Abstract
Purpose
An estimated 10% of breast and ovarian cancers result from hereditary causes. Current testing guidelines for germ line susceptibility genes in patients with breast carcinoma were developed to identify carriers of BRCA1/2 variants and have evolved in the panel-testing era. We evaluated the capability of the National Comprehensive Cancer Network (NCCN) guidelines to identify patients with breast cancer with pathogenic variants in expanded panel testing.
Methods
An institutional review board–approved multicenter prospective registry was initiated with 20 community and academic sites experienced in cancer genetic testing and counseling. Eligibility criteria included patients with a previously or newly diagnosed breast cancer who had not undergone either single- or multigene testing. Consecutive patients 18 to 90 years of age were consented and underwent an 80-gene panel test. Health Insurance Portability and Accountability Act–compliant electronic case report forms collected information on patient demographics, diagnoses, phenotypes, and test results.
Results
More than 1,000 patients were enrolled, and data records for 959 patients were analyzed; 49.95% met NCCN criteria, and 50.05% did not. Overall, 8.65% of patients had a pathogenic/likely pathogenic (P/LP) variant. Of patients who met NCCN guidelines with test results, 9.39% had a P/LP variant. Of patients who did not meet guidelines, 7.9% had a P/LP variant. The difference in positive results between these groups was not statistically significant (Fisher’s exact test P = .4241).
Conclusion
Our results indicate that nearly half of patients with breast cancer with a P/LP variant with clinically actionable and/or management guidelines in development are missed by current testing guidelines. We recommend that all patients with a diagnosis of breast cancer undergo expanded panel testing.
INTRODUCTION
Approximately 330,000 patients are diagnosed with breast cancer every year in the United States.1 An estimated 10% of these cancers likely result from hereditary causes.2 Studies have estimated that less than 10% of all BRCA1 and BRCA2 carriers have been identified.3 Moreover, 50% to 80% of individuals at risk have not received genetic testing, in part because they do not meet the family history criteria of current testing guidelines,4,5 and insurance seldom reimburses testing in such cases. An estimated 35,000 patients with breast cancer have pathogenic BRCA1/2 variants; however, only 30% have been identified.3,4 A recent study of Medicare patients with breast cancer found no significant difference in the germ line pathogenic/likely pathogenic (P/LP) rate from multigene panel tests between patients who did and did not meet genetic testing guidelines.6
In addition to enhanced cancer screening, risk reduction, and surgical treatment, germ line genetic results are increasingly relevant to systemic therapy,7-9 and results in affected individuals also yield valuable information for cascade family variant testing and thus efficient discovery of unaffected carriers, facilitating prevention of breast and other cancers.
Recent studies have identified the clinical actionability associated with expanded multigene panels, and these management strategies are now an accepted part of clinical guidelines.6,10 For example, a study of 488 patients with breast cancer (stage I to III) at a single center yielded a P/LP variant in 10.7% of women, only 6.1% of whom were BRCA1/2.11 In other studies, up to 10% of women who tested negative for BRCA1/2 had P/LP variants in other cancer predisposition genes, providing additional information that might change management of these patients.12
National Comprehensive Cancer Network (NCCN) guidelines for genetic testing were established approximately 20 years ago to identify patients with the highest likelihood of carrying BRCA1/2 variants to reduce the number needed to test at a time when BRCA1/2 genetic testing cost $2,000 to $5,000 per test and in line with known management implications at the time. However, with the landmark Supreme Court case on BRCA1/2 in 2013, which overturned patents on naturally occurring genes, and with next-generation sequencing (NGS) spurring competition, the availability and cost of testing has dropped (as low as $250 per test13). Simultaneously management guidelines are developing rapidly and have been revised multiple times. However, testing guidelines have become more complicated and have not been sufficiently reevaluated in the panel-testing era. Testing guidelines remain limited for BRCA1/2, TP53, and PTEN. The use of multigene panels is pointing to additional genes beyond BRCA1/2 that may be implicated in breast cancer (and other cancers) and for which management guidelines have been proposed or are in development.
Accumulating evidence suggests that the rate of germ line pathogenic variants in the US population is higher than originally suspected and that BRCA1/2 mutation prevalence may be as high as one in 200.14 A recently published study in which the exomes of 50,000 patients were sequenced found that close to 50% of patients with BRCA1/2 variants did not meet published guidelines for clinical testing.14 A study by Buys et al15 of 35,000 patients with breast cancer who were tested with a 25-gene panel found a P/LP variant rate of 9.3%, with more than 50% of these variants in genes other than BRCA1/2; a study by Susswein et al16 reporting NGS testing of 10,000 patients—two thirds of whom had a cancer diagnosis—found a similar P/LP variant rate.
