Abstract
The Ca2+-dependent human S100A4 (Mts1) protein is part of the S100 family. Here, we studied the interactions of S100A4 with S100A1 using nuclear magnetic resonance (NMR) spectroscopy. We used the chemical shift perturbed residues from HSQC to model S100A4 and S100A1 complex with HADDOCK software. We observed that S100A1 and the RAGE V domain have an analogous binding area in S100A4. We discovered that S100A4 acts as an antagonist among the RAGE V domain and S100A1, which inhibits tumorigenesis and cell proliferation. We used a WST-1 assay to examine the bioactivity of S100A1 and S100A4. This study could possibly be beneficial for evaluating new proteins for the treatment of diseases.
1. Introduction
The family of human S100 proteins are Ca2+-dependent, slightly acidic proteins including more than 20 family members with molecular weights of 9−13 kDa in vertebrates [1]. S100 proteins are used as a biomarker to identify the malignant tumor, has been found repeatedly in human diseases and some of them have been proposed as medical targets or predictors of therapeutic response" or "predictive biomarkers [2–5]. Interactions of S100 protein play a role in the regulation of enzyme action, cell development and discrimination; many S100 proteins exhibit chemotactic and neurotrophic activities [3,6,7]. S100 proteins are known to be possible markers of various cancers such as breast and colorectal cancer pancreatic, thyroid, gastric bladder, and melanoma [3]. It is also found as a bunch on the human chromosome 1q21 [8]. The family of EF-hand Ca2+-binding proteins is familiar to science, but intracellular Ca2+ mediates signals in an unknown fashion [9,10]. The S100 family has hydrophobic residues that facilitates interactions of the protein [11–13].
The S100A4 protein is a part of the S100 superfamily, which includes the leading EF-hand Ca2+-binding proteins and regulates many proteins engaged in various cellular functions such as apoptosis, differentiation, proliferation, two-calcium ion (Ca2+) homeostasis, and energy metabolism [14–16]. The S100 superfamily controls a large variety of essential cellular developments via protein-protein interaction [9]. EF-hand motif calcium binding initiates the action of the S100 proteins with structural changes and allows them to interact via selectivity [17,18]. The S100A4 protein was first deduced from stromas and tumors. In solution, the S100A4 protein takes the form of a homo-dimeric and acts as a metastasis-supporting protein [19,20]. The presence of S100A4 has now been demonstrated in cancers (e.g., pancreatic gastric, colorectal, bladder, and breast). The S100A4 protein acts as a part of angiogenesis and tumor establishment [19–21]. The EF-hand hinge area and the C terminus of the Mts1 protein are specifically related to another S100 protein. However, the majority of S100 proteins are related to target protein-protein binding. Calcium ion binding results in conformational changes in proteins to expose the hydrophobic pocket in helices 3 and 5 of the C-terminal EF-hand and the hinge region [22–24].
S100A1 is a part of the S100 family—it is expressed the most in cardiomyocytes [25]. S100A1 has been noted in the heart, brain, skin, ovaries, thyroid gland, breasts, salivary glands, skeletal muscles, and kidneys. It is the source of various endometrial cancers such as melanoma, breast, thyroid, renal, endometrioid, and it is responsible for neurodegenerative disorders [25,26,35–39,27–34]. Due to the helix 3 and 4 conformation, S100A1 creates a large hydrophobic region between this helix, and many Ca2+-dependent target protein interactions take place in this region [40]. Previously studies have demonstrated the interaction of S100A1 with other proteins such as ATP2A2, RyR1, TRPM3, RyR2, and RAGE [41–46]. The conformational changes or activities of S100A1 support particular physiological roles. The S100A1 protein plays a crucial role in gene therapies, and it was recently used in human clinical trials related to heart failure [47].
Interaction between the S100 protein have in reported which have the ability to form the hetero- and homo-dimers [1,48]. In this report we have found that the interactions site of S100A1 and S100A4 on the molecular level, this binding is also reported in vitro using gel overlay, yeast two-hybrid system and affinity column chromatography [49,50]. We also studied the S100A4 as an inhibitor—it blocking the interface of the V domain and S100A1 [51] to stop the cell proliferation [52] and could be used as the treatment of cancer [53] and RAGE related disease [54–57]. We also used the WST-1 assays, suggest that the inhibition properties arise from the bioactivity of S100A4 [58]. We also present putative models of the S100A4-S100A1 complex. This study could possibly be beneficial for the development of new proteins useful for the treatment of diseases.
2. Materials and methods
2.1 Materials
Ninety-nine percent 15NH4Cl (15N-ammonium chloride) and 99% D2O (isotopic-labeled deuterium oxide) were purchased from Cambridge Isotope Laboratories. All of the buffers and solutions were prepared using milli-Q water. The buffers for the NMR spectroscopy sample were filtered using a 0.22-μm antiseptic filter. The SW480 assay cells were bought from CCL-288 (American Type Culture Collection).
