SUMMARY
One of the features that distinguishes modern humans from our extinct relatives and ancestors is a globular shape of the braincase [1–4]. As the endocranium closely mirrors the outer shape of the brain, these differences might reflect altered neural architecture [4, 5]. However, in the absence of fossil brain tissue, the underlying neuroanatomical changes as well as their genetic bases remain elusive. To better understand the biological foundations of modern human endocranial shape, we turn to our closest extinct relatives: the Neandertals. Interbreeding between modern humans and Neandertals has resulted in introgressed fragments of Neandertal DNA in the genomes of present-day non-Africans [6, 7]. Based on shape analyses of fossil skull endocasts, we derive a measure of endocranial globularity from structural MRI scans of thousands of modern humans and study the effects of introgressed fragments of Neandertal DNA on this phenotype. We find that Neandertal alleles on chromosomes 1 and 18 are associated with reduced endocranial globularity. These alleles influence expression of two nearby genes, UBR4 and PHLPP1, which are involved in neurogenesis and myelination, respectively. Our findings show how integration of fossil skull data with archaic genomics and neuroimaging can suggest developmental mechanisms that may contribute to the unique modern human endocranial shape.
In Brief
Gunz, Tilot et al. combine paleoanthropology, archaic genomics, neuroimaging, and gene expression to study biological foundations of the characteristic modern human endocranial shape. They find introgressed Neandertal alleles that associate with reduced endocranial globularity and affect expression of genes linked to neurogenesis and myelination.
RESULTS AND DISCUSSION
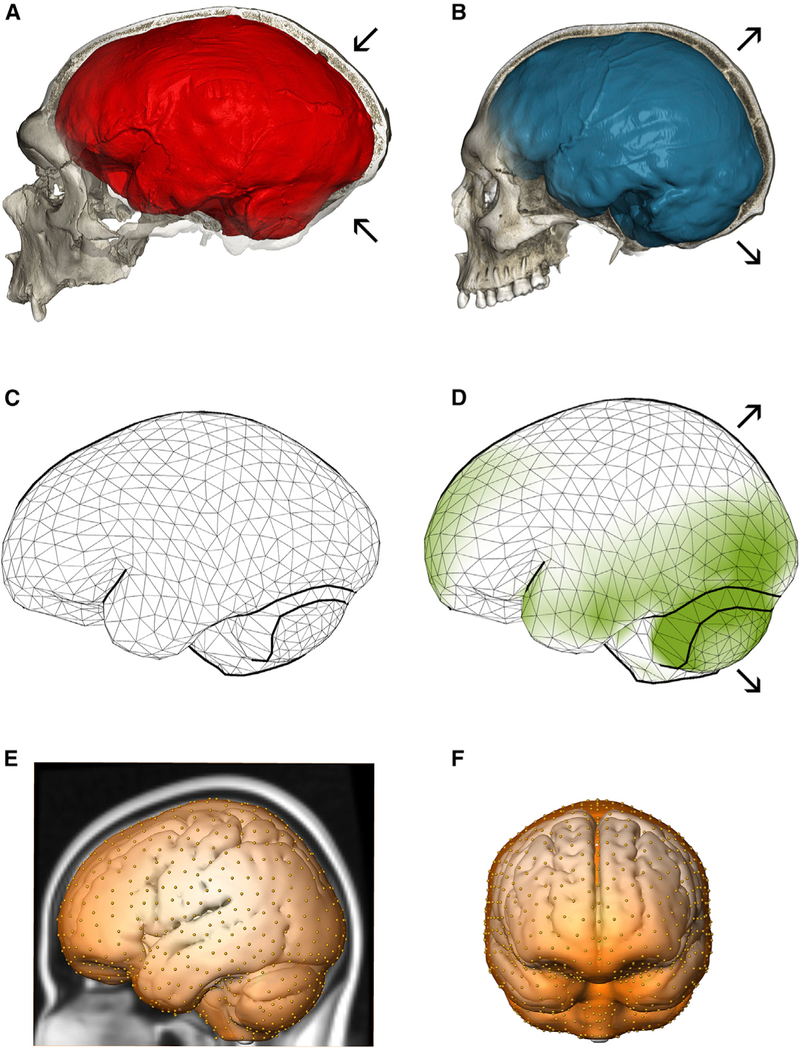
Neandertals (Figure 1A) and modern humans (Figure 1B) have similar endocranial volumes but distinct endocranial shapes. Modern humans have a globular-shaped braincase, with a round and expanded posterior cranial fossa housing the cerebellum, and more bulging parietal bones [2–4]. Contrasting the average endocranial shapes of modern humans and Neandertals, Kochiyama et al. [5] proposed that the biggest differences between these groups are found in the cerebellum. Comparative analyses suggest rapid evolutionary changes of this brain structure in great apes and humans [8, 9]. Our analysis of endocranial shape based on data from [4] demonstrates that other regions beyond the cerebellum are relatively larger in modern humans than in Neandertals, including parts of the prefrontal cortex and the occipital and temporal lobes (Figures 1C and1D). In contrast, parietal bulging [2–4] is not linked to an increased surface area, suggesting that the parietal lobe is ‘‘displaced’’ by reorganization of other—presumably subcortical—parts of the brain.
Figure 1. Endocranial Shape Differences between Neandertals and Modern Humans.
(A) CT scan of the Neandertal fossil from La Chapelle-aux-Saints with a typical elongated endocranial imprint (red).
(B) CT scan of a modern human showing the characteristic globular endocranial shape (blue). Arrows highlight the enlarged posterior cranial fossa (housing the cerebellum) as well as bulging of parietal bones in modern humans compared to Neandertals.
(C) Average endocranial shape of adult Neandertals; each vertex of the surface corresponds to a semilandmark.
(D) Average endocranial shape of modern humans. Areas shaded in green are relatively larger in modern humans than in Neandertals.
(E and F) The semilandmarks used to quantify overall endocranial shape from MRI scans of living people shown on the MNI 152 template in lateral and frontal views, respectively.
The evolutionary history of our species can currently be traced back to fossils from Jebel Irhoud (Morocco) dated to about 300,000 years ago [4, 10, 11]. While their faces and teeth look modern, their elongated braincases appear more like older human species and Neandertals [4, 10]. Together with crania from South Africa and Ethiopia, these fossils document an early evolutionary phase of Homo sapiens on the African continent [10, 11]. The globular endocranial shape emerged gradually in the Homo sapiens lineage, evolving independently of brain size: reorganization of cerebellar and lateral parietotemporal areas was followed by continued gradual changes in the organization of cerebellar and occipital areas [4]. From the perspective of ontogeny, braincase shape depends on a complex interplay between cranial bone growth, facial size, and the tempo and mode of neurodevelopment [1, 4, 12–14]. In present-day humans, globularity emerges during perinatal development [12, 13] in a period when the rapidly expanding brain is the main driver of braincase shape. It has therefore been proposed that endocranial globularity reflects evolutionary changes in early brain development [4, 12]. However, endocranial imprints only capture outer brain shape and cannot provide direct information about underlying features of neural reorganization.