We created a cohort of patients with breast cancer seen in practice and who agreed to participate in largely community-based clinics using a multigene panel. The primary objective was to determine whether there was a difference in the incidence of actionable variants between patients who met 2017 NCCN testing guidelines and those who did not.
METHODS
Investigator Selection
An institutional review board (IRB)–approved multicenter prospective registry was initiated with 20 community and academic breast practices experienced in cancer genetic risk assessment and management. IRB approval and oversight were provided by Western Institutional Review Board (Puyallup, WA) or via a local IRB. Sites were selected with the aim of having the ethnicity of participants commensurate with US ethnicity demographics and in the interest of generating hereditary genetic testing data for traditionally underrepresented and underserved ethnic populations.
Participant Accrual
Patients were eligible to participate if they were 18 years of age or older, had a personal diagnosis of breast cancer, were either currently being treated or had previously been treated, and had not previously undergone either single- or multigene germ line testing. Patients were enrolled in two equal cohorts—those who met NCCN genetic testing guidelines17 and those who did not. Sites identified consecutive patients meeting enrollment criteria (breast cancer, no previous germ line genetic testing), with each site enrolling equivalent numbers of patients who met guidelines and who did not, per study design. Site clinicians classified patients as meeting or not meeting NCCN (version 2.2017) guidelines based on their responses to a multipart question developed with the assistance of a cancer genetic counselor. If a clinician answered no to this question for a particular patient, the patient did not meet guidelines for testing. Sites stopped enrolling when they met their individual enrollment goals. All enrolled patients consented to have their deidentified data included in the registry.
Genetic Testing
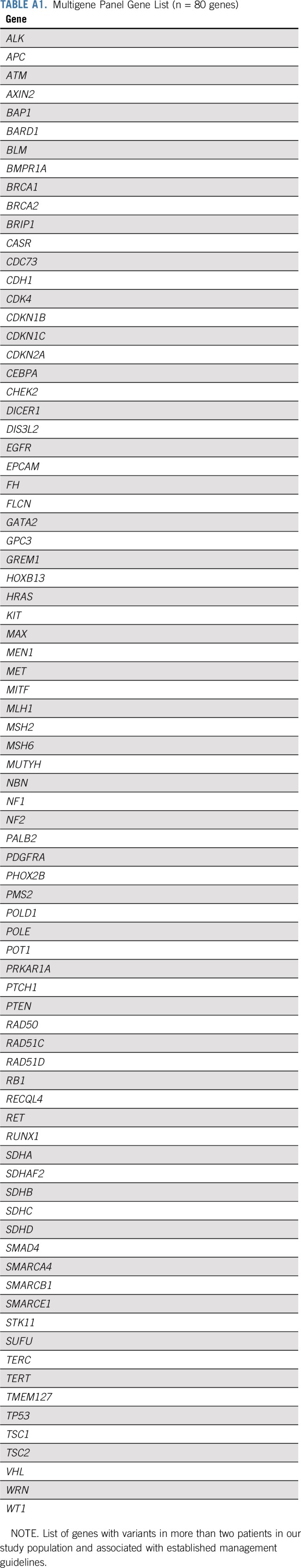
All patients underwent germ line genetic testing with a multicancer panel of 80 genes (Appendix Table A1, online only). Among these genes, 11 (BRCA1, BRCA2, ATM, CDH1, CHEK2, NBN, NF1, PALB2, PTEN, STK11, TP53) are referenced in NCCN management guidelines and commonly included in diagnostic breast cancer panels. Full-gene sequencing, deletion/duplication analysis, and variant interpretation were performed at Invitae (San Francisco, CA), as previously described.12,18
Test results were deidentified and recorded in a Health Insurance Portability and Accountability Act–compliant electronic study registry (Genae, Belgium), along with other relevant clinical data including demographic and clinical data and whether and how patients met 2017 NCCN criteria. All reported patients had their variant findings source verified and confirmed by independent review of the test results by a medical geneticist. Patients were informed of their test results, and physicians initiated appropriate management actions if indicated by current guidelines and as they saw fit.
Statistical Analysis
This noninferiority study was powered to detect a difference in P/LP variant rate of 4 percentage points between those meeting and not meeting testing guidelines with statistical significance (Fisher’s exact test P < .05). Participant characteristics and genetic testing results were tabulated, with descriptive statistics including medians, means, and standard deviations for continuous data and proportions with 95% CIs for categorical data. All P values are two tailed.