2.2 Expression procedures of S100A4 and S100A1
The S100A4 protein includes four free cysteine residues. We used dithiothreitol (DTT) as a reductant in the buffer for the NMR experiments. The cDNA of the S100A4 was obtained from Mission Biotech Company. The S100A4 protein contains the vector pET21b, and E. coli was used for the transformed and over-expressed in the host cell BL21 (DE3). M9 medium was used for bacterial growth, and 15NH4Cl was the source of 15N labelled. The cultures were grown at a temperature 37°C until they obtained an optical density (OD) of 0.75–1.00. 1.0 mM of IPTG used to induce the culture, which was grown at 25°C for 15–18 h. The cells were lysed and harvested using the buffer composition (300 mM KCl, 1 mM DTT, 2 mM CaCl2, 20 mM Tris, 1 M (NH4)2SO4 and 1 mM EDTA at pH 7.5). Sample was sonicated again for 30 min and centrifuged at 11000 rpm. S100A4 was obtained in soluble form, and purification was carried out using a Hi-Prep 16/60 Phenyl FF column with hydrophobic interaction chromatography (HIC, GE Healthcare). The HIC column was rinsed with buffer-1 containing 300 mM KCl, 1 mM DTT, 2 mM CaCl2, 20 mM Tris, 1 M (NH4)2SO4 and 1 mM EDTA at pH 7.5 and then washed with buffer-2 containing 1 mM EDTA, 20 mM Tris, 300 2 mM CaCl2, mM KCl, and 1 mM DTT at pH 7.5. Finally, it was eluted with a buffer containing 10 mM EDTA, 20 mM Tris, and 1 mM DTT at pH 7.5. The S100A4 portion was placed in the buffer with 20 mM Tris-HCl, 1 mM DTT and 0.02% NaN3 at pH 7.5 and was purified with a Q-Sepharose (XL 16/10) column on a AKTA FPLC system. S100A4 was eluted by a gradient with a buffer containing 1 mM DTT, 20 mM Tris-HCl, 0.02% NaN3 and 1.5M NaCl at pH 7.5. The S100A4 protein was exchanged with an NMR buffer containing 8 mM NaCl, 0.1 mM EDTA, 16 mM Tris, 6 mM CaCl2, and 0.34 mM NaN3 at pH 6.0 using a Millipore centrifuge tube. The purity of S100A4 was confirmed using SDS-PAGE (S1 Fig).
The cDNA of S100A1 (1–93 amino acids) was inserted into XhoI or NdeI restriction sites. The expression vector pET20b was used for cloning, and host cell Rosetta (DE3) was used for the transformation and expression. Details of the purification of the S100A1 protein have been reported [51]. The purity of S100A1 was identified using SDS-PAGE (S2 Fig). These two purified protein S100A4 and S100A1 was dissolved in 8M urea separately to make them to be monomer. Then we mixed them together to generate mixture of S100A1 dimer, S100A4 dimer and heterodimer S100A4-S100A1. At the end we exchange with the NMR buffer for the experiments.
2.3 1H-15N HSQC NMR titration
A Varian 700 MHz NMR spectrometer was used for the HSQC titrations at 25°C. All of the samples for the titration contained the same buffer at pH 6.0: 0.1 mM EDTA, 8 mM NaCl, 6 mM CaCl2, 16 mM Tris, and 0.34 mM NaN3. The HSQC titrations were carried out by adding S100A1 to the 15N S100A4 in proportions of 1:0 and 1:1 molar. A second titration was also performed with 15N labeled S100A1 by adding S100A4 in proportions of 1:0 and 1:1 molar. The NMR HSQC spectra were purposely overlapped to decide whether the intensities should be decreased or the cross peaks shifted.
2.4 Docking study of S100A4 and S100A1 (HADDOCK)
The HADDOCK program (version 2.2) [59–61] was used to create the structure of the S100A4-S100A1 complex. The structures of S100A4 and S100A1 were selected from the PDB (ID: 2MRD, 2LP3, separately). The docking study and successive refinement were carried out using a number of ambiguous-interaction restraints (AIRs) and residues that represented at the HSQC spectra, peaks with decreased intensities or considerable chemical shifts [51]. The reported S100A4-S100A1 complex was selected from the cluster with the lowest energy, and the complex was illustrated and displayed using software called PyMOL [62].
2.5 Dissociation constant (Kd) measurements based on fluorescence
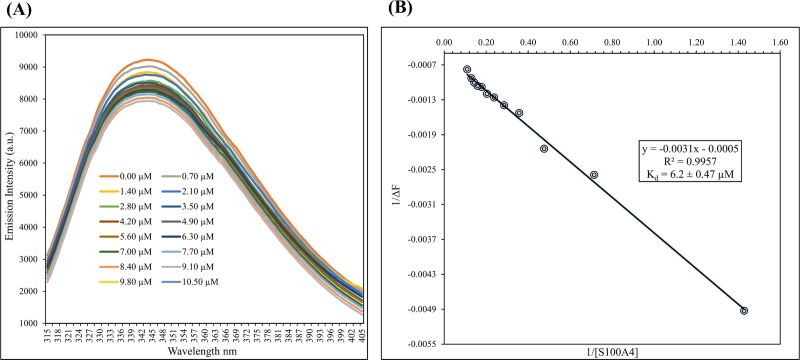
For binding constant measurement of protein-ligand and protein-protein interaction, fluorescence is a widely used method [63,64]. A F-2500 fluoro-spectrophotometer (Hitachi) was used for the experiments. In S100A4, there is no tryptophan. The tryptophan residue in the S100A1 protein excites and causes the protein to fluoresce. For S100A1, the excitation frequency of the tryptophan absorption band was found at 295 nm, and wavelength of 295 nm was used for the excitation. At a wavelength of 344 nm, S100A1 exhibits an emission band. Over the range of 315–405 nm, we noted the emission wavelengths. For the fluorescence measurements, we used a buffer containing 0.1 mM EDTA, 8 mM NaCl, 0.34 mM NaN3, 6 mM CaCl2, and 16 mM Tris at pH 6.0. The S100A4 concentrations were increased from 0 to 10.50 μM in increments of roughly 0.70 μM to the S100A1 which consist of 3.50 μM concentration. Variation in emission spectra was observed as the S100A4 concentration was increased in the complex solution. The curve was fitted and the following equations [65] were used to calculate the Kd of the S100A1-S100A4 complex [66].
| Eq (1) |
| Eq (2) |
In Eq 1, I0 refers to the intensity of the free S100A4 in the solution; I and I1 refer to the emission intensities at the middle and maximum concentration of S100A4, respectively. F is the fluorescence intensity change between I0 and I. Fmax is the maximum fluorescence difference and Kd is the dissociation constant. The actual curve was processed further as curve fitting and slope was calculated to get the Kd.
