Abstract
Early identification of high-risk septic patients in the emergency department (ED) may guide appropriate management and disposition, thereby improving outcomes. We compared the performance of machine learning models against conventional risk stratification tools, namely the Quick Sequential Organ Failure Assessment (qSOFA), National Early Warning Score (NEWS), Modified Early Warning Score (MEWS), and our previously described Singapore ED Sepsis (SEDS) model, in the prediction of 30-day in-hospital mortality (IHM) among suspected sepsis patients in the ED.
Adult patients who presented to Singapore General Hospital (SGH) ED between September 2014 and April 2016, and who met ≥2 of the 4 Systemic Inflammatory Response Syndrome (SIRS) criteria were included. Patient demographics, vital signs and heart rate variability (HRV) measures obtained at triage were used as predictors. Baseline models were created using qSOFA, NEWS, MEWS, and SEDS scores. Candidate models were trained using k-nearest neighbors, random forest, adaptive boosting, gradient boosting and support vector machine. Models were evaluated on F1 score and area under the precision-recall curve (AUPRC).
A total of 214 patients were included, of whom 40 (18.7%) met the outcome. Gradient boosting was the best model with a F1 score of 0.50 and AUPRC of 0.35, and performed better than all the baseline comparators (SEDS, F1 0.40, AUPRC 0.22; qSOFA, F1 0.32, AUPRC 0.21; NEWS, F1 0.38, AUPRC 0.28; MEWS, F1 0.30, AUPRC 0.25).
A machine learning model can be used to improve prediction of 30-day IHM among suspected sepsis patients in the ED compared to traditional risk stratification tools.
Keywords: electrocardiography, emergency service, hospital, machine learning, sepsis, triage
1. Introduction
Sepsis is increasing in incidence and has a 10% to 20% in-hospital mortality (IHM) rate.[1–3] Risk stratification of septic patients in the Emergency Department (ED) may help to guide appropriate management and disposition, thereby reducing morbidity and mortality.[4–6] A number of clinical tools have been developed to risk stratify septic patients in the ED, where certain clinical information, such as laboratory investigations, is not readily available. The Quick Sequential Organ Failure Assessment (qSOFA) score was externally validated among septic patients presenting to the ED using the worst level of the 3 components observed during the ED stay and showed good prognostic accuracy for IHM.[7] A recent study showed that commonly used early warning scores such as the National Early Warning Score (NEWS) and the Modified Early Warning Score (MEWS) were more accurate than qSOFA in predicting mortality in patients with suspected infection presenting to the ED.[8]
Several studies have also reported the prognostic value of heart rate variability (HRV) parameters in septic patients presenting to the ED.[9–11] Septic patients have reduced sympatho-vagal balance and impaired sympathetic activity, which lead to varying degrees of cardiac autonomic dysfunction.[12] This can be detected by HRV analysis, a quick, non-invasive technique of evaluating the beat-to-beat variation in heart rate. HRV analyses are divided into linear and non-linear methods.[13] Linear methods include HRV parameters measured in time or frequency domains. Time domain HRV parameters are statistical calculations of consecutive R-R time intervals and how they correlate with each other. Frequency domain HRV parameters are based on spectral analysis. Studies have suggested that regulators of the cardiovascular system interact in a non-linear way[14,15] and HRV analysis using non-linear methods reflect these mechanisms.[16]
We previously described a 5-variable Singapore ED Sepsis (SEDS) model to predict the risk of 30-day IHM among septic patients in the ED.[17] The SEDS model was the first risk stratification tool to incorporate HRV parameters with other traditional prognosticators such as patient demographics and vital signs. It was developed via stepwise logistic regression and had improved predictive performance over existing tools that only utilize vital signs in their scoring criteria.[17]
With the widespread adoption of electronic medical records in healthcare and availability of high-resolution data particularly in the intensive care unit (ICU) setting, machine learning algorithms have become popular for modelling patient health status. Machine learning models have shown good performance in early detection of sepsis among ICU patients[18,19] and prediction of progression to septic shock among patients with sepsis.[20] A randomised controlled trial also showed that the use of a machine learning-based severe sepsis surveillance and alert system improved patient outcomes such as length of stay and IHM.[21]
To date, only 1 study has demonstrated the use of machine learning for risk prediction of septic patients in the ED. However, it did not explore modern algorithms such as boosting and support vector machine, and did not incorporate HRV measures.[22] In this study, we aimed to compare the performance of HRV-based machine learning models against the SEDS model and other conventional risk stratification tools, namely the qSOFA, NEWS and MEWS, in the prediction of 30-day IHM among suspected sepsis patients in the ED setting. We are interested in the use of these models for early risk stratification based on clinical information that are quickly obtainable during triage and without chart review.