RESULTS
Participant Characteristics
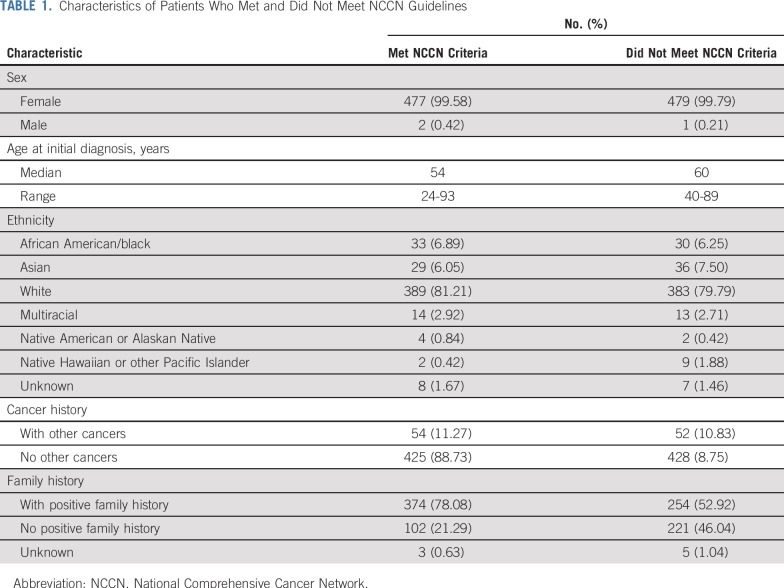
Participating sites enrolled 1,001 patients with breast cancer from April 2017 to September 2018. Forty-two patients were excluded from analysis because their data were incomplete or could not be source verified. A total of 959 patients meeting all study requirements and having all required data collected and available were included in our evaluation. Among the 959 unselected patients with breast cancer in this cohort, 479 (49.95%) met established 2017 NCCN germ line genetic testing guidelines, and 480 (50.05%) did not meet these guidelines for genetic testing. Age, sex, ethnicity, and personal and family cancer history information of the in-guideline and out-of-guideline patient groups are listed in Table 1. The cohort included 650 patients who were recently diagnosed (within 12 months of consent) and 309 who were not recently diagnosed. There were 106 patients with a previous cancer other than breast cancer.
TABLE 1.
Characteristics of Patients Who Met and Did Not Meet NCCN Guidelines
Pathogenic Variants in Cancer Genes
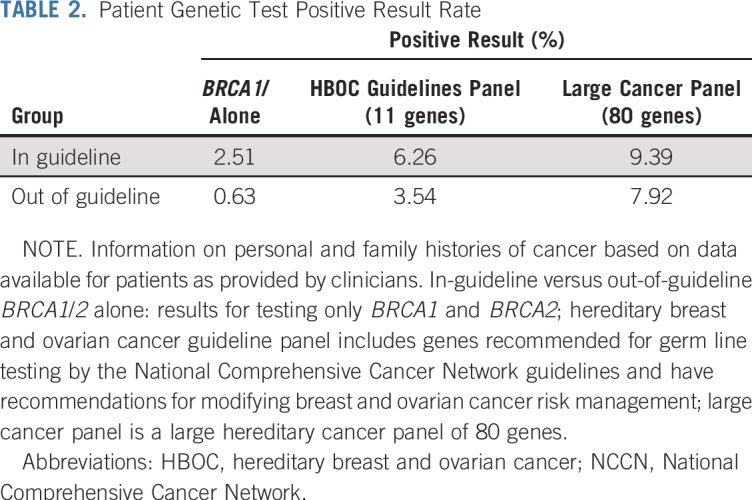
Overall, 83 (8.65%) of 959 patients had a P/LP variant. Of these, 45 (9.39%) of 479 patients who met NCCN testing guidelines and 38 (7.9%) of 480 patients who did not meet guidelines had a P/LP variant. The difference of positive cases between the two groups was not statistically significant (P = .4241). Overall, 47 patients (4.9%) had a P/LP variant if only an 11-gene breast cancer panel was considered, and only 15 patients (1.56%) had a P/LP variant if only BRCA1/2 mutation were considered (Table 2). When only results from BRCA1 and BRCA2 testing were considered, the positive rate of the in-guideline group was four-fold that of the out-of-guideline group (2.51% v 0.63%; P = .0201; Table 2). Variant of uncertain significance (VUS) rates were virtually identical between the two groups. The overall VUS rate was 54.22% for the entire patient population. Accordingly, almost half of the patients with breast cancer tested had either a P/LP variant or a completely negative test with no P/LP variant or VUS found in 80 genes.
TABLE 2.
