Abstract
Accumulation of protein aggregates with age was first described in aged human tissue over 150 years ago and has since been described in virtually every human tissue. Ubiquitin modifications are a canonical marker of insoluble protein aggregates; however, the composition of most age-related inclusions remains relatively unknown. To examine the landscape of age-related protein aggregation in vivo, we performed an antibody-based pulldown of ubiquitinated proteins coupled with metabolic labeling and mass spectrometry on young and old mice on calorie restriction (CR), rapamycin (RP)-supplemented, and control diets. We show increased abundance of many ubiquitinated proteins in old mice and greater retention of preexisting (unlabeled) ubiquitinated proteins relative to their unmodified counterparts—fitting the expected profile of age-increased accumulation of long-lived aggregating proteins. Both CR and RP profoundly affected ubiquitinome composition, half-live, and the insolubility of proteins, consistent with their ability to mobilize these age-associated accumulations. Finally, confocal microscopy confirmed the aggregation of two of the top predicted aggregating proteins, keratins 8/18 and catalase, as well as their attenuation by CR and RP. Stable-isotope labeling is a powerful tool to gain novel insights into proteostasis mechanisms, including protein aggregation, and could be used to identify novel therapeutic targets in aging and protein aggregation diseases.
Keywords: Bioinformatics, Caloric restriction, Proteomics, Rapamycin, mTOR
Maintenance of proper protein homeostasis (proteostasis) is essential to cellular and organismal health. A wealth of research has shown that age-related diseases and conditions are associated with the inability of the cell to maintain healthy proteins or remove defective proteins (1). These include neurodegenerative diseases (2), cardiac dysfunction (3,4), cataracts (5), and sarcopenia (6). Loss of proteostasis can manifest at the cellular level in a number of ways: protein aggregation (7), unfolding (8), oxidative damage (9,10), posttranslational modification (11–14), and age-related altered rates of protein synthesis and turnover (15–17). The age-dependent accumulation of proteins and other macromolecular “junk” was first described over a hundred years ago (18), and has since been reported to occur in virtually every human tissue with age (1,19–21). Insoluble inclusions have been extensively cataloged based on histological appearance, macromolecular structure, or origin, and have been described by names such as lipofuscin (22), amyloid (5), aggresome-like-induced structures (23,24), advanced glycation end-products (AGEs) (25), plaques (26), dendritic cell aggresome-like-induced structures (DALIS) (27), particle-rich cytoplasmic structure (PaCS) (28), inclusion bodies (29), ceroid (18), and aggresomes (19). In some disease-associated inclusions, particularly those of neurodegenerative diseases, the primary protein constituents involved are well known, such as amyloid beta and alpha-synuclein in Alzheimer’s disease and Parkinson’s disease, respectively (30,31). Most other protein inclusions, however, are identified by histological or biochemical features, as the identities of the protein components are not known.
Ubiquitin modification is most commonly associated the ubiquitin–proteasome system, a robust and highly precise system of maintaining proteome homeostasis by the recognition of damaged, unfolded, overly abundant, or otherwise dysfunctional proteins, marking them for degradation by the proteasome. In addition to this well-known association with proteosomal degradation, ubiquitination signals mitochondrial protein turnover through autophagy (mitophagy) (32), promotes autophagic clearance of protein aggregates (aggrephagy) (33–35), localizes to viral stress granules (36), and decorates pathogen containing vacuoles to facilitate host-cell defense (37). Ubiquitination is also closely associated with insoluble protein inclusions characteristic of aged tissue (38), in which case the aggregated proteins appear to have escaped degradation.
Using a combination of stable isotope labeling and proteomics, we recently reported that a large number of age-related changes in protein abundances, in vivo protein turnover rates, total protein ubiquitination, and protein quality could be mitigated or reversed in mice treated with short-term calorie restriction (CR) or rapamycin (RP) (15,16). To examine how ubiquitin-mediated proteostasis may underlie these changes, we used a similar stable isotope approach to more specifically examine two subproteome fractions: the ubiquitinated (UB) proteome (ubiquitinome) and the urea soluble and insoluble proteomes. We demonstrate that aging and CR and RP treatments have widespread effects on the landscape of ubiquitin-modified and insoluble proteins. With age, there is an accumulation of preexisting ubiquitin-modified proteins relative to their unmodified counterparts. Additionally, a number of proteins become increasingly insoluble with age. Confocal microscopy confirms the presence of two predicted in vivo aggregating proteins, and their reduction by CR and RP, demonstrating the utility of this proteomics approach for identifying novel aggregating proteins
Materials and Methods
Animals
C57BL/6 female mice, which have previously shown a larger response to RP than males (39), were obtained at 3 and 25 months of age (Figure 1A) from the NIA Charles River colony. Mice were housed at 20°C with a 12-hour light and dark cycle. All animals were handled according to the guidelines of the Institutional Animal Care Committee of the University of Washington and the National Institutes of Health. One week after arrival, all mice were started on a synthetic diet (Harlan Teklad diet #TD.99366) that was nutritionally similar to the NIH-31 standard for rodents. The use of this diet facilitated the subsequent substitution of heavy-labeled [5,5,5-2H3]-leucine for light leucine, which enabled the protein turnover measurements. Mouse weights and food intake were recorded weekly, with results previously reported (16). The young and old mice were individually housed after 3 weeks of acclimation to the synthetic chow and were randomly assigned to three groups: (i) an ad libitum synthetic food regimen (40); (ii) RP-containing synthetic diet (RP); and (iii) calorie restriction (CR).
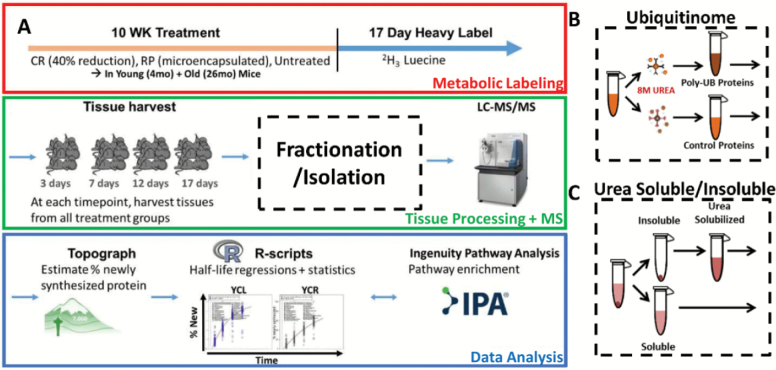
Figure 1.
Experimental workflow. (A) Young and old mice are treated for 10 weeks with CR, RP, or ad libitum diet, then dietary leucine is replaced with deuterated “heavy” leucine for 17 days while mice remain on treatments. Tissues are harvested from each treatment group over four time points over the 17-day labeling period, followed by processing of livers into protein fractions and analysis by nLC-MS/MS. Topograph software is used to calculate precursor-pool corrected estimates of percentage of newly synthesized protein as well as perform peak area integration. Statistical analysis and visualizations are performed by in-house R-scripts. Pathway enrichments are performed using commercially available Ingenuity Pathway Analysis (IPA) software. (B) A portion of each liver was homogenized in a highly denaturing buffer for ubiquitinome enrichment. The resulting lysates were then split in half—the first half was used for enrichment of ubiquitin-modified proteins, and the remaining half underwent enrichment with an isotope control antibody. (C) An additional portion of each liver was used to prepare the soluble and insolublomes. The supernatant of a low-speed centrifugation was kept as the soluble portion. The remaining pellet was re-solubilized in urea and both portions were then processed for MS analysis. CR = calorie restriction; RP = rapamycin.