Study Design and Hypothesis
In this study, we combine paleoanthropological data from Neandertal fossils with neuroimaging and genomic data from thousands of present-day humans, as well as gene expression data, to interrogate the molecular basis of endocranial globularity. As overall endocranial shape is a complex trait, we expect that it is influenced by many genetic loci, each with only a small effect. We therefore use both phenotypic and genetic differences between modern humans and Neandertals as filters to constrain our search space. Analyses of the genomes of Neandertals and modern humans show that they encountered each other outside Africa and interbred [6, 15, 16]. Introgressed fragments of Neandertal DNA can be reliably identified in modern humans and are estimated to account for 1%–2% of the genomes of non-African individuals, such that collectively ~40% of the Neandertal genome is represented in people living today [17–19]. Here, we quantify the endocranial shape differences between Neandertals and modern humans and study how Neandertal introgressed fragments affect this phenotype.
Metric for Endocranial Globularity
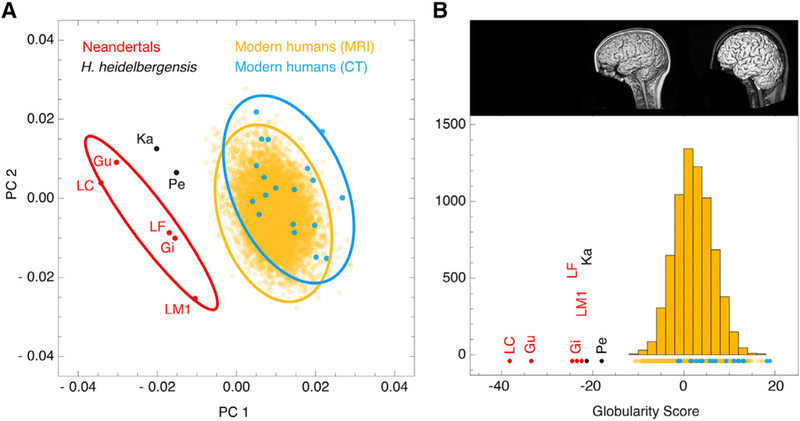
Using computed tomographic (CT) scans of fossil and extant skulls, we generated virtual imprints of the interior braincase (endocasts) and quantified endocranial shape differences between modern humans from Europe (n = 19) and Neandertals (n = 7) based on a dense mesh of semilandmarks (Figures 1C and 1D). We placed the same mesh on the endocranial surface segmented manually from the MNI-152 template—an average of 152 registered structural MRI scans of living humans (Figures 1E and 1F). A principal component analysis of endocranial shape shows that there is no overlap between the more globular endocrania of modern humans and the more elongated endocrania of Neandertals (Figure 2A). We therefore used these distinctive group differences to develop a summary metric for endocranial globularity. This ‘‘globularity score’’ quantifies overall endocranial shape by projecting the endocranial measurements of each individual onto the vector between the average shape of Neandertals and that of present-day humans (Figure 2B). By applying this morphometric approach to structural MRI scans of thousands of healthy human adults (n = 6,575) from the general population (predominantly of European origin), we obtained an evolutionarily derived quantitative index of globularity, which we showed with repeat scans to be robust and reliable. We replicated the findings shown in Figure 2 in a more diverse endocranial dataset, building on data from [4] (Figure S1). As the sample composition differs from the one shown in Figure 2, the values of the derived globularity scores also differ—the overall pattern, however, remains highly consistent. Endocranial shapes extracted from CT and MRI scans largely overlap in both analyses (Figures 2, S1B, and S1C).
Figure 2. Globularity Scores of CT and MRI Scans.
(A) Principal component analysis of endocranial shape. 99% confidence ellipses are shown for modern human CT scans from Europe (blue; n = 19), MRI scans of present-day humans (yellow; n = 6,575), and Neandertal CT scans (red; n = 7); two Homo heidelbergensis individuals are plotted in black.
(B) Frequency plot of globularity scores computed for data shown in (A). This globularity score quantifies overall endocranial shape by projecting each individual onto the vector between the elongated average shape of Neandertals and the globular average shape of present-day humans. Inset shows example MRI scans associated with low (left) and high (right) globularity scores among present-day humans.
See also Figure S1.
Voxel-Based Morphometry
We explored potential underlying structural contributions to interindividual variation in the globularity phenotype in healthy present-day humans using voxel-based morphometry (VBM) of MRI scans in two large European cohorts from Germany (SHIP and SHIP-Trend, total n = 2,929). These analyses revealed multiple significant (p(peak,FWE) % 0.025) clusters where globularity was positively or negatively associated with interindividual variability in gray matter (GM). In both hemispheres, greater values of globularity were associated with higher GM volumes in temporal regions, vermis and adjacent parts of the cerebellum, and in subcortical structures including the hippocampus, thalamus, amygdala, caudate, and putamen (Figure S2; Table S1). A significant inverse relationship of globularity and GM (Table S2) was detected in large parts of the frontal, temporal, and occipital gyri; parts of the cerebellum; and several subcortical regions (thalamus, putamen, hippocampus).
Effect of Introgressed Neandertal Alleles
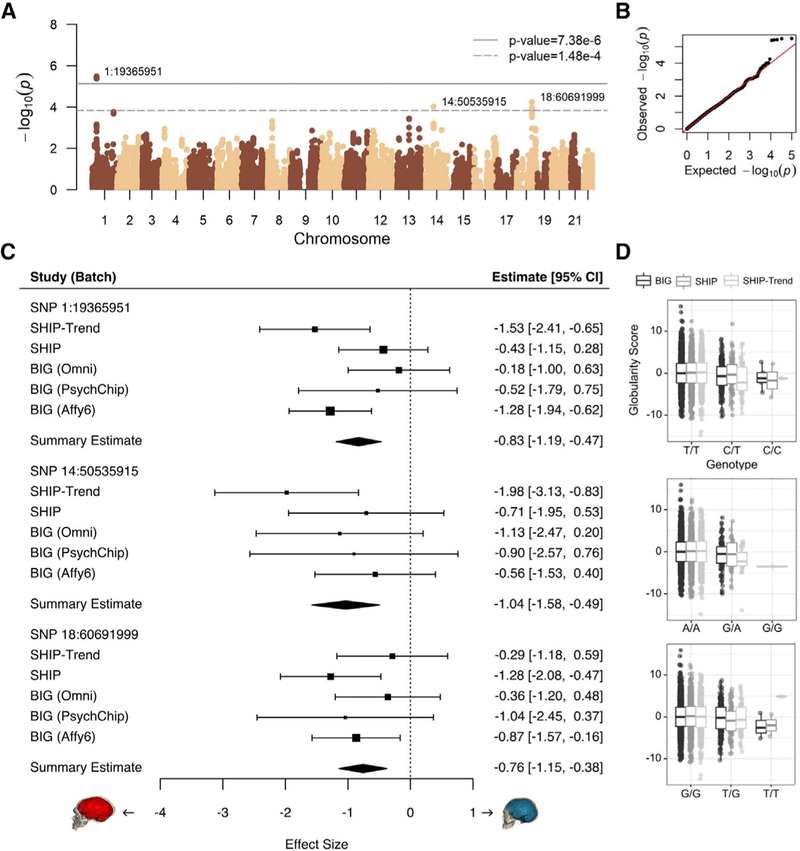
To investigate molecular correlates of the fossil-based globularity score, we used genotype data from European individuals to identify introgressed Neandertal alleles [18, 20] and studied their association with variability in this phenotype. We analyzed five datasets comprising 4,468 individuals with European ancestry for whom both MRI and genotype data were available: three batches of the Dutch BIG cohort (total n = 2,433) and the genotyped subsets of the German SHIP (n = 1,139) and SHIP-Trend (n = 896) cohorts, mentioned above. We analyzed each of 50,057 archaic SNPs with a minor allele frequency (MAF) of at least 0.01, testing the hypothesis that Neandertal-introgressed fragments would promote elongation of endocranial shape in modern humans (controlling for age, sex, ancestry, and scanning parameters). These SNPs cover 42% of the known, high-confidence Neandertal haplotypes [20] (Figures 3A and 3B). Although the cohorts were all European, we used multidimensional scaling (MDS) to exclude outlier individuals and, for extra rigor, accounted for residual effects of population stratification using principal components (PCs). Prior to association analysis, we designated significance thresholds that appropriately account for the extent of multiple testing, based on the linkage disequilibrium structure within the set of Neandertal alleles being tested (6,778 independent tests, estimated using the Genetic Type 1 error calculator [21]). Thus, our multiple-testing corrected thresholds were p < 7.38 × 10−6 for significant association and p < 1.48 × 10−4 for suggestive association, following standard guidelines of the field.