2. Methods
Ethics approval for the study was obtained from SingHealth's Centralised Institutional Review Board (CIRB, Reference Number 2016/2858), with waiver of patient consent. We conducted secondary analysis of electronic health data from patients above 21 years old who presented to the Singapore General Hospital (SGH) ED between September 2014 and April 2016 with suspected sepsis, and who met at least 2 of the 4 Systemic Inflammatory Response Syndrome (SIRS) criteria.[23] The SIRS criteria are temperature (<36°C or >38°C), heart rate (>90 beats/min), respiratory rate (>20 breaths/min) and total white count (<4000/mm3 or >12,000/mm3).
All patients presenting to the SGH ED are triaged by a trained nurse on arrival. The first set of vital signs recorded and routine 5-minute one-lead electrocardiogram (ECG) tracings performed at triage were used for analysis. Patient demographics and vital signs were obtained from the hospital's electronic medical records. The ECGs were obtained from X-Series Monitor (ZOLL Medical Corporation, Chelmsford, MA) and subsequently loaded into Kubios HRV software version 2.2 (Kuopio, Finland) for computation of HRV parameters.[24]
The program automatically detected QRS complexes, but each ECG was also manually screened to ensure QRS detection was correct, and their positions were adjusted if misplaced. The R-R interval time series was then screened for rhythm, artifacts and ectopic beats. If artifacts or ectopic beats were few (<5), they were removed from the R-R interval time series. Patients with non-sinus rhythm or >5 artifacts and/or ectopic beats were excluded.
The outcome of interest was IHM within 30 days of ED admission. Objective variables quickly obtainable during triage and without chart review were considered as predictors, namely 3 demographic variables (age, gender, ethnicity), 6 vital signs (temperature, heart rate, respiratory rate, systolic and diastolic blood pressures, and Glasgow Coma Scale (GCS) score), and 22 HRV parameters in time, frequency and non-linear domains. These variables were also compared between patients who met the outcome and patients who did not using the Mann-Whitney U test for continuous variables, and the chi-square test or Fisher exact test as appropriate for categorical variables.
One-hot encoding was applied to categorical variables (such as ethnicity) and all variables were scaled prior to modeling. We randomly selected 60% of the observations to train the models, holding the remaining 40% as a test set for subsequent model evaluation. Baseline models were created using qSOFA, NEWS and MEWS scores. Their scoring criteria and thresholds for predicting positive outcome (>=2 for qSOFA, >=7 for NEWS, >=5 for MEWS) were taken from their original articles.[25–27] Two sets of qSOFA scores were computed, one using initial vital signs recorded at triage, and another using worst vital signs recorded during the entire ED stay as described by Freund et al.[7] Candidate models were trained using k-nearest neighbors (KNN), random forest (RF), adaptive boosting (ADA), gradient boosting (GB) and support vector machine (SVM). Class imbalance was addressed by applying class weights. Parameter tuning was performed via grid search 5-fold cross-validation with the aim of optimizing F1 score.
We used each model to predict on the test set and calculated its precision (equivalent to positive predictive value) and recall (equivalent to sensitivity) from its confusion matrix. For each model, we also generated a precision-recall curve (PRC), and calculated its F1 score, which is the harmonic mean of precision and recall, as well as area under the PRC (AUPRC). We chose these performance metrics as they are more informative and less misleading than specificity and Receiver Operating Characteristics (ROC) plots for evaluating binary classifiers on imbalanced datasets.[28] We computed 95% confidence intervals (CI) for the F1 scores by sampling from 1000 bootstrapped test sets. We used F1 as our main evaluation metric since it takes both precision and recall into account and we believe both are important in this context.