Immunoaffinity Enrichment of Ubiquitinated Proteins
Portions of harvested mouse livers were used for MS analysis to determine abundance and proportion of newly synthesized and preexisting ubiquitin-modified proteins. Livers were removed at necropsy, rinsed in cold saline, and homogenized in cold isolation buffer (250 mM sucrose, 1 mM EGTA, 10 mM HEPES, 10 mM Tris–HCl pH 7.4). Lysates were centrifuged at 800g for 10 minutes to eliminate debris. To purify ubiquitinated proteins, lysates were immunopurified with FK2 antibody, which recognizes ubiquitinated proteins but not free ubiquitin (41). Each liver lysate was split into two samples for incubation on filtration columns with either the FK2 antibody or an isotype control antibody. To prevent enrichment of interacting proteins, lysates contain highly denaturing 8M urea which does not affect the FK2 antibody binding. Columns are washed several times and eluted with 100 mM glycine, pH 2.8. Protein from each sample will be methanol-chloroform precipitated and re-suspended into a trypsin compatible buffer followed by trypsin digestion, neutral ion removal, and MS analysis as described earlier.
Using the software Topograph (described below), we measured abundance and the proportion of new and existing proteins within ubiquitinated proteins. To further control for nonspecific contaminants, the values obtained in the FK2 enrichments are later corrected to the control enrichments (protein by protein).
Enrichment and Analysis of Ubiquitinated and Insoluble Proteins
Portions of harvested mouse livers were used for MS analysis to determine abundance and proportion of newly synthesized and preexisting ubiquitin-modified proteins. Livers were removed at necropsy, rinsed in cold saline, and homogenized in cold isolation buffer (50 mM ammonium bicarbonate). Lysates were centrifuged at 8,000g for 10 minutes to separate the soluble and insoluble fractions. Soluble fractions were removed and suspended with Rapigest (Waters Corporation, Milford, MA) to a final concentration of 0.1% and boiled for 5 minutes. Insoluble pellets were re-solubilized in 8M urea in RIPA buffer, sonicated, cooled in ice for 30 minutes, centrifuged at 10,000g for 15 minutes, and transferred to new tubes. Both soluble and insoluble fractions were treated with 5 mM DTT at 60°C for 30 minutes to reduce disulfide bonds. The free sulfhydryls were then alkylated with treatment of 15 mM iodoacetamide at room temperature for 30 minutes. The insoluble fractions were then transferred to Amicon Ultra-0.5 Centrifugal Filter Units (EMD Millipore, Billerica, MA), spun at 14,000g to concentrate, followed by three washes with 50 mM ammonium bicarbonate, and resuspension in buffer containing 50 mM ammonium bicarbonate and 0.2% Rapigest. Trypsin was added to both soluble and insoluble fractions to a final concentration of 1:100 (trypsin/protein) and the sample was digested at 37°C for 2 hours. The trypsin and Rapigest were hydrolyzed by 200 mM HCl, at 37°C for 30 minutes. The samples were then centrifuged for 10 minutes at 20,000g and the supernatant was washed using a Waters Oasis MCX sample extraction column, according to the manufacturer’s protocol (Waters Corporation). The digested samples were then loaded to the ultra-performance liquid chromatography and mass spectrometry (UPLC-MS/MS), using a Waters nanoAcquity LC system and a Thermo Scientific LTQ-FT Ultra. The LC mobile phase consisted of buffer A (water, 0.1% formic acid) and buffer B (acetonitrile, 0.1% formic acid).
Data Repository
The raw data from MS/MS and extended supplementary files are available at https://chorusproject.org/pages/blog.html#/961. In order to view the data, a free account must be obtained by following the instructions on the Chorus Project website. For R-scripts, spreadsheets, and other data, please contact the corresponding author.
MS Data Analysis
MS data were processed with the Hardklor (v1.33) and Bullseye (v1.25) algorithms to refine precursor mass measurements, followed by database search against all mouse entries of the UniProt database (UniProt release 2013_02) with the SEQUEST algorithm (vUW2012.01.7)—searching a total of 74,888 protein entries that were designated Mus musculus (Mouse). A dynamic modification of 3.0188325 for leucine was set to account for [5,5,5-2H3]-leucine and a static modification of 57.021461 for cysteine was set for carbamidomethyl modifications. The precursor monoisotopic mass tolerance was set to ±10 ppm and the fragment mass tolerance window was set to 0.36 m/z. Enzyme specificity was set to semi-tryptic, allowing for up to two missed cleavage sites per peptide. The false discovery rate for spectrum matches was determined by the Percolator algorithm (v2.04) using a reversed copy of the UniProt database as a decoy (42). Only results with a q-value less than 0.01 were kept for further analysis. This filter was used as it allows a small number (1%) of false positives to the next portion of analysis while providing enough data for categorization and statistical analysis of subsets of proteins. In addition, less than 5% of accepted false positives are expected to pass a subsequent turnover score filter, described below.
Estimation of Protein Turnover
Topograph (v1.1.0.297) software performs estimation of peptide turnover rates in stable-isotope labeling experiments. It was designed with the unique ability to calculate the enrichment of amino acid precursor pool, allowing an accurate estimation of peptide turnover rates when the amino acid precursor pool is not fully labeled (43). (http://proteome.gs.washington.edu/software/topograph/). Determining precursor relative isotope abundance (RIA) is a critical step in accurately estimating the fraction of newly synthesized and preexisting peptides.
Prior to analysis of abundance and turnover, peptides with a turnover score less than 0.98 were filtered out of the data. Turnover score is a metric internal to Topograph, ranging from 0 to 1, which describes the closeness of each observed isotopologue distribution with its closest matching theoretical distribution. A cutoff score of 0.98 was derived by plotting a receiver operator curve of true positive results versus true negative results (not shown), where true positive was defined as any peptide measurement that fell within 2 SDs of the mean label enrichment for all peptides, and true negatives were results that did not meet these criteria.
Relative peptide abundances were determined by integrating MS1 peaks. For peptides that were identified in one sample, the regression of the identified peptide’s MS/MS scan number is used to estimate a window for the same peptide in the other samples and a matching chromatographic peak was identified within that time range. This method allows peaks areas to be measured even in samples in which they are low and otherwise difficult to identify (44).
Only peptides that uniquely mapped to a single UniProt protein accession for Mus musculus (UniProt release 2013_02), consisting of 74,888 entries from UniProtKB/Swiss-Prot and UniProtKB/TrEMBL, were used for quantification of abundance and turnover. First, sequences were searched against Swiss-Prot (reviewed) entries and accepted in a unique match is found. If no match was found, a second search was performed on TrEMBL (unreviewed) entries and the unique matches were retained. All remaining peptides, consisting of peptides with either no matching proteins or greater than one matching protein, were filtered out. For the cases where a protein consisted of more than one peptide, statistical models were modified to account for the multiple peptides by using a blocking factor. For each protein we applied nonlinear regression fits of first-order exponential curves to the percent newly synthesized protein using: y = 100 − β1e−αΔt. To determine whether the rates of turnover (slopes, α) were statistically different between experimental groups, ANCOVA was used. Half-lives are calculated directly from slopes, where t1/2 = −ln(2)/slope. For details see the methods supplement of Hsieh and colleagues (43).
