2017.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 49.Grubert F, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–65. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A. 2010;107:139–44. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 53.Carter H, et al. Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer. Cancer Discov. 2017;7:410–423. doi: 10.1158/2159-8290.CD-16-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Litchfield K, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun. 2015;6 doi: 10.1038/ncomms6973. 5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drier Y, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–35. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–3. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.