Abstract
Macrophages enforce anti-tumor immunity by engulfing and killing tumor cells. Although these functions are determined by a balance of stimulatory and inhibitory signals, the role of macrophage metabolism is unknown. Here, we study the capacity of macrophages to circumvent inhibitory activity mediated by CD47 on cancer cells. We show that stimulation with CpG, a TLR9 agonist, evokes changes in the central carbon metabolism of macrophages that enable anti-tumor activity, including engulfment of CD47+ cancer cells. CpG activation engenders a metabolic state, that requires fatty acid oxidation and shunting of tricarboxylic acid cycle intermediates for de novo lipid biosynthesis. This integration of metabolic inputs is underpinned by carnitine palmitoyltransferase 1A and ATP citrate lyase, which together, impart macrophages with anti-tumor potential capable of overcoming inhibitory CD47 on cancer cells. Our findings identify central carbon metabolism to be a novel determinant and potential therapeutic target for stimulating anti-tumor activity by macrophages.
Macrophages govern the immune landscape of many cancers and are key proponents of tumor growth1. However, macrophages can also enact anti-tumor functions. These opposing roles are explained by the phenotypic polarity of macrophages, which are often classified as either pro-inflammatory M1 macrophages that enforce anti-tumor immunity or immunosuppressive M2 macrophages that promote tumor progression2. While macrophages most commonly adopt a phenotype that is supportive of tumor growth3, their biology is pliable. As a result, under the appropriate conditions, macrophages can be redirected with anti-tumor activity4–6. The mechanisms that determine pro- versus anti-tumor functions of macrophages, though, are still being elucidated.
One mechanism governing pro- and anti-tumor roles of macrophages is the balance of stimulatory and inhibitory signals. For example, a key negative regulator of macrophage activity is CD47, a membrane-bound protein overexpressed by many cancers7, 8. CD47 is a “don’t eat me” signal that suppresses the phagocytic activity of macrophages upon binding SIRPα (signal regulatory protein α)-receptor present on phagocytes9. Blocking CD47-SIRPα binding promotes macrophage engulfment of tumor cells and induces anti-tumor responses in multiple xenograft models7, 10. However, in models of pancreatic ductal adenocarcinoma (PDAC), CD47-blockade as a monotherapy has shown modest anti-tumor efficacy11, which may be explained by the limited pro-phagocytic effect of CD47-blockade seen in non-hematopoietic tumor models12. These findings suggest that additional stimuli are required to potentiate anti-tumor activity by macrophages.
Macrophage stimulation is directed by cytokines and agonists of pathogen recognition receptors, such as Toll-like receptors (TLRs), which together determine macrophage phenotype13. Unique combinations of stimuli have been used in vitro to define classical phenotypic states of macrophages, such as M1 and M2. However, in pathological settings, such as cancer, macrophages more commonly acquire phenotypes that span a spectrum of differentiation states14, 15.
When examined using systems-based approaches, macrophage phenotypes can be distinguished by their core metabolic processes15, 16. For example, M1 macrophages rely on glycolytic metabolism and reduced oxidative phosphorylation, whereas M2 macrophages perform de novo lipogenesis and glutaminolysis to support fatty acid oxidation (FAO)17–19. Additional studies support an association between M2-macrophage polarity and FAO, but indicate that these can also occur independently20. In particular, FAO and lipid metabolism underpin the anti-tumor functions of multiple myeloid subsets (e.g. dendritic cells and myeloid-derived suppressor cells)21–23. Similarly, lipid availability can modulate macrophage engulfment of red blood cells and macromolecules24, 25. Together, these findings underscore the potential role of metabolism in defining myeloid cell biology and suggest that lipid metabolism may likewise coordinate macrophage function in cancer.
To understand the metabolic determinants that govern macrophage anti-tumor function, we utilized metabolomic approaches and a syngeneic model of PDAC to study macrophage engulfment of PDAC cells upon TLR stimulation. Further, we leveraged targeted knockout of CD47 in PDAC cells to understand how macrophage activation acts in concert with inhibition of anti-phagocytic signals present in cancer. Our studies reveal a novel role for metabolic pathways in regulating macrophage anti-tumor functions and underscore the potential of targeting macrophage metabolism for overriding inhibitory signals used by cancer cells to evade elimination by innate immunity.
Results
Macrophage activation, but not loss of inhibitory CD47, is sufficient for anti-tumor activity in PDAC
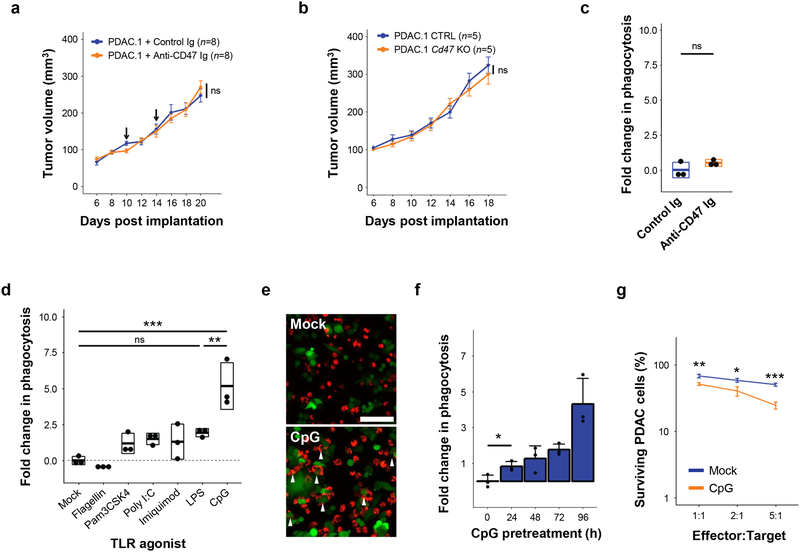
Macrophages can be induced with therapeutic and anti-tumor functions by activating pro-inflammatory signaling pathways such as CD40 and TLRs26. However, macrophage biology is ultimately determined by a balance of stimulatory and inhibitory signals that are sensed within the tumor microenvironment (Supp Fig 1a). One major inhibitory signal that is involved in suppressing macrophage anti-tumor activity is CD47, which is overexpressed on cancer cells across a wide range of solid malignancies27. Elevated CD47 can be detected in both mouse (Supp Fig 1b–c) and human PDAC11. Therefore, we initially studied the role of CD47 as a macrophage-inhibitory signal in a murine model of PDAC. To do this, we administered a CD47-blocking antibody via intratumoral delivery to mice implanted with KrasG12D/+ Trp53R172H/+ mutant PDAC tumors. In this fully syngeneic and immunocompetent model, antibody blockade of CD47 did not alter tumor growth (Fig 1a). To address possible limitations in bioavailability or insufficient blockade, expression of Cd47 was ablated in PDAC cells using transient expression of CRISPR/Cas9 (Supp Fig 1d–e). Unlike models of leukemia where CD47 overexpression is a key determinant of immune escape7, deletion of Cd47 in PDAC cells did not impact tumor engraftment or growth (Fig 1b). This observation suggested that mechanisms other than CD47 can regulate macrophage-dependent anti-tumor responses in PDAC, consistent with xenograft models of this disease and other non-hematopoietic malignacies11, 12. In support of this finding, we also found that antibody blockade of CD47 on PDAC cells did not enhance in vitro phagocytosis by murine bone-marrow derived macrophages (BMDMs) (Fig 1c).
Figure 1. Macrophage activation via TLR agonists, but not disruption of CD47, induces anti-tumor activity in a model of pancreatic cancer.