To better understand how the GB model worked, we also visualized feature importance in terms of the total decrease in node impurity (indicated by Gini index) due to branching over a given predictor, averaged over all trees.
Univariate statistical analysis was carried out in Stata version 13 (StataCorp 2013, College Station, TX). Machine learning models were developed in Python 3.6 (Python Software Foundation, Wilmington, DE) using the scikit-learn library.[29]
3. Results
Figure 1 shows the cohort selection process. A total of 214 patients were included in the study, of whom 40 (18.7%) met the outcome. One hundred eight (50.5%) of them were male, with median age of 67.5 years (inter-quartile range, IQR 57–79). The most commonly identified sources of infection were respiratory (33.2%), urinary tract (17.8%), gastrointestinal (7.0%), musculoskeletal (5.6%), and hepatobiliary (5.6%). There were no significant differences in the sources of infection between those who did and did not meet the outcome.
Figure 1.
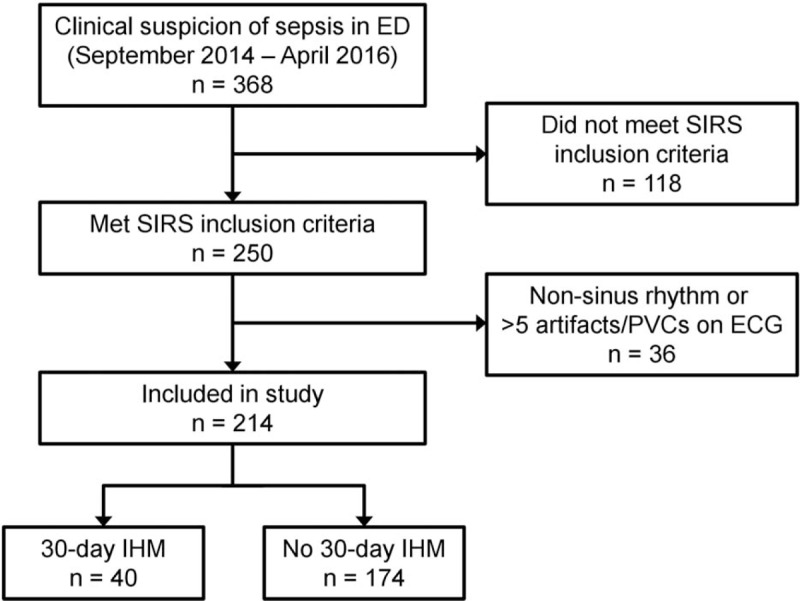
Cohort selection flow with breakdown of outcome.
Table 1 compares the patient demographics, vital signs and HRV parameters of the 2 patient groups. Patients who met the outcome were older (median age 76 years; IQR 68–83 years) than those who did not (median age 66 years; IQR 56–77 years). There were no significant differences in gender and ethnicity distributions between the 2 groups. In terms of vital signs, patients who met the outcome had higher respiratory rates, as well as lower temperatures and GCS scores, compared to patients who did not meet the outcome. Most of the HRV parameters across time, frequency and non-linear domains showed significant differences between the 2 groups.
Table 1.
Summary of predictor variables by presence of outcome.
