Supplemental Digital Content is available in the text
Keywords: biomarkers, expression profiling, integrative analysis, long non-coding RNA, osteosarcoma
Abstract
Transcriptome profiling of osteosarcoma (OS) by next generation sequencing technology (NGS) has been broadly performed by previous researches, which uncovers a large number protein-coding driver genes, facilitates our understanding of the molecular mechanisms of OS formation, progression and metastasis. Recently, more and more researchers realize the importance of long non-coding RNAs (lncRNAs) on the development of OS. However, few studies focus on discovering driver lncRNAs.
Here we collected somatic copy number alterations (SCNAs) and gene expression profiles of 84 samples from Therapeutically Applicable Research to Generate Effective Treatments (TARGET) project. The RNA sequencing data detected 13,903 expressed lncRNAs, 157 of which were previously reported to be associated with cancer based on the annotations from Lnc2Cancer database.
By analyzing the SNP array data, several significant SCNAs were detected, such as the amplifications on chromosomes 1q, 4q, 17p, 17q, and 19q, and deletions on 1q, 3q, 9p, 10q, and 15q. With the SCNA and gene expression profiles, we identified 167 driver genes by integrative analysis, including 162 novel driver lncRNAs, 2 lncRNAs reported to be associated with OS, and another 3 associated with other cancers. Furthermore, functional characterization and survival analysis revealed that RP11-241F15.10 may function as a tumor suppressor in OS, and loss of function may contribute to activation of Wnt signaling pathway.
This study not only facilitates our understanding of the oncogenic or tumor-suppressor role of lncRNAs in OS, but also provides potential therapies for the patients with OS with metastasis or relapse.
1. Introduction
Osteosarcoma (OS) is the most common primary cancer of bone, which can destroy tissue and weaken the bone, and mostly occurs in childhood and adolescence. The incidence of osteosarcoma in the general population is 1 per million per year, but is higher in adolescence, in which the annual incidence peaks at about 10 per million per year at 15 to 19 years of age.[1,2] There are several imaging tests for OS diagnosis, such as X-ray, magnetic resonance imaging (MRI),[3] bone scan, and CT.[4] These imaging tests play important roles in diagnosis and characterization, which will help guide osteosarcoma therapy.[5,6]
In most patients, the etiology of OS remains unclear. Risk factors include height,[7–9] birth weight,[8] and genetic background,[10] pubertal hormones and other factors related to bone growth.[11] The treatment for primary osteosarcoma is surgery. However, the survival of patients with OS treated with only surgery is approximately 15 to 17%.[12,13] In the early 1970 s, high-dose methotrexate and vincristine followed by folinic acid, was introduced as adjuvant chemotherapy for patients with non-metastatic disease.[14] Current therapies incorporate surgical resection and combinational chemotherapy, which cures ∼70% of patients. However, survival for patients with metastatic or relapsed osteosarcoma has remained unchanged over the past four decades, with a 5-year overall survival rate of about 20%,[13,15] suggesting that new therapies are urgently needed for the patients with metastatic or relapsed osteosarcoma.
The latest developments in next generation sequencing (NGS) technologies have profiled mutational spectrums, deregulated expression and epigenetic changes of several cancers by Therapeutically Applicable Research to Generate Effective Treatments (TCGA) studies.[16] Whole genome or exome sequencing of OS have identified some recurrently mutated genes, such as TP53, RB1, CDKN2A, PTEN, and YAP1.[17] Transcriptome sequencing of cancers shows remarkable potential to identify both novel biomarkers and uncharacterized aspects of tumor biology, particularly some long non-coding RNAs. The LncRNAs are transcripts that are unable to translate proteins in the intracellular space. It has been widely accepted that more than 90% of the human genome DNA is thought to be transcribed, while only about 2% of it can encode proteins. The LncRNAs can participate in tumorigenesis or progression by a variety of ways. For instance, TUG1,[18] H19,[19] LINC00161,[20] and LOC285194[21] can act as miRNA sponges, thereby indirectly regulating the miRNA targets. Moreover, lncRNAs can also directly regulate gene transcription in OS, such as MEG3[22] and ZEB1-AS1.[23]
In the present study, we collected somatic copy number alterations and RNA-seq based gene expression profiles of 84 samples from TARGET project. The RNA-seq based gene expression profiles include more lncRNAs than microarray data, which is more beneficial for us to detect novel driver lncRNAs. To identify driver lncRNAs in OS, integrative analysis of somatic copy number alterations (SCNAs) and gene expression profiles was performed. Pathway enrichment analysis highlighted some candidate driver lncRNAs that may participate in cancer-related pathways. The present study not only facilitated our understanding of the oncogenic or tumor-suppressor role of lncRNAs in OS, but also provided potential prognostic biomarkers and therapies for the patients with OS with metastasis or relapse.