(a) Syngeneic mice with established PDAC tumors were treated with 50 μg isotype or anti-CD47 Ig on days 10 and 14 after tumor implantation (n=8). Arrows indicate intratumoral delivery of antibody; shown is mean ± standard error. (b) Syngeneic mice were challenged with PDAC tumors in which CD47 was knocked out (Cd47–/–) versus control (CTRL) using CRISPR-Cas9 gene editing (n=5); shown is mean ± s.e.m. (c) In vitro phagocytosis of PDAC cells by BMDMs in the presence of isotype control or anti-CD47 Ig. (d) In vitro phagocytosis of PDAC cells by BMDMs stimulated with indicated TLR agonists for 24 hours. (e) Representative immunofluorescence images showing phagocytosis of PDAC cells (green) by macrophages (BMDM, red) pretreated with vehicle (mock) or CpG for 24 hours. Arrows indicate phagocytic events. Scale bar, 100μm. (f) In vitro phagocytosis of PDAC cells by BMDMs stimulated with CpG for increasing lengths of time (n=3), error bars represent standard error. (g) PDAC cell survival following 48-hour co-culture with macrophages pretreated with vehicle (Mock) or CpG for 96 hours (n=3), mean ± s.e.m. Boxplots (c, d) represent mean ± s.d. Statistical significance determined using two-tailed Student’s t-test with Hochberg correction: ns (not significant). Results are representative of at least two independent experiments (a-g). *, p<0.05; **, p<0.01; ***,p<0.001.
We next hypothesized that PDAC cells might not provide sufficient activating signals to stimulate macrophages with anti-tumor functions, and that delivery of discrete stimuli may be necessary to induce macrophages with anti-tumor activity. We tested a panel of Toll-like receptor (TLR) pathway agonists for their capacity to stimulate macrophages with anti-tumor functions, such as phagocytosis. In the absence of TLR stimulation, mock-treated BMDMs lacked the capacity to phagocytose tumor cells upon co-culture with PDAC cells (Fig. 1d). Further, the phagocytic capacity of BMDMs increased only modestly with 24-hour pretreatment with Pam3CSK4, Poly(I:C), lipopolysaccharide (LPS), flagellin, and imiquimod – which stimulate TLR1 & TLR2, TLR3, TLR4, TLR5, and TLR7, respectively (Fig 1d). In contrast, ODN1826, a Class B CpG oligonucleotide that preferentially stimulates TLR9 expressed by macrophages and B cells, was found to be a potent activator of macrophage phagocytosis of PDAC cells (Fig 1d–e). Upon increasing the duration of CpG pretreatment, CpG-activated macrophages (CpG-BMDM) exhibited enhanced phagocytic capacity relative to mock-stimulated macrophages (mock-BMDM) (Fig 1f). To ascertain the potential of CpG-activation to produce anti-tumor activity, we also performed an extended co-culture of pretreated macrophages with PDAC cells for 48 hours. We found that CpG-activation rendered macrophages with potent anti-tumor activity leading to a decrease in tumor cell survival in comparison to co-culture with mock-BMDMs (Fig 1g). Together, these data show loss of inhibitory CD47 alone to be insufficient for unleashing anti-tumor activity by macrophages in PDAC, and that an activated phenotype is also necessary for macrophages to engage in anti-tumor functions.
Tumor-associated macrophages are essential for CpG-activated anti-tumor responses
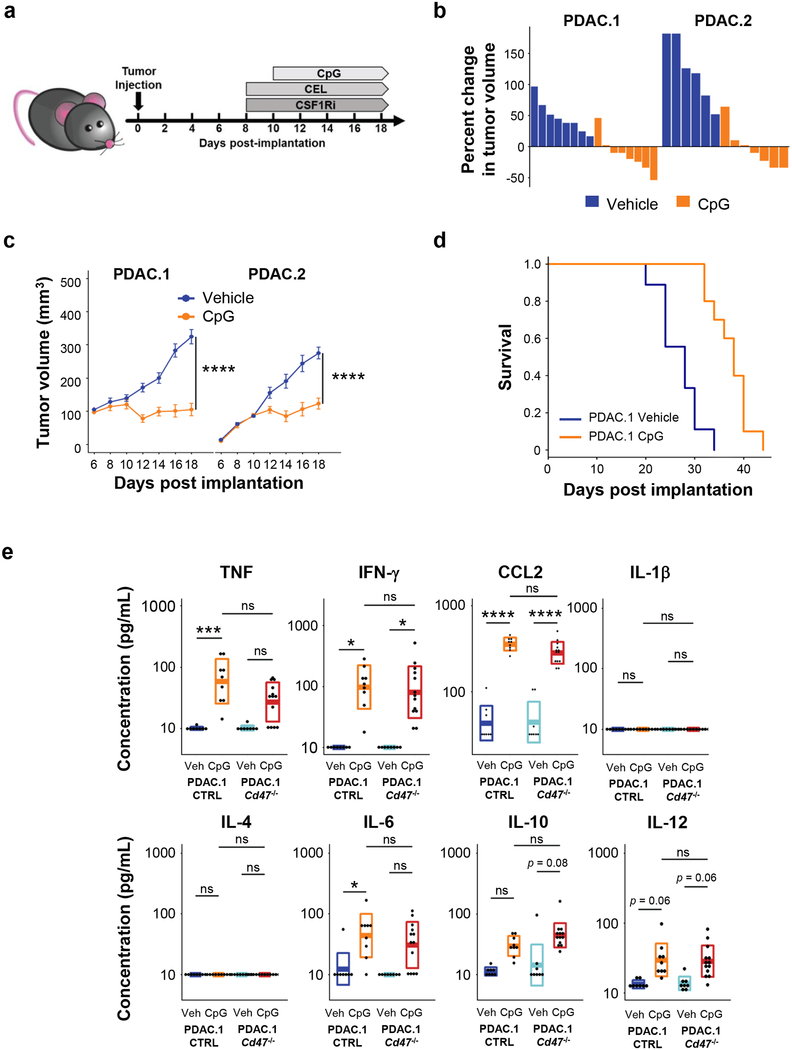
We next evaluated the in vivo anti-tumor activity of CpG on established murine PDAC tumor growth. Treatment was initiated with intraperitoneal injection of vehicle (PBS) or CpG on day 10 with repeated administration every other day for a total of five doses (Fig 2a). CpG treatment potently suppressed tumor growth in two independent PDAC tumor models (Fig. 2b–c). Although tumors ultimately relapsed, CpG significantly prolonged overall survival (Fig. 2d) without inducing gross toxicity or lethality. This effect was also independent of any direct cytotoxic activity of CpG on tumor cells, as treatment of PDAC cells in vitro with supratherapeutic concentrations of CpG did not affect tumor cell survival (Supp Fig 2).
Figure 2. CpG induces anti-tumor activity in vivo.
(a) Schematic showing implantation of PDAC tumor cells (day 0) and subsequent treatment schedule (beginning on day 8) for CpG and methods of macrophage depletion using clodronate encapsulated liposomes (CEL) and a CSF1R-inhibitor (CSF1Ri). (b) Two distinct syngeneic KPC-derived PDAC tumors (PDAC.1 and PDAC.2) were treated with vehicle or CpG beginning on day 10. Waterfall plots show percent change in tumor volume at day 14 relative to baseline prior to treatment (day 10). (c) Longitudinal growth curves shown as mean ± s.e.m. for PDAC tumors treated with vehicle or CpG (n=6–8); (d) Kaplan-Meier curve showing survival of tumor bearing mice (n=10) treated with vehicle of CpG (p=5.3×10−5). Treatment was initiated beginning on day 10. Significance was determined using log-rank analysis. (e) Syngeneic mice bearing PDAC.1 control (CTRL) or PDAC.1 Cd47–/– tumors were treated with vehicle or CpG beginning on day 10 after tumor implantation. Shown are box plots displaying mean ± s.d. of cytokine concentration detected in peripheral blood on day 14. Significance determined using two-tailed Student’s t-test with Hochberg correction: ns (not significant). Results are representative of at least two independent experiments (b-e). *, p<0.05; ****, p<0.0001.