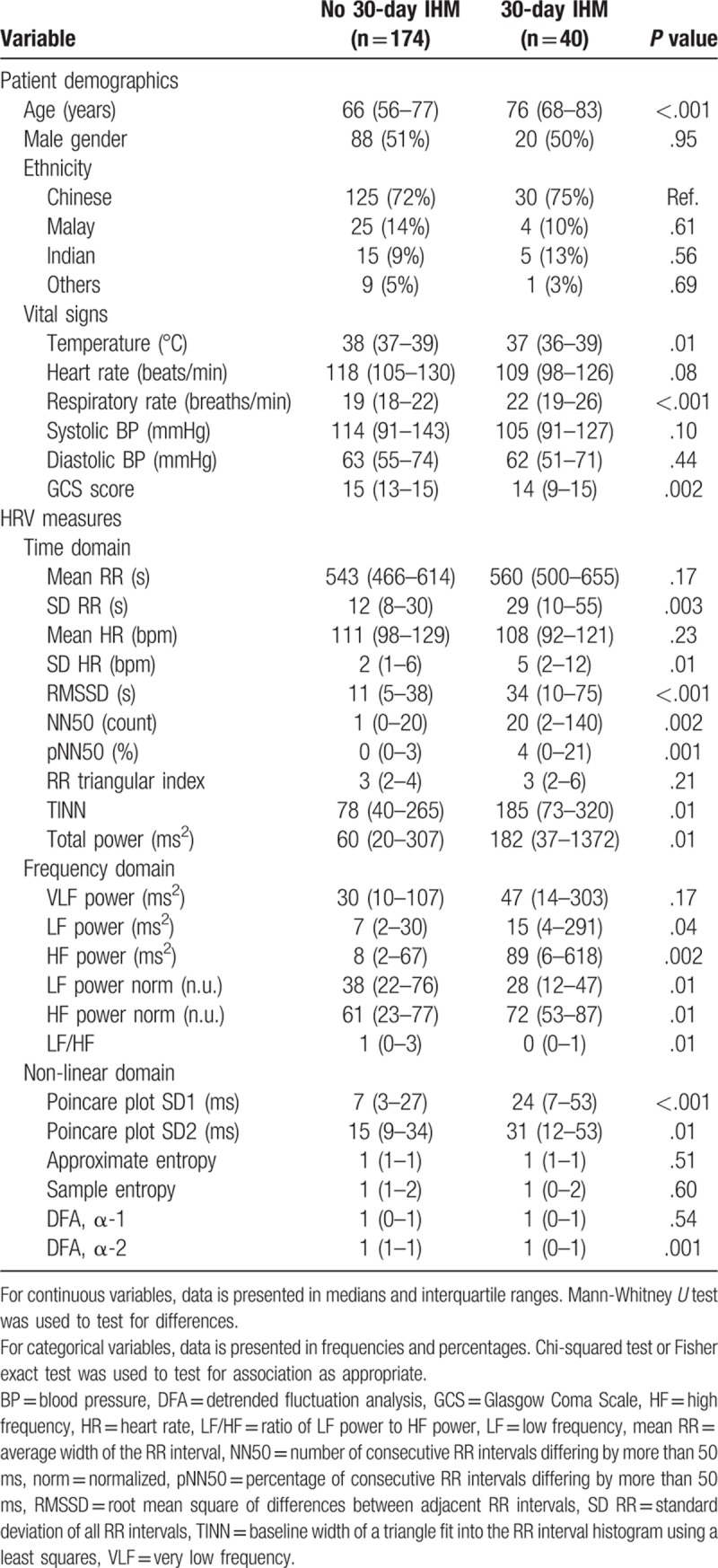
Table 2 summarizes the precision, recall, F1 score and AUPRC of the baseline and candidate models. Gradient boosting (GB) was the best candidate model with a F1 score of 0.50 and AUPRC of 0.35, and performed better than all the baseline models. Figure 2 shows the precision-recall curves of the GB model and baseline comparators.
Table 2.
Results of model evaluation.