2. Materials and methods
2.1. Data sources and osteosarcoma samples
The SCNA, gene expression data, and clinical information were downloaded from the publicly available website of the National Cancer Institute TARGET Data Matrix (https://ocg.cancer.gov/programs/target/data-matrix), which deposits multi-omics datasets of some pediatric cancers, such as somatic mutations, SCNA, structural variations (SV), DNA methylation, miRNA expression, and gene expression. The SCNA and gene expression data of OS have been preprocessed and quantified, and could be directly used for downstream analysis. To meet the requirement for data analysis, we only collected 84 osteosarcoma samples with paired SCNA and gene expression data.
2.2. Significantly amplified and deleted regions
Before identifying the significantly altered regions, the segment mean of each CNA call was transformed as log2-ratio:
The identification of significantly altered regions was implemented by GISTIC2 on GenePattern server (https://genepattern.broadinstitute.org/gp). Notably, the thresholds for amplifications and deletions were set as 0.4 and -0.4, and other parameters were left as their defaults.
2.3. Identification of driver genes by integrative analysis
To evaluate whether expression of these genes was related to CNV, patients were classified into 2 groups according to their CNV status: 1 group is for copy number variated (gain/loss), and the other group for copy number neutral. For each gene, a Wilcoxon rank-sum test was applied to the gene expression levels between copy number variated tumor tissues and copy number neutral tumor tissues. To identify CNV-driven genes, only genes with concordant changes in copy number and gene expression were collected for further analyses.
2.4. Pathway enrichment analysis
The enrichment analysis was implemented in fgsea package with pre-rank mode in R programming language. The genes were ranked based on their correlation with each lncRNA. Pathway database was downloaded from MsigDB (http://software.broadinstitute.org/gsea/index.jsp). We only retained NCI-PID[24] for enrichment analysis.
2.5. Cox-regression based survival analysis
The survival analysis based on Cox-regression model was implemented in R with package survival. The comparison of survival curves for high and low expression groups was performed using the G-rho family of tests with survdiff function. Kaplan–Meier curve was used to visualize the survival probability for each group. The hazard ratios and corresponding p-values were calculated by hazard.ratio function in survcomp package.
3. Results
3.1. Landscape of expressed lncRNAs in osteosarcoma
We analyzed the gene expression profiles of 84 osteosarcomas from TARGET project, which included a total of 39,615 genes with ENSEMBL gene annotation (GRCh37.71 version). Following the previous study,[25] 6 biotypes, as well as pseudogenes, were defined as long non-coding RNAs: lincRNA, processed_transcript, sense_intronic, sense_overlapping, antisense, and 3prime_overlapping_ncrna. In total, 33,214 genes were deemed as expressed genes that have expression (TPM > 1) in at least 1 sample, including 18,868 protein-coding genes, 13,903 lncRNAs, and 443 other ncRNAs (Fig. 1A). Compared with the protein-coding genes, lncRNAs were expressed at lower levels (Fig. 1B, Wilcoxon rank-sum test, P-value < 2.2e-16), which is consistent with previous studies.[25–27]
Figure 1.

Overview of the protein-coding genes and long non-coding RNAs in osteosarcoma (OS). (A) The proportions of protein-coding genes, long non-coding RNAs (lncRNAs), and other RNAs detected by RNA sequencing are illustrated in the pie chart. The specific numbers are presented on the bottom right. (B) The estimated probability density functions for the expression levels (log2-TPM) of protein-coding genes and lncRNAs. (C) The top-10 most highly expressed cancer lncRNAs in OS. The expression values were normalized as TPM, and log2-transformed. OS = osteosarcoma, lncRNAs = long non-coding RNAs.