2.6 Bioactivity study with the WST-1 assay
We used the WST-1 assay to determine the physiological condition of S100A1 proteins [67,68]. In living cells, WST-1 is split to a soluble formazan via mitochondrial dehydrogenases. The number of formazan created is directly proportional to the dehydrogenase enzymatic activity. Therefore, the difference in OD (optical density) values at a suitable wavelength is correlated with the number of active metabolically cells in the culture. The WST-1 assay was conducted in the same manner as the Roche method. We placed the SW480 cells at a density of 5×103 cells/well in a 96-well plate one day prior to the experiments [69]. Next, in a serum medium consisting of bovine serum albumin (0.1%), the cells were incubated for one day. The serum-starved cells were titrated with 100 nM S100A1 with S100A4 and left free of S100A4 protein for an additional 48 h to detect the exchange of the proportional cell number. Prior to collecting the 1/10 volume of the WST-1 chemical agent was added to all of the wells. This agent consisted of 100 μL of culture, and the cells were incubated for an additional 4 h at 37°C. The culture cell plate was then incubated with slight agitation for 10 min on an incubator. The absorbance was determined at a wavelength of 450 nm using a synergy 2 micro-plate reader [51].
3. Results and discussion
3.1 The binding site of S100A4 and S100A1
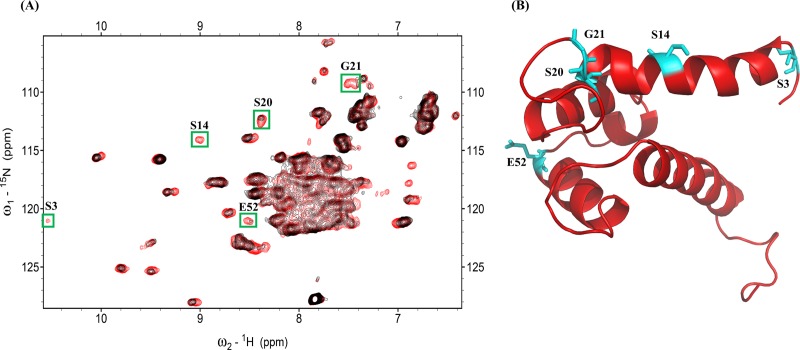
1H-15N NMR HSQC is generally used to determine the binding site between a ligand and a protein. Interacting residues of the S100A4 and S100A1 proteins can be determined by calculating the resonances on the HSQC NMR spectra of S100A4 and correlated them with S100A4 in the complex with S100A1. Superimposed HSQC NMR spectra of free 15N S100A4 and the 15N S100A4-S100A1 complex are shown in Fig 1A. Portions of the NMR signals were decreased after the addition of unlabeled S100A1 to 15N S100A4. The NMR HSQC signals of an 15N labeled S100A4 protein complex with unlabeled S100A1 residues at the interaction site were lower than those acquired with free 15N labeled S100A4. The assignment of S100A4 (BMRB under accession number 25069) was previously reported [70]. This finding is likely due to the affected nuclei at the interface between the S100A4 and S100A1 proteins.
Fig 1.
(A) Superimposed HSQC of the 15N S100A4 complex with unlabeled S100A1 (red) and free 15N S100A4 (black). Peaks indicating reduced intensity appear in green boxes. (B) Ribbon diagram showing the structure of S100A4; intensity decreasing residues are labeled on the structure in cyan.
The adjacent nuclei at the interacting site were reformed by another protein, which brought together and led to a decrease in the strength of peaks in the HSQC spectrum. When the S100 protein formed a complex with its target protein, the residues of the S100 protein were affected. Previous NMR HSQC studies noted changes in HSQC resonance among protein complex interfaces. The spectra of free 15N labeled S100A4 and the 15N labeled S100A4 complex with unlabeled S100A1 were superimposed to classify the decreased intensity of S100A4 residues (S3, S14, S20, G21, E52) at the interface.
3.2 The interface site of S100A1 and S100A4
We also performed a reverse titration experiment on S100A4 titration with 15N S100A1 to define the residues of S100A1 binding to S100A4. The HSQC spectra of free 15N labeled S100A1 (The assignment of S100A1 in BMRB Entry 18101) [71] and 15N labeled S100A1 complexed with unlabeled S100A4 (Fig 2A). Residues that exhibited a low intensity were selected for plotting on a cartoon model of S100A1 (Fig 2B): G1, E3, N13, H16, G43, F44, D52, A53, V76, L77, A79 and W90. We found the some common interaction residues (G43, V76, L77 and A79) which is reported in the binding HSQC spectra of S100A1-RAGE V domain [51].
Fig 2.
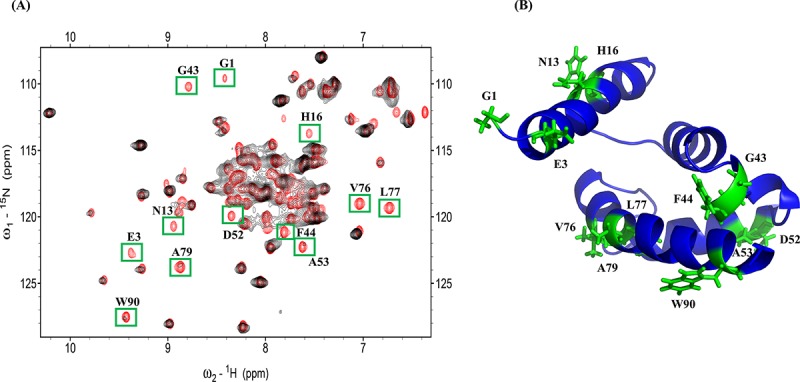
(A) Overlapping 15N S100A1 HSQC spectra (black) and the 15N S100A1 complex with unlabeled S100A4 (red). The cross peaks, which exhibited decreased intensity, are boxed in green. (B) Ribbon diagram showing the structure of S100A1—decreasing residues are labeled on the structure in green.
3.3 The S100A4-S100A1 complex model
The complex structure of S100A4 and S100A1 (Fig 3) was developed using the HADDOCK program to compute the protein complex. Ambiguous interaction restraints were developed based on the difference in resonance between the NMR HSQC of S100A4 and S100A1. Input parameters for the HADDOCK program were selected based on those residues that exhibited perturbations from the spectra (NMR HSQC). We also observed that some of the residues disappeared at the N-terminal of S100A1, which may be due to the flexibility of the N-terminal being high when a complex is formed.
Fig 3. Model of the S100A1-S100A4 complex.