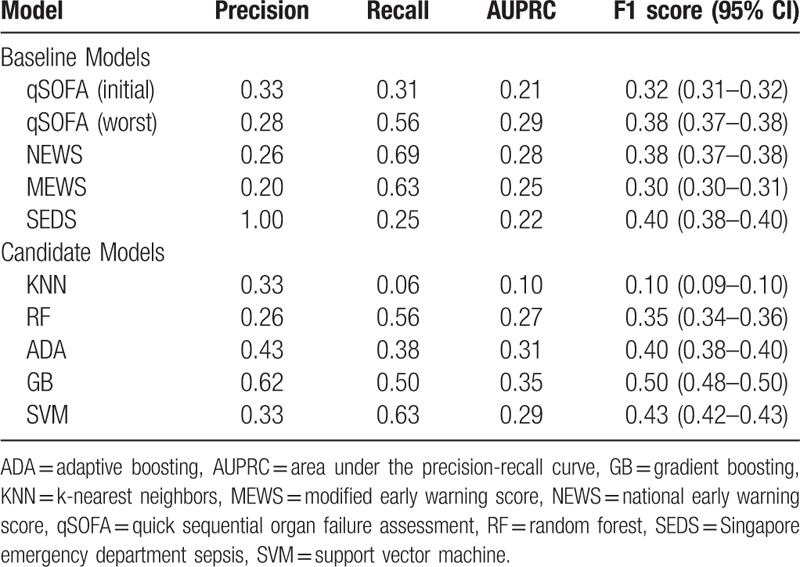
Figure 2.
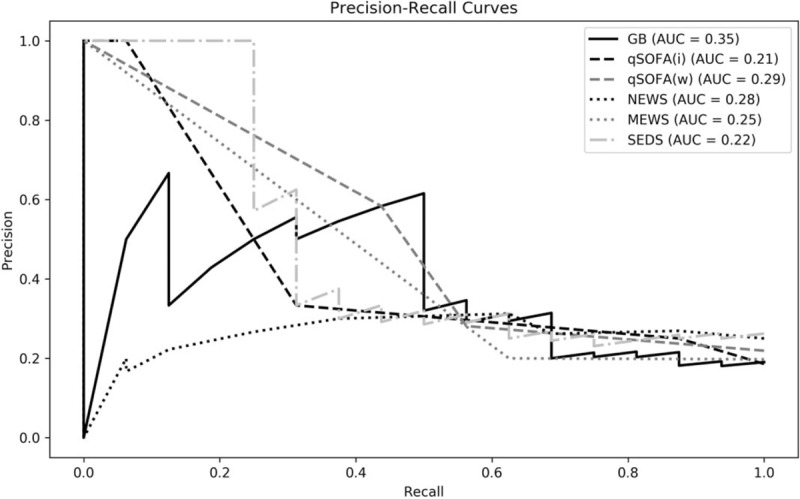
Precision-recall curves of gradient boosting model and baseline comparators.
Figure 3 shows the most predictive features in the GB model and their relative importance. Top predictors for 30-day IHM included temperature, detrended fluctuation analysis (DFA) α-2, heart rate, Glasgow Coma Scale (GCS) score and approximate entropy.
Figure 3.
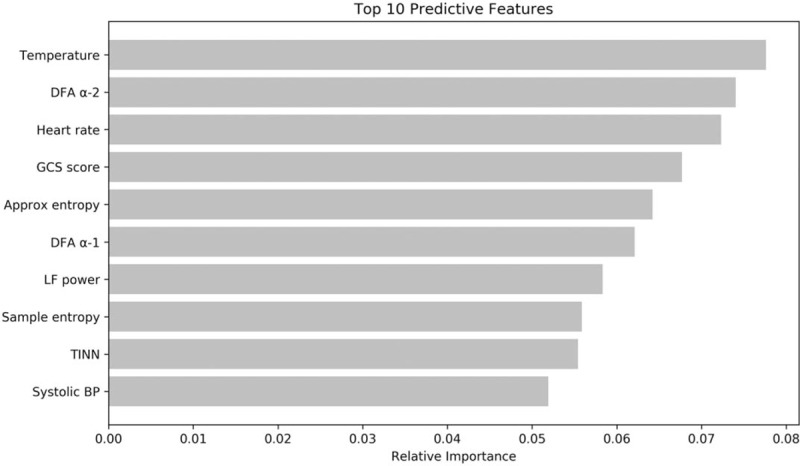
Relative importance of top 10 predictive features in the gradient boosting model.