Patient Genetic Test Positive Result Rate
Clinical Implications of P/LP Variants
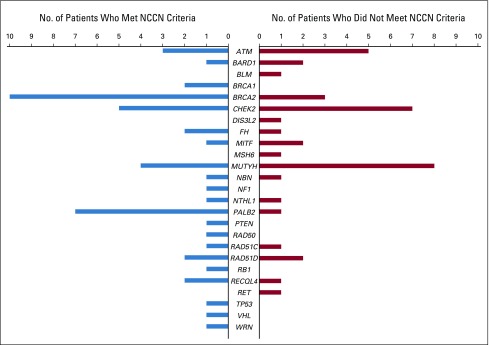
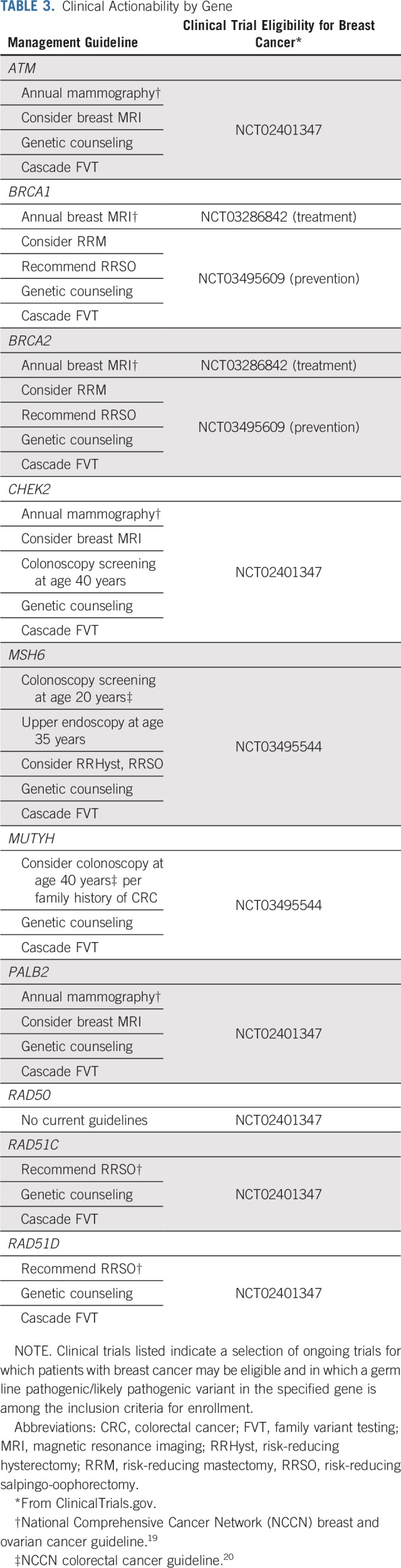
The P/LP variants identified for both patient groups occurred in genes associated specifically with breast cancer, as well as in known cancer genes traditionally associated with other hereditary cancers (Fig 1). The spectrum of P/LP variants differed somewhat between the two groups, with some overlap. The yield of P/LP variants from the 80-gene panel was greater than that with an 11-gene panel (8.65% v 4.90%), and with few exceptions, the genes in which P/LP variants were identified have published management recommendations and are clinically actionable for treatment of a patient’s cancer, post-treatment surveillance and prophylaxis, or cascade testing for at-risk family members (Table 3).
FIG 1.
Pathogenic/likely pathogenic variants identified by gene. Includes BRCA1 and BRCA2 and other genes associated with breast cancer, genes associated with breast and/or gynecologic cancers, and genes associated with other cancers. NCCN, National Comprehensive Cancer Network.
TABLE 3.
Clinical Actionability by Gene
DISCUSSION
The rate of P/LP variants in a large 80-gene panel was similar among patients with breast cancer who did and did not meet 2017 NCCN guidelines for genetic testing; the difference was not statistically significant. In fact, the results of our study suggest that a strategy that simply tests all patients with a personal history of breast cancer would almost double the number of patients identified as having a clinically actionable genetic test result.
Advances in NGS technologies have dramatically reduced the cost of BRCA1/2 testing and enabled simultaneous sequencing and deletion/duplication testing of BRCA1/2 concomitantly with dozens of additional risk genes for breast, ovarian, and/or other cancers (eg, PALB2, PTEN, ATM, CHEK2). Furthermore, current testing guidelines do not adequately account for the full range of clinical presentations described to date as associated with breast cancer, and carriers of clinically actionable variants in genes other than BRCA1/2 are likely to fall outside of the current guidelines.