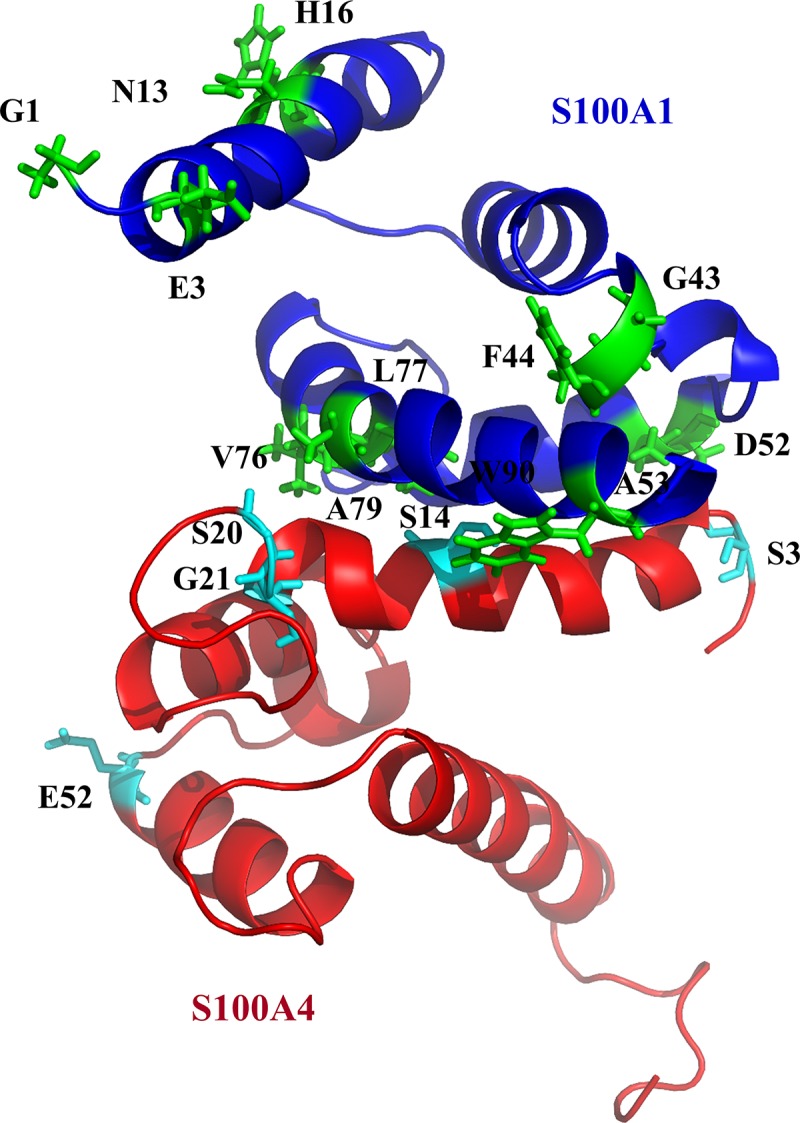
S100A1 and S100A4 are shown in blue and red, respectively. The interacting residues are shown in cyan and green.
Based on the HADDOCK results, we generated a heterodimeric complex of S100A4 and S100A1. The three-dimensional S100A1 and the S100A4 structures were achieved from PDB (ID: 2MRD,2LP3). Nearly 5000 complex structures were produced by applying rigid-body minimization with HADDOCK. The top 200 structures with lower energies after water refinement were used in this study. The structure of the heterodimer S100A4-S100A1 complex is shown in Fig 3. We observed that, due to segmental motion after S100A4-S100A1 complex formation, residues G1, E3, N13 and H16 disappeared at the N-terminal of S100A1. These residues are far away from the core of complex, which are experienced high degree of flexibility and mobility of the N-terminal when a complex is formed.
The Ramachandran plot analysis of the acquired complex S100A1-S100A4 had fair bond angles ψ and φ in the stereochemistry of the proteins. The Ramachandran plot (S3 Fig) reveals that 91.6% of the complex residues were glycine free in the favorite region. Six residues (3.2%) were glycine free in the outlier area.
3.4 Dissociation constant measurements based on fluorescence of the complex S100A4-S100A1
S100A1 protein consist one tryptophan residue at the sequence W90 and it is exposed. Tryptophan 90 is situated at the binding region of the S100A1. However, the excitation frequency of the tryptophan absorption band was found at 295 nm, and drop in fluorescence intensity shows around W90. Decrease in intensity was observed after addition of S100A4 at the titration curve. These data were processed to get linear curve fitting and slope was used to calculated to get the Kd. Dissociation constant for S100A4-S100A1 complex was calculated which is in the range of about 6.2 ± 0.47 μM. In Fig 4 we show the data graphed as 1/[S100A4] versus 1/(I—I0). Dissociation constant of S100A1-RAGE V domain was reported as 6.13 ± 1.29 μM [51], which indicating that binding of S100A1 is stronger with S100A4 than RAGE V domain.
Fig 4.
(A) Emission of fluorescence spectra of S100A1 titrations showing a decrease in intensity with the addition of S100A4 at μM-level concentrations. (B) Change in fluorescence intensity versus 1/[S100A4] obtained at a wavelength of 344 nm. Kd was measured 6.2 ± 0.47 μM.
3.5 Functional bioactivity study with the WST-1 assay
A WST-1-based cell proliferation assay was used to describe the downstream signaling activity transmitted by the RAGE V domain through S100A1 [16,46,72–76]. We selected SW480 cells (which contains RAGE) for the bioactivity functional assays because they have epithelial-like structure and are frequently used to study cancer in vitro. The SW480 cells were treated for 2 days with S100A1 prior to the variability analysis with the WST-1 assay.