Pathway Analysis
Top pathways were determined using QIAGEN’S Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA; www.qiagen.com/ingenuity) on all proteins which were significantly changed (p-value < .05) in abundance or half-life by aging (OWT vs YWT) or after treatment (old CR or RP vs old controls) expression. IPA determines the p-values of enrichment into canonical pathways by Fisher exact test. All significantly changed proteins were then grouped by IPA canonical pathway and z-scores were visualized on a heatmap created in R using the gplots package. Values within each pathway category were clustered by Ward’s method.
Immunoblotting and ELISA
A portion of each tissue was aliquoted into separate tubes containing cold isolation buffer (250 mM sucrose, 1 mM EGTA, 10 mM HEPES, 10 mM Tris–HCl pH 7.4) as well as protease and phosphatase inhibitors (Pierce #87786 and #78420, Waltham, MA) at the time of harvest, and stored at −80°C to be used for immunoblotting and other bench assays as necessary. Western blotting was done on the NuPAGE Bis-Tris gel system (Life Technologies #WG1403BOX, Carlsbad, CA) according to the manufacturer protocols with primary antibody concentrations of 1,000× and secondary antibody concentrations of 10,000×. Western blots were imaged on a FluorChem Q imager and quantified using AlphaView software (ProteinSimple). Statistical comparisons of treatment groups were performed using two-tailed t tests. The following primary antibodies were used: multi-ubiquitin FK2 (D058-3, MBL), agarose-conjugated multi-ubiquitin FK2 (D058-3, MBL), FK2 isotype control (M075-3), agarose-conjugated FK2 isotype control (M075-8); secondary antibodies: Donkey anti Rabbit (42PI31458, Waltham, MA), Goat anti Mouse (42PI31432, Waltham, MA), Rabbit anti Goat (42AP106P, Waltham, MA), Qdot 705 (Q-11061MP, Thermo Fisher), Qdot 585 (Q-11011MP, Thermo Fisher).
Confocal Microscopy
Liver tissue was fixed in 4% paraformaldehyde overnight at 4°C, followed by paraffin embedding. Prior to deparaffinization, liver sections were photobleached 2 hours under a 30-W UV lamp. Sodium citrate buffer at 99°C was used for antigen retrieval. Slides were blocked in 5% BSA in PBS and incubated at 4°C overnight with the first of two primary antibodies in blocking buffer, followed by three washes with PBST, 1 hour incubation with secondary antibody, and three further PBST washes. Sections were then incubated on the second primary antibody at 4°C overnight, washed three times, incubated in secondary antibody for 1 hour, and washed three times again. Sections were immersed in 0.1% Sudan Black B in 70% ethanol for 30 minutes, washed three times with PBST, and nuclei were stained using Hoechst 33342. Slides were dehydrated through serial ethanol and mounted in Vectashield (Vector Laboratories, Burlingame, CA). Images were taken on a Leica TCS SP8 X. For controls, we prepared slides containing only secondary antibodies as well as a no antibody control. Analysis of confocal images was performed using the Costes automatic threshold method provided in the JACoP plugin (45) in Fiji (ImageJ) (46). Costes randomization indicates whether observed co-localization is statistically significant. This method creates random images by shuffling pixels in the green channel and then calculating a new Pearson correlation for each randomized image in the green channel (the image in the red channel is kept unchanged). The correlation of the original image in the green and red channel is then compared with the correlations of the randomized green images and the significance (p-value) is calculated. This p-value (expressed as percentage) is inversely proportional to the probability that the correlation with the original image is obtained by chance.
Results
Experimental Workflow
The experimental workflow, summarized in Figure 1, utilizes a previously described metabolic labeling proteomic workflow (16,43) in combination with an antibody-based enrichment of the ubiquitin-modified proteome. Briefly, 4- and 26-month-old mice were subjected to one of three treatments for 10 weeks (six groups total): ad libitum synthetic chow control diet (CL), caloric-restricted diet (CR), or ad libitum diet plus rapamycin (RP). RP was used at the concentration and formulation previously shown by the NIA Interventions Testing Program to extend mouse life span (47). Following 10 weeks of treatment, deuterated “heavy” leucine was substituted into the diet and maintained along with the treatments for 3, 7, 12, or 17 days before tissues were harvested and frozen. In order to enrich the ub proteome, antibody enrichment of proteins performed from liver lysates using the FK2 multi-ubiquitin antibody in a highly denaturing 8M urea buffer to avoid nonspecific binding (Figure 1B). This antibody has been previously shown to be effective in pulldowns under these conditions, which we were able to confirm (Supplementary Figure 1). To further control for nonspecific binding, a portion of each lysate was subjected to a pulldown with an isotype control antibody under the same conditions. All ubiquitin-enriched and control samples were analyzed by nLC-MS/MS followed by an analysis of abundances and percentage of newly synthesized proteins using Topograph software as previously described (43) (see Materials and Methods section), and ubiquitin-enriched fractions were corrected using the isotype control pulldowns to exclude any possible nonspecifically binding proteins from further analysis. For isolation of the urea soluble and insoluble proteome (Figure 1C), a low-speed spin was performed and the supernatant was prepared as the soluble portion. The insoluble portion was prepared with the remaining pellet by solubilizing it in 8M urea solution prior to preparation for mass spectrometry.
Accumulation of Increasingly Insoluble Ubiquitinated Proteins With Age
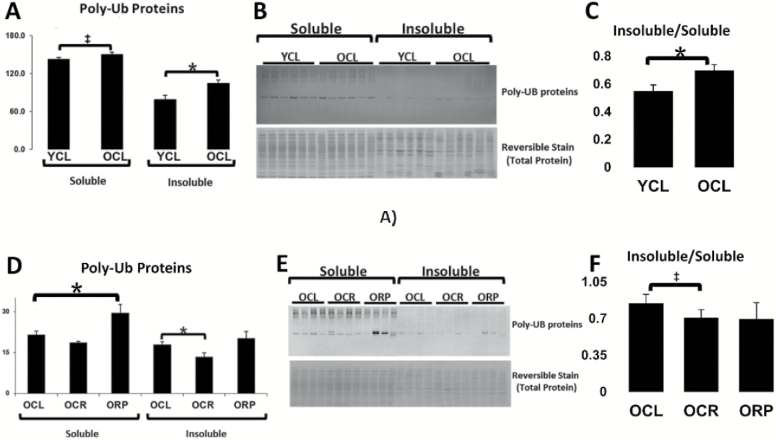
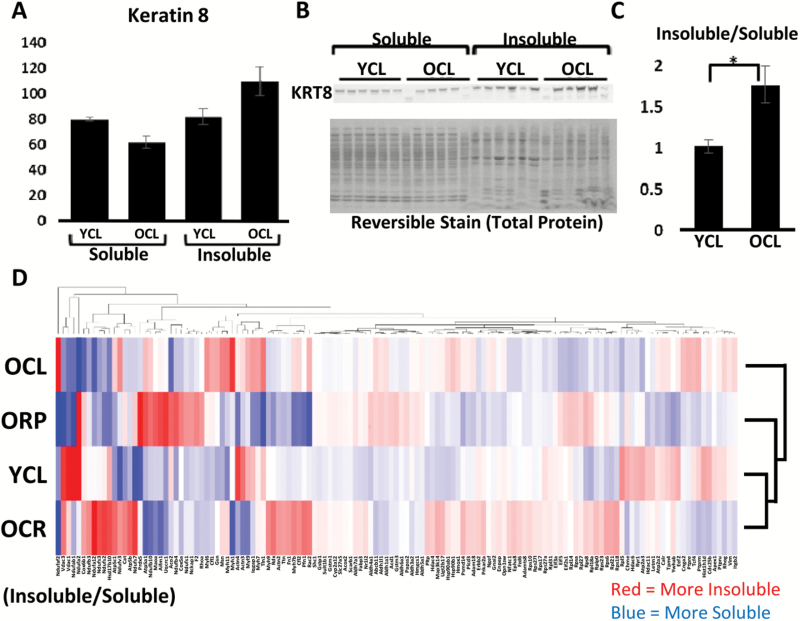
To assess whether the age-dependent increase in UB proteins that we previously reported (16) could be due to accumulation and retention of insoluble protein aggregates, we performed Western blotting of UB proteins in the soluble and insoluble proteomes with age (Figure 2). There was a significant increase in insoluble UB proteins with age, as well as a trending increase in soluble UB proteins (Figure 2A). Due to differences in buffer conditions insoluble and soluble UB proteins cannot be compared directly; however, quantification of the insoluble/soluble ratios in young and old mice suggests that there is an increasing proportion of insoluble proteins in the aged ubiquitinome (Figure 2C). We also assessed whether these aging changes were reversed by CR and RP treatment (Figure 2D–F). A significant reduction was seen in insoluble UB proteins in old mice on CR. Interestingly, aged mice fed RP showed a significant increase in soluble UB proteins (Figure 2D). However, when taking the ratio of insoluble/soluble proteins in each treatment, both CR and RP show a decreased proportion of insoluble UB proteins on average, though this is only statistically significant with CR (Figure 2F).