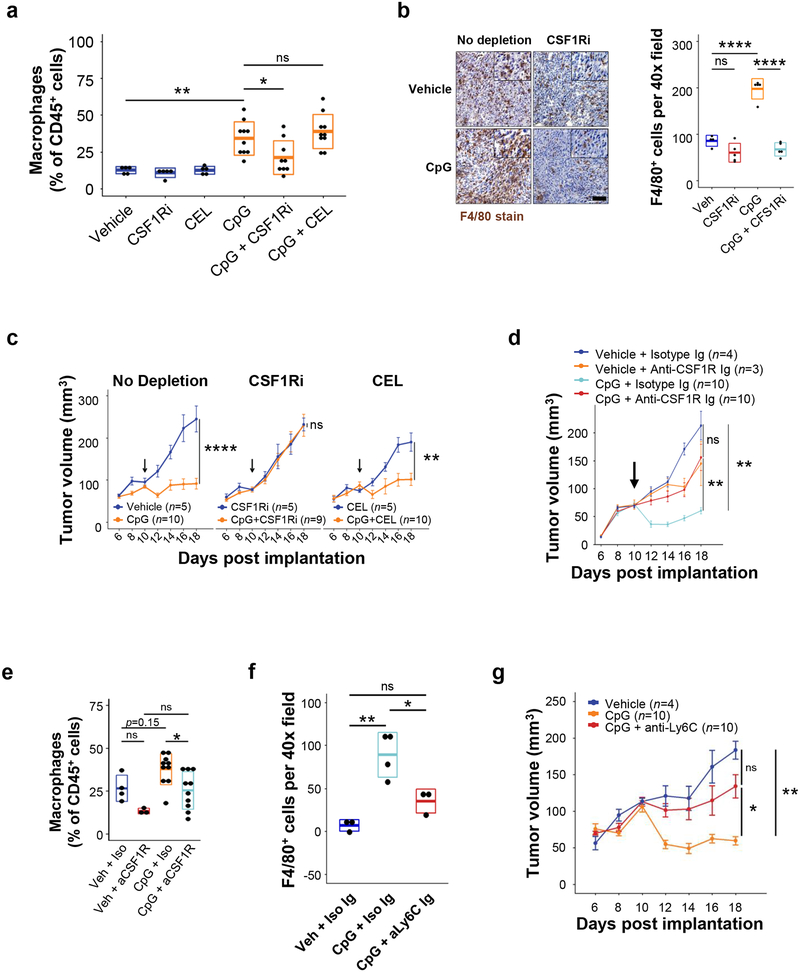
Repeated dosing of CpG to non-tumor bearing mice has been found to stimulate a macrophage activation syndrome28. Therefore, we examined the impact of treatment on the systemic release of inflammatory cytokines. We found that in vivo delivery of CpG to tumor-bearing mice increased pro-inflammatory cytokines in the serum, including TNF, IFN-γ, and CCL2 (Fig 2e). Consistent with this increase in inflammatory and chemotactic factors, we observed an increase in tumor-associated macrophages following CpG treatment (Fig 3a–b). In addition, we detected an increase in F4/80+ macrophage phagocytosis of tumor cells in CpG-treated tumors (Supp Fig 3). To ascertain the role of tumor-associated macrophages in the response to CpG treatment, we depleted distinct macrophage populations using GW2580, an inhibitor of colony stimulating factor 1-receptor (CSF1Ri), and clodronate encapsulated liposomes (CEL), which target phagocytes residing outside of the tumor microenvironment29. Depletion with either CEL or CSF1Ri alone did not affect tumor outgrowth in the vehicle-treated groups (Fig 3c). In contrast, CSF1Ri abrogated the anti-tumor effect of CpG, whereas CEL did not (Fig 3c). Similarly, we found that administration of an anti-CSF1R antibody attenuated the CpG-induced anti-tumor response (Fig 3d). These findings implicated a CSF1R+ population of macrophages, which are not targeted by liposomes, in mediating the anti-tumor response by CpG. Consistent with this observation, we found that CEL did not alter macrophage presence within tumors (Fig 3a), whereas both CSF1Ri (Fig 3a–b) and anti-CSF1R antibody (Fig 3e) treatment decreased the abundance of tumor-associated macrophages.
Figure 3. CpG induced anti-tumor activity requires macrophages.
(a) Shown is frequency of CD11b+ F4/80+ macrophages among total CD45+ cells detected by flow cytometry in PDAC tumors at day 18, in mice treated with CpG, CSF1Ri, or CEL. (b) Representative images (left) and quantification (right) of F4/80+ macrophages detected in PDAC tumors by immunohistochemistry after treatment with vehicle or CpG with or without CSF1Ri. (c) Longitudinal growth curves of PDAC.1 tumors treated with CpG and vehicle (no depletion), CSF1Ri, or clodronate encapsulated liposomes (CEL); Arrows indicate the initiation of CpG treatment. (d) Longitudinal growth curves of PDAC.1 tumors in mice treated as indicated. (e) Frequency of F4/80+ CD11b+ macrophages among total CD45+ cells detected by flow cytometry in PDAC tumors at day 18 in mice treated as indicated. (f) Quantification by immunohistochemistry of F4/80+ cells in tumors of mice on day 18 after treatment as indicated. Representative of one experiment. (g) Longitudinal growth curves of PDAC.1 tumors in mice treated with vehicle, CpG, or CpG + anti-Ly6C antibody (Monts1). Boxplots (e, f) represent mean ± s.d. Growth curves (c, d, g) show mean ± s.e.m. Significance determined using two-tailed Student’s t-test with Hochberg correction: ns (not significant). Results are representative of at least two independent experiments (a-c, g). *, p<0.05; **, p<0.01; ****, p<0.0001.
To ascertain the possible contribution of other immune effectors, we considered a role for lymphocytes in CpG-induced anti-tumor activity. We found that CpG did not significantly alter the infiltration of T cell subsets (CD8+, CD4+, and CD4+ Foxp3+) into tumors (data not shown). In addition, we detected no significant change in the expression of immune regulatory markers, including PD-1 and Tim3, by T cells after CpG treatment (data not shown). CpG-induced anti-tumor activity was also preserved in Rag2-deficient mice bearing PDAC.1 tumors, thereby excluding a role for lymphocytes (Supp Fig 4a). Anti-tumor activity induced with CpG was also preserved in mice depleted of natural killer (NK) cells using the anti-NK1.1 antibody (Supp Fig 4b). In addition, we found that depletion of dendritic cells (DC) using the CD11c-DTR-eGFP mouse model did not alter the anti-tumor response stimulated by CpG (Supp Fig 4c). Collectively, these data indicate that T and B lymphocytes as well as DC and NK cells are not required for the anti-tumor response induced by CpG.
We have previously shown a role for peripheral blood myeloid cells in mediating anti-tumor activity against PDAC4. Thus, we investigated the expression of CSF1R on myeloid cell populations in the peripheral blood. We found that CSF1R was expressed by a subset of CD11b+ F4/80+ myeloid cells bearing high levels of the monocyte marker Ly6C (data not shown), which marks a population of myeloid cells that have been previously shown to be recruited to PDAC tumors4, 30. Further, administration of an anti-CSF1R antibody significantly reduced the Ly6Chi myeloid population in the peripheral blood (data not shown). We also found, similar to CSF1Ri and anti-CSF1R treatment, that anti-Ly6C antibodies, which deplete the Ly6Chi monocyte population in vivo4, blocked both the accumulation of F4/80+ macrophages in tumors (Fig 3f) and the CpG-induced anti-tumor response (Fig 3g). Together, these data implicate myeloid cells marked by expression of Ly6C and CSF1R in the anti-tumor activity induced by CpG31.