The rate of BRCA1/2 P/LP variants we observed was lower among both groups of patients with breast cancer than the 6% to 9% rate estimates of previous reports. This is not surprising for our in-guideline patient cohort, because this registry specifically excluded patients who had previously been tested. Previously diagnosed patients at our research sites with clearly identifiable personal and family histories consistent with NCCN testing guidelines (ie, BRCA1/2 positive) were likely to have already undergone testing and therefore would have been excluded from this study. Our study population was tested using a comprehensive multigene panel strategy, which identified a substantial number of patients with P/LP variants in genes such as PALB2, ATM, CHEK2, MSH6, MUTYH, RAD50, RAD51C, and RAD51D that would have been missed by a restrictive testing strategy (Fig 1). Furthermore, of the patients with P/LP variants who did not meet NCCN germ line testing guidelines, 56% were potentially eligible for precision therapeutic clinical treatment trials, 76% for established clinical management recommendations, and 82% overall for clinical treatment trials and/or established management recommendations based in part on their germ line test results.
These management implications are invaluable to clinicians caring for these patients, including breast specialists who observe their patients over many years and are in position to test and counsel patients and make recommendations for next steps; this is critical as evidence emerges on cancer genes, guidelines change, and information about VUSs evolves. This proposed expansion of testing will require surgeons and other physicians who order testing to be comfortable either counseling patients appropriately about their risk profiles or referring them to genetic counselors.
The rate of VUSs observed in our study is not surprising given the number of genes included in the panel. Given the lack of definitive clinical significance for these variants, participating physicians counseled their patients not to be concerned about VUSs, and they did not use VUSs to alter patient management with regard to cancer treatment or the testing of family members. This does represent a future opportunity to have patients return to be counseled as evidence emerges and supports the reclassification of an identified VUS. This requires that diagnostic laboratories provide VUS updates to clinicians over time, which is already the practice of some commercial laboratories. It also necessitates that clinicians stay current with management guidelines and access reliable information resources to implement these updates effectively for their patients (eg, cancer genetic counselors, resources such as ASK2me21).
The results of this study suggest that a substantial modification of the scope and intent of existing genetic testing guidelines is critically overdue. We conclude that guidelines should be expanded immediately to include genetic testing of all patients with breast cancer. This conclusion is not as radical as it may seem. Our proposal is analogous to published studies and recommendations supporting universal germ line genetic testing of patients with pancreatic or metastatic prostate cancer, which have been incorporated into NCCN guidelines, as well as recently published data calling for tumor sequencing of all colorectal cancers.22-24 Furthermore, geneticist Mary Claire King has suggested that all women older than 30 years of age have BRCA1/2 testing.24a
Testing would facilitate informed decision making for patients with breast cancer and identify family members through cascade testing before they develop cancer, thereby activating surveillance and risk-reduction options. Any conversation about universal germ line genetic testing for all patients with breast cancer needs to acknowledge the implementation burden such testing would create. Because of the persisting shortage of genetic professionals, consideration and research must be devoted to developing novel genetics service delivery models for pretest counseling, delivery of the associated volume of test results, and coordination of appropriate specialty follow-up as indicated. These are issues with which the American Society of Breast Surgeons, the National Society of Genetic Counselors, the American College of Medical Genetics and Genomics, and independent researchers are actively engaged. For example, a variety of models for delivering genetic counseling with improved access have been proposed, including novel genetic counselor extender models.25-31
The scope of this study did not allow for patient-reported outcomes to document patient experiences in receiving test results, which is a limitation. These are important aspects of testing and merit additional research. Determination of the economic implications and cost effectiveness of germ line genetic testing in patients with breast cancer, including testing under broadened criteria, was also outside the scope of this study but warrants additional study and dedicated research efforts. Studies investigating these crucial issues have already generated evidence supporting the cost effectiveness of germ line genetic testing and cascade family variant testing for patients with breast cancer,32,33 and additional research is needed.
Although the additional cost of germ line testing for all patients with breast cancer is not negligible, it is approximately 10 times less than it was at the time the genetic testing guidelines were originally established, and the cost will continue to decrease. Naturally, this cost needs to be weighed in the context of current practices considered standard of care. For example, insurers currently pay for mammography/three-dimensional mammograms (current standard of care regionally in the United States) and routine population screening using advanced lipid panels that are repeated yearly. By contrast, hereditary testing is a one-time cost with information that may be valuable over the lifetime of a patient—a value likely to increase over time commensurate with the ever-advancing understanding of medical genetics by the medical community. Furthermore, the finite cost of genetic testing for all patients with breast cancer must be considered against the potentially dramatic cost of such patients undergoing prolonged/failed treatments because genetic information necessary to apply the most effective treatment/clinical trial was not generated. It must also be considered in the context of weighing the opportunity for preventive management, including screening patients with breast cancer for other cancers for which they have identified genetic risk, against the high cost of treating the catastrophic presentation of a late-stage malignancy in patients with a genetic risk that was not identified. Additionally, the opportunity for true prevention, which is cost effective, may be afforded to affected patients’ family members through cascade testing and subsequent risk-reduction decisions.