Figure 2.
Increasingly insoluble UB proteins with age. (A) A significant increase in UB proteins in the insoluble fraction with age and a trending increase in the soluble fraction. (B) Representative Western blot and total protein stain. (C) The ratio of insoluble/soluble protein increases with age. (D) Increased soluble UB-modified proteins in old RP-treated mice compared with untreated controls and a decrease in insoluble UB proteins in mice on CR compared with controls. (E) Representative Western blot and total protein reversible stain. (F) The proportion of insoluble/soluble proteins significantly decreases with CR, and RP treatment appears to move the ratio in the same direction. Values are mean ± SEM. Two-tailed t tests: *p < .05, ‡p < 0.1. CR = calorie restriction; RP = rapamycin; UB = ubiquitinated.
Abundance Changes in the Ubiquitinome
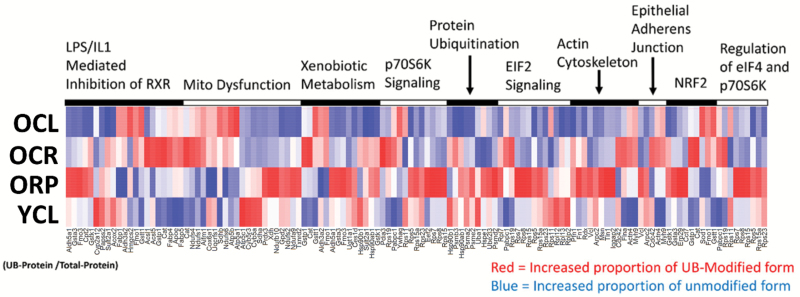
To identify the ubiquitinated proteins that are altered by aging, CR, and RP, we performed mass spectrometry analysis of the antibody-enriched ubiquitinome as described above (Figure 1A and B). To account for the possibility that an increase in a given UB protein might simply be a reflection of a general increase in the abundance of the total protein, areas of UB proteins were normalized to their unmodified signals in a total (non-enriched) fraction. All UB proteins that significantly increased or decreased in abundance with age after normalization were analyzed using Ingenuity Pathway Analysis (IPA) software to determine canonical pathways that were most significantly enriched. These results were visualized on a heatmap of the top 10 enriched pathways (Figure 3A). An increase (red) indicates a greater accumulation of a UB protein than is seen in the protein overall, whereas blue represents a decrease in ubiquitination. It is interesting to note the similarity in pathways enriched here and in our previously reported protein abundance and turnover analyses in liver (16). Young mice showed clearly lower levels of UB protein (blue) compared with aged control mice. Aged mice treated with CR or RP showed patterns that were different from either young or old controls and from each other; these included subsets of proteins that both increased (red) and decreased (blue) in UB protein abundance.
Figure 3.
Heatmap of top pathways altered in ubiquitination. Heatmap depicting ubiquitin-modified proteins that were significantly altered with age, arranged by the top 10 most significantly enriched pathways and shown for OCL, OCR, ORP, and YCL. Each ubiquitin-modified protein was normalized to changes in its unmodified form in the total protein fraction. A high quality PDF of Figure 3 is available as a supplementary file.
Turnover Changes in the Ubiquitinome
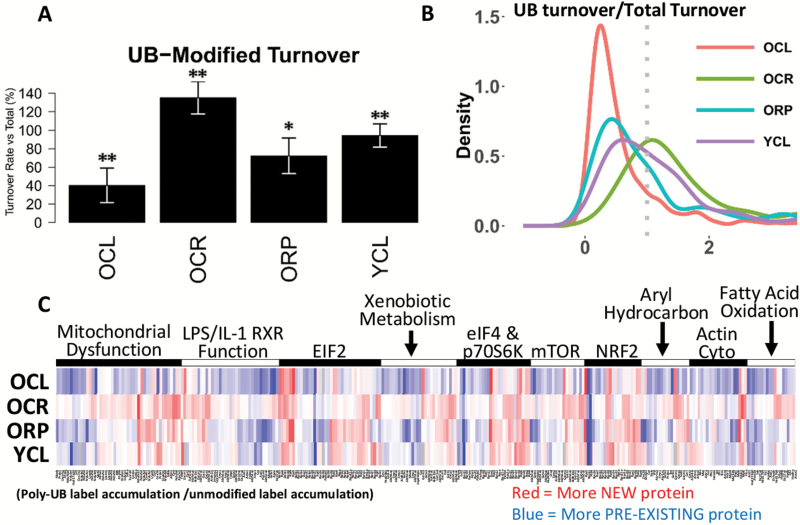
Ubiquitin-modified protein aggregates, particularly large aggregates observed in histological studies, are typically insoluble and difficult to degrade. We therefore sought to utilize our metabolic labeling strategy to measure the turnover of proteins within the ubiquitinome to build a list of candidates which are primarily composed of long-lived, preexisting proteins. We therefore compared the proportion of newly synthesized and preexisting proteins in the UB fraction to the unmodified fraction, with the results summarized in Figure 4A and B. In old control mice, the ubiquitin-modified proteome had less than half the percentage of newly synthesized protein observed in the total fraction (Figure 4A), whereas in young mice there was a nearly equivalent proportion of new proteins among both modified and unmodified proteins. Both CR and RP increased the proportion of new proteins within the UB fractions in old mice (Figure 4A). The results for individual proteins were plotted on a heatmap (Figure 4C; Supplementary Figure 2). Although old untreated mouse livers were largely composed of preexisting proteins (blue), both CR and RP treatments showed higher proportions of newly synthesized UB proteins (redder) in comparison. Interestingly, the majority of red-shaded areas of the heatmap in OCR and ORP overlap with the red areas in YCL, whereas few of these areas in OCL overlap with YCL. Hierarchical clustering of heatmap data clustered YCL most closely with OCR, followed by ORP, then OCL (Supplementary Figure 2).
Figure 4.
