Abstract
Background:
Nonsmall cell lung cancer (NSCLC) is a serious leading cause of death worldwide. Recently, multiple researches have identified that microRNA (miRNA) in sputum could be a useful tool for NSCLC diagnosis. The objective of this study was to assess whether aberrant miRNA expression could be regarded as a useful biomarker in sputum specimen for the diagnosis of NSCLC.
Methods:
Eligible studies were searched in PubMed, Web of Science, China National Knowledge Infrastructure (CNKI), Wanfang, and VIP databases up to June 2018. We calculated the pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the curve (AUC) to investigate the diagnostic value of miRNA in sputum for NSCLC. MetaDisc1.4 and STATA12.0 were used to analyze the retrieved data.
Results:
Finally, a total of 14 articles were included in this meta-analysis involving 1009 NSCLC patients and 1006 controls. The results were as followed: the pooled sensitivity, specificity, PLR, NLR, DOR, were 0.75 (95%CI:0.72–0.78), 0.88 (95%CI:0.86–0.90), 5.70 (95%CI:4.82–6.75), 0.30 (95%CI:0.26–0.34), 22.43 (95%CI:17.48–28.79), respectively. The AUC of overall summary receiver operator characteristic curve (SROC) was 0.8917.
Conclusion:
Our comprehensive analysis indicated that miRNAs in sputum specimen may be noninvasive diagnostic biomarkers for NSCLC. However, much more studies should be conducted before clinical application.
Keywords: diagnosis, meta-analysis, miRNAs, NSCLC, sputum
1. Introduction
Lung cancer, which includes small cell lung cancer (SCLC) and nonsmall cell lung caner (NSCLC), is the second most common malignancies and the number one cancer killer worldwide.[1] The morbidity and mortality of NSCLC are increasing year after year despite great improvements in treatment strategies including surgery, chemoradiotherapy and immunotherapy. NSCLC is the predominant subtype of lung cancer accounting for approximately 80% of all incidences, while SCLC accounts for 20%. NSCLC is also divided into 4 main histological subtypes: adenocarcinoma (AC), squamouscell carcinoma (SCC), large cell carcinoma and other (neuroendocrine cancers, carcinoids etc).[2,3] The 5-year survival rate is approximately 10% for advanced stage of lung cancer, however, it is almostly 80% for stage I lung cancer. Furthermore, the median survival time of lung cancer with treatment is 18 to 24 months. The median survival of extensive-stage lung cancer is 6 to 12 months with treatment, and only 2 to 4 months without treatment.[3,4] The unpromising phenomenon is due to the lack of early diagnostic methods. Hence, it is urgent to find potential markers for diagnosis to improve the early detection of lung cancer patients.
Up to now, pathological biopsy is still the diagnostic “golden standard” of lung cancer, however, the approaches may have risks of pneumothorax, hemorrhage and false-negative results.[5] Chest x-ray has been used to screen the high risk population. In addition, high-resolution CT (HRCT) and the widespread use of computed tomography (CT) is applied to screen varieties of cancers, such as breast cancer and gastrointestinal cancer, this is certainly an encouraging and exciting challenge, however, none of them is truly optimal, either with low sensitivity and specificity or the methods are expensive and invasive.[6,7] Sputum cytology has been performed for diagnosis of lung cancer, but it depends on the skills required for identifying subtle cellular morphological abnormalities in bronchial epithelial cells, thus the sensitivity is very low.[8] Therefore, it is necessary to explore and seek the relevant molecular mechanisms of lung cancer to identify potential diagnostic biomarkers.
Compared with other specimen, sputum is one of the most conveniently and noninvasively accessible biological fluids. Sputum consists of many components, such as salivary amylase, lysozyme, mucoprotein, immune globulin (Ig) and airway epithelial cells. Analysis of sputum can find the specific source of the abnormal airway epithelial cells in the lung and then provide an organ-specific way for lung cancer diagnosis.[9] Mounting studies have shown that sputum could be a promising “remote medium” for early detection of NSCLC. For example, Bagheri et al[10] found that altered miR-223 expression in sputum could discriminated NSCLC patients from cancer-free individuals and might be potential noninvasive marker for diagnosis of NSCLC patients. In addition, P16 hypermethylation was found in sputum collected from patients with lung cancer and could be a clinical diagnostic biomarker of NSCLC.[11,12] Furthermore, Varella-Garcia and his colleagues[13] found that the assessment of chromosomal aneusomy in exfoliated cells of sputum could help diagnose lung cancer with 76% sensitivity and 88% specificity.