Among the expressed lncRNAs, 157 were previously reported to be associated with cancer based on the annotation from Lnc2Cancer database[28] (Supplementary Table S1). The top-ten most highly expressed cancer lncRNAs were GAS5, linc-ITGB1, H19, SNHG5, SNHG16, ZFAS1, UFC1, DANCR, CRIP2, and SNHG1 (Fig. 1C). Particularly, recent studies revealed that ZFAS1,[29] H19,[30] DANCR,[31] and SNHG1[32] could function as miRNA sponges in osteosarcoma, thereby promoting tumorigenesis or tumor progression. These results indicated that lncRNAs may act as a crucial role in initiation or progression of osteosarcoma.
3.2. Significant somatic copy number alterations in patients with osteosarcoma
The significant somatic copy number alterations were identified by GISTIC2 on the GenePattern server. As shown in Figure 2, several significant SCNAs were detected, such as the amplifications on chromosomes 1q, 4q, 17p, 17q, and 19q, and deletions on 1q, 3q, 9p, 10q, and 15q. In general, some oncogenes and tumor suppressors were located within the amplified and deleted regions, respectively. Some well-recognized driver genes, such as MCL1 (1q21.3), MYC and PVT1 (8q23.21), 19q12 (CCNE1), IGF1R (15q26.3), PDGFRA (4q12), NCOR1 (17p12), JAK1 (1p31.2), TERT (5p15.33), and KDR and KIT (4q12), were frequently amplified in patients with osteosarcoma. Notably, MYC and NCOR1 were regulators in gene transcription, and the other oncogenes like IGFR1, PDGFRA, JAK1, KDR, and KIT were involved in signaling transduction pathways. As expected, tumor suppressors, such as TP53 (17p13.1), RB1 (13q14.2), CDKN2A/CDKN2B (9p21.3), PTEN (10q23.31), and NF1 (17q11.2) were frequently deleted. Particularly, loss of TP53, RB1, and CDKN2A/CDKN2B may result in uncontrolled cell cycle progression, and loss of PTEN and NF1 may activate PI3K/AKT/mTOR and Ras signaling. Although some of the significant SCNA regions were characterized by oncogenes or tumor suppressors, most of these regions were still lack of specific cancer driver genes to associate with the disease, giving us a hint that some lncRNAs located within these regions may function as cancer drivers.
Figure 2.
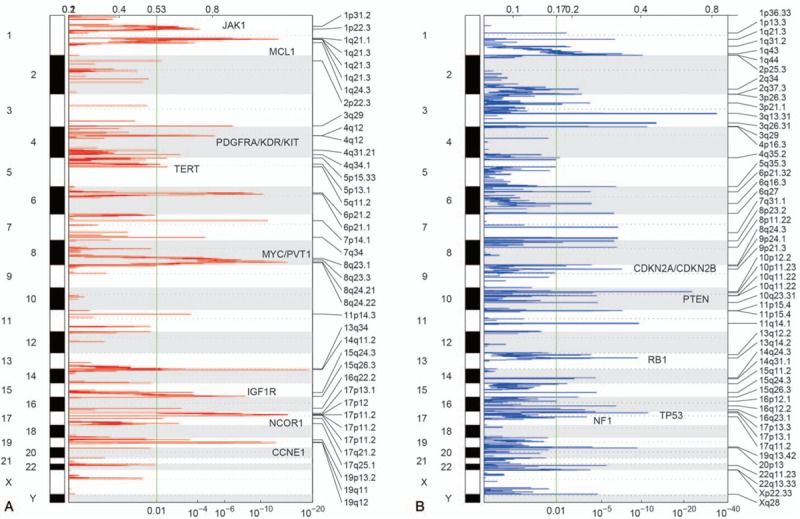
Significantly amplified and deleted regions in osteosarcoma (OS). The significantly amplified (A) and deleted (B) regions are identified by GISTIC2. The red and blue represent the copy number gains and losses, respectively. The numbers on the top and bottom represent the mutated frequency and the significance level of Q-value. OS = osteosarcoma.