Figure 3. Endocranial Globularity in Modern Humans Is Associated with Introgressed Neandertal Haplotypes.
(A) Association statistics for introgressed Neandertal SNPs (solid line, significance, adjusted for multiple testing; dashed line, suggestive significance).
(B) Quantile-quantile plot of association p values, showing the expected uniform distribution, with the outliers representing significant associations.
(C) Forest plots depict the effects of the top Neandertal SNPs, for each study and genotyping batch. Boxes are proportional to weight, with whiskers representing the 95% confidence interval; diamonds represent a linear mixed-effect model incorporating all five datasets.
(D) Covariate-corrected globularity scores by genotype. All data points are shown; boxes represent 25th and 75th percentiles; whiskers represent 1.5 times the interquartile range.
Five SNPs within a ~200 kilobase (kb) archaic haplotype on chromosome 1 spanning 1:19244479–19453365 passed the significance threshold (top SNP 1:19365951, p = 3.26 × 10−6). A second Neandertal haplotype on chromosome 18 (500 kb, 18:60279290–60776578) contains six SNPs that passed the suggestive significance threshold (top SNP 18:60691999, p = 5.66 × 10−5). A single SNP, representing a 170 kb haplotype on chromosome 14 (14:50535915, p = 9.29 × 10−5), also passed suggestive significance. All haplotypes showed consistent directions of effect across all five datasets from the three cohorts, with the Neandertal-like haplotypes showing association with more oblong endocranial shape (Figure 3C) and with the top SNPs each showing an additive effect (i.e., heterozygous carriers were intermediate in score between homozygous groups; Figure 3D). We checked the top SNP at each locus to further discount residual effects of population stratification by increasing the number of PCs from 2 to 15. In this conservative follow-up analysis, there was a slight attenuation of signals: the chromosome-1 association remained significant (p = 6.94 × 10−6), and the chromosome-18 association remained suggestive (p = 9.54 × 10−5), but the isolated chromosome-14 SNP dropped below the threshold (p = 0.00019). The MAFs for the top SNPs were low in all three European cohorts, and very few individuals were homozygous for the Neandertal alleles (1:19365951, MAF = 0.0436, 8 homozygotes; 18:60691999, MAF = 0.052, 6 homozygotes; 14:50535915, MAF = 0.0204, 1 homozygote; Figure 3D). As expected for a genetically complex neuroanatomical phenotype [22], effect sizes of individual SNPs were small (Figure 3C) but detectable here due to inclusion of cohorts comprising thousands of individuals.
A recent study explored Neandertal DNA variants and brain shape in a substantially smaller sample of present-day humans [23]. The authors looked for associations between features of brain shape and a composite score reflecting the overall percentage of Neandertal DNA that a person carries. However, because individual introgressed fragments are rare, two people can have the same total amount of Neandertal-derived DNA, and thus an identical summary score, but share few (if any) Neandertal variants. Thus, it is difficult to draw biologically informative conclusions from overall genomic percentages, limiting the interpretation of the prior study. In their analysis of only 146 people, the authors highlighted Neandertal SNPs in the GPR26 gene as being of particular interest [23]; we assessed these markers in our sample of 4,468 individuals but saw no association with variability in endocranial globularity (all p > 0.05).
Gene Expression
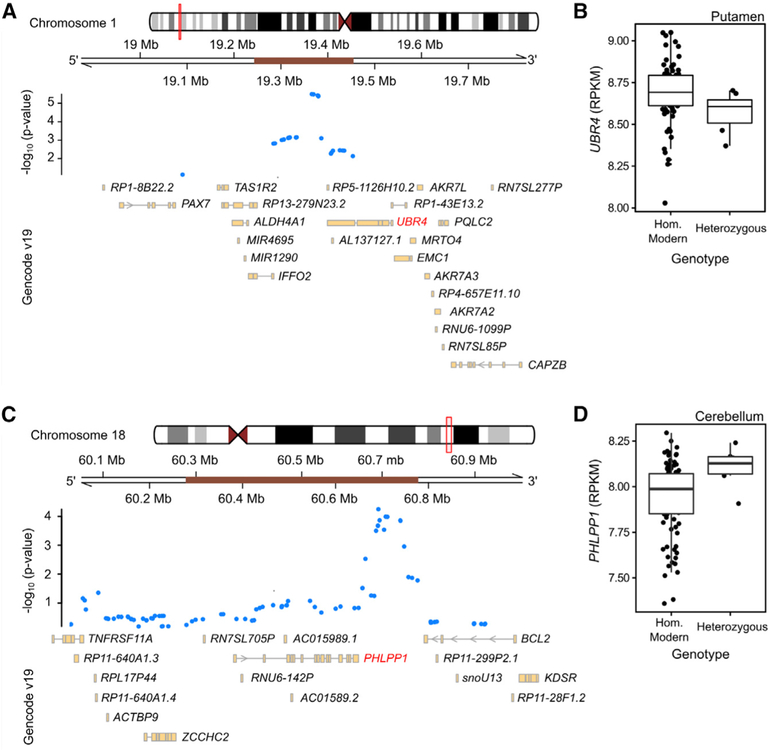
The associated SNPs lie outside protein-coding regions, but could potentially affect expression of nearby genes, in the brain and other tissues. We tested this hypothesis for the Neandertal haplotypes tagged by the two top SNPs on chromosome 1 and 18 using the GTEx database of expression quantitative trait loci (eQTL) data from over 400 donors [24]. To increase our power to detect associations with Neandertal introgressed alleles, which tend to be at lower frequency in present-day genomes, we used a set of Neandertal eQTLs that were previously defined, based on the top 5% of genes in each GTEx tissue showing association between gene expression and the presence of a nearby introgressed archaic haplotype [25]. The Neandertal variants that were associated with lower globularity scores showed modest but consistent effects on the expression in brain tissues of nearby genes that regulate multiple aspects of brain growth. Among the top 5% eQTL associations, we found that the chromosome-1 SNP, 1:19365951 (rs28445963), which had the strongest association with endocranial shape in our study, showed significant eQTLs in multiple tissues, including downregulation of UBR4 in the putamen (part of the basal ganglia) in carriers of the Neandertal allele (Figures 4A and 4B; Spearman correlation p = 0.031). The Neandertal allele of the most highly associated chromosome-18 SNP, 18:60691999 (rs72931809), was associated with upregulation of PHLPP1 in the cerebellum (Figures 4C–4D, Spearman correlation p = 0.024). Additional eQTL associations of these SNPs in other tissues are shown in Figures S3 and S4.