The availability of germ line genetic information for the routine clinical care of patients with cancer is one of the most significant opportunities to revolutionize the practice of medicine today. Genetic information expands clinicians’ ability to apply precision treatments and personalize risk assessment and management options for patients and their families. Key to accomplishing this is increasing access to germ line genetic testing. As the cost of testing falls, it becomes more accessible to all patients. Current testing guidelines represent another barrier that, if relaxed, would increase access to genetic testing, particularly as more insurance companies cover the delivery of this clinically vital information and commercial laboratories offer testing programs to facilitate access for Medicare recipients, low-income patients, and other underserved populations. Although broader clinical implementation of genetic testing requires additional clinical education to inform patient management, it also holds the promise of greater access to targeted interventions and increased cost effectiveness from interventional and preventive care tailored to individuals and their families. Universal genetic testing of patients with breast cancer, including currently underserved populations, with a comprehensive gene panel has the potential to reveal the full component that genetics play in breast cancer development regardless of the age or family history of the patient.
Expanded panel testing can provide information that may present additional treatment and follow-up options, including clinical trials, for all patients with breast cancer, not just those who meet testing guidelines.
Additional research is needed to quantify the impact on clinical outcomes when all patients with a personal history of breast cancer are eligible to undergo genetic testing (ie, elimination of additional guidelines), including multigene panel testing; however, our results suggest that approximately 45% of patients with breast cancer with clinically actionable germ line variants are being missed when testing is restricted to patients meeting current NCCN guidelines and when testing strategies are limited to panels containing only BRCA1/2 or to less comprehensive panels. We propose that testing criteria be expanded to include all patients with breast cancer.
ACKNOWLEDGMENT
We thank Nancy Jacoby for manuscript editing assistance, Chloe Wernecke (TME) for database management, and Carole Leung and Alla Zarifyan for graphic design assistance. Genetic counselor Erin O'Leary developed a multipart questionnaire to help the investigators determine whether patients met NCCN guidelines
Appendix
TABLE A1.
Multigene Panel Gene List (n = 80 genes)
Footnotes
Processed as a Rapid Communication manuscript.
See accompanying Editorial on page 445
AUTHOR CONTRIBUTIONS
Conception and design: Peter D. Beitsch, Pat W. Whitworth, Rakesh Patel, Barry Rosen, Paul Baron, Cynara Coomer, Dennis R. Holmes, Patricia Clark, Shan Yang, Ed D. Esplin, Robert L. Nussbaum
Administrative support: Cynara Coomer, Patricia Clark, Robert L. Nussbaum
Provision of study material or patients: Peter D. Beitsch, Rakesh Patel, Barry Rosen, Gia Compagnoni, Rache Simmons, Linda Ann Smith, Karen Barbosa, Samuel Lyons, Sadia Khan, Heather MacDonald, Lisa Curcio
Collection and assembly of data: Peter D. Beitsch, Pat W. Whitworth, Rakesh Patel, Barry Rosen, Gia Compagnoni, Paul Baron, Rache Simmons, Linda Ann Smith, Ian Grady, Michael Kinney, Cynara Coomer, Karen Barbosa, Eric Brown, Linsey Gold, Patricia Clark, Lee Riley, Samuel Lyons, Antonio Ruiz, Heather MacDonald, Lisa Curcio, Mary Kay Hardwick, Shan Yang, Robert L. Nussbaum
Data analysis and interpretation: Peter D. Beitsch, Pat W. Whitworth, Kevin Hughes, Rakesh Patel, Barry Rosen, Paul Baron, Cynara Coomer, Patricia Clark, Antonio Ruiz, Sadia Khan, Mary Kay Hardwick, Shan Yang, Ed D. Esplin, Robert L. Nussbaum
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST AND DATA AVAILABILITY STATEMENT
Underdiagnosis of Hereditary Breast Cancer: Are Genetic Testing Guidelines a Tool or an Obstacle?