3.3. Identification of potential driver lncRNAs in osteosarcoma
To explore the genes in the significant SCNAs, we focused on 16,310 genes with at least 10% (9/84) of samples showing copy number changes and having expression (TPM > 1) in the following analyses. To identify the potential driver lncRNAs, we evaluated whether the expression levels of both protein-coding genes and lncRNAs were associated with SCNAs. For each gene, the patients were divided into 2 groups based on the copy number statuses (Methods), and a Wilcoxon rank-sum test was applied to the 2 groups, by which we identified 1162 differentially expressed genes (DEGs) (false discovery rate [FDR] < 0.05 and |log2 fold change|>1), including 995 protein-coding genes and 167 lncRNAs (Supplementary Table S2), which may be candidate driver genes in OS. The 995 protein-coding driver genes were overrepresented in driver genes from COSMIC Cancer Gene Census (CGC) database (Fisher test, P-value < .05), indicating that our integrative analysis was effective to detect driver genes. Overrepresentation enrichment analysis revealed that the up-regulated DEGs due to copy number gains were significantly enriched in regions such as 13q34, 17p11.2, 17p13.1, 19q13.11, and 6p21.1, while the down-regulated DEGs due to copy number deletion were mostly located within regions like 13q34, 16q24.3, 17p13.1, 17p12, and 1q32.1 (Fig. 3, FDR < 0.05).
Figure 3.
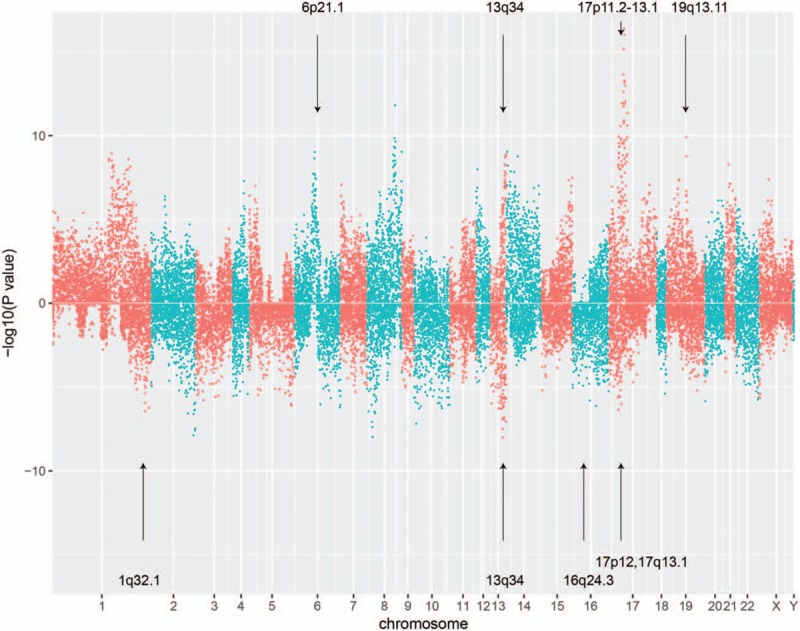
The potential driver genes in osteosarcoma (OS) by integrative analysis. The top and bottom panels represent the copy number gain and loss, respectively. The overrepresented regions that contain more candidate driver genes were highlighted by the arrows. OS = osteosarcoma.
Of the 167 candidate driver lncRNAs, PVT1,[33] and ZFAS1[29] have been reported to promote osteosarcoma metastasis or invasion. Three antisense RNAs, CBR3-AS1,[34,35] FOXD2-AS1,[36] and HOXA-AS2,[37–39] were associated with other cancers by previous studies. For the 3 antisense RNAs and the remaining 162 candidate driver lncRNAs, it is necessary to further investigate molecules or pathways that they may regulate or participate in.
3.4. Prediction of dysregulated pathways of candidate driver lncRNAs
To clarify the dysregulated pathways of three antisense RNAs and remaining 162 candidate driver lncRNAs, correlation analysis and gene set enrichment analysis (GSEA) were performed (Methods). Firstly, Song W, et al[40] found that CBR3-AS1 could promote colorectal cancer cell progression by activating PI3K/Akt signaling pathway. The GSEA revealed that highly correlated genes with CBR3-AS1 were enriched in MET signaling pathway (Q-value < 0.05), which could activate multiple signal transduction pathways like PI3K/Akt signaling pathway[35] (Fig. 4A), and further confirmed the association of CBR3-AS1 with PI3K/Akt signaling pathway in OS. Secondly, FOXD2-AS1 was positively correlated with genes from beta-catenin nuclear pathway (Q-value < 0.05), which was consistent with the results that FOXD2-AS1 could promote non-small cell lung cancer progression via Wnt/β-catenin signaling by Rong L, et al[36] (Fig. 4B). Thirdly, HOXA-AS2 overexpression was found in colorectal cancer[38] and hepatocellular carcinoma,[39] and could promote tumor cell proliferation. Further analysis found that HOXA-AS2 may be involved in mTOR signaling pathway based on GSEA (Fig. 4C, Q-value < 0.05), thereby leading to uncontrolled cell proliferation of OS.