CpG activation of macrophages bypasses inhibitory CD47 on PDAC cells
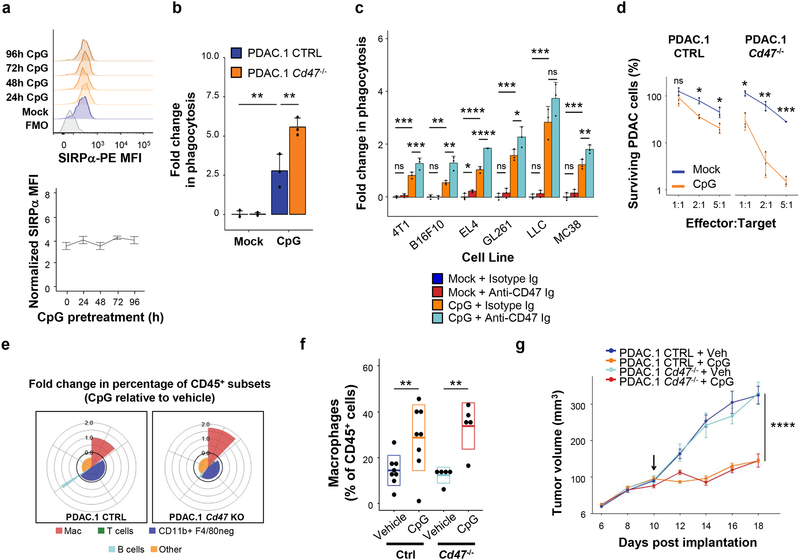
We next sought to understand the mechanism by which CpG stimulates macrophages with enhanced phagocytic and anti-tumor activity. We hypothesized that CpG might alter the capacity of macrophages to respond to CD47 as a negative regulatory signal. To test this, we assessed the impact of CpG on SIRPα expression. However, we observed no changes in SIRPα surface expression with increasing duration of CpG-activation (Fig 4a). We also examined the effect of CpG-activation on macrophage-inhibitory signals present in vitro. We found that CpG-BMDMs phagocytosed PDAC cells that either expressed or lacked CD47, indicating that CpG stimulates macrophages with anti-tumor activity that is independent, at least in part, of CD47 as a macrophage-inhibitory signal (Fig 4b). This finding was generalizable as CpG-activation enabled macrophage engulfment of CD47-expressing syngeneic tumor cells derived from breast cancer, melanoma, colorectal cancer, glioblastoma, lung cancer and lymphoma (Fig 4c). CD47-blockade alone also showed limited capacity to enhance macrophage phagocytosis in these tumors, except for EL4 lymphoma cells. However, despite limited activity by itself, we found that genetic ablation of CD47 in tumor cells (Fig 4b) as well as CD47 blockade (Fig 4c) significantly enhanced the capacity of CpG-stimulated macrophages to phagocytose tumor cells in vitro, implying that appropriate macrophage-activation signals are critical for overcoming CD47 in non-hematopoietic tumors.
Figure 4. CpG stimulates macrophage anti-tumor activity in vitro and in vivo that is independent of the anti-phagocytic signal CD47 expressed on PDAC cells.
(a) Representative histogram plots of SIRPα expression (top) and normalized SIRPα expression (bottom) detected by flow cytometry on BMDMs after activation with CpG for 24–96 hours (n=3). (b) Vehicle (mock) versus CpG stimulated macrophage phagocytosis of PDAC cells in which CD47 was knocked out (Cd47–/–) using CRISPR-Cas9 gene editing versus control (CTRL) (n=3). (c) In vitro phagocytosis of the indicated syngeneic CD47+ murine tumor cell lines treated with isotype or anti-CD47 antibody, and co-cultured with BMDMs stimulated with vehicle of CpG (n=3). (d) Survival of PDAC.1 CTRL and Cd47–/–cells following 48-hour co-culture with vehicle (mock) or CpG activated macrophages pretreated for 72 hours, using the indicated effector (macrophage) to target (tumor) ratios (n=3). (e) Coxcomb plots showing distribution and fold change in leukocyte subsets (as a percentage of CD45+ cells on day 18), for CpG treatment relative to vehicle treatment in control (left) and Cd47–/– (right) PDAC tumors. Radial axis represents relative fold change in leukocyte subset percentage (n=5–8). (f) Percentage of CD11b+ F4/80+ macrophages in control and Cd47–/–tumors following CpG treatment. Boxplot represents mean ± s.d. (g) Tumor volume of control and Cd47–/– tumors treated with CpG (n=5–8); arrow indicates initiation of CpG; Box plots (f) show mean + s.d. Plots in b-d and g show mean + s.e.m. Two-tailed Student’s t-test with Hochberg correction: ns (not significant). Results are representative of at least two independent experiments (a-g). *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001.
Prior studies have suggested that TLR agonists might promote phagocytic and anti-tumor activity by stimulating macrophages to produce calreticulin32. To test this possibility, we quantified calreticulin expression – including both membrane-bound and intracellular levels – but detected no differences between mock-BMDMs and CpG-BMDMs (Supp Fig 5). Further, upon extended co-culture, CpG-BMDMs effectively eliminated PDAC cells, with loss of CD47 expression in tumor cells providing an additive benefit in anti-tumor activity (Fig 4d). However, the ability of CpG to promote anti-tumor activity by macrophages despite CD47 expression on tumor cells is in contrast with other TLR agonists, for which blockade of CD47 is critical for macrophage activity32.
We then asked whether disruption of CD47 enhances the in vivo anti-tumor activity of macrophages. Delivery of CpG to mice bearing control or CD47 knockout (Cd47–/–) tumors produced similar increases in the relative abundance of tumor-associated macrophages (Fig 4e–f). The impact of CpG on tumor growth was also similar in tumors that expressed or lacked CD47 (Fig 4g). These findings underscore the dominant response elicited by CpG activation of macrophages in vivo that can overcome CD47 expressed by PDAC cells.
CpG induces a shift in macrophage metabolism
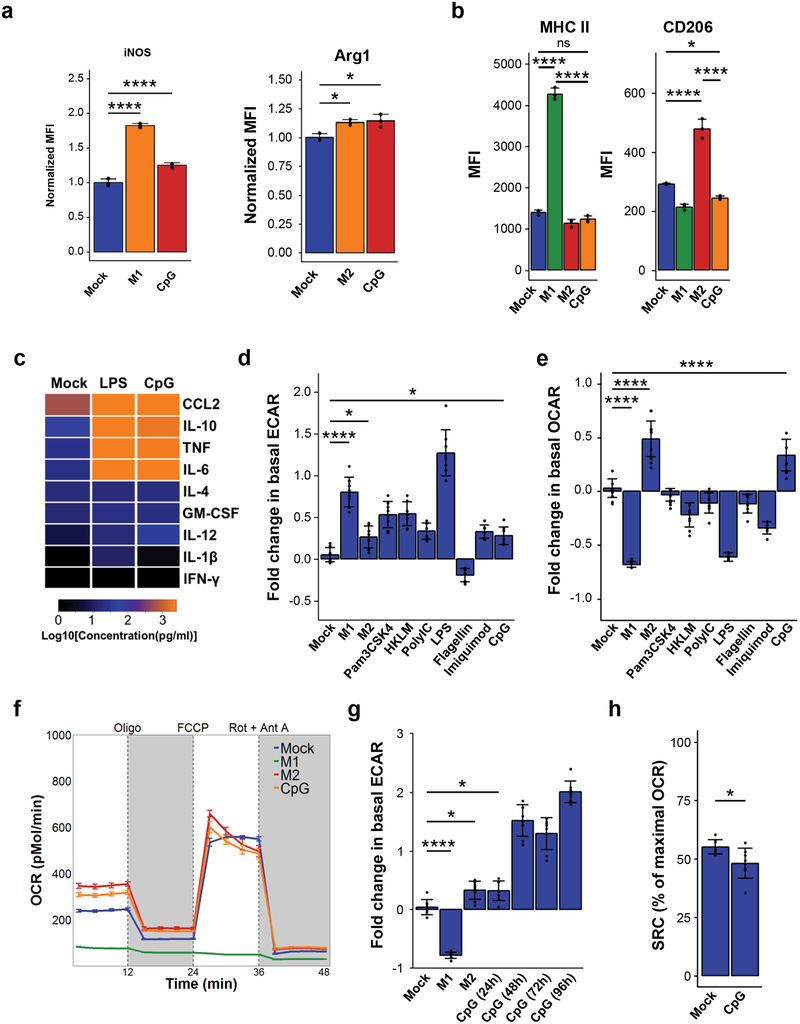
Microenvironmental cues can induce distinct phenotypes in macrophages. For example, the combination of LPS and IFN-γ promote an M1 phenotype in macrophages, which is associated with pro-inflammatory and anti-tumor activity in cancer1, 2. In contrast, the combination of IL-4 and IL-13 endows macrophages with M2-polarity, which suppresses inflammation and promotes tumor growth. To study the direct effect of CpG on macrophage polarity, we evaluated CpG-BMDMs for markers associated with M1 and M2 macrophages, including arginase 1 (Arg1), CD206, MHC-II, inducible nitric oxide synthase (iNOS), IL-6 and IL-1233. We found that CpG-activation increased the expression of both iNOS and Arg1 which are associated with M1(LPS) and M2(IL-4) macrophages, respectively (Fig 5a). However, CpG did not significantly alter expression of MHC-II or CD206, markers increased by M1(LPS) and M2(IL-4) polarization, respectively (Fig 5b). CpG also induced production of both pro-inflammatory cytokines (i.e. IL-6, IL-12, CCL2, and TNF) and anti-inflammatory cytokines (i.e. IL-10) relative to mock-treatment (Fig 5c). Further examination of additional markers revealed that CpG did not modulate IL-4R alpha, CD80 or CD86, but upregulated expression of FcγRIII and PD-L1 (data not shown).