MiRNA, which was found in 1990, is one of the small noncoding (19∼25 nucleotides in length) single stranded RNA and plays an important role in regulating gene and protein expression.[14] In recent years, miRNAs might function as tumor suppressors or oncogenes and dysregulated miRNA expressions have been confirmed to participate in many pathological and physiological processes in various cancers. Another characteristic of miRNAs is prominent stability in different kinds of biological samples, such as urine, plasma, serum, sputum, formalin-fixed, paraffin-embedded clinical tissues, fresh snap-frozen materials and even in very harsh conditions, this is due to their resistance to endogenous or exogenous RNA enzyme, extreme temperatures and pH, long storage in frozen conditions, and repeated freezethaw cycles.[15] These features introduce miRNA as a great target for different aspects of biological and medical investigations, which can be regarded as diagnostic and prognostic biomarkers in multiple diseases. In the passed decades, a number of studies[16–18] have verified the diagnostic significance of sputum miRNAs in NSCLC. However, there are still inconsistencies about the results, which may account for stage, region, sample size and so on. Therefore, we performed this meta-analysis to assess the diagnostic value of miRNA expression in sputum for NSCLC patients.
2. Methods
2.1. Publication search
Systematic comprehensive search was carried out to find relevant studies for this meta-analysis. We searched PubMed, Web of Science, CNKI, Wanfang and VIP databases from articles published to June 2018 with languages in English or Chinese. The search terms are as follows: microRNAs or miRNAs, nonsmall cell lung cancer or NSCLC, sputum or flema, diagnosis or diagnostic, sensitivity or specificity. At the same time, additional bibliographies were also retrieved in the selected literatures in order to prevent omitting relevant articles. Because this is a systematic review and meta-analysis, the ethical approval and patient written informed consent are not required.
2.2. Inclusion and exclusion criteria
The suitable studies must satisfied the following criteria: studies were conducted on humans; and studies were case-control investigations; researches involved in miRNAs and NSCLC; specimen was sputum or bronchoalveolar lavage (BAL); researches had sufficiently retrieved data. Articles were excluded if they: investigated about cell lines or animals; reviews, case reports, letters, meeting records; without enough data.
2.3. Data extraction and quality assessment
Two investigators (Wang and Zhang) independently extracted data and information from the selected studies. Any discrepancies were resolved by the third author or discussion. We retrieved the following information: first author's name, publication year, country, the number of cases and controls, miRNAs, detection method, sensitivity, specificity and AUC. The quality assessment of all the eligible studies was conducted according to the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2), which is a useful tool to evaluate the quality assessment of diagnostic accuracy studies.[19] The QUADAS-2 consists of 4 domains: patient selection, the index test, the reference standard and flow and timing. The questions will be answered as “yes” with one point, and “no” or “unclear” with zero point. The maximum score is 7 point, any studies obtained more than 5 point would be regarded as eligible candidates.
2.4. Statistic analysis
The analysis software we used were MetaDisc1.4 (XI Cochrane Colloquium, Barcelona, Spain) and STATA12.0 (StataCorp LP, College Station, TX). The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and area under curve (AUC) were calculated to assess diagnostic value of sputum miRNAs in distinguishing NSCLC patients from healthy individuals. Heterogeneity analysis was conducted by Cochranc's Q test and Higgins I-squared test. If P < .1 or I2 > 50%, a random effect model was applied, while P > .1 or I2 < 50%, the fixed effect model was used. Subgroup analysis was conducted to investigate the potential sources of heterogeneity. Besides, we also performed Deek's funnel plot to assess publication bias.
3. Results
3.1. Data selection and study characteristics
A total of 230 articles were searched from PubMed, Web of Science, CNKI, and VIP databases initially. After wiping out duplicates, there remained 148 studies. According to reading the titles and abstracts, 128 studies were removed, which contained 96 studies with other specimen (blood, plasma, serum and tissue), 20 non-NSCLC patients, 3 meeting records, 7 letters and 2 reviews. After browsing the full-texts, 6 articles were excluded without sufficient data. Finally, there were 14 publications[10,16–18,20–29] satisfying our meta-analysis. The flow chart of searching process was shown in Figure 1.
Figure 1.