Figure 4.
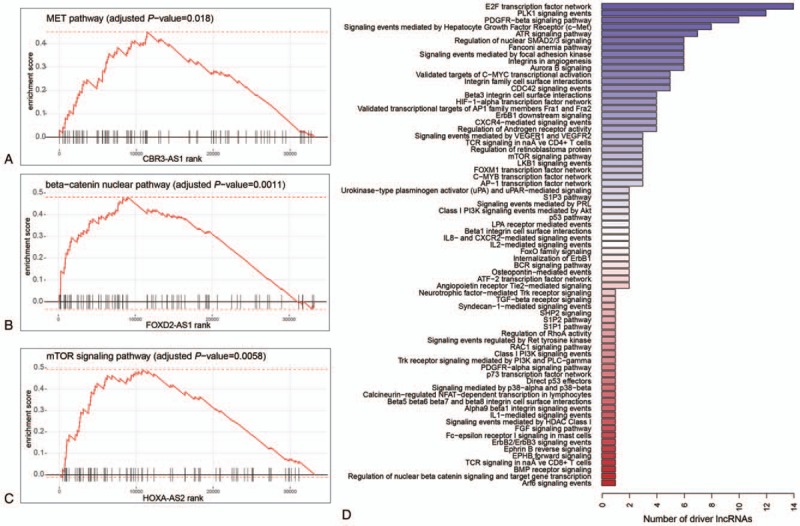
The functional characterization of the candidate driver lncRNAs. The CBR3-AS1, FOXD2-AS1, and HOXA-AS2 were positively correlated with genes from MET pathway (A), beta-catenin nuclear pathway (B), and mTOR signaling pathway (C). The distribution of predicted pathways across the candidate driver lncRNAs. The x-axis represents the number of lncRNAs that were characterized by each pathway. lncRNAs = long non-coding RNAs.
Similarly, the dysregulated pathways of the remaining 162 candidate driver lncRNAs were also predicted, of which, 66 lncRNAs were successfully annotated with 5 most significantly enriched pathways based on GSEA (Q-value < 0.05, Fig. 4D). Overall, most of these lncRNAs were closely associated with signaling transduction pathways, such as PLK1 signaling, PDGFR-beta signaling, signaling events mediated by c-Met, and ATR signaling, which were frequently activated in multiple cancers. Moreover, highly correlated genes with lncRNAs were also significantly enriched in target genes of proliferation-related transcription factors, such as E2F, MYC, FOXM1, AP-1, and C-MYB, indicating that the candidate driver lncRNAs may be associated with excessive proliferation. In addition, integrins in angiogenesis and HIF1-alpha transcription factor network were also identified, suggesting that some lncRNAs may contribute to angiogenesis, thereby promoting tumor metastasis. In summary, the prediction of dysregulated pathways of the candidate driver lncRNAs revealed that these lncRNAs may participate in several cancer-related pathways, and be closely associated with tumor initiation, progression, or metastasis.