Figure 5. CpG evokes metabolic changes in macrophages without polarization to M1 or M2.
(a) Normalized mean fluorescence intensity of iNOS (left) and Arg1 (right) in mock, M1 or M2 as indicated, and CpG-activated BMDMs (n=3). (b) Mean fluorescence intensity for BMDMs pretreated with vehicle (mock) or CpG for 96 hours, compared with M1 and M2 BMDMs (n=3). (c) Heatmap of cytokine concentrations produced in vitro by BMDMs treated with vehicle (mock), LPS, or CpG for 48 hours (n=3), scaled to Log10(Concentration in pg/ml). (d) Extracellular acidification rate (ECAR) of BMDMs after 24-hour activation with indicated TLR agonists (n=8), relative to mock-treated baseline. (e) Basal oxygen consumption rate (OCR) of BMDMs after 24-hour activation with indicated TLR agonists (n=8), relative to mock-treated baseline. (f) Profile of OCR in BMDMs pretreated for 24 hours with vehicle (mock) or CpG, in comparison to M1 and M2 BMDMs (n=8). OCR was measured as picomoles of O2 per minute upon sequential administration with oligomycin, FCCP, and Rotenone + Antimycin A. (g) Basal OCR of BMDMs following activation with CpG for increasing durations (n=8), relative to mock-treated baseline. (h) Spare respiratory capacity of BMDMs treated with vehicle (mock) or CpG for 96 hours (n=8), shown as the difference between basal OCR and maximal OCR (after addition of FCCP). Significance determined using two-tailed Student’s t-test with Hochberg correction: ns (not significant). Plots (a-b, d, f-h) show mean ± s.d. Plots (e) show mean ± s.e.m. Results are representative of at least two independent experiments (a-h). *, p<0.05; ****, p<0.0001.
Pro-inflammatory stimuli can also alter macrophage metabolism15, 34. Therefore, we sought to understand the impact of CpG-activation on the metabolic state of macrophages by assessing their ECAR (extracellular acidification rate) and OCR (oxygen consumption rate). We found that CpG and several other TLR agonists increased ECAR relative to mock-BMDMs, indicating an increase in glycolytic flux (Fig 5d, Supp Fig 6). However, distinct from other TLR agonists, CpG elevated the basal OCR of macrophages, signifying higher rates of oxidative phosphorylation (Fig 5e–f, Supp Fig 6). This metabolic change seen in CpG-stimulated macrophages is consistent with metabolic activation seen in dendritic cells in response to CpG35. Further, we observed that the basal OCR of macrophages increased with prolonged CpG stimulation (Fig 5g) and corresponded with reduced spare respiratory capacity (Fig 5h). Collectively, these findings demonstrate that CpG-activation confers a unique metabolic shift in macrophages and that CpG-activated macrophages do not fully conform to the classical M1(LPS) or M2(IL-4) classification.
Lipid metabolism is critical for anti-tumor functions induced by CpG
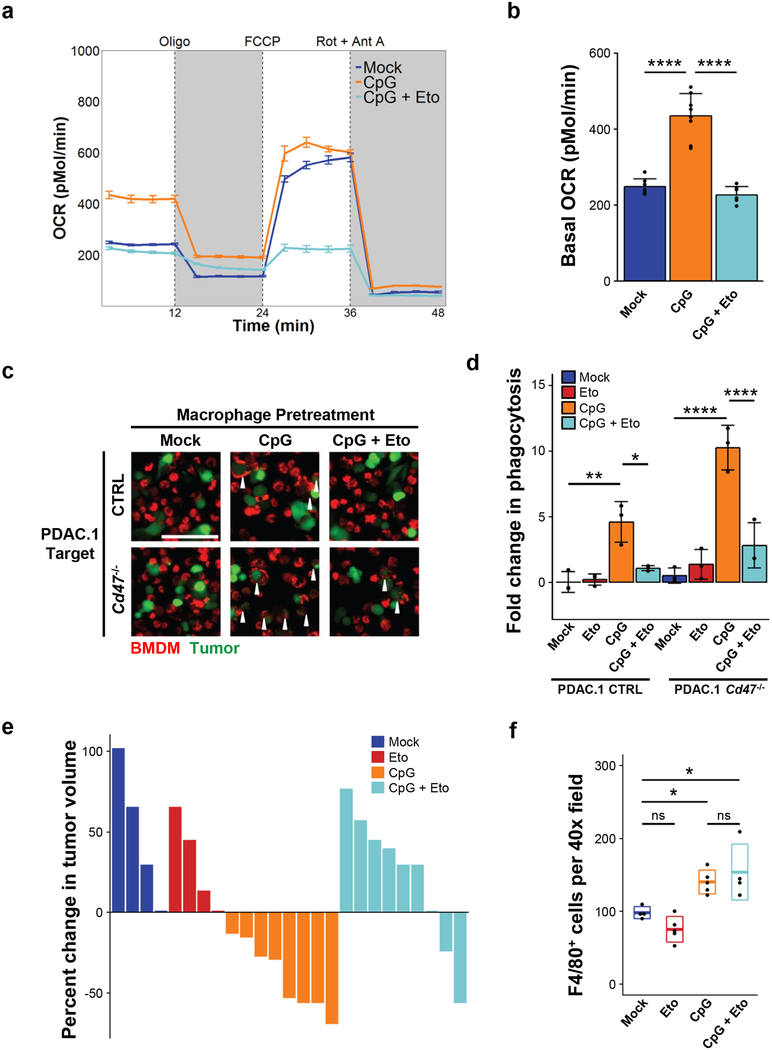
We next investigated the significance of elevated oxygen consumption in response to CpG activation by first assessing mitochondrial abundance in macrophages. CpG-BMDMs exhibited an increase in total mitochondria, in comparison to mock-BMDMs (Supp Figure 7a). To further evaluate changes in mitochondrial function, we tested the contribution of FAO toward elevated oxygen consumption under normal culture conditions, in the presence of serum and without other exogenous lipids. FAO was restricted using etomoxir, an irreversible inhibitor of the enzyme carnitine palmitoyltransferase 1A (CPT1A) that is involved in fatty acid breakdown. We found that FAO-inhibition blocked the CpG-induced increase in macrophage OCR without affecting the maximal respiratory capacity (Fig 6a–b). Furthermore, FAO inhibition with etomoxir attenuated the capacity of CpG-BMDMs to phagocytose PDAC cells, whether they expressed inhibitory CD47 or not (Fig 6c–d).