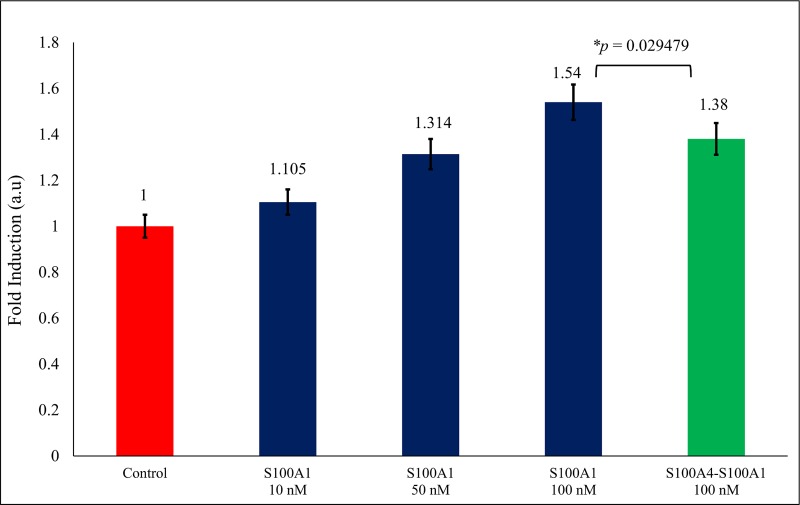
SW480 cells containing serum were treated with S100A1 (concentration: 10nM, 50nM and 100nM). We absorbed the 1.54-fold growth (Fig 5, Lane 4) in the viable cell count over control (Fig 5, Lane 1) which is free from serum. This result can be clarified by the point that S100A1 interact with the RAGE V domain and hence induce the cell proliferation [51]. A 1.38-fold decrease was observed in cells when treated with the complex of S100A4-S100A1 (100 nM) protein (Fig 5, Lane 5). These findings indicate that the S100A4 protein potentially inhibits contact between S100A1 and the RAGE V domain and inhibits cell proliferation activity.
Fig 5. The SW480 cells were treated with the S100A1 (blue, Lanes 2, 3 and 4).
Decrease in fold was observed when treated with the complex of 100 nM S100A4-S100A1 (green) and value of p is considering as p ≤ 0.05.
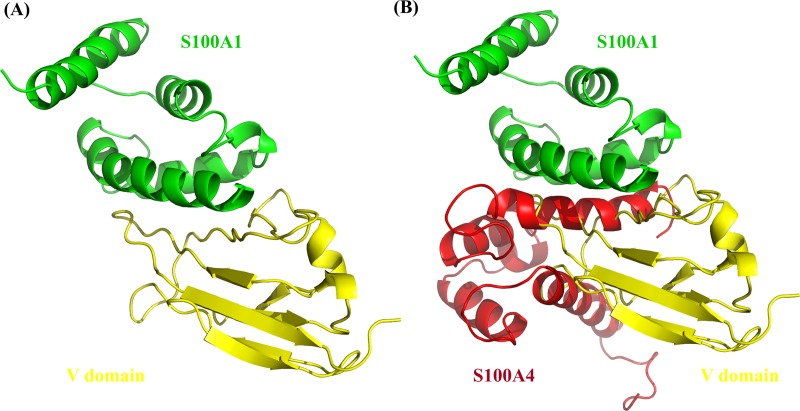
The interactions between S100A1 and the V domain suggest that their binding may activate an auto-phosphorylation route that leads to several signal transduction cascades that regulate migration and cell proliferation. In this study, S100A1-RAGE V domain complex was used which is previously reported (Fig 6A) [51].
Fig 6.
(A) The structure of S100A1-RAGE V domain complex (yellow and green respectively). (B) S100A4 (in red) blocking the interaction between S100A1-RAGE V domain.
4. Conclusions
The paracrine, endocrine, and autocrine aspects of S100 family proteins are unknown. These proteins can be present in sufficient concentrations to determine cellular signaling in diverse cell types. We used NMR titration experiments to describe the binding between S100A1 and S100A4. Using HSQC chemical shift perturbation, we have chosen the amino acid residue to run the HADDOCK to generate the S100A4-S100A1 complex model (Fig 3). We can see the S100A4 blocking the interaction between RAGE V domain and S100A1 (Fig 6B). WST-1 analysis is also supporting our finding, that S100A4 inhibit the cell proliferation (Sherbet and Lakshmi, 1998) when S100A4-S100A1 complex were added (Fig 5 lane 5 in green) [49,50]. Based on our results, we can conclude that S100A4 plays an essential role as an antagonist between the RAGE V domain and S100A1 [51]. Furthermore, our findings are essential to designing S100A4 analogs that may possibly act as effective blockers to inhibit the RAGE V domain-S100A1 route for the treatment of many kinds of human cancer [53] and inflammation [54–57]. The complex structure of S100A1-V domains [51] is shown in Fig 6A. The overlapping complexes S100A4-S100A1 are shown Fig 6B.
The S100A4 molecule acts as an inhibitor and stop the cell proliferation to block the interaction of the RAGE V domain and S100A1. This complex model may be valuable for improving antagonists and may potentially benefit protein-design studies that target S100A1 and the RAGE V domain.
Supporting information
(TIF)
(TIF)
Ninety-one percent of the residues were in the favored area, 5.3% were in the allowed area, and 3.2% were in the disallowed region.
(TIF)
(XLSX)
(XLSX)
Acknowledgments
We acknowledge the Department of Chemistry, National Tsing Hua University, for allowing us to use their Varian NMR 700 MHz spectrometer.