4. Discussion
In this study, we applied machine learning to improve the 30-day IHM prediction of suspected sepsis patients in the ED. Baseline comparators were the qSOFA, NEWS, MEWS, and SEDS scoring systems. Our gradient boosting model outperformed all of them in terms of F1 score and AUPRC.
Compared to a previous study by Taylor et al which employed all available clinical variables collected during the entire ED stay,[22] our study only used predictors that were objective and quickly attainable in the first 5 minutes of patient presentation, namely demographics, vital signs and HRV parameters derived from routine ECGs. This allows risk stratification to be done at triage, facilitating early recognition of high-risk patients for allocation of care resources in the ED.
Our outcome of interest was in-hospital mortality within 30 days during the same admission where the vitals and ECG were taken. Some studies did not specify a time period for mortality[8] or if it was strictly within the same admission or not.[30] We chose this endpoint as it is more likely to be sepsis-related compared to an out-of-hospital mortality or mortality from a subsequent admission. It is also more meaningful for physicians in terms of administering possible interventions such as closer monitoring and less conservative management of high-risk patients.[7]
Among the top predictors in our machine learning model are temperature and heart rate, which are also part of the NEWS and MEWS scoring criteria, as well as GCS score, which is part of qSOFA, and similar to the AVPU scale used in NEWS and MEWS. The most important HRV predictor is DFA, which is a non-linear parameter quantifying the self-similarity of signals using the fractal property.[16,31,32] In other words, it measures the long-range correlation patterns of the R-R interval time series, which includes a short-term and long-term fractal scaling exponent, α-1 and α-2, respectively. The degree of fractal correlation has been shown to reflect sympathetic and parasympathetic tone.[33] Nonetheless, more research is needed to understand the physiological significance and normal range of values for each of the HRV parameters.
Our study had several limitations. Firstly, this was a single-institution study with a small sample size. Therefore, the results might not be generalizable to other settings and larger multi-centre prospective studies are required to validate our results. Secondly, we had included patients in our study based on clinical suspicion of sepsis and meeting at least 2 of the 4 SIRS criteria. Sepsis largely remains a clinical diagnosis and there is no gold standard to determine whether a patient is septic. Other studies have attempted to address this issue by including only patients with administered intravenous antibiotics, blood culture investigations or confirmed source of infection.[7,8] We acknowledge that our cohort definition reflects suspected sepsis rather than confirmed sepsis. However, given the aim of early risk stratification during triage where laboratory testing and confirmed diagnoses are not available, we believe this is suitable and does not detract from the model's value. In addition, while the SIRS criteria has recently been replaced with a new state of sepsis, defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection, the usefulness of the SIRS criteria in diagnosis of sepsis was still emphasized by the same task force.[25,34] Lastly, even though HRV measures are predictive of adverse outcomes in suspected sepsis patients as shown in this study, they cannot be manually calculated from a patient's ECG. Currently, we are developing a portable hardware device which can be used at the bedside to perform HRV analysis.
We acknowledge that the use of a machine learning model requiring computational resources on the ground may be challenging. However, many modern EDs already employ electronic data collection systems, on which predictive machine learning models could be deployed, making them even more convenient than traditional manual scoring tools. Future studies should implement the clinical use of such models and evaluate whether they translate into improved outcomes for septic patients.
In conclusion, a machine learning model incorporating HRV analysis can be used to improve prediction of 30-day IHM among suspected sepsis patients in the ED compared to traditional risk stratification tools. This model could be used at triage as a clinical decision support tool to identify high-risk septic patients for early, appropriate management.
Acknowledgments
We would like to thank all doctors, nurses and research assistants from the Department of Emergency Medicine, Singapore General Hospital, who contributed towards this project.
Author contributions
Conceptualization: Calvin J Chiew, Nan Liu, Marcus EH Ong.
Data curation: Zhi Xiong Koh.
Formal analysis: Calvin J Chiew, Nan Liu, Takashi Tagami, Ting Hway Wong, Zhi Xiong Koh.
Methodology: Calvin J Chiew, Nan Liu, Takashi Tagami, Ting Hway Wong, Zhi Xiong Koh, Marcus EH Ong.