Figure 4. Introgressed Neandertal Fragments Associated with Globularity.
(A and C) Detailed views of the kilobase surrounding the Neandertal SNPs within the introgressed haplotypes (brown bars) on chromosomes 1 (A) and 18 (C).
(B and D) Expression quantitative trait loci data from the GTEx resource, showing the impact of Neandertal alleles on gene expression for UBR4 (B) and PHLPP1 (D), as defined by [24].
See also Figures S3 and S4.
UBR4 encodes a ubiquitin ligase that regulates neurogenesis in the developing neocortex and promotes neuronal migration, among other roles [26]. Loss of the mouse ortholog (called p600) in the developing brain impairs neurogenesis, resulting in microcephaly [27]. In humans, UBR4 is intolerant to loss-of-function mutations (pLI score = 1.0), suggesting that even small expression changes may have functional consequences [28].
PHLPP1 encodes a negative regulator of the PI3K/Akt growth-factor signaling pathway that drives myelination. Overexpression of Akt in transgenic mice leads to hypermyelination compared to wild-type controls [29]. With a high pLI score (0.92), PHLPP1 is a tumor suppressor in humans and mice [30, 31]. The PI3K/ AKT/mTOR signaling pathway broadly promotes brain growth and development of the myelin sheath [32–35]. In carriers of the Neandertal allele, PHLPP1 expression is slightly higher in the cerebellum, which would be predicted to have dampening effects on Akt-driven cerebellar myelination, consistent with the less globular endocranial shape associated with this allele in our study. However, comparative studies indicate that the increased endocranial globularity of present-day humans as compared to Neandertals is not merely the result of absolute cerebellar enlargement [4, 5], suggesting that endocranial shape variation involves complex shifts in relative, rather than absolute, volumes of different brain structures. This viewpoint is supported also by our VBM analyses of interindividual differences in globularity, which found that such variation involves both positive and negative GM changes, distributed across various brain regions. In one of the cohorts that we studied here, more globular modern human endocrania have slightly smaller absolute cerebellar volumes (Table S3), but this subtle effect was not observed in the other cohorts. The VBM analyses suggest that, at least within present-day Europeans, higher endocranial globularity is associated with increased GM in some parts of the cerebellum but also with decreased GM in others (Figure S2; Tables S1 and S2).
Conclusions
We have integrated evidence from paleoanthropology, comparative genomics, neuroimaging, and gene expression to begin identifying genes associated with variation in endocranial globularity, a defining feature of modern humans. The directions of effect were consistent in five separate batches of data from three independent cohorts and consistent with our hypothesis that Neandertal alleles would push the endocranium toward a more elongated shape. The associated variants were connected to genes involved in neurogenesis and myelination pathways in putamen and cerebellum, respectively. The eQTL data suggest that Neandertal alleles near UBR4 and PHLPP1 are linked to lower levels of neural proliferation. We speculate that this may contribute to altered neuroanatomical morphology of some subcortical structures and the cerebellum and thereby to lower overall globularity. The Neandertal haplotypes may thus be associated with developmental gene expression patterns that influence endocranial globularity through effects on neurogenesis and myelination during brain development. Functional impacts of Neandertal alleles on neural properties and brain development can in future be empirically tested [36], for example by using gene-editing techniques to insert changes into human induced pluripotent stem cells, which can be differentiated into distinct types of neurons or organoids.
Globularity is a multifactorial trait, involving combined influences of many different loci, and the effects of individual SNPs on overall endocranial shape are small. It is likely that future genome-wide studies in sufficiently large high-powered samples will reveal additional relevant genes and associated pathways. The potential links between evolutionary changes in endocranial globularity and mechanisms affecting the basal ganglia and cerebellum are nevertheless intriguing, because both brain structures receive direct input from the motor cortex and are involved in the preparation, learning, and sensorimotor coordination of movements. Expanded cerebellar interconnections with prefrontal, premotor, and superior-posterior parietal cortices, which also project densely to the putamen, may be particularly relevant to cognitive abilities of modern humans [9, 37, 38]. In addition to their involvement in sensorimotor coordination, the basal ganglia also contribute to diverse cognitive functions in memory, attention, planning, skill learning [39], and potentially to speech and language evolution [40, 41].
STAR*METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical Commercial Assays | ||
| 1.5 Tesla Siemens Sonata and Avanto scanners, 3 Tesla Siemens Trio and Trio TIM scanners | Siemens Medical Systems, Erlangen, Germany | N/A |
| 3 Tesla GE MR750 scanner | General Electric, USA | N/A |
| Affymetrix 6.0 genotyping array | Affymetrix, USA | N/A |
| PsychChip genotyping array | Psychiatric Genomics Consortium, USA | RRID:SCR_004495 |
| OmniExpress genotyping array | Illumina, USA | N/A |
| Omni 2.5 genotyping array | Illumina, USA | N/A |
| Deposited Data | ||
| Introgressed Neandertal SNPs | [18, 20] | N/A |
| GTEx eQTL data | gtexportal.org | RRID:SCR_013042 |
| BrainSpan Atlas | http://www.brainspan.org/static/download.html | RRID:SCR_008083 |
| Software and Algorithms | ||
| Globularity measurement script (Mathematica) | This Study | N/A |
| FreeSurfer (v5.3) | http://surfer.nmr.mgh.harvard.edu/ | RRID:SCR_001847 |
| FSL FIRST (v5.0, v5.0.9), FLIRT (v6.0) | https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/ | RRID:SCR_002823 |
| Avizo | Thermo Scientific | RRID:SCR_014431 |
| Mathematica | Wolfram | RRID:SCR_014448 |
| SPM12 | Wellcome Trust Centre for Neuroimaging, University College London | RRID:SCR_007037 |
| CAT12 toolbox | Christian Gaser, University of Jena, Germany, http://www.neuro.uni-jena.de | N/A |
| xjview toolbox | http://www.alivelearn.net/xjview | RRID:SCR_008642 |
| PLINK | https://www.cog-genomics.org/plink2/ | RRID:SCR_001757 |
| minimac (release 2012–05-29) | https://genome.sph.umich.edu/wiki/Minimac | RRID:SCR_009292 |
| mach2qtl | https://www.unc.edu/~yunmli/software.html | RRID:SCR_009621 |
| IMPUTE (v2.2.2) | http://mathgen.stats.ox.ac.uk/impute/impute.html | RRID:SCR_009245 |
| QUICKTEST (v0.99b) | https://wp.unil.ch/sgg/quicktest/ | N/A |
| easyQC | https://www.uni-regensburg.de/medizin/epidemiologie-praeventivmedizin/genetische-epidemiologie/software/ | N/A |
| METAL | https://genome.sph.umich.edu/wiki/METAL_Documentation | RRID:SCR_002013 |
| Genomic Type 1 Error Calculator | http://grass.cgs.hku.hk/gec/ | N/A |
| DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | RRID:SCR_015687 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Simon E. Fisher (simon.fisher@mpi.nl).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Participants
The Nijmegen ‘‘Brain Imaging Genetics’’ (BIG) cohort is a Dutch population-based sample of healthy, unrelated volunteers. Established in 2007, BIG is part of Cognomics, a joint initiative by the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud University Medical Centre, and the Max Planck Institute for Psycholinguistics. MRI and genetic data were collected from individuals at the Donders Center for Cognitive Neuroscience, in Nijmegen, the Netherlands. The BIG dataset has been described in several previous studies [42, 43]. The MRI data in the present study come from 2,913 participants (53% female), with an average age of 27 years (range 17–82). The genetic data presented come from a subset of 2,433 individuals (53% female), with an average age of 25 years (range 18–82). All participants gave written informed consent for analysis of both their DNA and MRI scans, and the regional ethics committee approved the study.