3.5. Clinical significance of expression of driver lncRNAs in osteosarcoma
To determine the clinical significance of expression of driver lncRNAs, we investigated whether their expression levels correlated with event-free survival (EFS) or overall survival (OS). In total, 17 and 11 were identified as EFS- and OS-related driver lncRNAs, respectively (log-rank test, P-value < .05), and 7 of these lncRNAs, including EMG1, AC068831.10, RP11-360D2.2, ASMTL-AS1, AC004019.13, RP11-241F15.10, and CTD-2319I12.1, were associated with both EFS and OS (Table 1). To validate the associations between lncRNA expression and prognosis, we collected another 17 tumor samples from TARGET, and performed Wilcoxon rank sum test to examine the differential expression of these lncRNAs between samples with and without event, and between dead and alive samples, respectively. In accordance with the results above, RP11-360D2.2 and RP11-241F15.10 were still significantly associated with both EFS and OS (P < .1 and fold change > 1 or < 1/2, Supplementary Table S3), further suggesting their key roles in OS progression. Remarkably, the potential tumor suppressor, RP11-241F15.10, with ENSEMBL identifier ENSG00000250753, was frequently deleted in OS (15/84, 19%), which resulted in significant decrease of its expression (Fig. 5A, FDR < 0.05). Survival analysis demonstrated that it was negatively correlated with both event-free survival (Fig. 5B, P-value = .0014) and overall survival (Fig. 5C, P-value = .0013). Moreover, we found that down-regulation of RP11-241F15.10 was associated with activation of components from WNT signaling, such as beta-catenin nuclear pathway, and MYC activity pathway based on GSEA (Fig. 5D–E). The predicted functionality of RP11-241F15.10 suggested that it is a potential tumor suppressor by inhibiting the activity of nuclear transcription factors, such as TCF7 and LEF1 (Fig. 5F–G), thereby down-regulating WNT-targets. In summary, dysregulated driver lncRNAs could promote OS metastasis or relapse by activating some cancer-related signaling pathways.
Table 1.
The summary for the 7 long non-coding RNAs (lcnRNAs) associated with both event-free survival (EFS) and overall survival (OS).

Figure 5.

The clinical significance and functional annotation of RP11-241F15.10. (A) The expression levels of RP11-241F15.10 between wild and deleted OS samples. (B) and (C) display the event-free and overall survival time between OS samples with high and low expression of RP11-241F15.10. (D) and (E) display the negatively correlated pathways with RP11-241F15.10, Beta-catenin nuclear pathway and MYC activity pathway. (F) and (G) The TCF7 and LEF1 expression levels in RP11-241F15.10 wild and deleted samples. OS = osteosarcoma.
4. Discussion
The molecular basis of OS about protein-coding genes has largely been studied in the context of tumorigenesis, progression and metastasis. Despite extensive researches about the function of protein-coding genes in OS, the lack of effective biomarkers for OS therapies is still not thoroughly solved. Meanwhile, a majority of long non-coding RNAs are characterized to act as cancer driver RNAs, and understanding their deregulation and regulatory roles can facilitate the development of new diagnostic or therapeutic strategies.
The present study aims to uncover the candidate driver lncRNAs, and characterize their functionality. In the cohort of 84 OS, RNA sequencing data detected 13,903 expressed lncRNAs, which was more beneficial for us to carry out this research. The analysis of SCNA data found several recurrently gained or deleted regions, which contained some well-recognized driver genes, such as MCL1, MYC and PVT1, 19q12, IGF1R, PDGFRA, NCOR1, JAK1, TERT, and KDR, KIT, TP53, RB1, CDKN2A/CDKN2B, PTEN, and NF1. However, most of these regions were not characterized by any protein-coding driver genes, giving us a hint that some lncRNAs located within these regions may function as cancer drivers. With the paired SCNA data for the 84 samples, integrative analysis was performed, and successfully identified 995 driver protein-coding genes, and 167 candidate driver lncRNAs. Moreover, the 995 protein-coding driver genes were overrepresented in driver genes from COSMIC CGC database (Fisher test, P-value < .05). The functionalities of candidate driver lncRNAs were subsequently annotated based on GSEA. For the three lncRNAs that were reported to associate with other cancers, the GSEA analysis also provided the evidence about their regulatory roles in OS. Furthermore, most of the candidate driver lncRNAs were predicted to participate in some cancer-related pathways, further illustrating their important roles in OS. In addition, the dysregulated candidate driver lncRNA, RP11-241F15.10, could also be potentially used to predict event-free survival and overall survival of patients with OS, suggesting that its aberrant expression may promote tumor progression, metastasis, relapse, and even death for patients with OS. However, the present study also has some limitations. For example, as we aim to uncover some potential driver lncRNAs by integrative analysis, molecular experiments are very necessary to further validate their biological function in tumor formation and development. Moreover, independent RNA sequencing datasets of tumor tissues can also be used to validate their clinical significance.