Supervision: Nan Liu, Marcus EH Ong.
Writing – original draft: Calvin J Chiew, Nan Liu, Takashi Tagami, Ting Hway Wong, Zhi Xiong Koh, Marcus EH Ong.
Writing – review & editing: Calvin J Chiew, Nan Liu, Takashi Tagami, Ting Hway Wong, Zhi Xiong Koh, Marcus EH Ong.
Footnotes
Abbreviations: ADA = adaptive boosting, AUPRC = area under the precision-recall curve, BP = blood pressure, CI = confidence intervals, DFA = detrended fluctuation analysis, ED = emergency department, GB = gradient boosting, GCS = Glasgow Coma Scale, HF = high frequency, HR = heart rate, HRV = heart rate variability, ICU = intensive care unit, IHM = in-hospital mortality, KNN = k-nearest neighbors, LF = low frequency, MEWS = Modified Early Warning Score, NEWS = National Early Warning Score, NN50 = number of consecutive RR intervals differing by more than 50 ms, pNN50 = percentage of consecutive RR intervals differing by more than 50 ms, PRC = precision-recall curve, qSOFA = quick sequential organ failure assessment, RF = random forest, RMSSD = root mean square of differences between adjacent RR intervals, ROC = receiver operating characteristics, SD = standard deviation, SEDS = Singapore Emergency Department Sepsis, SGH = Singapore general hospital, SIRS = systemic inflammatory response syndrome, SVM = support vector machine, TINN = baseline width of a triangle fit into the RR interval histogram using least squares, VLF = very low frequency.
The authors received no specific funding for this work. Nan Liu and Marcus Ong have a patent filing that is not directly related to this study (System and method of determining a risk score for triage, Application Number: US 13/791,764). Marcus Ong has a similar patent filing unrelated to this study (Method of predicting acute cardiopulmonary events and survivability of a patient, Application Number: US 13/047,348). Marcus Ong also has a licensing agreement with ZOLL Medical Corporation for the above patented technology. There are no further patents, products in development or marketed products to declare. All the other authors do not have either commercial or personal associations or any sources of support that might pose a conflict of interest in the subject matter or materials discussed in this manuscript.
The authors report no conflicts of interest to disclose.
References
- [1].Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med 2013;369:840–51. [DOI] [PubMed] [Google Scholar]
- [2].Gaieski DF, Edwards JM, Kallan MJ, et al. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med 2013;41:1167–74. [DOI] [PubMed] [Google Scholar]
- [3].Lagu T, Rothberg MB, Shieh MS, et al. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Crit Care Med 2012;40:754–61. [DOI] [PubMed] [Google Scholar]
- [4].Liu B, Ding X, Yang J. Effect of early goal directed therapy in the treatment of severe sepsis and/or septic shock. Curr Med Res Opin 2016;32:1773–82. [DOI] [PubMed] [Google Scholar]
- [5].Nguyen HB, Rivers EP, Havstad S, et al. Critical care in the emergency department: a physiologic assessment and outcome evaluation. Acad Emerg Med 2000;7:1354–61. [DOI] [PubMed] [Google Scholar]
- [6].Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 2017;43:304–77. [DOI] [PubMed] [Google Scholar]
- [7].Freund Y, Lemachatti N, Krastinova E, et al. Prognostic accuracy of Sepsis-3 criteria for in-hospital mortality among patients with suspected infection presenting to the emergency department. JAMA 2017;317:301–8. [DOI] [PubMed] [Google Scholar]
- [8].Churpek MM, Snyder A, Han X, et al. Quick sepsis-related organ failure assessment, systemic inflammatory response syndrome, and early warning scores for detecting clinical deterioration in infected patients outside the intensive care unit. Am J Respir Crit Care Med 2017;195:906–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Barnaby D, Ferrick K, Kaplan DT, et al. Heart rate variability in emergency department patients with sepsis. Acad Emerg Med 2002;9:661–70. [DOI] [PubMed] [Google Scholar]
- [10].Chen WL, Chen JH, Huang CC, et al. Heart rate variability measures as predictors of in-hospital mortality in ED patients with sepsis. Am J Emerg Med 2008;26:395–401. [DOI] [PubMed] [Google Scholar]