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Peter D. Beitsch
Stock and Other Ownership Interests: Targeted Medical Education
Pat W. Whitworth
Stock and Other Ownership Interests: Reverse Medical (I), Rebound Medical (I), Lazarus (I), Cerebrotech (I), Targeted Medical Education
Honoraria: Medtronic
Consulting or Advisory Role: Medtronic, Lumicell, ImpediMed
Research Funding: Invitae, Intact Medical
Travel, Accommodations, Expenses: ImpediMed
Kevin Hughes
Stock and Other Ownership Interests: Hughes riskApps
Honoraria: Focal Therapeutics, 23andMe
Consulting or Advisory Role: Health Beacons
Barry Rosen
Speakers’ Bureau: Cianna, Zeiss, Hologic
Paul Baron
Honoraria: Myriad Genetics
Speakers’ Bureau: Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Ian Grady
Honoraria: GE Healthcare, Bard Medical
Speakers’ Bureau: GE Healthcare, Bard Medical
Research Funding: GE Healthcare
Michael Kinney
Stock and Other Ownership Interests: Cianna Medical
Consulting or Advisory Role: CMR Naviscan
Karen Barbosa
Stock and Other Ownership Interests: Genomic Health
Consulting or Advisory Role: Lumicell
Travel, Accommodations, Expenses: Allergan
Eric Brown
Research Funding: Invitae
Patricia Clark
Honoraria: Medtronic
Consulting or Advisory Role: Lumicell, Medtronic
Research Funding: Targeted Medical Education
Lisa Curcio
Employment: Agendia, bioTheranostics, Ice Cure
Honoraria: Ice Cure
Consulting or Advisory Role: Ice Cure, bioTheranostics, Agendia
Research Funding: Ice Cure
Expert Testimony: NorthGauge, MD Review
Travel, Accommodations, Expenses: Ice Cure
Mary Kay Hardwick
Honoraria: Provista
Travel, Accommodations, Expenses: Provista
Shan Yang
Employment: Invitae
Stock and Other Ownership Interests: Invitae
Travel, Accommodations, Expenses: Invitae
Ed D. Esplin
Employment: Invitae
Stock and Other Ownership Interests: Invitae
Robert L. Nussbaum
Employment: Invitae
Leadership: Invitae
Stock and Other Ownership Interests: Invitae
Honoraria: Third Rock Ventures
Consulting or Advisory Role: Pfizer, Genome Medical
Patents, Royalties, Other Intellectual Property: Royalties from National Institutes of Health and the University of California for mouse model
Travel, Accommodations, Expenses: Pfizer
No other potential conflicts of interest were reported.
REFERENCES
- 1. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2017-2018.pdf American Cancer Society: Breast Cancer Facts & Figures 2017-2018.
- 2.Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drohan B, Roche CA, Cusack JC, Jr, et al. Hereditary breast and ovarian cancer and other hereditary syndromes: Using technology to identify carriers. Ann Surg Oncol. 2012;19:1732–1737. doi: 10.1245/s10434-012-2257-y. [DOI] [PubMed] [Google Scholar]
- 4.Levy-Lahad E, Lahad A, King M-C. Precision medicine meets public health: Population screening for BRCA1 and BRCA2. J Natl Cancer Inst. 2014;107:420. doi: 10.1093/jnci/dju420. [DOI] [PubMed] [Google Scholar]
- 5.Childers CP, Childers KK, Maggard-Gibbons M, et al. National estimates of genetic testing in women with a history of breast or ovarian cancer. J Clin Oncol. 2017;35:3800–3806. doi: 10.1200/JCO.2017.73.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Leary E, Iacoboni D, Holle J, et al. Expanded gene panel use for women with breast cancer: Identification and intervention beyond breast cancer risk. Ann Surg Oncol. 2017;24:3060–3066. doi: 10.1245/s10434-017-5963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo F, Hirth JM, Lin YL, et al. Use of BRCA mutation test in the US, 2004-2014. Am J Prev Med. 2017;52:702–709. doi: 10.1016/j.amepre.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans DGR, Ingham SL, Baildam A, et al. Contralateral mastectomy improves survival in women with BRCA1/2-associated breast cancer. Breast Cancer Res Treat. 2013;140:135–142. doi: 10.1007/s10549-013-2583-1. [DOI] [PubMed] [Google Scholar]
- 9.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Axilbund JE, O’Leary E, et al. Underdiagnosis of hereditary breast and ovarian cancer in Medicare patients: Genetic testing criteria miss the mark. Ann Surg Oncol. 2018;25:2925–2931. doi: 10.1245/s10434-018-6621-4. [DOI] [PubMed] [Google Scholar]
- 11.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32:2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. https://www.ncbi.nlm.nih.gov/gtr Genetic Testing Registry. [Google Scholar]
- 14.Manickam K, Buchanan AH, Schwartz MLB, et al. Exome sequencing–based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open. 2018;1:e182140. doi: 10.1001/jamanetworkopen.2018.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buys SS, Sandbach JF, Gammon A, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 16.Susswein LR, Marshall ML, Nusbaum R, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med. 2016;18:823–832. doi: 10.1038/gim.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Comprehensive Cancer Network https://www.nccn.org/professionals/physician_gls/default.aspx NCCN clinical practice guidelines in oncology, genetic/familial high-risk assessment: Breast and ovarian (version 2.2017)
- 18.Nykamp K, Anderson M, Powers M, et al. Sherloc: A comprehensive refinement of the ACMG-AMP variant classification criteria. Genet Med. 2017;19:1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Comprehensive Cancer Network https://www.nccn.org/professionals/physician_gls/default.aspx : NCCN clinical practice guidelines in oncology, genetic/familial high-risk assessment: Breast and ovarian (version 2.2019).