The ‘‘Study of Health in Pomerania’’ (SHIP), established in 1997, is a prospective cohort study, part of the Community Medicine Research net of the University of Greifswald. SHIP is a population-based project in West Pomerania, a region in the northeast of Germany, that consists of two independent prospectively collected cohorts (SHIP and SHIP-Trend) assessing the prevalence and incidence of common population-based diseases and their risk factors. The study design has been previously described in detail [44]. For SHIP, baseline examinations were carried out from 1997 until 2001, and the sample finally comprised 4,308 participants. Baseline examinations for SHIP-Trend were carried out between 2008 and 2012, finally comprising 4,420 participants. We conducted a voxel-based morphometry (VBM) analysis [45] on individuals from SHIP and SHIP-TREND. Complete datasets (including MRI, globularity score, and covariates for adjustments) were available for 3,309 subjects. After exclusion of medical conditions (e.g., a history of cerebral tumor, stroke, Parkinson’s diseases, multiple sclerosis, epilepsy, hydrocephalus, enlarged ventricles, pathological lesions) or technical reasons (e.g., severe movement artifacts or inhomogeneity of the magnetic field) 2,952 subjects were available. We finally performed the homogeneity check and excluded 23 extreme outliers. Our final sample for the VBM analysis consisted of 2,929 individuals (53% female). So as to study the effect of Neandertal alleles on globularity we used samples from the second five-year follow-up of SHIP (SHIP-2, n = 1,139) and the baseline of SHIP-Trend (n = 896) for which both MRI and genotype data were available. The samples had an average age of 56 (range 30–90) years for SHIP-2, and 50 (range 22–81) years for SHIP-Trend. The cohorts included 51% and 56% females for SHIP-2 and SHIP-Trend, respectively. The medical ethics committee of the University of Greifswald approved the study protocol, and oral and written informed consents were obtained from each of the study participants.
The ‘‘Function Biomedical Informatics Research Network’’ (FBIRN) Phase 3 cohort included 186 individuals with schizophrenia (average age 38 years, 22% female) and 176 healthy volunteers (average age 37.5 years, 28% female) with an age range of 18–62 years. Written informed consent, including permission to share de-identified data between the centers, approved by the University of California Irvine, Los Angeles, and San Francisco, Duke University, University of North Carolina, New Mexico, Iowa, and Minnesota Institutional Review Boards, was obtained from all study participants. The study methods have been previously described in detail [46]. Data from FBIRN was included in the initial description of brain globularity (see Methods - Structural MRI), however, the cohort was not included in the genetic association analyses (see Methods - Genotyping, Statistical analyses).
METHOD DETAILS
Structural MRI
In the BIG cohort, MRI data were obtained using either a 1.5 Tesla Siemens Sonata or Avanto scanner, or a 3 Tesla Siemens Trio or Trio TIM scanner (Siemens Medical Systems, Erlangen, Germany). Image processing was completed using FreeSurfer version 5.3 and FSL FIRST version 5.0 with FLIRT version 6.0, as described previously [42].
In the SHIP cohorts, participants were scanned using a 1.5 Tesla Siemens Avanto scanner, and image processing was completed using FreeSurfer 5.3 and FSL FIRST version 5.0.9 (with FLIRT version 6.0).
The FBIRN Phase 3 cohort was scanned at 7 sites using six 3 Tesla Siemens Trio TIM (Siemens, Erlangen, Germany) and one 3 Tesla GE MR750 scanner (General Electric, USA). Image processing was completed using FreeSurfer 5.3 and FSL FIRST version 5.0.9 (with FLIRT version 6.0). In all cohorts, visual inspection was used to resolve potential outliers.
Computed tomography
Original fossil human crania (n = 7) and a European sample of recent modern human adults (n = 19) were scanned using computed tomography (CT). The sample shown in Figure 2 comprises the Homo neanderthalensis specimens Gibraltar 1 (labeled as Gi), Guattari (Gu), La Chapelle-aux-Saints (LC), La Ferrassie 1 (LF), Le Moustier 1 (LM1), and the Homo heidelbergensis specimens Kabwe (Ka) and Petralona (Pe).
For all CT specimens, virtual endocasts of the braincase were created using segmentation in Avizo (Thermo Scientific) following [4]. Any missing data in fossil specimens were reconstructed using established protocols [47–49].
Globularity score computation for MRI scans
To quantify the endocranial shape differences between modern humans and Neandertals, we combined the methods of geometric morphometrics [48] using scripts in Mathematica (Wolfram Inc.) with standard neuroimaging data processing protocols (FSL 5.0 and FreeSurfer 5.3). On each virtual endocast, we first distributed a dense mesh of semilandmarks [4, 47–49]. The same mesh of semilandmarks was distributed on an endocast segmented manually from the MNI 152 brain template (MNI 152 T1 2 mm). Semilandmarks are a geometric morphometric technique for quantifying relatively smooth curves and surfaces based on the same number 3D coordinates on each individual [4]. One starts by distributing the same number of 3D coordinates in approximately corresponding locations on each individual. Subsequently these semilandmarks are allowed to slide along the surface so as to remove the influence of the arbitrary initial point spacing. Our iterative semilandmark algorithm allows the surface semilandmarks to slide along tangent planes until the Procrustes distance between each individual and a template shape is minimal (this template shape can be sample average shape, or a single individual as detailed below), and projects the semilandmarks back onto the surface. These iterative sliding steps establish geometric homology among the semilandmarks within a sample [48, 49].
Next, we registered the FreeSurfer segmentation of each MRI scan to the MNI 152 template: we used ‘‘mri_label2vol’’ to register wmparc.mgz to the respective individual’s native anatomical space (rawavg.mgz), and then the command ‘‘flirt’’ to create a transformation matrix between this image and the MNI 152 template. Subsequently, we used ‘‘convert_xfm -omat’’ to create an inverse of this transformation matrix.
In Mathematica, we then applied the inverse of each transformation matrix to the 3D coordinates of the dense mesh of semilandmarks on the MNI 152 template, thereby bringing the semilandmarks into the native anatomical space of each individual. Next, we allowed the semilandmarks on all specimens (i.e., CT scans and MRI scans) to slide so as to minimize the Procrustes distance between each individual and the MNI 152 template (following ref. [49]). This sliding step establishes geometric homology of the semilandmarks [4, 47–49].