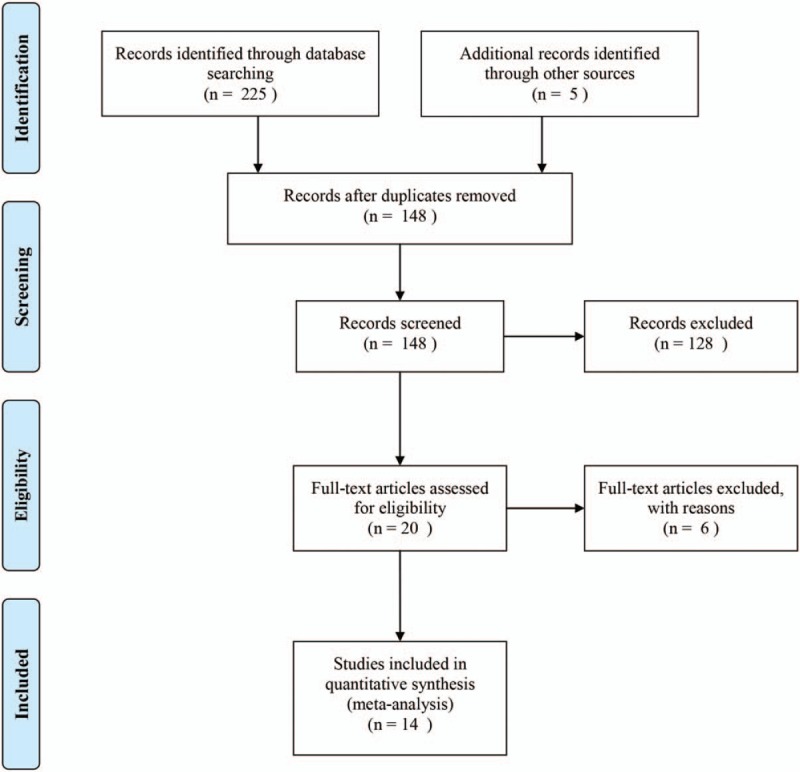
The flow chart of searching eligible articles process in this meta-analysis.
The characteristics of the included studies were described in Table 1. Among the 14 eligible articles, there were 1009 NSCLC patients and 1006 controls, which included 11 kinds of miRNAs. The methods to detect the level of miRNAs included real-time polymerase chain reaction (RT-PCR),[16,29] quantitative real-time polymerase chain reaction (qRT-PCR),[10,17,20–28] and digital polymerase chain reaction (Digital PCR).[18] The sample size of these studies ranged from 30 to 291 individuals. Three studies[10,16,29] evaluated a single miRNA in sputum as diagnostic biomarker, while eleven studies researched multiple miRNAs. QUADAS-2 was used to assess the quality of the included studies. All the eligible literatures obtained had satisfying scores. The quality of included studies was assessed by QUADAS-2 and most studies had moderately high scores. The risk of bias and applicability concerns diagram were shown in Figure 2.
Table 1.
Characteristics of the 14 included studies.
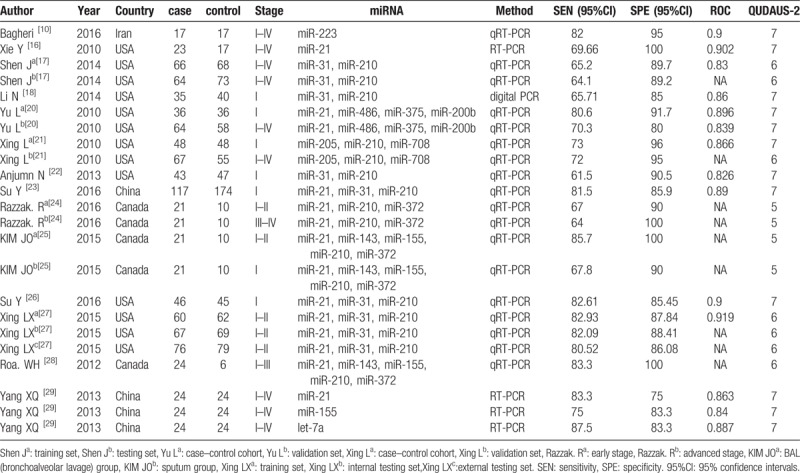
Figure 2.
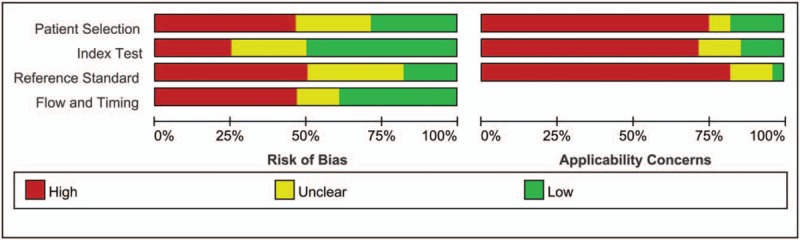
Bar charts of the quality assessment of included studies using the tool of Quality Assessment of Diagnostic Accuracy Studies 2 (QUADUA-2). (Left) Risk of Bias. (Right) Applicability Concerns.
3.2. Pooled diagnosis accuracy of miRNAs in NSCLC
The heterogeneity analysis was conducted by Cochranc's Q test and I2 test. I2 value of sensitivity and specificity were 35.5%, 24.1%, respectively, so the fixed effect model was used to assess the pooled diagnosis accuracy of miRNAs in NSCLC.
The pooled sensitivity was 0.75 (95%CI:0.72–0.78), pooled specificity 0.88 (95%CI:0.86–0.90), positive likelihood ratio (PLR) 5.70 (95%CI:4.82–6.75), negative likelihood ratio (NLR) 0.30 (95%CI:0.26–0.34), diagnostic odds ratio (DOR) 22.43 (95%CI:17.48–28.79). The area under the curve (AUC) was 0.8917, which indicated that miRNAs in sputum samples had a high diagnostic efficiency for NSCLC. The overall forest plots of sensitivity and specificity, DOR, SROC were presented in Figures 3–6.