Figure 6. Fatty acid oxidation induced by CpG is essential for macrophage anti-tumor activity.
(a) Profile of oxygen consumption rate (OCR) in BMDMs treated with vehicle (mock), CpG, or CpG + etomoxir for 96 hours (n=8). Shown is OCR measured as picomoles of O2 per minute upon sequential administration with oligomycin, FCCP, and Rotenone + Antimycin A. (b) Basal OCR of BMDMs treated with vehicle (mock), CpG, and CpG + etomoxir for 96 hours (n=8). (c) Representative immunofluorescence images showing phagocytosis of PDAC.1 control (CTRL) or Cd47–/– cells (green) after co-culture with macrophages (BMDM, red) pretreated with vehicle (mock), CpG, or CpG + etomoxir for 96 hours. Arrows indicate phagocytic events. Scale bar, 100μm. (d) In vitro phagocytosis of PDAC.1 CTRL or Cd47–/– cells following co-culture with BMDMs pretreated with vehicle (mock), CpG, or CpG + etomoxir for 96 hours. (e) Treatment of PDAC.1 CTRL tumors with vehicle, etomoxir, CpG, or CpG + etomoxir beginning on day 10 after tumor implantation. Waterfall plot shows percent change in tumor volume at day 14 relative to baseline prior to CpG-treatment (day 10). (f) Quantification by immunohistochemistry of F4/80+ macrophages in PDAC tumors (n=4) on day 14, following treatment with vehicle (mock), etomoxir, CpG, or CpG + etomoxir. Two-tailed Student’s t-test with Hochberg correction: ns (not significant). Boxplots represent mean ± s.d. Plots (a-b, d) show mean ± s.e.m. Results are representative of at least two independent experiments (a-f). *, p<0.05; **, p<0.01; ****, p<0.0001.
To test the role of FAO for the in vivo anti-tumor activity induced by CpG, etomoxir was administered daily to mice beginning on day 9 prior to initiating CpG treatment on day 10. We found that etomoxir treatment blocked the anti-tumor effect of CpG, whereas treatment with etomoxir alone had no significant impact on tumor outgrowth (Fig 6e). Importantly, etomoxir treatment also did not alter the abundance of F4/80+ macrophages in the tumor microenvironment, indicating that the inhibitory effect observed was not a result of depletion or exclusion of macrophages from the tumor microenvironment (Fig 6f). Rather, the inhibitory effect of etomoxir was associated with decreased engulfment of tumor cells by F4/80+ macrophages (Supp Fig 3). Together, these data demonstrate a key role for FAO in macrophage-dependent anti-tumor activity induced with CpG.
De novo lipogenesis, but not exogenous fatty acids, are critical for CpG-activated anti-tumor activity
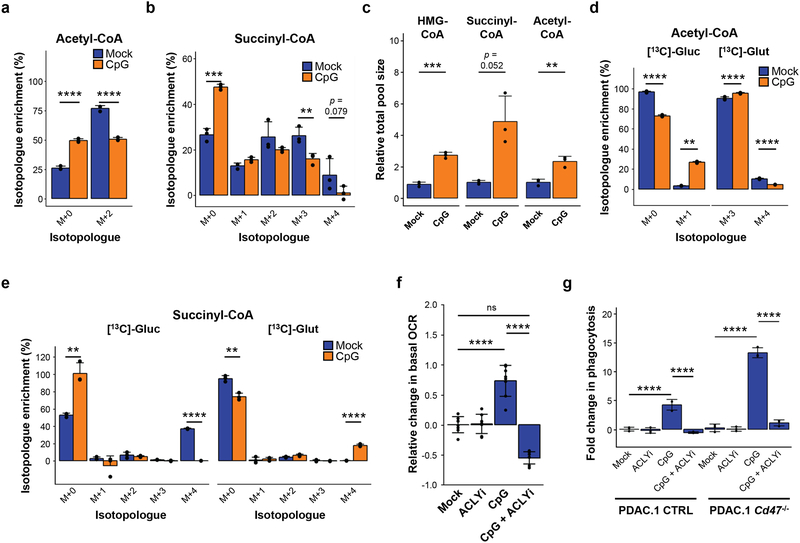
To define the metabolic changes induced by CpG, we assessed the abundance of short chain coenzyme A (CoA) species in mock- and CpG-treated macrophages. We hypothesized that an increase in FAO in CpG-BMDMs would require breakdown and incorporation of exogenous fatty acids into acetyl-CoA and TCA substrates. Surprisingly, we found that the incorporation of labeling from 13C-palmitate into acetyl-CoA and succinyl-CoA was decreased in CpG-BMDMs, indicating that substrates other than exogenous palmitate likely serve as precursors for generating acetyl-CoA and succinyl-CoA in response to CpG stimulation (Fig 7a–b). We next investigated the impact of CpG on increased fatty acid uptake which is a key feature of M2 macrophages. We found that CpG-BMDMs did not upregulate fatty acid transporters such as CD36 which instead, was significantly downregulated in response to CpG stimulation (Supp Fig 7b). In addition, CpG-BMDMs showed a limited increase in the uptake of BODIPY-labeled palmitate, indicating that utilization of exogenous fatty acids was not significantly altered in macrophages by CpG stimulation (Supp Fig 7c). Together, these findings illustrate that while CpG-macrophages share several metabolic features with M2 macrophages, such as their oxidative phenotype, they differ significantly in their utilization of exogenous fatty acids.
Figure 7. Metabolic rewiring of TCA cycle supports the oxidative phenotype and anti-tumor activity of CpG-activated macrophages.
BMDMs were pretreated with CpG or vehicle for 96 hours, plated, and then labeled (i) with the addition of 100 μM 13C-palmitate for 4 hours (n=3) (a,b) or (ii) in the presence of 25 mM 13C-glucose or 4 mM 13C-glutamine for 2 hours (n=3) (c-e). (a) Percent isotopologue enrichment of acetyl-CoA with addition of 13C-palmitate. (b) Percent isotopologue enrichment of succinyl-CoA with addition of 13C-palmitate. (c) Relative total pool size of HMG-CoA, succinyl-CoA, and acetyl-CoA after labeling with tracers (13C-glucose or 13C-glutamine). (d) Percent isotopologue enrichment of acetyl-CoA from 13C-glucose and 13C-glutamine. (e) Percent isotopologue enrichment of succinyl-CoA from 13C-glucose and 13C-glutamine. (f) Basal oxygen consumption of BMDMs pretreated as indicated (n=8). (g) In vitro phagocytosis of PDAC.1 CTRL or Cd47–/– cells by BMDMs pretreated as indicated (n=3). Two-tailed Student’s t-test with Hochberg correction: ns (not significant). Plots (a-g) show mean ± s.d. Results are representative of at least two independent experiments (f,g). **, p<0.01; ***, p<0.001; ****, p<0.0001.