We then used Procrustes superimposition on the slid semilandmarks to standardize location and orientation and to scale them to the same centroid size [48]. These Procrustes shape variables were analyzed using principal component analysis (Figure 2A). To quantify globularity, we computed the difference between the Procrustes mean shape of the Neandertal endocrania and the average shape of all modern European endocrania extracted from CT scans. We then projected the Procrustes shape coordinates of all endocrania (i.e., CT and MRI data) onto this multivariate group-difference vector (Figure 2B). This final step yields a ‘‘globularity score’’ for each individual, a reliable continuous trait with a normal distribution, which we used to quantify the phenotype. A subsample (n = 399) of repeated MRI scans (from the BIG cohort) of the same individual on different occasions shows that this ‘‘globularity score’’ has a high test-retest repeatability, with a correlation of 0.97 after correcting for scanning covariates.
Complementary shape analysis and globularity score
We replicated the findings shown in Figure 2 in a more diverse endocranial dataset (Figure S1). This complementary shape analysis is based on endocranial data published in [4], with two additional Neandertal specimens (Saccopastore 1 and Le Moustier 1). This dataset comprises coordinate measurements on computer-generated endocasts of a geographically diverse sample of recent modern human adults (n = 89; the European specimens overlap with the crania used in the main text Figure 2) and fossil humans (n = 20): 8 fossil Homo erectus specimens — KNM-ER 3733 (labeled as ER3733 in Figure S1), KNM-ER 3883 (ER3883), KNM-WT 15000 (WT15k), OH 9, Ngandong 14 (Ng14), Ngawi (Nga), Sambungmacan 3 (Sam3), and Sangiran 2 (Sa2), and the Homo heidelbergensis specimens Kabwe (Ka), and Petralona (Pe). The Neandertal sample includes Amud 1 (labeled as A1), Feldhofer 1 (Fe), Gibraltar 1 (Gi), Guattari 1 (Gu), La Chapelle-aux-Saints (LC), La Ferrassie 1 (LF), Le Moustier 1 (LM1), Saccopastore 1 (SAC1), Spy 1 (S1), and Spy 2 (S2). A manual endocranial segmentation of the MNI 152 template (Figures 1E and1F) was measured following the measurement and data processing protocol of [4]. We then used the same transformation matrices as for the computations in the main text (Figure 2) to transform these landmarks and semilandmarks to the native space of each MRI scan. After a Procrustes superimposition these shape coordinate data were analyzed using between-group principal component analysis based on the group mean endocranial shapes of Homo erectus, Homo neanderthalensis, and recent Homo sapiens (Figure S1B). Globularity scores for this dataset were computed as described above for Figure 2B. Homo erectus, Neandertals, and modern humans have distinct endocranial shapes (Figure S1B). Whereas the globularity scores of Neandertals and Homo erectus overlap completely (indicating that both groups have elongated endocranial shapes), modern humans form a distinct cluster without overlap (Figure S1C).
Characterization of the phenotype
Aspects of brain shape variation within modern humans have recently been linked to naturally occurring brain size variation [50]. Our evolutionarily-derived measure of brain globularity, however, is not related to brain size, as we found no significant correlation between globularity and intracranial volume (Table S3). Moreover, the evolutionary trajectory of globularity within Homo sapiens is not related to endocranial volume, and the adult endocranial volumes of Homo sapiens and Neandertals overlap [4].
We found that endocranial globularity subtly changes with age in modern adult Europeans, with older adults having slightly higher globularity scores (i.e., more rounded endocranial shapes) (Table S3). Notably, however, not a single modern human individual overlapped with Neandertals in overall endocranial shape (Figure 1F, Figure 2B) and thus globularity score (Figure 1G, Figure 2C). Longitudinal MRI data may be able to identify the factors contributing to this subtle age-effect, whether it be due to regional differences in brain volume loss or other processes.
Several studies have associated Neandertal-introgressed genetic variants with variability in phenotypes in modern humans, such as immunity, metabolism, and adaptation to environmental conditions, like temperature and sunlight, as well as neurological and behavioral phenotypes related to depression and addiction [17–20, 51]. To understand whether endocranial globularity is associated with other evolutionarily-linked traits, we tested their partial correlations in the SHIP-2 and SHIP-TREND cohorts (Table S3). Controlling for the effect of age, we saw no significant correlation between the globularity measure and any trait previously associated with Neandertal introgressed alleles.
Voxel-based morphometry analysis
MRI scans were processed for voxel-based analysis (Figure S2, Tables S1 and S2) with SPM12 (Wellcome Trust Centre for Neuroimaging, University College London) and the CAT12 toolbox (developed by Christian Gaser, University of Jena, Germany, www.neuro.uni-jena.de). The images were bias-corrected, spatially normalized by using the high-dimensional DARTEL normalization, segmented into the different tissue classes, modulated for non-linear warping and affine transformations and smoothed by a Gaussian kernel of 8 mm FWHM. The homogeneity of gray matter images was checked using the covariance structure of each image with all other images (outlier R 3 standard deviations from the mean), as implemented in the check data quality function in the CAT12 toolbox. We ran a linear regression model in every voxel of the gray matter segmentations with the globularity score as the exposure variable and adjusted for the following covariates: age (modeled continuously using restricted cubic splines), sex, ICV, and cohort (SHIP, SHIP-Trend). The statistical threshold for significant voxels was set to family wise error (FWE)-corrected peak-level p values ppeak,FWE<0.025 as we were testing two-sided for positive and negative associations with globularity. The labeling of the significant clusters was done within the xjview toolbox (http://www.alivelearn.net/xjview) on the basis of the Anatomical Automatic Labeling atlas (AAL) [52].
Genotyping
Samples from the BIG cohort were genotyped in three batches over a period of several years, using the Affymetrix 6.0, Psychiatric Genomics Consortium PsychChip, or Illumina OmniExpress arrays. The SHIP samples were genotyped using the Affymetrix 6.0 (SHIP) and Illumina Omni 2.5 (SHIP-Trend) arrays.
Sample and SNP-level quality control was performed in PLINK in accordance with the ENIGMA consortium protocol, described previously [43]. Briefly, sample-level quality control included missingness (SHIP: > 8%, BIG: > 5%) and identity-by-descent estimation (removing duplicates). Multidimensional scaling (MDS) components, as calculated in PLINK [53], were used to exclude any outliers resulting from population stratification.
SNPs passing initial quality control measures (SHIP: pHWE > 0.0001 and CallRate > 0.8; SHIP-Trend: pHWE > 0.0001 and CallRate > 0.9; BIG: pHWE > 0.000001, CallRate > 0.95) were imputed to the 1000 Genomes Project (phase 1, version 3, ALL populations) reference panel using IMPUTE v2.2.2 (SHIP) or minimac (release 2012–05-29, BIG) [54–56]. Variants with imputation quality scores (R2) higher than 0.6 were carried forward in the analyses. The reference human genome for SNP annotation was the hg19 (GRCh37) human genome assembly.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Partial correlations between the globularity score and other covariates were performed in R using the ‘ppcor’ package. Scaled globularity scores were corrected for the scanner parameters (BIG only, since the SHIP cohorts used more homogeneous acquisition), and participants’ age and sex (Table S3). As noted above, based on MDS quality control checks in genotype processing, we are confident of minimal population substructure in these European cohorts. Nonetheless, for additional rigor we included the first two principal components to correct for any remaining subtle population stratification. With the model residuals as the trait, association statistics were generated with mach2qtl (BIG) or QUICKTEST version 0.99b (SHIP cohorts) using an additive model. The five sets of summary statistics were aligned using easyQC with the 1000 Genomes Project European phase 1 version 3 reference files.