Figure 3.
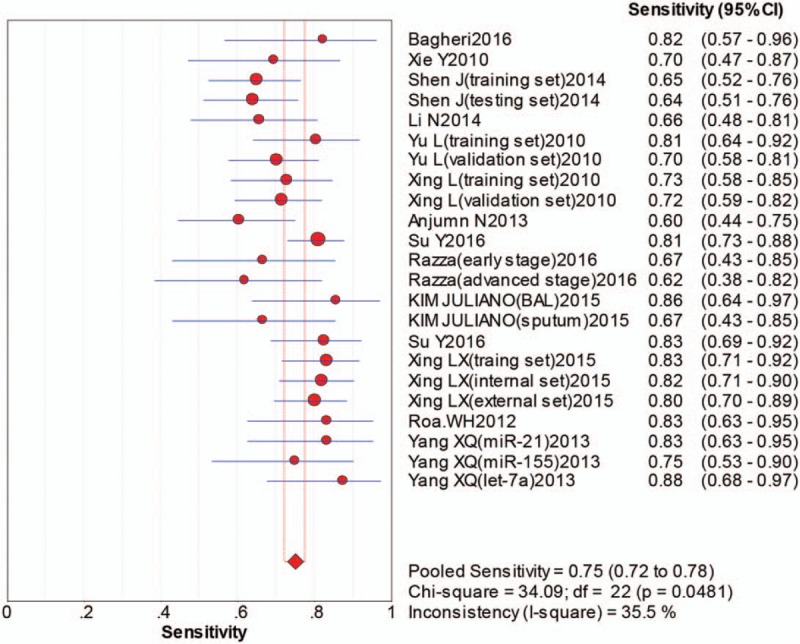
Pooled sensitivity forest plot of sputum miRNAs in diagnosing of nonsmall cell lung cancers.
Figure 6.
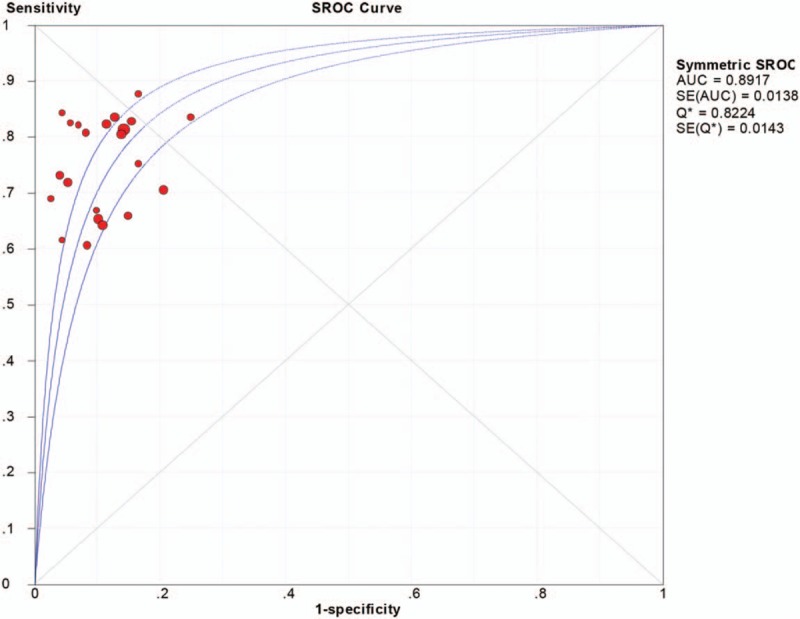
Summary receiver operator characteristic curve (SROC) with area under curve (AUC) of sputum miRNAs in diagnosing of nonsmall cell lung cancers. AUC = area under curve, SROC = summary receiver operator characteristic curve.
Figure 4.