In this study, our analyses demonstrated that lncRNAs could function as oncogenic or tumor suppressor RNAs. Although further characterization of their molecular mechanism remains necessary, these lncRNAs may play functionally vital roles in OS formation or progression, and provide some resources for the biological researchers. In summary, our integrative analysis of lncRNAs further illustrated the important roles of lncRNAs in OS, and generates novel insight into cancer biology.
Author contributions
Conception and design: ZL, LX and CW; Development of methodology: ZL, JL, BD and CW; Data collection: ZL, LX, JL and BD; Analysis and interpretation of data: J ZL, LX, JL and BD; Writing, review, and/or revision of the manuscript: ZL, LX, JL BD, CW. All authors read and approved the final manuscript.
Conceptualization: Zhenguo Luo, Li Xiao, Jing Li, Chunsheng Wang.
Data curation: Zhenguo Luo, Li Xiao, Jing Li, Buhuai Dong, Chunsheng Wang.
Formal analysis: Buhuai Dong.
Methodology: Chunsheng Wang.
Writing – original draft: Zhenguo Luo, Li Xiao, Jing Li, Chunsheng Wang.
Writing – review & editing: Jing Li, Buhuai Dong, Chunsheng Wang.
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: CGC = Cancer Gene Census, DEGs = differentially expressed genes, EFS = event-free survival, FDR = false discovery rate, GSEA = gene set enrichment analysis, NGS = next generation sequencing, OS = osteosarcoma, OS = overall survival, SCNAs = somatic copy number alterations, TARGET = Therapeutically Applicable Research to Generate Effective Treatments.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Stiller CA, Bielack SS, Jundt G, et al. Bone tumours in European children and adolescents, 1978-1997. Report from the Automated Childhood Cancer Information System project. Eur J Cancer 2006;42:2124–35. [DOI] [PubMed] [Google Scholar]
- [2].Stiller CA, Craft AW, Corazziari I, et al. Survival of children with bone sarcoma in Europe since 1978: results from the EUROCARE study. Eur J Cancer 2001;37:760–6. [DOI] [PubMed] [Google Scholar]
- [3].Razek A, Nada N, Ghaniem M, et al. Assessment of soft tissue tumours of the extremities with diffusion echoplanar MR imaging. Radiol Med 2012;117:96–101. [DOI] [PubMed] [Google Scholar]
- [4].Abdel Razek AAK, Samir S. Diagnostic performance of diffusion-weighted MR imaging in differentiation of diabetic osteoarthropathy and osteomyelitis in diabetic foot. Eur J Radiol 2017;89:221–5. [DOI] [PubMed] [Google Scholar]
- [5].Abdel Razek AA, Castillo M. Imaging appearance of primary bony tumors and pseudo-tumors of the spine. J Neuroradiol 2010;37:37–50. [DOI] [PubMed] [Google Scholar]
- [6].Surov A, Nagata S, Razek AA, et al. Comparison of ADC values in different malignancies of the skeletal musculature: a multicentric analysis. Skeletal Radiol 2015;44:995–1000. [DOI] [PubMed] [Google Scholar]
- [7].Mirabello L, Pfeiffer R, Murphy G, et al. Height at diagnosis and birth-weight as risk factors for osteosarcoma. Cancer Causes Control 2011;22:899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen S, Yang L, Pu F, et al. High birth weight increases the risk for bone tumor: a systematic review and meta-analysis. Int J Environ Res Public Health 2015;12:11178–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Arora RS, Kontopantelis E, Alston RD, et al. Relationship between height at diagnosis and bone tumours in young people: a meta-analysis. Cancer Causes Control 2011;22:681–8. [DOI] [PubMed] [Google Scholar]
- [10].Kansara M, Teng MW, Smyth MJ, et al. Translational biology of osteosarcoma. Nat Rev Cancer 2014;14:722–35. [DOI] [PubMed] [Google Scholar]
- [11].Musselman JR, Bergemann TL, Ross JA, et al. Case-parent analysis of variation in pubertal hormone genes and pediatric osteosarcoma: a Children's Oncology Group (COG) study. Int J Mol Epidemiol Genet 2012;3:286–93. [PMC free article] [PubMed] [Google Scholar]
- [12].Bernthal NM, Federman N, Eilber FR, et al. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer 2012;118:5888–93. [DOI] [PubMed] [Google Scholar]