Abbreviations
- NMR
Nuclear Magnetic Resonance
- HSQC
Heteronuclear Single Quantum Coherence
- HADDOCK
High Ambiguity Driven biomolecular DOCKing
- AIRs
Ambiguous Interaction Restraints
- PDB
Protein Data Bank
- DTT
Dithiothreitol
- Kd
Dissociassion Constant
- TFA
Trifluoroacetic acid
- RAGE
Receptor for Advanced Glycation Endproducts
- OD
optical density
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Ministry of Science and Technology (MOST) of Taiwan (grant number MOST 104–2113-M-007-019-MY3), MOST 105-2320-B-039-059-MY3 and MOHW107-TDU-B-212-114025 from Ministry of Health and Welfare (MOHW), and the Drug Development Center, China Medical University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Santamaria-Kisiel L, Rintala-Dempsey AC, Shaw GS. Calcium-dependent and -independent interactions of the S100 protein family. Biochem J. 2006;396: 201–214. 10.1042/BJ20060195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugam T, Logsdon CD. S100P: A novel therapeutic target for cancer. Amino Acids. 2011. p. 893–899. 10.1007/s00726-010-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34: 357–64. 10.1016/j.ejso.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 4.Yang WS, Moon H-G, Kim HS, Choi E-J, Yu M-H, Noh D-Y, et al. Proteomic Approach Reveals FKBP4 and S100A9 as Potential Prediction Markers of Therapeutic Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. J Proteome Res. 2012;11: 1078–1088. 10.1021/pr2008187 [DOI] [PubMed] [Google Scholar]
- 5.Penumutchu SR, Chou RH, Yu C. Structural insights into calcium-bound S100P and the V domain of the RAGE complex. PLoS One. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochemical and Biophysical Research Communications. 2004. p. 1111–1122. 10.1016/j.bbrc.2004.07.096 [DOI] [PubMed] [Google Scholar]
- 7.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60: 540–551. 10.1002/jemt.10296 [DOI] [PubMed] [Google Scholar]
- 8.Engelkamp D, Schäfer BW, Mattei MG, Erne P, Heizmann CW. Six S100 genes are clustered on human chromosome 1q21: identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc Natl Acad Sci U S A. 1993;90: 6547–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schäfer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: Functions and pathology. Trends Biochem Sci. 1996;21: 134–140. [DOI] [PubMed] [Google Scholar]
- 10.Davey GE, Murmann P, Heizmann CW. Intracellular Ca2+ and Zn2+ Levels Regulate the Alternative Cell Density-dependent Secretion of S100B in Human Glioblastoma Cells. J Biol Chem. 2001;276: 30819–30826. 10.1074/jbc.M103541200 [DOI] [PubMed] [Google Scholar]
- 11.Rintala-Dempsey AC, Santamaria-Kisiel L, Liao Y, Lajoie G, Shaw GS. Insights into S100 target specificity examined by a new interaction between S100A11 and annexin A2. Biochemistry. 2006;45: 14695–14705. 10.1021/bi061754e [DOI] [PubMed] [Google Scholar]
- 12.Sastry M, Ketchem RR, Crescenzi O, Weber C, Lubienski MJ, Hidaka H, et al. The three-dimensional structure of Ca2+-bound calcyclin: Implications for Ca2+-signal transduction by S100 proteins. Structure. 1998;6: 223–231. [DOI] [PubMed] [Google Scholar]
- 13.Ma L, Sastry M, Chazin WJ. A Structural Basis for S100 Protein Specificity Derived from Comparative Analysis of Apo and Ca 2. J Mol Biol. 2002; 279–290. 10.1006/jmbi.2002.5421 [DOI] [PubMed] [Google Scholar]
- 14.Donato R, R. Cannon B, Sorci G, Riuzzi F, Hsu K, J. Weber D, et al. Functions of S100 Proteins. Curr Mol Med. 2013;13: 24–57. [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann A, Donato R, Weiger TM, Chazin WJ. S100 calcium binding proteins and ion channels. Front Pharmacol. 2012;3: 1–10. 10.3389/fphar.2012.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclerc E, Heizmann CW. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front Biosci (Schol Ed). 2011;3: 1232–62. [DOI] [PubMed] [Google Scholar]
- 17.Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins Struct Funct Genet. 1999;37: 499–507. [DOI] [PubMed] [Google Scholar]
- 18.Weber DJ, Zimmer DB. The calcium-dependent interaction of S100B with its protein targets. Cardiovascular Psychiatry and Neurology. 2010. p. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambartsumian N, Grigorian M, Lukanidin E. Genetically modified mouse models to study the role of metastasis-promoting S100A4(mts1) protein in metastatic mammary cancer. J Dairy Res. 2005;72: 27–33. [DOI] [PubMed] [Google Scholar]
- 20.Ismail TM, Zhang S, Fernig DG, Gross S, Martin-Fernandez ML, See V, et al. Self-association of calcium-binding protein S100A4 and metastasis. J Biol Chem. 2010;285: 914–922. 10.1074/jbc.M109.010892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford HL, Zain SB. Interaction of metastasis associated Mts1 protein with nonmuscle myosin. Oncogene. 1995;10: 1597–1605. [PubMed] [Google Scholar]
- 22.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, et al. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47: 5111–5126. 10.1021/bi702537s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra SK, Siddique HR, Saleem M. S100A4 calcium-binding protein is key player in tumor progression and metastasis: Preclinical and clinical evidence. Cancer and Metastasis Reviews. 2012. p. 163–172. 10.1007/s10555-011-9338-4 [DOI] [PubMed] [Google Scholar]
- 24.Semov A, Moreno MJ, Onichtchenko A, Abulrob A, Ball M, Ekiel I, et al. Metastasis-associated protein S100A4 induces angiogenesis through interaction with annexin II and accelerated plasmin formation. J Biol Chem. 2005;280: 20833–20841. 10.1074/jbc.M412653200 [DOI] [PubMed] [Google Scholar]
- 25.Remppis A, Greten T, Schäfer BW, Hunziker P, Erne P, Katus HA, et al. Altered expression of the Ca2+-binding protein S100A1 in human cardiomyopathy. Biochim Biophys Acta—Mol Cell Res. 1996;1313: 253–257. [DOI] [PubMed] [Google Scholar]