Turnover of ubiquitin-modified proteins. (A) Bar plot showing, on average, the percentage of ubiquitin-modified proteins which are newly synthesized when compared with the corresponding unmodified proteins (100%) for OCL, OCR, ORP, and YCL. (B) A probability density plot showing the distribution of percentage of newly synthesized ubiquitinated proteins compared with the equivalent unmodified proteins. The dotted line represents an equivalent percentage of newly synthesized proteins in the ubiquitin-modified and unmodified forms. (C) Heatmap depicting the ratio of newly synthesized ubiquitin-modified proteins over equivalent unmodified proteins, arranged by the top 10 most significantly enriched pathways and shown for OCL, OCR, ORP, and YCL. Red indicates a greater proportion of newly synthesized, labeled protein in the modified form and blue indicated a greater proportion of preexisting protein. **p-value < .001 for all pairwise comparisons. *p-value < .05 versus YCL and p-value < .001 versus OCL and OCR. A high quality PDF of Figure 4C is available as a supplementary file.
Urea Soluble and Insoluble Proteomes
The data above suggest that the accumulation of UB proteins in old livers may be in part due to an inability to degrade such proteins. We hypothesized that proteins following this pattern would be likely to be forming insoluble aggregates, as aggregates are less efficiently degraded and removed from cells, if they are degraded at all. The UB proteins that were increasing in abundance with age were filtered for those that also retained a high proportion of preexisting proteins. However, these data from the ubiquitinome do not conclusively indicate that a protein is forming an insoluble aggregate—as protein may also be accumulating in solution. In order to further identify candidate aggregating proteins, we performed another round of MS analysis on the lysates from the same mice; however, for each mouse we prepared the insoluble fraction in addition to the soluble fraction (Figure 1A and C). The insoluble pellets from a low-speed spin of whole lysates followed were resolubilized in an 8M urea-containing buffer. The ratio of intensities of each identified peptide in the insoluble fractions over the soluble fraction was then determined for each liver sample, and a q-value threshold of 0.05 was applied to identify proteins significantly increasing in the insoluble to soluble ratio with age, as determined by two-tailed t testing (see Materials and Methods section). The insoluble/soluble ratios of these proteins in YCL, OCL, OCR, and ORP livers were visualized in a heatmap, shown in Figure 5D. It is interesting to note that the overall insolubility of proteins globally does not appear to change dramatically with age (Figure 5B). Rather, the spectrum of insoluble proteins is changed with some becoming increasingly and others decreasingly soluble with age. Aged mice treated with CR or RP have a profile more similar to that of young mice, as indicated by clustering on the heatmap (Figure 5D). To refine this list to those proteins most likely to be present in age-related aggregates, we filtered for only proteins which also significantly increased in the ratio of insoluble proteins with age, as determined by the analysis of the urea soluble and insoluble fractions. The 10 top predicted “aggregator” proteins are shown in Table 1. We then compared this list against proteins well established in the literature as components of aggregates in the liver.
Figure 5.
Soluble and insoluble proteomes. (A) Quantification of keratin 8 in the soluble and insoluble fractions of young and old mice by Western blotting (normalized to total protein reversible stain). (B) Representative Western blot of krt8 and total protein stain. (C) Quantification of the insoluble to soluble intensity ratio of keratin 8 in young and old mice. (D) Heatmap depicting the relative proteomic abundance of proteins in the insoluble fractions versus the soluble fractions for each treatment group, with column and row clustering and dendrograms. Red indicates a greater proportion of insoluble protein and blue indicates a greater proportion of soluble protein. Values are mean ± SEM. Two-tailed t tests: *p < .05. A high quality PDF of Figure 5D is available as a supplementary file.
Table 1.
Top Candidate Aggregating Proteins
| Relative Abundance (Log2 Fold Change) | New Protein (Log2 Fold Change) | ||||||
|---|---|---|---|---|---|---|---|
| Entry | Gene names | OCL/YCL | OCR/OCL | ORP/OCL | OCL/YCL | OCR/OCL | ORP/OCL |
| P41216 | Acsl1 | 5.19 | −3.09 | −3.02 | −1.65 | 2.88 | 1.71 |
| P11679 | Krt8 | 3.01 | −1.82 | −2.95 | −6.26 | 9.08 | −2.00 |
| P57780 | Actn4 | 1.53 | −2.18 | −5.12 | −1.83 | 3.63 | 1.88 |
| Q8BWF0 | Aldh5a1 | 3.84 | 2.22 | −1.41 | −1.42 | 2.70 | −1.89 |
| Q03265 | Atp5a1 | 2.46 | 1.04 | −2.31 | −2.00 | 3.07 | 2.26 |
| P56480 | Atp5b | 1.71 | 1.62 | −2.46 | −1.75 | 1.98 | 1.67 |
| P24270 | Cat | 3.02 | −3.29 | −5.10 | −1.39 | 2.46 | −1.05 |
| P26041 | Msn | 2.64 | −1.01 | −1.48 | −3.65 | 2.44 | 2.07 |
| Q8VDD5 | Myh9 | 1.22 | −1.99 | −4.74 | −1.59 | 2.29 | 1.84 |
| P26039 | Tln1 | 3.03 | −1.15 | −2.00 | −1.23 | 2.46 | 1.25 |
Note: OCL = old control; OCR = old calorie restriction; ORP = old rapamycin; YCL = young calorie restriction. q-value < 0.05 for all OCL/YCL ratio comparisons.
Mallory-Denk bodies (MDBs) are a highly studied and well-characterized protein aggregates in the liver associated with fatty liver disease (48). These aggregates are known to be highly modified with ubiquitin (49) and are composed primarily of a single protein—keratins 8/18 (48). There is also an increased risk of having MDBs with age in both mice and humans (50,51). Interestingly, keratins 8/18 was among the proteins showing the largest abundance increases in the UB fraction in addition to being highly composed of preexisting proteins (Table 1). In addition, Western blotting for keratins 8/18 in the insoluble and soluble fractions showed that keratins 8/18 became increasingly insoluble with age (Figure 5A–C), further suggesting heavy-label MS may be an effective method of uncovering the identities of novel aggregate-prone proteins.
Confirmation of “Aggregators” by Confocal Microscopy
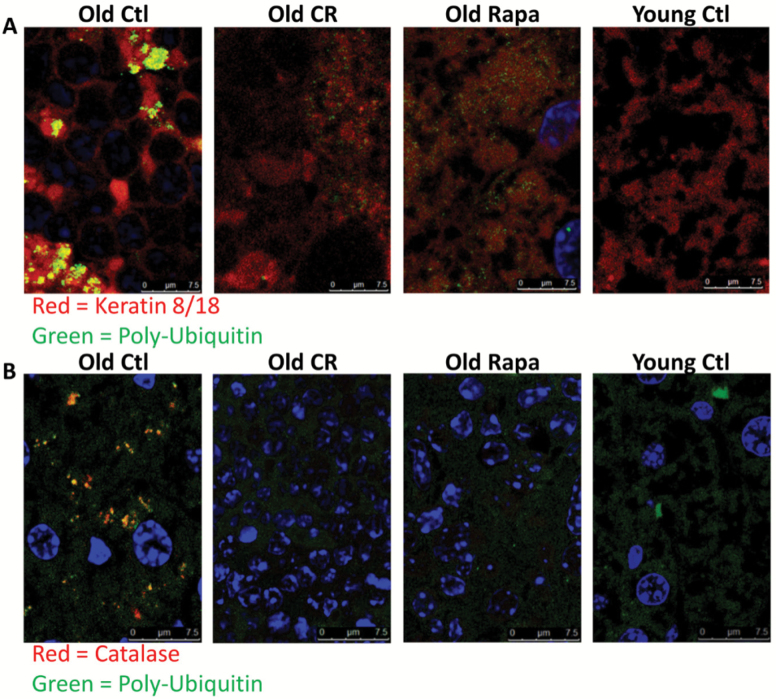
To visually confirm the presence of ubiquitinated keratin 8/18-containing MDB aggregates, as predicted by (i) the accumulation of preexisting, or “old,” ubiquitin-modified krt8 as well as (ii) the increased ratio of insoluble/soluble krt8 with age, we performed confocal microscopy on liver sections probed with keratins 8/18 and ubiquitin antibodies. Consistent with predictions, we observed the appearance of co-localized aggregates of krt8 and ubiquitin in old mouse tissue (Figure 6A, R = 0.63, p = 1, Costes automatic thresholding (45)). Consistent with the predictions from proteomic analysis (Table 1), confocal microscopy showed that these aggregates were diminished in both CR and RP mice (Figure 6A, R = 0.41 and 0.44 for old CR and RP, respectively).