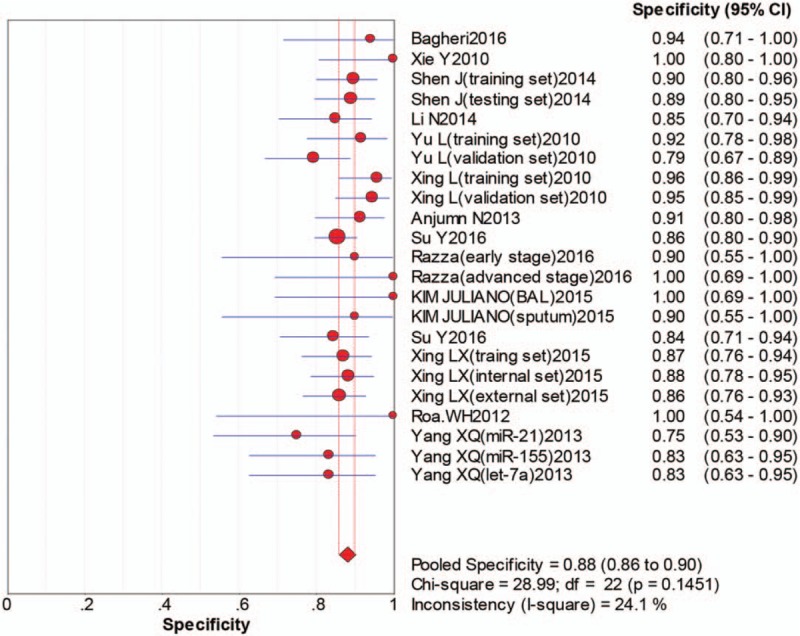
Pooled specificity forest plot of sputum miRNAs in diagnosing of nonsmall cell lung cancers.
Figure 5.
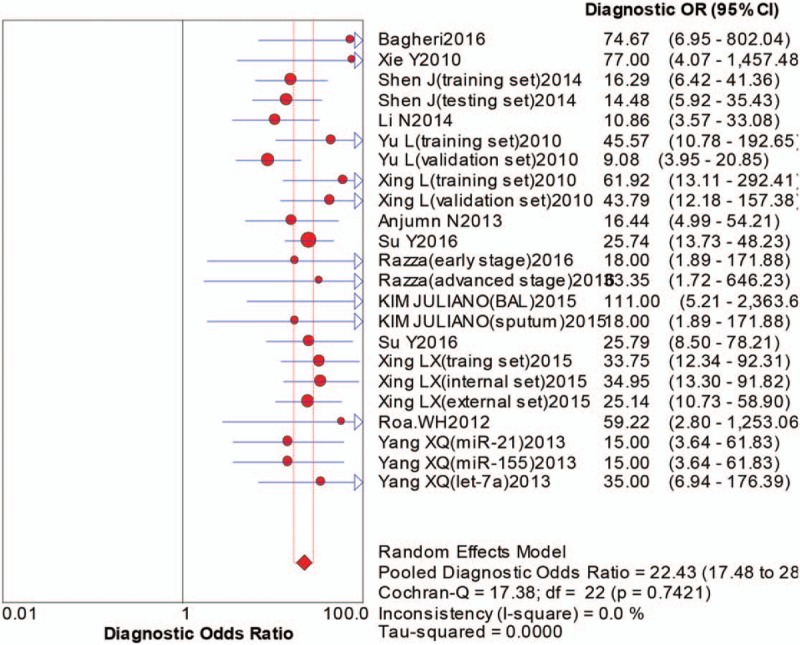
The forest plot of diagnostic odds ratio of sputum miRNAs in diagnosing of nonsmall cell lung cancers.
3.3. Subgroup analysis
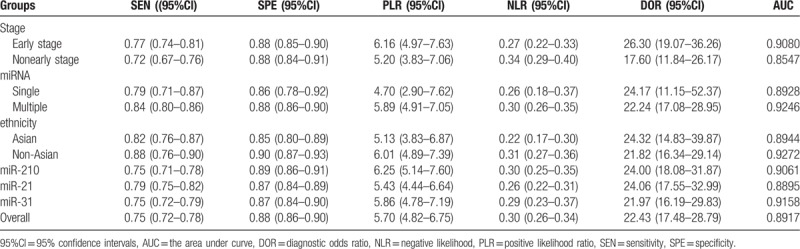
In order to find the origins of the heterogeneity, we performed subgroup analysis (Table 2). The results demonstrated that the accuracy of sputum miRNAs test in early stage produced sensitivity of 0.77 (95%Cl: 0.74–0.81), specificity of 0.88 (95%Cl: 0.85–0.90) and AUC of 0.9080. The corresponding values of nonearly stage were 0.72 (95%Cl: 0.67–0.76) for sensitivity, 0.88 (95%Cl: 0.84–0.91) and 0.8547 for AUC. This result indicated that the sensitivity of miRNAs in early stage was more significant than in nonearly stage. Then, we implemented subgroup analysis based on miRNA profiling (single miRNA vs multiple miRNAs). We found that multiple miRNAs had a higher diagnostic value with sensitivity 0.84 (95%Cl: 0.80–0.86), specificity 0.88 (95%Cl: 0.86–0.90) and AUC 0.9246 than single miRNA with sensitivity 0.79 (95%Cl: 0.71–0.87), specificity 0.86 (95%Cl: 0.78–0.92) and AUC 0.8928, respectively. Furthermore, we also conducted subgroup analysis on the basis of ethnicity. Through comparison, the miRNAs obtained a better diagnostic value in non-Asian population with sensitivity 0.88 (95%Cl: 0.76–0.90), specificity 0.90 (95%Cl: 0.87–0.93) and AUC 0.9272, respectively, while Asian population with sensitivity 0.82 (95%Cl: 0.77–0.87), specificity 0.85 (95%Cl: 0.80–0.89) and AUC 0.8944, respectively. Compared with other miRNAs, miR-210, miR-21 and miR-31 were more often used as diagnostic markers. However, they were usually associated with other miRNAs. The sensitivity, specificity, and AUC were, respectively, 0.75 (95%CI: 0.71–0.78), 0.89 (95%CI: 0.86–0.91) and 0.9061 for miR-210 with other miRNAs. The sensitivity, specificity and AUC of miR-21 with other miRNAs were, respectively, 0.79 (95%CI: 0.75–0.82), 0.87 (95%CI: 0.84–0.89) and 0.8895. The sensitivity, specificity and AUC of miR-31 with other miRNAs were, respectively, 0.75 (95%CI: 0.72–0.79), 0.87 (95%CI: 0.84–0.90) and 0.9158. Outcomes of subgroup analysis confirmed that the stage of NSCLC, miRNAs profiling, and ethnicity might be the sources of heterogeneity among the literatures.