We restricted our meta-analysis to the list of SNPs originating from Neandertal introgression, which was first presented [18] and further refined as described [20]. The full set included 132,296 variants that differ from the Homo sapiens–Neandertal common ancestor and match the Neandertal sequence. The meta-analysis was performed in METAL, using the standard error based method, a minimum minor allele frequency of 0.01, and including the genomic control option [57]. In follow-up tests of the three top SNPs, assessing how the number of genetic principal components in the model affected association statistics, the genomic control option was not used as the meta-analysis involved only three markers. Any SNPs missing from one or more of the cohorts were excluded, leaving 50,057 variants in the final analysis. As we tested a pre-defined, directional hypothesis that Neandertal alleles would be associated with a shift toward more archaic endocranial shapes, we calculated p values based on a one-tailed association test.
Neighboring SNPs on introgressed fragments are often in linkage disequilibrium with each other, and hence show varying degrees of non-independence in association testing. To appropriately adjust for the multiple testing of many SNPs, accounting for the existing linkage disequilibrium structure, the effective number of independent tests was calculated using the Genetic Type-1 Error Calculator (GEC), with the 1000 Genomes Project VCF as input [21]. Significance was assessed using the significant (7.38 3 10—6) and suggestive thresholds (1.48 3 10—4) provided by GEC.
GTEx eQTL Analysis
The GTEx dataset was obtained from dbGaP (accession number phs000424.v6.p1.c1, accessed on 05/23/2016). The processing of GTEx v6 data for 48 tissues, for which > 50 individuals had available genotype data, has been described in detail elsewhere [25]. We retained protein-coding genes for a given tissue if there were at least two individuals in the dataset that had a read count greater than 0. We used DESeq2 to normalize all expressed genes in each tissue between individuals. For genes within 50 kilobases of a Neandertal SNP, we calculated Spearman correlations between the SNP and the normalized gene expression, if there were at least two genotypes with a minimum of two individuals each. As the top 5% of genes associated with Neandertal SNPs contained an excess of low p values, these were defined as a significant gene expression associations for each tissue, following the practice of the prior published work using this approach [25]: instead of simply using the eQTLs identified by GTEx consortium we re-evaluated the full dataset in order to increase power to detect associations with Neandertal introgressed alleles, which tend to be at lower frequency in present day genomes. Because of this low frequency, the power to detect associations with Neandertal alleles is more limited than for the higher frequency variants (for which the cut-offs used in the GTEx study were optimized). Additionally, the power to detect associations differs substantially between tissues as a result of the differences in sample size. We therefore used a quantile cut-off to avoid discrimination against low frequency alleles, and tissues with lower sample sizes. As a result it is possible that we identify rare Neandertal-introgressed variants that are excluded in the GTEx browser due to frequency filters. Sample sizes are listed in Data S1.
DATA AND SOFTWARE AVAILABILITY
Association summary statistics are available in Data S1. The Mathematica script used to compute the globularity scores is available from the authors upon request.
Supplementary Material
Highlights.
We use fossil skull data to derive an index of endocranial shape in human MRI scans
In 4,468 Europeans, we screen introgressed Neandertal SNPs for association with the index
Lead SNPs consistently associate with reduced globularity in five separate subsamples
These SNPs affect neural expression of two genes linked to neurogenesis and myelination
ACKNOWLEDGMENTS
BIG is part of Cognomics, supported by the Donders-Centre for Cognitive Neuroimaging, Departments of Human-Genetics and Cognitive Neuroscience (Radboud University Medical Centre), MPI for Psycholinguistics. Additional funding: BBMRI-NL, Hersenstichting-Nederland, Netherlands Organisation for Scientific Research (NWO), FP7/2007–2013: 602450 (IMAGEMEND) and 602805 (Aggressotype), NIH-U54-EB020403. SHIP funded by BMBF Germany (01ZZ9603, 01ZZ0103, 01ZZ0403, 03IS2061A, 03ZIK012), Federal State, Ministry of Cultural Affairs & Social Ministry of Mecklenburg-West Pomerania, Siemens Healthineers. Thanks to InterSystems (Cache´-Campus), German Research Foundation (GR-1912/5–1), Max Planck Society, Nomis Foundation (to S.P.), and NIH 1-U24-RR021992 and 1-U24-RR025736–01. Funding sources had no role in study design, data collection, analysis, and interpretation. We thank the anonymous referees for detailed and constructive feedback that helped improve the manuscript.
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing interests. B.F. received educational speaking fees from Shire and Medice.
REFERENCES
- 1.Lieberman DE, McBratney BM, and Krovitz G (2002). The evolution and development of cranial form in Homo sapiens. Proc. Natl. Acad. Sci. USA 99, 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruner E, Manzi G, and Arsuaga JL (2003). Encephalization and allometric trajectories in the genus Homo: evidence from the Neandertal and modern lineages. Proc. Natl. Acad. Sci. USA 100, 15335–15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruner E (2004). Geometric morphometrics and paleoneurology: brain shape evolution in the genus Homo. J. Hum. Evol 47, 279–303. [DOI] [PubMed] [Google Scholar]
- 4.Neubauer S, Hublin JJ, and Gunz P (2018). The evolution of modern human brain shape. Sci. Adv 4, eaao5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochiyama T, Ogihara N, Tanabe HC, Kondo O, Amano H, Hasegawa K, Suzuki H, Ponce de León M.S., Zollikofer CPE, Bastir M, et al. (2018). Reconstructing the Neanderthal brain using computational anatomy. Sci. Rep 8, 6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. (2010). A draft sequence of the Neandertal genome. Science 328, 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyrégne S, et al. (2017). A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton RA (2012). Embodied cognitive evolution and the cerebellum. Philos. Trans. R. Soc. Lond. B Biol. Sci 367, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton RA, and Venditti C (2014). Rapid evolution of the cerebellum in humans and other great apes. Curr. Biol 24, 2440–2444. [DOI] [PubMed] [Google Scholar]
- 10.Hublin JJ, Ben-Ncer A, Bailey SE, Freidline SE, Neubauer S, Skinner MM, Bergmann I, Le Cabec A, Benazzi S, Harvati K, and Gunz P (2017). New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature 546, 289–292. [DOI] [PubMed] [Google Scholar]
- 11.Scerri EML, Thomas MG, Manica A, Gunz P, Stock JT, Stringer C, Grove M, Groucutt HS, Timmermann A, Rightmire GP, et al. (2018). Did Our Species Evolve in Subdivided Populations across Africa, and Why Does It Matter? Trends Ecol. Evol 33, 582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunz P, Neubauer S, Maureille B, and Hublin JJ (2010). Brain development after birth differs between Neanderthals and modern humans. Curr. Biol 20, R921–R922. [DOI] [PubMed] [Google Scholar]
- 13.Neubauer S, Gunz P, and Hublin JJ (2010). Endocranial shape changes during growth in chimpanzees and humans: a morphometric analysis of unique and shared aspects. J. Hum. Evol 59, 555–566. [DOI] [PubMed] [Google Scholar]