Figure 6.
Increasingly insoluble UB proteins with age. (A) Representative images of confocal microscopy of a top candidate “aggregator” keratin 8, shown in red, and ubiquitin, shown in green, in mouse liver sections. Old control mice showed significant co-localization of red and green fluorescence (R = 0.63, p = 1, Costes automatic thresholding (45)), whereas old treated mice showed diminished co-localization (R = 0.41 and 0.44 for old CR and Rapa-treated mice, respectively). (B) Representative confocal images in mouse liver sections probed for catalase (red) and ubiquitin (green) and DNA (DAPI, blue). Old control mice showed significant co-localization of red and green fluorescence (R = 0.42, p = 1, Costes automatic thresholding), whereas other treatment groups did not (R = 0.12, 0.02, and 0.04 for young controls, old CR-treated mice, and old rapamycin-treated mice, respectively). Fluorescence bleed-through from DAPI into the red channel was removed from catalase images by removing red staining within nuclei in Photoshop. The Costes’ p-value from Costes’ randomized analysis was p = 1, or 100%, for all images. This indicates a high probability that the co-localization observed was not the co-localization of pixels due to chance (see Materials and Methods section). CR = calorie restriction; UB = ubiquitinated.
Although MDBs are a well-studied protein aggregates known to be composed of krt8, the majority of the proteins among our top candidates are not known to aggregate in the liver with age. We probed for another candidate “aggregator” protein, catalase, and examined its co-localization with ubiquitin by confocal microscopy. In agreement with our prediction, the ratio of insoluble to soluble catalase was increased with age by Western blot (Supplementary Figure 3). Additionally, co-localized aggregates of catalase and ubiquitin were seen in aged but not young livers (Figure 6B, R = 0.42 and 0.12 for old and young controls, respectively), and were reduced in livers from old mice treated with CR or RP (R = 0.02 and 0.04 for old CR and RP, respectively), all consistent with the predictions shown in Table 1.
Discussion
In this study we sought to form a more complete picture of the dynamics of ubiquitin-mediated proteostasis and its relative contribution to protein aggregation during aging, CR, and RP treatment. In order to perform a comprehensive survey of the ubiquitin-modified proteins and their homeostasis, we utilized a combination of metabolic labeling and mass spectrometry analysis to simultaneously measure abundances and turnover of proteins enriched by a ubiquitin-specific antibody pulldown. We performed these analyses on the total proteome, insoluble protein fraction (insolublome), and the solublome of all samples to identify proteins potentially contributing to age-related accumulation of insoluble protein aggregates. Compared with young mice, old mice showed a bulk increase in protein ubiquitination which is mirrored on the individual protein level by increases in ubiquitination across nearly all proteins. Bulk levels of ubiquitination in old mice given either treatment was reduced as well; however, among individual proteins there was a mixture of proteins with both increased and decreased ubiquitination and solubility. Among proteins increasing in ubiquitination with age, a large portion was decreased after either treatment. Interestingly, CR, and RP to a lesser extent, altered ubiquitination and solubility in an even greater number of proteins with no age-related changes, and many of these, particularly in CR, were increases. It is unclear what role these CR- and RP-induced increases might play or whether they could potentially form aggregates.
Strikingly, we found that the majority of ubiquitin-modified proteins had a significantly greater proportion that were preexisting (unlabeled) than their unmodified counterparts, suggesting that UB proteins were specifically retained or perhaps aggregating. To further narrow the focus on the latter, we identified proteins that were both ubiquitinated and that were enriched in the insoluble protein fraction. Choosing two of the top predicted protein aggregates, keratins 8/18 and catalase, we performed confocal microscopy on liver tissues and confirmed the presence of ubiquitin-positive aggregates of both proteins with age, demonstrating that this approach has predictive power.
CR and RP treatment are interventions known to extend life span and improve health, as well as to slow or reverse the age-related decline in proteostasis (15,47,52–54). Previous work from our lab and others has described changes in proteome-wide abundances and turnover rates in the liver (16,53,55–58) and other tissues (15,57–60) with age and with CR or RP treatment (15,16,59). In the current study, we observed that compared with aged control mice, old mice treated for 10 weeks with CR or RP had significantly fewer preexisting proteins across most of the top enriched canonical pathways; 10-week CR mice showed the greater reversal that was also most uniform across pathways. In addition, old mice on either treatment showed a reduction in co-localized aggregates of keratins 8/18 and catalase compared with old untreated mice. These changes could be explained by activation of protein turnover through autophagy, which is known to occur after CR or treatment with RP. Aggregates specifically containing ubiquitin modification may also be more specifically targeted by autophagy. For example, mitochondrial-specific autophagy (mitophagy) is ubiquitin-dependent and therefore activation of autophagy by CR or RP may therefore increase turnover of ubiquitin-modified mitochondrial proteins through this pathway (13,32,61,62). Consistent with such a mechanism, we have recently reported that RP treatment induces mitochondrial remodeling (63). In addition, several studies have shown that ubiquitination of protein aggregates promotes association with the aggresome and facilitates recruitment of protein complexes participating in the formation of autophagosomes, thereby promoting their clearance through autophagy (33–35,62). Finally, a reduction of total protein synthesis rates also takes place after CR and RP (16) and this may decrease the rate at which new aggregates form, even if the rate of aggregate removal is unchanged.
MDBs are cytoplasmic inclusions of liver cells most commonly associated with alcoholic hepatitis and cirrhosis (64), and have a strong age-related component (51). Ubiquitin-modified keratin 8 has been established to be the major protein component of MDBs (65). In agreement with this, modified keratin 8 was a top predicted age-related aggregate based on our proteomic criteria, which we were able to confirm by confocal microscopy. In old CR or RP-fed mice, there was significantly reduced accumulation of ubiquitin-modified keratin 8 in both proteomic and imaging data, suggesting that CR and RP may be potentially effective treatments for the elimination of MDBs. This result is consistent with a prior study which found RP to be effective in reducing or eliminating MDBs in an autophagy-dependent manner (66).