Table 2.
Subgroup analysis of all studies and diagnostic results.
3.4. Publication bias
In this meta-analysis, Deek's Funnel plot asymmetry test was used to evaluated the publication bias of the included studies. As was shown in Figure 7, there was no significant evidence of publication bias in our study. It demonstrated the final results were not affected by the individual study.
Figure 7.
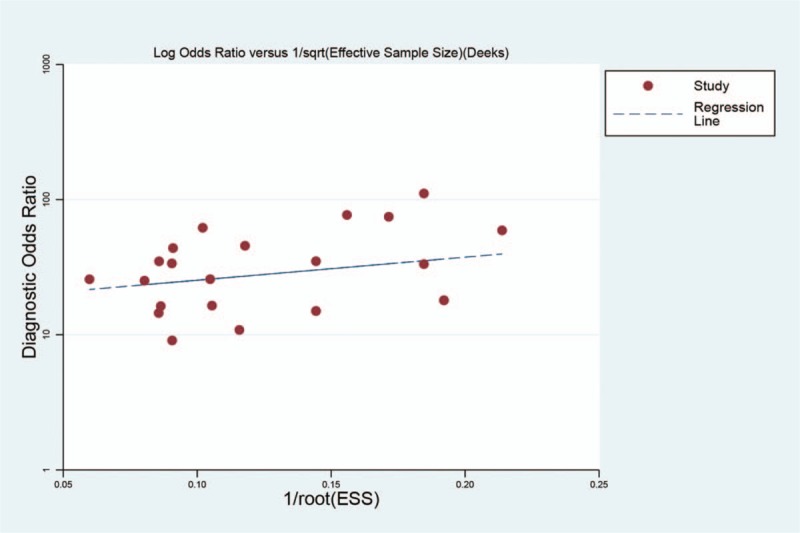
Deek's funnel graph of the diagnostic role of sputum miRNAs in nonsmall cell lung cancers.
4. Discussion
It is reported that nonsmall cell lung cancer (NSCLC) is the second malignant disease and one of the cancer killers in the world.[1] The five-year survival rate is as low as 10% for late stage of NSCLC. The poor statistics is unpromising and it is urgent to find accurate and reliable method to diagnose NSCLC as early as possible.[2–4] Recently, the methods to screen NSCLC patients are computed tomography (CT) and bronchoscopy. However, National Lung Screening Trail (NLST) manifested that the specificity of CT screening is only 60% and bronchoscopy is invasive technique.[17] Therefore, it is essential to look for novel noninvasive biomarkers to diagnose lung cancer.
MiRNA is a class of small noncoding RNA that can regulate gene expression post-transcriptionlly through binding to the 3’ untranslational region (3’UTR) of target mRNAs.[15] In addition, miRNA may regulate tumor occurrence and development process, such as proliferation, differentiation and apoptosis.[24,25] The different expression level of miRNAs in human cancers and its potential diagnostic values have been previously investigated. The recent proof showed that endogenous miRNAs were present in sputum in a dramatically stable form and could easily be detected by real-time reverse transcription quantitative PCR (qRT-PCR).[26] So far, emerging articles suggest that the sputum miRNAs can be used as diagnostic markers for NSCLC patients. Xing et al[21] used different techniques to validate sputum miRNAs as biomarkers for early stage NSCLC, with sensitivity of 73% and specificity of 96%. Anjuman et al[22] found that the use of Lung Flute could conveniently and effectively obtain sputum from low reparatory track of individuals who are not able to expectorate spontaneous sputum, and the sputum miRNAs (miRs-31 and miR-210) produced a significantly higher sensitivity (61.5% vs 35.9%) but a lower specificity (90.5% vs 95.2%) compared with cytology approach for NSCLC diagnosis.