- 20.National Comprehensive Cancer Network doi: 10.6004/jnccn.2016.0108. https://www.nccn.org/professionals/physician_gls/default.aspx : NCCN clinical practice guidelines in oncology, genetic/familial high-risk assessment: Colorectal (version 1.2018). [DOI] [PubMed]
- 21.ASK2ME https://ask2me.org : All Syndromes Known to Man Evaluator.
- 22.Hampel H, Pearlman R, Beightol M, et al. Assessment of tumor sequencing as a replacement for Lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol. 2018;4:806–813. doi: 10.1001/jamaoncol.2018.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shindo K, Yu J, Suenaga M, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol. 2017;35:3382–3390. doi: 10.1200/JCO.2017.72.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Helwick C. Dr. Mary-Claire King proposes population screening in all young women for BRCA mutations. The ASCO Post, 2015. http://www.ascopost.com/issues/february-10-2015/dr-mary-claire-king-proposes-population-screening-in-all-young-women-for-brca-mutations/ [Google Scholar]
- 25.McCuaig JM, Stockley TL, Shaw P, et al. Evolution of genetic assessment for BRCA-associated gynaecologic malignancies: A Canadian multisociety roadmap. J Med Genet. 2018;55:571–577. doi: 10.1136/jmedgenet-2018-105472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen SA, Marvin ML, Riley BD, et al. Identification of genetic counseling service delivery models in practice: A report from the NSGC Service Delivery Model Task Force. J Genet Couns. 2013;22:411–421. doi: 10.1007/s10897-013-9588-0. [DOI] [PubMed] [Google Scholar]
- 27.Wham D, Vu T, Chan-Smutko G, et al. Assessment of clinical practices among cancer genetic counselors. Fam Cancer. 2010;9:459–468. doi: 10.1007/s10689-010-9326-9. [DOI] [PubMed] [Google Scholar]
- 28.Cohen SA, Nixon DM. A collaborative approach to cancer risk assessment services using genetic counselor extenders in a multi-system community hospital. Breast Cancer Res Treat. 2016;159:527–534. doi: 10.1007/s10549-016-3964-z. [DOI] [PubMed] [Google Scholar]
- 29.Cohen SA, Gustafson SL, Marvin ML, et al. Report from the National Society of Genetic Counselors service delivery model task force: A proposal to define models, components, and modes of referral. J Genet Couns. 2012;21:645–651. doi: 10.1007/s10897-012-9505-y. [DOI] [PubMed] [Google Scholar]
- 30.Bernhardt BA, Geller G, Doksum T, et al. Evaluation of nurses and genetic counselors as providers of education about breast cancer susceptibility testing. Oncol Nurs Forum. 2000;27:33–39. [PubMed] [Google Scholar]
- 31.Gronwald J, Huzarski T, Byrski T, et al. Direct-to-patient BRCA1 testing: The Twoj Styl experience. Breast Cancer Res Treat. 2006;100:239–245. doi: 10.1007/s10549-006-9261-5. [DOI] [PubMed] [Google Scholar]
- 32.Patel S, Legood R, Evans DG, et al. Cost effectiveness of population based BRCA1 founder mutation testing in Sephardi Jewish women. Am J Obstet Gynecol. 2018;218:431.e1–431.e12. doi: 10.1016/j.ajog.2017.12.221. [DOI] [PubMed] [Google Scholar]
- 33.Tuffaha HW, Mitchell A, Ward RL, et al. Cost-effectiveness analysis of germ-line BRCA testing in women with breast cancer and cascade testing in family members of mutation carriers. Genet Med. 2018;20:985–994. doi: 10.1038/gim.2017.231. [DOI] [PubMed] [Google Scholar]