We have also confirmed a previously unknown age-related aggregate in the liver: ubiquitinated catalase. Catalase is a conserved antioxidant enzyme, normally localized to peroxisomes, which protects cells from ROS by catalyzing the decomposition of hydrogen peroxide to water and oxygen. Although the mechanisms leading to its reduced solubility, aggregation, and ubiquitination in liver remain unclear, accumulation of insoluble catalase has been reported in autophagy-deficient or heat-stressed plants (67–70). It is not known whether this accumulation in plants is associated with ubiquitin modification; however, studies of pexophagy (peroxisome autophagy) have reported ubiquitin modification of defective peroxisomes (69) and it is possible a similar mechanism occurs in mammals. During aging in both human and yeast cells, peroxisomal protein import is disproportionately impaired for catalase compared with other proteins, and leads to its accumulation in the cytosol (71,72), a plausible mechanism contributing to its aggregation.
The age-related and disease-related accumulation of insoluble aggregates of protein and other macromolecular components has been a widely known and well-documented phenomenon for many years, and has been described to occur in virtually all tissues with age. Although several commonly studied aggregates such as amyloid beta associated with Alzheimer’s disease have known protein constituents, the majority of age-related aggregates are described by histological characteristics, such as amyloid and the “age pigment” lipofuscin. We have shown that the use of a multifaceted proteomic approach which combines global measurement of protein abundance, turnover rate, relative ubiquitination, and solubility can be used to identify many candidates at once. We propose that the use of this approach and similar strategies integrating proteome abundances and turnover can be used to assess the effect of therapeutic interventions on protein aggregation dynamics as well as to identify the protein composition of many disease- or age-related aggregates that presently remain unknown.
Supplementary Material
Supplementary data is available at The Journals of Gerontology, Series A: Biomedical Sciences and Medical Sciences online.
Funding
Grant support was provided by NIH P30 AG0132280, RO1 HL101186, P01 AG001751, and RO1 AG038550, by the Ellison Medical Foundation grant AG-SS-2535-10, an American Federation for Aging Research Breakthroughs in Gerontology award to P.S.R., and a Glenn/AFAR Scholarship for Research in the Biology of Aging awarded to N.B.B. We also acknowledge the support from the NIH to the UW W. M. Keck Microscopy Center (S10 OD016240).
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Material
References
- 1. Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res Rev. 2011;10:205–215. doi: 10.1016/j.arr.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010;190:719–729. doi:10.1083/jcb.201005144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christians ES, Benjamin IJ. Proteostasis and REDOX state in the heart. Am J Physiol Heart Circ Physiol. 2012;302:H24–H37. doi:10.1152/ajpheart.00903.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hedhli N, Pelat M, Depre C. Protein turnover in cardiac cell growth and survival. Cardiovasc Res. 2005;68:186–196. doi:10.1016/j.cardiores.2005.06.025 [DOI] [PubMed] [Google Scholar]
- 5. Surguchev A, Surguchov A. Conformational diseases: looking into the eyes. Brain Res Bull. 2010;81:12–24. doi:10.1016/j.brainresbull.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 6. Vinciguerra M, Musaro A, Rosenthal N. Regulation of muscle atrophy in aging and disease. Adv Exp Med Biol. 2010;694:211–233. doi:10.1007/978-1-4419-7002-2_15 [DOI] [PubMed] [Google Scholar]
- 7. Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi:10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 8. Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nat Cell Biol. 2015;17:829–838. doi:10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannizzo ES, Clement CC, Morozova K, et al. . Age-related oxidative stress compromises endosomal proteostasis. Cell Rep. 2012;2:136–149. doi:10.1016/j.celrep.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones DP. Redox theory of aging. Redox Biol. 2015;5:71–79. doi:10.1016/j.redox.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsakiri EN, Sykiotis GP, Papassideri IS, et al. . Proteasome dysfunction in Drosophila signals to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. Aging Cell. 2013;12:802–813. doi:10.1111/acel.12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baskin KK, Taegtmeyer H. AMP-activated protein kinase regulates E3 ligases in rodent heart. Circ Res. 2011;109:1153–1161. doi:10.1161/CIRCRESAHA.111.252742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan JM, Wong ES, Kirkpatrick DS, et al. . Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi:10.1093/hmg/ddm320 [DOI] [PubMed] [Google Scholar]
- 14. Brehm A, Krüger E. Dysfunction in protein clearance by the proteasome: impact on autoinflammatory diseases. Semin Immunopathol. 2015;37:323–333. doi:10.1007/s00281-015-0486-4 [DOI] [PubMed] [Google Scholar]
- 15. Dai DF, Karunadharma PP, Chiao YA, et al. . Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13:529–539. doi:10.1111/acel.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karunadharma PP, Basisty N, Dai DF, et al. . Subacute calorie restriction and rapamycin discordantly alter mouse liver proteome homeostasis and reverse aging effects. Aging Cell. 2015;14:547–557. doi:10.1111/acel.12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kruse SE, Marcinek DJ, Karunadharma PP, et al. . Age modifies respiratory complex I and protein homeostasis in a muscle type-specific manner. Aging Cell. 2016;15:89–99. doi:10.1111/acel.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jolly RD, Dalefield RR, Palmer DN. Ceroid, lipofuscin and the ceroid-lipofuscinoses (Batten disease). J Inherit Metab Dis. 1993;16:280–283. doi:10.1007/BF00710265 [DOI] [PubMed] [Google Scholar]
- 19. Corboy MJ, Thomas PJ, Wigley WC. Aggresome formation. Methods Mol Biol. 2005;301:305–327. doi:10.1385/1-59259-895-1:305 [DOI] [PubMed] [Google Scholar]
- 20. Cuanalo-Contreras K, Mukherjee A, Soto C. Role of protein misfolding and proteostasis deficiency in protein misfolding diseases and aging. Int J Cell Biol. 2013;2013:638083. doi:10.1155/2013/638083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jana NR. Protein homeostasis and aging: role of ubiquitin protein ligases. Neurochem Int. 2012;60:443–447. doi:10.1016/j.neuint.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 22. Rotteveel JJ, Mullaart RA. [Antioxidative therapy in ceroid lipofuscinosis]. Tijdschr Kindergeneeskd. 1989;57:181–186. [PubMed] [Google Scholar]
- 23. Calise J, Powell SR. The ubiquitin proteasome system and myocardial ischemia. Am J Physiol Heart Circ Physiol. 2013;304:H337–H349. doi:10.1152/ajpheart.00604.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mi L, Gan N, Chung FL. Aggresome-like structure induced by isothiocyanates is novel proteasome-dependent degradation machinery. Biochem Biophys Res Commun. 2009;388:456–462. doi:10.1016/j.bbrc.2009.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Semba RD, Nicklett EJ, Ferrucci L. Does accumulation of advanced glycation end products contribute to the aging phenotype?J Gerontol A Biol Sci Med Sci. 2010;65:963–975. doi:10.1093/gerona/glq074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferreira ME, de Vasconcelos AS, da Costa Vilhena T, et al. . Oxidative stress in Alzheimer’s disease: should we keep trying antioxidant therapies?Cell Mol Neurobiol. 2015;35:595–614. doi:10.1007/s10571-015-0157-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herter S, Osterloh P, Hilf N, et al. . Dendritic cell aggresome-like-induced structure formation and delayed antigen presentation coincide in influenza virus-infected dendritic cells. J Immunol. 2005;175:891–898. doi:10.4049/jimmunol.175.2.891 [DOI] [PubMed] [Google Scholar]