We proceeded to examine alternative pathways that may support metabolic activation in CpG-BMDMs. Upon substitution of glucose or glutamine with 13C-glucose or 13C-glutamine in the media, the relative total pool sizes for acetyl-CoA, succinyl-CoA and HMG (hydroxymethylglutaryl)-CoA increased in CpG-BMDMs compared to mock-BMDMs (Fig 7c). The percent of acetyl-CoA derived from glucose (M+2 isotopologue) increased in CpG-BMDMs relative to mock-BMDMs (Fig 7d). This shift was accompanied by a decreased percent of acetyl-CoA derived from glutamine (M+2 isotopologue) in CpG-BMDMs (Fig 7d). Conversely, incorporation of 13C-glucose into succinyl-CoA (M+4 isotopologue) decreased upon CpG-stimulation, and was accompanied by an increase in labeled, glutamine-derived succinyl-CoA (M+4 isotopologue) in CpG-BMDMs compared to mock-BMDMs (Fig 7e). Together, these data show that CpG activation of macrophages promotes a shift away from complete utilization of carbon from glucose and toward glutamine anaplerosis for generating TCA cycle intermediates such as succinyl-CoA.
The incorporation of glucose into acetyl-CoA and HMG-CoA (a precursor for the cholesterol synthesis pathway) seen in CpG-stimulated BMDMs was consistent with a shunting by ATP citrate lyase (ACLY) and de novo biosynthesis of cholesterol and/or lipids16. Lipid metabolism can impact multiple biological processes in macrophages, including cellular organelle production, cytokine secretion, and lipid membrane properties36, 37. Therefore, we examined a role for secretory capacity in CpG-induced macrophage phagocytosis. We found that blocking endoplasmic reticulum (ER) secretion using brefeldin A inhibited CpG-induced phagocytosis of tumor cells (Supp Fig 7d). This finding was consistent with previous work showing a role for the ER as a source of membrane for phagocytosis38. Therefore, we next hypothesized that disruption of ACLY would alter membrane properties and directly impede the anti-tumor activity of CpG-activated macrophages. We found that inhibition of ACLY during CpG-induced macrophage activation attenuated the oxidative phenotype (Fig 7f) and reversed membrane fluidity (Supp Fig 7e), an established regulator of macrophage phagocytosis. Consistent with this, we also found that inhibition of ACYL abrogated the anti-tumor phagocytic activity of CpG-stimulated macrophages even with combined CD47 ablation (Fig 7g). Together, our findings demonstrate that CpG imparts macrophages with a unique metabolic state, defined by enhanced oxidative phosphorylation and a shunting of TCA cycle intermediates, which determines their anti-tumor potential and capacity to circumvent inhibitory signals received by CD47 expressed on malignant cells.
Discussion
Macrophages dominate the microenvironment of many cancers, wherein they are directed toward a pro-tumor role and restricted in their anti-tumor potential. The capacity of macrophages to engulf and kill tumor cells is countervailed by their phenotype as well as inhibitory signals present within their surrounding microenvironment, including the anti-phagocytic molecule CD47, which is overexpressed by malignant cells7, 8. Our study identifies the central carbon metabolism of macrophages as a regulator of their anti-tumor functions, including the capacity to phagocytose tumor cells. We found that redirection of macrophages toward an oxidative phenotype is essential for anti-tumor activity induced by CpG stimulation in vitro and in vivo. CpG-stimulated alterations in macrophage metabolism were necessary for circumventing inhibitory signals mediated by CD47. Together, our findings highlight rewiring macrophage metabolism – by promoting FAO and redirecting the use of TCA intermediates – as a mechanism to circumvent negative regulatory signals present on malignant cells and to endow macrophages with anti-tumor potential.
Inhibitory signals expressed by tumor cells may not always be dominant in regulating anti-tumor activity by macrophages. Unlike findings in other cancers, we found that disruption of CD47 activity via antibody-blockade was not sufficient for inciting anti-tumor responses in an immunocompetent tumor model of PDAC. Deletion of tumor-expressed CD47 was also not sufficient for inducing in vitro phagocytosis of tumor cells or in vivo anti-tumor responses. The absence of macrophage activity in this setting was not due to a lack of pro-phagocytic signals as treatment with CD47-blocking antibodies, which contain Fc-domains for driving Fc receptor dependent pro-phagocytic signals in macrophages, also failed to elicit macrophage anti-tumor activity in vitro and in vivo. Further, in the absence of macrophage activation via CpG stimulation, we found that CD47-blocking antibodies were insufficient for stimulating macrophage phagocytosis of multiple solid tumor types including colorectal, glioblastoma, melanoma, breast cancer, and lung cancer. Our data support a conceptual model in which inhibitory signals, such as CD47, can be overcome by the activation state of macrophages, which is defined by core metabolic pathways that govern their anti-tumor potential. Consistent with this, we observed a significant role for CD47 in regulating macrophage anti-tumor activity in vitro, but only when macrophages were activated with CpG, which provides rationale for combining CD47 antagonists with myeloid agonists. However, this finding was not appreciated in vivo where CpG-induced anti-tumor activity was indistinguishable between tumors that expressed or lacked CD47. This may reflect the emergence of additional anti-phagocytic signals that are redundant with CD47 expressed by tumor cells39 or even the capacity of CpG to more effectively activate macrophages in vivo to circumvent inhibitory signals mediated by CD47.
The functional consequences of metabolic changes in macrophages has remained poorly defined in cancer, despite findings to suggest that lipid metabolism has a pivotal role in governing the anti-tumor functions of several other immune cell types (e.g. myeloid-derived suppressor cells, dendritic cells, and T-cells)21, 22, 40. We showed that, similar to M1 macrophages, CpG-activated macrophages redirected exogenous fatty acids and glucose toward acetyl-CoA generation and de novo lipid biosynthesis. We also found, as seen in M2 macrophages, that FAO and glutaminolysis were essential for the oxidative phenotype of CpG-activated macrophages17, 18. Notably, M2 macrophages are recognized to possess superior abilities than M1 macrophages in phagocytosing antibody-opsonized tumor cells41, suggesting an association between FAO and phagocytic capacity. Functionally, FAO in anti-tumor macrophages may enable the breakdown of large lipid loads after engulfment of target cells or fulfill the metabolic demands for phagocytosis42. Together, our findings substantiate the importance of lipid metabolism as a dominant regulator of anti-tumor activity by macrophages.
Changes in lipid metabolism can alter plasma membrane properties in macrophages and in doing so, confer them with an ability to phagocytose more effectively25. Our data show enhancement of membrane fluidity induced by CpG, that is dependent on acetyl-CoA shunting for de novo lipogenesis. In turn, membrane fluidity can modulate CD47-SIRPα signaling and receptor clustering at phagocytic synapses43, 44. Macrophage activation by TLRs has been shown to promote lipid-modifying programs that alter ceramide and sphingolipid species which may influence the membrane properties of macrophages45. Analogously, CpG-activation enhances lipogenesis in macrophages, which consequently modulates secretory function37, 46. Consistent with this, we identified secretory pathways between the ER and Golgi to be critical for CpG-induced phagocytosis. In non-cancerous settings, lipogenesis supports macrophage engulfment of microparticles, due to its role in phospholipid synthesis and expansion of cellular organelles (e.g. ER and Golgi)36. However, the relevance of de novo lipogenesis for macrophage activity in cancer and for circumventing inhibitory signals of phagocytosis (i.e. CD47) has not been previously reported. We find that CpG increases mitochondrial abundance and enhances FAO, along with shunting of acetyl-CoA toward cholesterol biosynthesis. This mechanism is necessary for CpG to endow macrophages with the capacity to phagocytose tumor cells, without need for engaging Fc-receptors or blocking CD47 activity.
Most clinical studies evaluating CpG oligonucleotides have investigated subcutaneous delivery and weekly administration with the goal of stimulating dendritic cells and mobilizing T cell immunity. However, thus far, CpG has demonstrated limited efficacy47–49. In contrast to these studies, we administered CpG systemically and repeatedly to incite robust and sustained macrophage activation. Our data suggest that activation of adaptive immunity may not always be required for CpG-induced anti-tumor activity and support evaluating the potential of CpG to stimulate macrophage anti-tumor activity in the clinical setting.