- [13].Link MP, Goorin AM, Miser AW, et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med 1986;314:1600–6. [DOI] [PubMed] [Google Scholar]
- [14].Jaffe N, Frei E, 3rd, et al. Adjuvant methotrexate and citrovorum-factor treatment of osteogenic sarcoma. N Engl J Med 1974;291:994–7. [DOI] [PubMed] [Google Scholar]
- [15].Meyers PA, Healey JH, Chou AJ, et al. Addition of pamidronate to chemotherapy for the treatment of osteosarcoma. Cancer 2011;117:1736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xie CH, Cao YM, Huang Y, et al. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumour Biol 2016;37:15031–41. [DOI] [PubMed] [Google Scholar]
- [19].He P, Zhang Z, Huang G, et al. miR-141 modulates osteoblastic cell proliferation by regulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675. Am J Transl Res 2016;8:1780–8. [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Y, Zhang L, Zheng X, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett 2016;382:137–46. [DOI] [PubMed] [Google Scholar]
- [21].Pasic I, Shlien A, Durbin AD, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res 2010;70:160–71. [DOI] [PubMed] [Google Scholar]
- [22].Tian ZZ, Guo XJ, Zhao YM, et al. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol 2015;8:15138–42. [PMC free article] [PubMed] [Google Scholar]
- [23].Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res 2016;8:4095–105. [PMC free article] [PubMed] [Google Scholar]
- [24].Schaefer CF, Anthony K, Krupa S, et al. PID: the Pathway Interaction Database. Nucleic Acids Res 2009;37:D674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 2006;16:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ning S, Zhang J, Wang P, et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res 2016;44:D980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu G, Wang L, Han H, et al. LncRNA ZFAS1 promotes growth and metastasis by regulating BMI1 and ZEB2 in osteosarcoma. Am J Cancer Res 2017;7:1450–62. [PMC free article] [PubMed] [Google Scholar]
- [30].Li M, Chen H, Zhao Y, et al. H19 Functions as a ceRNA in promoting metastasis through decreasing miR-200 s Activity in Osteosarcoma. DNA Cell Biol 2016;35:235–40. [DOI] [PubMed] [Google Scholar]
- [31].Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett 2017;405:46–55. [DOI] [PubMed] [Google Scholar]
- [32].Jiang Z, Jiang C, Fang J. Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/beta-catenin pathway. Biochem Biophys Res Commun 2018;495:238–45. [DOI] [PubMed] [Google Scholar]
- [33].Zhou Q, Chen F, Zhao J, et al. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget 2016;7:82620–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fang Z, Xu C, Li Y, et al. A feed-forward regulatory loop between androgen receptor and PlncRNA-1 promotes prostate cancer progression. Cancer Lett 2016;374:62–74. [DOI] [PubMed] [Google Scholar]
- [35].Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev 2008;27:85–94. [DOI] [PubMed] [Google Scholar]
- [36].Rong L, Zhao R, Lu J. Highly expressed long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer progression via Wnt/beta-catenin signaling. Biochem Biophys Res Commun 2017;484:586–91. [DOI] [PubMed] [Google Scholar]
- [37].Ding J, Xie M, Lian Y, et al. Long noncoding RNA HOXA-AS2 represses P21 and KLF2 expression transcription by binding with EZH2, LSD1 in colorectal cancer. Oncogenesis 2017;6:e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tong G, Wu X, Cheng B, et al. Knockdown of HOXA-AS2 suppresses proliferation and induces apoptosis in colorectal cancer. Am J Transl Res 2017;9:4545–52. [PMC free article] [PubMed] [Google Scholar]
- [39].Wang F, Yang H, Deng Z, et al. HOX antisense lincRNA HOXA-AS2 promotes tumorigenesis of hepatocellular carcinoma. Cell Physiol Biochem 2016;40:287–96. [DOI] [PubMed] [Google Scholar]
- [40].Song W, Mei JZ, Zhang M. Long noncoding RNA PlncRNA-1 promotes colorectal cancer cell progression by regulating the PI3K/Akt signaling pathway. Oncol Res 2018;26:261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