- 26.Moore BW. A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun. 1965;19: 739–744. [DOI] [PubMed] [Google Scholar]
- 27.Maco B, Mandinova A, Dürrenberger MB, Schäfer BW, Uhrík B, Heizmann CW. Ultrastructural distribution of the S100A1 Ca2+-binding protein in the human heart. Physiol Res. 2001;50: 567–574. [PubMed] [Google Scholar]
- 28.Durussel I, Cox JA, Heizmann CW. S100A13. Biochemical characterization and subcellular localization in different cell lines. 2000;275: 8686–8694. [DOI] [PubMed] [Google Scholar]
- 29.Adhikari BB, Wang K. S100A1 modulates skeletal muscle contraction by desensitizing calcium activation of isometric tension, stiffness and ATPase. FEBS Lett. 2001;497: 95–98. [DOI] [PubMed] [Google Scholar]
- 30.Naim R, Hormann K. The role of S100A1 in external auditory canal cholesteatoma. Oncol Rep. 2006;16: 671–675. [PubMed] [Google Scholar]
- 31.Li G, Barthelemy A, Feng G, Gentil-Perret A, Peoc’h M, Genin C, et al. S100A1: A powerful marker to differentiate chromophobe renal cell carcinoma from renal oncocytoma. Histopathology. 2007;50: 642–647. 10.1111/j.1365-2559.2007.02655.x [DOI] [PubMed] [Google Scholar]
- 32.Mori M, Yamada K, Ohomura H, Wataru K, Takai Y, Ilg E, et al. Immunohistochemical localization of S100A1 and S100A6 in postnatally developing salivary glands of rats. Histochem Cell Biol. 1998;110: 579–587. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Zhang S, Fernig DG, Martin-Fernandez M, Rudland PS, Barraclough R. Mutually antagonistic actions of S100A4 and S100A1 on normal and metastatic phenotypes. Oncogene. 2005;24: 1445–1454. 10.1038/sj.onc.1208291 [DOI] [PubMed] [Google Scholar]
- 34.DeRycke MS, Andersen JD, Harrington KM, Pambuccian SE, Kalloger SE, Boylan KLM, et al. S100A1 expression in ovarian and endometrial endometrioid carcinomas is a prognostic indicator of relapse-free survival. Am J Clin Pathol. 2009;132: 846–856. 10.1309/AJCPTK87EMMIKPFS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus C, Rohde D, Weidenhammer C, Qiu G, Pleger ST, Voelkers M, et al. S100A1 in cardiovascular health and disease: Closing the gap between basic science and clinical therapy. Journal of Molecular and Cellular Cardiology. 2009. p. 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afanador L, Roltsch EA, Holcomb L, Campbell KS, Keeling DA, Zhang Y, et al. The Ca2+sensor S100A1 modulates neuroinflammation, histopathology and Akt activity in the PSAPP Alzheimer’s disease mouse model. Cell Calcium. 2014;56: 68–80. 10.1016/j.ceca.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 37.Diaconescu DE, Dima L, Marinescu DM, Ţânţu MM, Rogozea LM. S100-positive dendritic cells in squamous cell laryngeal cancer. Rom J Morphol Embryol. 2014;55: 1371–1375. [PubMed] [Google Scholar]
- 38.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nature Reviews Cancer. 2015. p. 96–109. 10.1038/nrc3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Xu C, Jin Q, Liu Z. S100 protein family in human cancer. Am J Cancer Res. 2014;4: 89–115. [PMC free article] [PubMed] [Google Scholar]
- 40.Streicher WW, Lopez MM, Makhatadze GI. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys Chem. 2010;151: 181–186. 10.1016/j.bpc.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prosser BL, Wright NT, Hernãndez-Ochoa EO, Varney KM, Liu Y, Olojo RO, et al. S100A1 Binds to the Calmodulin-binding Site of Ryanodine Receptor and Modulates Skeletal Muscle Excitation-Contraction Coupling. J Biol Chem. 2008;283: 5046–5057. 10.1074/jbc.M709231200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Most P, Remppis A, Pleger ST, Löffler E, Ehlermann P, Bernotat J, et al. Transgenic Overexpression of the Ca2+-binding Protein S100A1 in the Heart Leads to Increased in Vivo Myocardial Contractile Performance. J Biol Chem. 2003;278: 33809–33817. 10.1074/jbc.M301788200 [DOI] [PubMed] [Google Scholar]
- 43.Wright NT, Prosser BL, Varney KM, Zimmer DB, Schneider MF, Weber DJ. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J Biol Chem. 2008;283: 26676–26683. 10.1074/jbc.M804432200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiewitz R, Acklin C, Schäfer BW, Maco B, Uhrík B, Wuytack F, et al. Ca2+-dependent interaction of S100A1 with the sarcoplasmic reticulum Ca2+-ATPase2a and phospholamban in the human heart. Biochem Biophys Res Commun. 2003;306: 550–557. [DOI] [PubMed] [Google Scholar]
- 45.Holakovska B, Grycova L, Jirku M, Sulc M, Bumba L, Teisinger J. Calmodulin and S100A1 protein interact with N terminus of TRPM3 channel. J Biol Chem. 2012;287: 16645–16655. 10.1074/jbc.M112.350686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: An update. Biochimica et Biophysica Acta—Molecular Cell Research. 2009. p. 993–1007. [DOI] [PubMed] [Google Scholar]
- 47.Rohde D, Ritterhoff J, Voelkers M, Katus HA, Parker TG, Most P. S100A1: A multifaceted therapeutic target in cardiovascular disease. Journal of Cardiovascular Translational Research. 2010. p. 525–537. 10.1007/s12265-010-9211-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deloulme JC, Gentil BJ, Baudier J. Monitoring of S100 homodimerization and heterodimeric interactions by the yeast two-hybrid system. Microscopy research and technique. 2003. p. 560–568. 10.1002/jemt.10298 [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Rudland PS, White MR, Barraclough R. Interaction in vivo and in vitro of the metastasis-inducing S100 protein, S100A4 (p9Ka) with S100A1. J Biol Chem. 2000;275: 11141–11146. [DOI] [PubMed] [Google Scholar]
- 50.Tarabykina S, Kriajevska M, Scott DJ, Hill TJ, Lafitte D, Derrick PJ, et al. Heterocomplex formation between metastasis-related protein S100A4 (Mts1) and S100A1 as revealed by the yeast two-hybrid system. FEBS Lett. 2000;475: 187–197. [DOI] [PubMed] [Google Scholar]