- 14.Ponce de León M.S., Bienvenu T, Akazawa T, and Zollikofer CP (2016). Brain development is similar in Neanderthals and modern humans. Curr. Biol 26, R665–R666. [DOI] [PubMed] [Google Scholar]
- 15.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. (2014). The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, et al. (2016). Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530, 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, and Reich D (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vernot B, and Akey JM (2014). Resurrecting surviving Neandertal lineages from modern human genomes. Science 343, 1017–1021. [DOI] [PubMed] [Google Scholar]
- 19.Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. (2016). Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, et al. (2016). The phenotypic legacy of admixture between modern humans and Neandertals. Science 351, 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li MX, Yeung JM, Cherny SS, and Sham PC (2012). Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet 131, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, et al. ; Alzheimer’s Disease Neuroimaging Initiative; CHARGE Consortium; EPIGEN; IMAGEN; SYS (2015). Common genetic variants influence human subcortical brain structures. Nature 520, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory MD, Kippenhan JS, Eisenberg DP, Kohn PD, Dickinson D, Mattay VS, Chen Q, Weinberger DR, Saad ZS, and Berman KF (2017). Neanderthal-Derived Genetic Variation Shapes Modern Human Cranium and Brain. Sci. Rep 7, 6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium GTEx (2017). Genetic effects on gene expression across human tissues. Nature 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dannemann M, Prüfer K, and Kelso J (2017). Functional implications of Neandertal introgression in modern humans. Genome Biol 18, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons K, Nakatani Y, and Nguyen MD (2015). p600/UBR4 in the central nervous system. Cell. Mol. Life Sci 72, 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belzil C, Asada N, Ishiguro K, Nakaya T, Parsons K, Pendolino V, Neumayer G, Mapelli M, Nakatani Y, Sanada K, and Nguyen MD (2014). p600 regulates spindle orientation in apical neural progenitors and contributes to neurogenesis in the developing neocortex. Biol. Open 3, 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. ; Exome Aggregation Consortium (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flores AI, Narayanan SP, Morse EN, Shick HE, Yin X, Kidd G, Avila RL, Kirschner DA, and Macklin WB (2008). Constitutively active Akt induces enhanced myelination in the CNS. J. Neurosci 28, 7174–7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao T, Furnari F, and Newton AC (2005). PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell 18, 13–24. [DOI] [PubMed] [Google Scholar]
- 31.Brognard J, Sierecki E, Gao T, and Newton AC (2007). PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell 25, 917–931. [DOI] [PubMed] [Google Scholar]
- 32.Franke B, Stein JL, Ripke S, Anttila V, Hibar DP, van Hulzen KJE, Arias-Vasquez A, Smoller JW, Nichols TE, Neale MC, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; ENIGMA Consortium (2016). Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat. Neurosci 19, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryskalin L, Lazzeri G, Flaibani M, Biagioni F, Gambardella S, Frati A, and Fornai F (2017). mTOR-Dependent Cell Proliferation in the Brain. BioMed Res. Int 2017, 7082696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reijnders MRF, Kousi M, van Woerden GM, Klein M, Bralten J, Mancini GMS, van Essen T, Proietti-Onori M, Smeets EEJ, van Gastel M, et al. (2017). Variation in a range of mTOR-related genes associates with intracranial volume and intellectual disability. Nat. Commun 8, 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figlia G, Gerber D, and Suter U (2018). Myelination and mTOR. Glia 66, 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pääbo S (2014). The human condition-a molecular approach. Cell 157, 216–226. [DOI] [PubMed] [Google Scholar]
- 37.Fallon J, Opole I, and Potkin S (2003). The neuroanatomy of schizophrenia: circuitry and neurotransmitter systems. Clin. Neurosci. Res 3, 77–107. [Google Scholar]
- 38.Ramnani N (2012). Frontal lobe and posterior parietal contributions to the cortico-cerebellar system. Cerebellum 11, 366–383. [DOI] [PubMed] [Google Scholar]
- 39.Middleton FA, and Strick PL (2000). Basal ganglia and cerebellar loops: motor and cognitive circuits. Brain Res. Brain Res. Rev 31, 236–250. [DOI] [PubMed] [Google Scholar]
- 40.Enard W (2011). FOXP2 and the role of cortico-basal ganglia circuits in speech and language evolution. Curr. Opin. Neurobiol 21, 415–424. [DOI] [PubMed] [Google Scholar]
- 41.Fisher SE (2017). Evolution of language: Lessons from the genome. Psychon. Bull. Rev 24, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guadalupe T, Zwiers MP, Teumer A, Wittfeld K, Vasquez AA, Hoogman M, Hagoort P, Fernandez G, Buitelaar J, Hegenscheid K, et al. (2014). Measurement and genetics of human subcortical and hippocampal asymmetries in large datasets. Hum. Brain Mapp 35, 3277–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibar DP, Adams HHH, Jahanshad N, Chauhan G, Stein JL, Hofer E, Renteria ME, Bis JC, Arias-Vasquez A, Ikram MK, et al. (2017). Novel genetic loci associated with hippocampal volume. Nat. Commun 8, 13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Völzke H, Alte D, Schmidt CO, Radke D, Lorbeer R, Friedrich N, Aumann N, Lau K, Piontek M, Born G, et al. (2011). Cohort profile: the study of health in Pomerania. Int. J. Epidemiol 40, 294–307. [DOI] [PubMed] [Google Scholar]
- 45.Ashburner J, and Friston KJ (2000). Voxel-based morphometry–the methods. Neuroimage 11, 805–821. [DOI] [PubMed] [Google Scholar]
- 46.van Erp TG, Greve DN, Rasmussen J, Turner J, Calhoun VD, Young S, Mueller B, Brown GG, McCarthy G, Glover GH, et al. (2014). A multi-scanner study of subcortical brain volume abnormalities in schizophrenia. Psychiatry Res 222, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunz P, Mitteroecker P, Neubauer S, Weber GW, and Bookstein FL (2009). Principles for the virtual reconstruction of hominin crania. J. Hum. Evol 57, 48–62. [DOI] [PubMed] [Google Scholar]
- 48.Mitteroecker P, and Gunz P (2009). Advances in Geometric Morphometrics. Evol. Biol 36, 235–247. [Google Scholar]
- 49.Gunz P, and Mitteroecker P (2013). Semilandmarks: a method for quantifying curves and surfaces. Hystrix It. J. Mamm 24, 103–109. [Google Scholar]
- 50.Reardon PK, Seidlitz J, Vandekar S, Liu S, Patel R, Park MTM, Alexander-Bloch A, Clasen LS, Blumenthal JD, Lalonde FM, et al. (2018). Normative brain size variation and brain shape diversity in humans. Science 360, 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dannemann M, and Kelso J (2017). The Contribution of Neanderthals to Phenotypic Variation in Modern Humans. Am. J. Hum. Genet 101, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, and Joliot M (2002). Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289. [DOI] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, and Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howie BN, Donnelly P, and Marchini J (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuchsberger C, Abecasis GR, and Hinds DA (2015). minimac2: faster genotype imputation. Bioinformatics 31, 782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, and Abecasis GR; 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willer CJ, Li Y, and Abecasis GR (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Association summary statistics are available in Data S1. The Mathematica script used to compute the globularity scores is available from the authors upon request.