In the present meta-analysis, we found that miRNAs in sputum samples for the detection of NSCLC yielded an overall sensitivity of 75% and an overall specificity of 88%. The AUC was 0.8917, indicating an accuracy of high level. Furthermore, the PLR was 5.70, NLR was 0.30 and DOR value was 22.43. Taken all together, it indicated that overall accuracy of NSCLC detection using sputum miRNAs testing was good enough.
Furthermore, we performed subgroup analysis to investigate the potential heterogeneity among the included studies. One of the sources of the heterogeneity is the stage of disease. The result of the subgroup analysis showed that the diagnostic accuracy in early stage was relatively higher than in nonearly stage (Table 2), it indicated that sputum miRNAs might be biomarkers to diagnose early NSCLC. And the future use of sputum-based miRNA biomarkers with radiological imaging would improve the possibility to detect lung cancer at its early stage where therapeutic interventions have a curative potential. Among the miRNAs profiling, miR-210, miR-21, miR-31 were associated with other miRNAs could be used for the detection of NSCLC. In addition, we also performed subgroup analysis of miRNAs profiling and the result showed that the pooled sensitivity and specificity of the multiple miRNAs was higher than the single one (Table 2). This may be explained by the theory that lung tumor is a heterogeneous disease and develops from complex molecular mechanisms and multi-step processes, which involved many miRNAs. So, the combination of panel miRNAs may be a better way to diagnose NSCLC more precisely, and our results are consistent with previous meta-analysis.[30,31] Additionally, in the subgroup analysis based on ethnicity, the diagnostic accuracy of miRNAs in non-Asian population was significantly higher than in Asian population (Table 2). This indicated that sputum miRNAs may be more suitable to detect NSCLC in non-Asian population than in Asian population.
4.1. Limitations
This meta-analysis still exists limitations despite the encouraging results. First, the sample size of the eligible studies was limited, which could result in insufficient statistic analysis for the clinical application of sputum miRNAs for NSCLC detection. Second, in the process of selecting the eligible studies, some publications may be excluded due to lacking of sufficient data. Third, the cut-off value was different among the 14 studies because there was no uniform standard, which certainly influenced the outcome. Finally, the population of included studies were almost from non-Asian (Canada or USA), Asian population was too small, this may not be representative of all nonsmall cell lung cancers.
In the future, more high quality and larger prospective clinical studies with standard cut-off values are essential to accurately define the role of miRNAs in the diagnosis of NSCLC. The information of the included studies revealed that the heterogeneity could be attributed to the differences in the publication year, stage of the cancers, the sample size, and the cut-off values of sputum miRNAs. We conducted subgroup analysis and sensitive analysis to find out the possible sources of heterogeneity. So these factors and studies should be paid more attention to when the concerning subjects were taken into consideration.
5. Conclusion
Our results indicated that miRNAs in sputum can be promising noninvasive and cost-effective biomarkers for the diagnosis of NSCLC patients, especially in the early stage. Due to the different diagnostic values of miRNAs, the combination of miR-210, miR-21, miR-31 with other miRNAs could be a better way to diagnose NSCLC more accurately. Considering the limitations in our meta-analysis, however, clinical application of miRNA diagnosis for NSCLC still needs more studies and large-scale investigations to improve the diagnostic accuracy.
Author contributions
Data curation: Qian Wang.
Methodology: Shijie Zhang.
Software: Shijie Zhang.
Supervision: Shijie Zhang, Qian Wang.
Visualization: Qian Wang.
Writing – original draft: Xiaoyun Zhang.
Writing – review & editing: Xiaoyun Zhang.
Footnotes
Abbreviations: AC = adenocarcinoma, AUC = area under curve, BAL = bronchoalveolar lavage, CNKI = China National Knowledge Infrastructure, CT = computed tomography, DOR = diagnostic odds ratio, HRCT = high-resolution computed tomography, NLR = negative likelihood ratio, NLST = National Lung Screening Trail, NSCLC = nonsmall cell lung cancer, PLR = positive likelihood ratio, qRT-PCR = quantitative real-time polymerase chain reaction, QUADAS-2 = quality assessment of diagnostic accuracy studies-2, RT-PCR = real-time polymerase chain reaction, SCC = squamouscell carcinoma, SCLC = small cell lung cancer, SROC = summary receiver's operative characteristics.