- [11].Chen WL, Kuo CD. Characteristics of heart rate variability can predict impending septic shock in emergency department patients with sepsis. Acad Emerg Med 2007;14:392–7. [DOI] [PubMed] [Google Scholar]
- [12].Scheff JD, Griffel B, Corbett SA, et al. On heart rate variability and autonomic activity in homeostasis and in systemic inflammation. Math Biosci 2014;252:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Heart rate variability: Standards of measurement, physiological interpretation, and clinical use. Eur Heart J 1996;17:354–81. [PubMed] [Google Scholar]
- [14].Chialvo DR, Jalife J. Non-linear dynamics of cardiac excitation and impulse propagation. Nature 1987;330:749–52. [DOI] [PubMed] [Google Scholar]
- [15].Krogh-Madsen T, Christini DJ. Nonlinear dynamics in cardiology. Annu Rev Biomed Eng 2012;14:179–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sassi R, Cerutti S, Lombardi F, et al. Advances in heart rate variability signal analysis: Joint position statement by the e-Cardiology ESC Working Group and the European Heart Rhythm Association co-endorsed by the Asia Pacific Heart Rhythm Society. Europace 2015;17:1341–53. [DOI] [PubMed] [Google Scholar]
- [17].Samsudin MI, Liu N, Sumanth M, et al. A novel heart rate variability based risk prediction model for septic patients presenting to the emergency department. Medicine 2018;97:e10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Calvert JS, Price DA, Chettipally UK, et al. A computational approach to early sepsis detection. Comput Biol Med 2016;74:69–73. [DOI] [PubMed] [Google Scholar]
- [19].Nemati S, Holder A, Razmi F, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med 2018;46:547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Henry KE, Hager DN, Pronovost PJ, et al. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015;7:299ra122. [DOI] [PubMed] [Google Scholar]
- [21].Shimabukuro DW, Barton CW, Feldman MD, et al. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res 2017;4:e000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: A local big data-driven, machine learning approach. Acad Emerg Med 2016;23:269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis: American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992;101:1644–55. [DOI] [PubMed] [Google Scholar]
- [24].Tarvainen MP, Niskanen JP, Lipponen JA, et al. Kubios HRV--heart rate variability analysis software. Comput Methods Programs Biomed 2014;113:210–20. [DOI] [PubMed] [Google Scholar]
- [25].Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016;315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Smith GB, Prytherch DR, Meredith P, et al. The ability of the national early warning score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation 2013;84:465–70. [DOI] [PubMed] [Google Scholar]
- [27].Subbe CP, Kruger M, Rutherford P, et al. Validation of a modified early warning score in medical admissions. QJM 2001;94:521–6. [DOI] [PubMed] [Google Scholar]
- [28].Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10:e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- [30].Wang JY, Chen YX, Guo SB, et al. Predictive performance of quick sepsis-related organ failure assessment for mortality and ICU admission in patients with infection at the ED. Am J Emerg Med 2016;34:1788–93. [DOI] [PubMed] [Google Scholar]
- [31].Peng CK, Havlin S, Stanley HE, et al. Quantification of scaling exponents and crossover phenomena in nonstationary heartbeat time series. Chaos 1995;5:82–7. [DOI] [PubMed] [Google Scholar]
- [32].Pikkujamsa SM, Makikallio TH, Sourander LB, et al. Cardiac interbeat interval dynamics from childhood to senescence: comparison of conventional and new measures based on fractals and chaos theory. Circulation 1999;100:393–9. [DOI] [PubMed] [Google Scholar]
- [33].Tulppo MP, Kiviniemi AM, Hautala AJ, et al. Physiological background of the loss of fractal heart rate dynamics. Circulation 2005;112:314–9. [DOI] [PubMed] [Google Scholar]
- [34].Balk RA. Systemic inflammatory response syndrome (SIRS): where did it come from and is it still relevant today? Virulence 2014;5:20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