- 51.Khan I, Su Y, Zou J, Yang L, Chou R. S100B as an antagonist to block the interaction between S100A1 and the RAGE V domain. PLoS One. 2018; 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherbet G V., Lakshmi MS. S100A4 (MTS1) calcium binding protein in cancer growth, invasion and metastasis. Anticancer Research. 1998. p. 18 (4A)2415-2421. [PubMed] [Google Scholar]
- 53.Wang G, Zhang S, Fernig DG, Martin-Fernandez M, Rudland PS, Barraclough R. Mutually antagonistic actions of S100A4 and S100A1 on normal and metastatic phenotypes. Oncogene. 2005;24: 1445–1454. 10.1038/sj.onc.1208291 [DOI] [PubMed] [Google Scholar]
- 54.Yan SF, Ramasamy R, Schmidt AM. Soluble RAGE: Therapy and biomarker in unraveling the RAGE axis in chronic disease and aging. Biochem Pharmacol. 2010;79: 1379–1386. 10.1016/j.bcp.2010.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, Kuraoka K, et al. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol. 2002;196: 163–170. 10.1002/path.1031 [DOI] [PubMed] [Google Scholar]
- 56.Logsdon C, Fuentes M, Huang E, Arumugam T. RAGE and RAGE Ligands in Cancer. Curr Mol Med. 2007;7: 777–789. [DOI] [PubMed] [Google Scholar]
- 57.Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, et al. RAGE (Receptor for advanced glycation endproducts), RAGE ligands, and their role in cancer and inflammation. Journal of Translational Medicine. 2009. p. 1–21. 10.1186/1479-5876-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berridge M V., Herst PM, Tan AS. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnology Annual Review. 2005. p. 127–152. 10.1016/S1387-2656(05)11004-7 [DOI] [PubMed] [Google Scholar]
- 59.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125: 1731–1737. 10.1021/ja026939x [DOI] [PubMed] [Google Scholar]
- 60.Van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, et al. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol. 2016;428: 720–725. 10.1016/j.jmb.2015.09.014 [DOI] [PubMed] [Google Scholar]
- 61.van Zundert GCP, Bonvin AMJJ. Modeling protein–protein complexes using the HADDOCK webserver “modeling protein complexes with HADDOCK”. Methods Mol Biol. 2014;1137: 163–179. 10.1007/978-1-4939-0366-5_12 [DOI] [PubMed] [Google Scholar]
- 62.L.L.C. Schrodinger. The PyMOL Molecular Graphics System, Version 2.0. 2018; 2–9.
- 63.Burstein EA, Vedenkina NS, Ivkova MN. Fluorescence and the location of tryptophan residues in protein molecules. Photochem Photobiol. 1973;18: 263–279. [DOI] [PubMed] [Google Scholar]
- 64.Xue J, Ray R, Singer D, Böhme D, Burz DS, Rai V, et al. The receptor for advanced glycation end products (RAGE) specifically recognizes methylglyoxal-derived AGEs. Biochemistry. 2014;53: pp 3327–3335. 10.1021/bi500046t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lakowicz JR. Principles of fluorescence spectroscopy [Internet]. Lakowicz JR, editor. Springer; US; 2006. [Google Scholar]
- 66.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci. 2005;102: 4735–4740. 10.1073/pnas.0501459102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan AS, Berridge M V. Superoxide produced by activated neutrophils efficiently reduces the tetrazolium salt, WST-1 to produce a soluble formazan: A simple colorimetric assay for measuring respiratory burst activation and for screening anti-inflammatory agents. J Immunol Methods. 2000;238: 59–68. [DOI] [PubMed] [Google Scholar]
- 68.Tsukatani T, Suenaga H, Higuchi T, Akao T, Ishiyama M, Ezoe K, et al. Colorimetric cell proliferation assay for microorganisms in microtiter plate using water-soluble tetrazolium salts. J Microbiol Methods. 2008;75: 109–116. 10.1016/j.mimet.2008.05.016 [DOI] [PubMed] [Google Scholar]
- 69.Liang H, Zhong Y, Zhou S, Peng L. Knockdown of RAGE expression inhibits colorectal cancer cell invasion and suppresses angiogenesis in vitro and in vivo. Cancer Lett. 2011;313: 91–98. 10.1016/j.canlet.2011.08.028 [DOI] [PubMed] [Google Scholar]
- 70.Cho CC, Hung K-W, Gorja DR, Yu C. The solution structure of human calcium-bound S100A4 mutated at four cysteine loci. J Biomol NMR. Springer Netherlands; 2015;62: 233–238. 10.1007/s10858-015-9927-6 [DOI] [PubMed] [Google Scholar]
- 71.Gupta AA, Mohan SK, Chin Y. 1H,13C and15N backbone and side chain resonance assignments of human halo S100A1. Biomol NMR Assign. 2012;6: 213–215. 10.1007/s12104-012-9360-7 [DOI] [PubMed] [Google Scholar]
- 72.Tian T, Li X, Hua Z, Ma J, Liu Z, Chen H CZ. S100A1 promotes cell proliferation and migration and is associated with lymph node metastasis in ovarian cancer. 2017;23: 235–245. [PubMed] [Google Scholar]
- 73.Tsoporis JN, Izhar S, Leong-Poi H, Desjardins JF, Huttunen HJ, Parker TG. S100B interaction with the receptor for advanced glycation end products (RAGE): A novel receptor-mediated mechanism for myocyte apoptosis postinfarction. Circ Res. 2010;106: 93–101. 10.1161/CIRCRESAHA.109.195834 [DOI] [PubMed] [Google Scholar]
- 74.Donato R, Sorci G, Riuzzi F, Arcuri C, Bianchi R, Brozzi F, et al. S100B’s double life: Intracellular regulator and extracellular signal. Biochimica et Biophysica Acta—Molecular Cell Research. 2009. p. 1793 (6) 1008–22. [DOI] [PubMed] [Google Scholar]
- 75.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J Biol Chem. 2008;283: 36221–36233. 10.1074/jbc.M806322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leclerc E. Measuring binding of S100 proteins to rage by surface plasmon resonance. Methods in Molecular Biology. 2013. p. 963:201–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
Ninety-one percent of the residues were in the favored area, 5.3% were in the allowed area, and 3.2% were in the disallowed region.
(TIF)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.