The authors have no conflicts of interest to disclose.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Hattori A, Takamochi K, Oh S, et al. New revisions and current issues in the eighth edition of the TNM classification for non-small cell lung cancer. Jap J Clin Oncol 2019;49:3–11. [DOI] [PubMed] [Google Scholar]
- [3].Kozu Y, Maniwa T, Takahashi S, et al. Results of surgical treatment for non-small cell lung cancer with positive sputum cytology: experience from a single institution. Thorac Cardiovasc Surg 2014;62:588–92. [DOI] [PubMed] [Google Scholar]
- [4].Wang J, Zhang KY, Liu SM, et al. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules (Basel, Switzerland) 2014;19:1912–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van der Heijden EH, Casal RF, Trisolini R, et al. Guideline for the acquisition and preparation of conventional and endobronchial ultrasound-guided transbronchial needle aspiration specimens for the diagnosis and molecular testing of patients with known or suspected lung cancer. Respiration 2014;88:500–17. [DOI] [PubMed] [Google Scholar]
- [6].Wang Z, Ge M. Progress of lung margin during sublobar resection for early-staged non-small cell lung cancer. Chin J Lung Cancer 2018;21:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kodama K, Higashiyama M, Yokouchi H, et al. Prognostic value of ground-glass opacity found in small lung adenocarcinoma on high-resolution CT scanning. Lung Cancer (Amsterdam, Netherlands) 2001;33:17–25. [DOI] [PubMed] [Google Scholar]
- [8].Jiang F, Todd NW, Li R, et al. A panel of sputum-based genomic marker for early detection of lung cancer. Cancer Prev Res (Philadelphia, Pa) 2010;3:1571–8. [DOI] [PubMed] [Google Scholar]
- [9].Thunnissen FB. Sputum examination for early detection of lung cancer. J Clin Pathol 2003;56:805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bagheri A, Khorshid HRK, Mowla SJ, et al. Altered miR-223 expression in sputum for diagnosis of non-small cell lung cancer. Avicenna J Med Biotechnol 2017;9:189–95. [PMC free article] [PubMed] [Google Scholar]
- [11].Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res 2002;62:2370–7. [PubMed] [Google Scholar]
- [12].Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer 2004;4:707–17. [DOI] [PubMed] [Google Scholar]
- [13].Varella-Garcia M, Schulte AP, Wolf HJ, et al. The detection of chromosomal aneusomy by fluorescence in situ hybridization in sputum predicts lung cancer incidence. Cancer Prev Res (Phila) 2010;3:447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis 2015;35:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Acunzo M, Croce CM. MicroRNA in cancer and cachexia—a mini-review. J Infect Dis 2015;212suppl 1:S74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer (Amsterdam, Netherlands) 2010;67:170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shen J, Liao J, Guarnera MA, et al. Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J Thorac Oncol Cancer 2014;9:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li N, Ma J, Guarnera MA, et al. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. J Cancer Res Clin Oncol 2014;140:145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qu YJ, Yang ZR, Sun F, et al. Risk on bias assessment: (6) A Revised Tool for the Quality Assessment on Diagnostic Accuracy Studies (QUADAS-2). Zhonghua Liu Xing Bing Xue Za Zhi 2018;39:524–31. [DOI] [PubMed] [Google Scholar]
- [20].Yu L, Todd NW, Xing L, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 2010;127:2870–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xing L, Todd NW, Yu L, et al. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol 2010;23:1157–64. [DOI] [PubMed] [Google Scholar]
- [22].Anjuman N, Li N, Guarnera M, et al. Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. Clin Transl Med 2013;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Su Y, Fang H, Jiang F. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin Epigenetics 2016;8:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Razzak R, Bedard EL, Kim JO, et al. MicroRNA expression profiling of sputum for the detection of early and locally advanced nonsmall-cell lung cancer: a prospective case-control study. Curr Oncol (Toronto, Ont ) 2016;23:e86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim JO, Gazala S, Razzak R, et al. Non-small cell lung cancer detection using microRNA expression profiling of bronchoalveolar lavage fluid and sputum. Anticancer Res 2015;35:1873–80. [PubMed] [Google Scholar]
- [26].Su Y, Guarnera MA, Fang H, et al. Small non-coding RNA biomarkers in sputum for lung cancer diagnosis. Mol Cancer 2016;15:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xing L, Su J, Guarnera MA, et al. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin Cancer Res 2015;21:484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roa WH, Kim JO, Razzak R, et al. Sputum microRNA profiling: a novel approach for the early detection of non-small cell lung cancer. Clin Invest Med 2012;35:E271. [DOI] [PubMed] [Google Scholar]
- [29].Yang XQ, Zhang YH, Sun B, et al. Diagnostic value of the detection of MicroRNAs in sputum of patients with nonsmall cell lung cancer. J Clin Pulm Med 2013;18:226–9. [Google Scholar]
- [30].Wang H, Wu S, Zhao L, et al. Clinical use of microRNAs as potential non-invasive biomarkers for detecting nons-mall cell lung cancer: a meta-analysis. Respirology (Carlton, Vic ) 2015;20:56–65. [DOI] [PubMed] [Google Scholar]
- [31].Jiang M, Li X, Quan X. Clinically correlated MicroRNAs in the diagnosis of non-small cell lung cancer: a systematic review and meta-analysis. Biomed Res Int 2018;2018:5930951. [DOI] [PMC free article] [PubMed] [Google Scholar]