- 28. Solcia E, Sommi P, Necchi V, Vitali A, Manca R, Ricci V. Particle-rich cytoplasmic structure (PaCS): identification, natural history, role in cell biology and pathology. Biomolecules. 2014;4:848–861. doi:10.3390/biom4030848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palmer I, Wingfield PT. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Curr Protoc Protein Sci. 2012;Chapter 6:Unit6.3. doi:10.1002/0471140864.ps0603s70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harman D. Alzheimer’s disease: a hypothesis on pathogenesis. J Am Aging Assoc. 2000;23:147–161. doi:10.1007/s11357-000-0017-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rochet JC, Hay BA, Guo M. Molecular insights into Parkinson’s disease. Prog Mol Biol Transl Sci. 2012;107:125–188. doi:10.1016/B978-0-12-385883-2.00011-4 [DOI] [PubMed] [Google Scholar]
- 32. Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi:10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- 33. Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi:10.1161/CIRCRESAHA.111.244707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong E, Bejarano E, Rakshit M, et al. . Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun. 2012;3:1240. doi:10.1038/ncomms2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zaarur N, Meriin AB, Bejarano E, et al. . Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol. 2014;34:1336–1348. doi:10.1128/MCB.00103-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi:10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haldar AK, Foltz C, Finethy R, et al. . Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A. 2015;112:E5628–E5637. doi:10.1073/pnas.1515966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi:10.1073/pnas.1216197110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fok WC, Chen Y, Bokov A, et al. . Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS One. 2014;9:e83988. doi:10.1371/journal.pone.0083988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coker RH, Miller S, Schutzler S, Deutz N, Wolfe RR. Whey protein and essential amino acids promote the reduction of adipose tissue and increased muscle protein synthesis during caloric restriction-induced weight loss in elderly, obese individuals. Nutr J. 2012;11:105. doi:10.1186/1475-2891-11-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5:4145–4151. doi:10.1002/pmic.200401280 [DOI] [PubMed] [Google Scholar]
- 42. Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi:10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- 43. Hsieh EJ, Shulman NJ, Dai DF, et al. . Topograph, a software platform for precursor enrichment corrected global protein turnover measurements. Mol Cell Proteomics. 2012;11:1468–1474. doi:10.1074/mcp.O112.017699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Finney GL, Blackler AR, Hoopmann MR, Canterbury JD, Wu CC, MacCoss MJ. Label-free comparative analysis of proteomics mixtures using chromatographic alignment of high-resolution muLC-MS data. Anal Chem. 2008;80:961–971. doi:10.1021/ac701649e [DOI] [PubMed] [Google Scholar]
- 45. Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. doi:10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 46. Schindelin J, Arganda-Carreras I, Frise E, et al. . Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi:10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harrison DE, Strong R, Sharp ZD, et al. . Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi:10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crawford JM. Histologic findings in alcoholic liver disease. Clin Liver Dis. 2012;16:699–716. doi:10.1016/j.cld.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 49. Bardag-Gorce F, van Leeuwen FW, Nguyen V, et al. . The role of the ubiquitin-proteasome pathway in the formation of Mallory bodies. Exp Mol Pathol. 2002;73:75–83. doi:10.1006/exmp.2002.2451 [DOI] [PubMed] [Google Scholar]
- 50. Kato M. Mallory bodies in hepatocytes of alcoholic liver disease and primary biliary cirrhosis contain Nε-(carboxymethyl)lysine-modified cytokeratin, but not those in hepatic carcinoma cells. Yonago Acta Med. 2006;49:83–92. [Google Scholar]
- 51. Hanada S, Harada M, Abe M, et al. . Aging modulates susceptibility to mouse liver Mallory-Denk body formation. J Histochem Cytochem. 2012;60:475–483. doi:10.1369/0022155412441478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Bokov A, Gelfond J, et al. . Rapamycin extends life and health in C57BL/6 mice. J Gerontol A Biol Sci Med Sci. 2014;69:119–130. doi:10.1093/gerona/glt056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thompson AC, Bruss MD, Price JC, et al. . Reduced in vivo hepatic proteome replacement rates but not cell proliferation rates predict maximum lifespan extension in mice. Aging Cell. 2016;15:118–127. doi:10.1111/acel.12414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi:10.1038/nature11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Price JC, Khambatta CF, Li KW, et al. . The effect of long term calorie restriction on in vivo hepatic proteostatis: a novel combination of dynamic and quantitative proteomics. Mol Cell Proteomics. 2012;11:1801–1814. doi:10.1074/mcp.M112.021204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miller BF, Robinson MM, Reuland DJ, et al. . Calorie restriction does not increase short-term or long-term protein synthesis. J Gerontol A Biol Sci Med Sci. 2013;68:530–538. doi:10.1093/gerona/gls219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Claydon AJ, Thom MD, Hurst JL, Beynon RJ. Protein turnover: measurement of proteome dynamics by whole animal metabolic labelling with stable isotope labelled amino acids. Proteomics. 2012;12:1194–1206. doi:10.1002/pmic.201100556 [DOI] [PubMed] [Google Scholar]
- 58. Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci U S A. 2010;107:14508–14513. doi:10.1073/pnas.1006551107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Karunadharma PP, Basisty N, Chiao YA, et al. . Respiratory chain protein turnover rates in mice are highly heterogeneous but strikingly conserved across tissues, ages, and treatments. FASEB J. 2015;29:3582–3592. doi:10.1096/fj.15-272666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim TY, Wang D, Kim AK, et al. . Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics. 2012;11:1586–1594. doi:10.1074/mcp.M112.021162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Thomas RL, Gustafsson AB. Mitochondrial autophagy–an essential quality control mechanism for myocardial homeostasis. Circ J. 2013;77:2449–2454. doi:10.1253/circj.CJ-13-0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamano K, Matsuda N, Tanaka K. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016;17:300–316. doi:10.15252/embr.201541486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chiao YA, Kolwicz SC, Basisty N, et al. . Rapamycin transiently induces mitochondrial remodeling to reprogram energy metabolism in old hearts. Aging (Albany NY). 2016;8:314–327. doi:10.18632/aging.100881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782:764–774. doi:10.1016/j.bbadis.2008.08.008 [DOI] [PubMed] [Google Scholar]
- 65. Nakamichi I, Toivola DM, Strnad P, et al. . Keratin 8 overexpression promotes mouse Mallory body formation. J Cell Biol. 2005;171:931–937. doi:10.1083/jcb.200507093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47:2026–2035. doi:10.1002/hep.22294 [DOI] [PubMed] [Google Scholar]
- 67. Yoshimoto K, Shibata M, Kondo M, et al. . Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci. 2014;127(Pt 6):1161–1168. doi:10.1242/jcs.139709 [DOI] [PubMed] [Google Scholar]
- 68. Shibata M, Oikawa K, Yoshimoto K, et al. . Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013;25:4967–4983. doi:10.1105/tpc.113.116947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Avin-Wittenberg T, Fernie AR. At long last: evidence for pexophagy in plants. Mol Plant. 2014;7:1257–1260. doi:10.1093/mp/ssu029 [DOI] [PubMed] [Google Scholar]
- 70. Zhou J, Zhang Y, Qi J, et al. . E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. doi:10.1371/journal.pgen.1004116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arlia-Ciommo A, Leonov A, Piano A, Svistkova V, Titorenko VI. Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae. Microbial Cell. 2014;1:163–178. doi:10.15698/mic2014.06.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Koepke JI, Nakrieko KA, Wood CS, et al. . Restoration of peroxisomal catalase import in a model of human cellular aging. Traffic. 2007;8:1590–1600. doi:10.1111/j.1600-0854.2007.00633.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.