The mechanisms involved in shifting the role of macrophages from pro- to anti-tumor have remained poorly defined. We found that CSF1R+ macrophages were necessary for mounting an anti-tumor response upon CpG-activation, despite this subset of macrophages being associated with immunosuppression50. Our findings show a role for core metabolic processes, which act in concert as a key regulator of macrophage anti-tumor activity. Our findings also highlight the importance of the metabolic state of macrophages for circumventing immune checkpoint signals with implications for broadening the impact of immunotherapy and identify macrophage metabolism as a novel therapeutic target that must be appropriately wired to enable macrophages to carry out anti-tumor functions.
Materials and Methods
Mice.
C57BL/6J, Rag2–/– (B6(Cg)-Rag2tm1.1Cgn/J), CD11c-DTR/eGFP (B6.FVB-1700016L21RikTg(Itgax-DTR/EGFP)57Lan/J) mice were obtained from Jackson Laboratories. CD11c-DTR/eGFP heterozygotes were enrolled in tumor studies. Animal protocols were reviewed and approved by the Institute of Animal Care and Use Committee (IACUC) of the University of Pennsylvania and conducted in compliance with the guidelines for animal research by the National Institutes of Health.
Cell lines.
PDAC.1 (152 PDA) and PDAC.2 (69 PDA) cell lines were derived from PDAC tumors, as previously described4, which arose spontaneously in KrasLSL.G12D/+; Trp53R172H/+; Pdx-Cre mice backcrossed onto the C57BL/6 background (Jackson Labs). Cell line authentication was performed as previously described4. Isogenic PDAC lines were established by cloning single cells for in vitro and in vivo experiments. PDAC knockout lines were generated as previously described (Supplementary Table 3): Cell lines from American Type Culture Collection (ATCC) include 4T1 (breast cancer), B16F10 (melanoma), EL4 (T cell lymphoma), GL261 (glioblastoma), LLC (Lewis Lung Carcinoma), and MC38 (colorectal cancer). Cell lines were negative for mycoplasma testing. 1×106 tumor cells from isogenic lines were transiently transfected with guide RNAs targeting GFP (5’-GTGAACCGCATCGAGCTGAA-3’) or CD47 (5’-GGAGCCATCCTTCTCATCCC-3’) sequences. Guide RNAs were inserted into TOPO-plasmids (Qiagen) per manufacturer instructions, and mixed 1:1 with LentiCRISPR V2 plasmid (Addgene). The plasmid mix was combined with Lipofectamine 2000 (Thermo Scientific Fisher) and Opti-MEM (Thermo Scientific Fisher) for transfection mixture. Tumor cells were transfected for 6 hours; at 24 hours post-transfection, cells were treated with 1 μg/ml puromycin (Invivogen) for 2 days. Following selection, tumor cells were stained for CD47 and purified as CD47+ and CD47− populations by fluorescence-activated cell sorting. Reporter cell lines were generated for phagocytosis and anti-tumor assays by transduction with lentivirus encoding GFP and Click Beetle Green luciferase, joined by a T2A signal peptide. After expansion, GFP-expressing PDAC cells were isolated using fluorescence-activated cell sorting.
Reagents.
TLR agonists were resuspended in PBS (phosphate buffered saline, Thermo Scientific Fisher) prior to addition to cell culture media at final concentrations of: 1 μg/ml Pam3CSK4 (Invivogen), 1 μg/ml PolyI:C (Invivogen), 1 μg/ml E. coli LPS (Invivogen), 1 μg/ml flagellin (Invivogen), 1 μg/ml imiquimod (Invivogen), 100 μg/ml ODN1826 CpG DNA (Invivogen and Integrated DNA Technologies). Etomoxir (Sigma) was reconstituted in DMSO (Sigma) and supplemented into media at a 200 μM concentration; BMS 303141 (Tocris) was reconstituted in DMSO and added into media at 50 μM concentration. Thapsigargin (Sigma) and staurosporine (Sigma) were reconstituted in DMSO and added into media at 10 nM and 1 mM, respectively. Brefeldin A at 5 μg/ml was applied to activated macrophages for 12–24 hrs prior to assays. For mock-treatment, PBS or DMSO vehicles were utilized as controls when appropriate.
Tissue Culture.
Tumor cells were cultured in DMEM (Dulbecco’s Modified Eagle’s Media, Thermo Scientific Fisher) supplemented with 10% v/v FCS (fetal calf serum, Gemini and VWR), 2 mM glutamax (Thermo Scientific Fisher), and 10 ng/ml gentamicin (Thermo Scientific Fisher). Cell cultures and in vitro co-cultures were maintained at 37oC and 5% CO2. Cell counts were determined using a hemacytometer. For in vitro experiments, PDAC cells were below passage 15, and for in vivo experiments, cells were below passage 10.
Generation of BMDMs.
BMDMs were derived by isolating bone marrow from mice euthanized with CO2 overexposure. Following treatment with ACK lysis buffer (Thermo Scientific Fisher). Bone marrow cells were then cultured in Iscove’s Modified Eagle’s Media (Thermo Scientific Fisher) supplemented with 10% v/v FCS, 2 mM glutamax, 10 ng/ml gentamicin, and 10 ng/ml M-CSF (Peprotech) for 7–10 days. Prior to use in experiments, BMDMs were removed from M-CSF and pretreated with TLR agonists or inhibitors for indicated durations.
Flow Cytometry.
Tumor cell cultures were treated with trypsin (Thermo Scientific Fisher) and prepared as a single-cell suspension for flow cytometric analysis; BMDMs in culture were incubated in cell dissociation buffer (Thermo Scientific Fisher) prior to mechanical detachment and resuspension. For intracellular stains, cells were permeabilized in 0.1% Triton-X (Sigma) for 15 min. For tumors, samples were harvested upon necropsy, mechanically separated, and digested in 1 mg/ml collagenase (Sigma) and 100 μUnits/ml DNase I (Roche). Tumor digests were filtered and resuspended as single cell suspensions. Cells were treated with Fc-block (BD Pharmingen), then stained with Amcyan live/dead dye (Thermo Scientific Fisher). For antibody staining (Supplementary Table 1), cells were washed with PBS containing 2% FCS, and stained on ice. Flow analysis was performed using FACS Canto (BD Biosciences), and singlets were gated on using FSC-H versus FSC-A.
BODIPY-C16 labeling.
BMDMs were seeded at 1×105 cells/well into a 96-well plate and stimulated with vehicle or 10 μg/ml CpG for 18 hours. Following stimulation, media was replaced with fresh DMEM supplemented with 10% v/v FCS, 2 mM glutamax, 10 ng/ml gentamicin, and 1 μg/ml BODIPY-C16 (Thermo Scientific Fisher) for 1 hour. After labeling, cells were washed, detached, and analyzed by flow cytometry on a FACS Canto.
In vitro phagocytosis assay.
Pretreated BMDMs were mechanically detached and labeled with 5 μM DiI (Life Technologies), then seeded into a transparent 96-well tissue culture plate at 5×104 cells/well. GFP-labeled PDAC cells were harvested as a single cell suspension and seeded at 5×104 cells/well. For other tumor cell lines, cell suspensions were labeled with 5 μM DiO (Life Technologies) and washed prior to being seeded at 5×104 cells/well. Following 4-hour co-culture, samples were washed and fixed in 4% PFA (paraformaldehyde, Sigma) for 15 min, prior to image collection. For antibody blockade, tumor cell suspensions were incubated with 10 μg/ml isotype (2A3, BioXcell) or rat-anti-mouse anti-CD47 (Miap301, BioXcell) antibodies for 20 min, prior to being washed and co-incubated with macrophages. Multiple random fields were collected per replicate, using an Olympus IX83 microscope, and the number of phagocytic events were scored and averaged for each replicate.
In vivo phagocytosis.
Frozen sections of PDAC tumors were fixed for 20 min in 4% PFA, followed by permeabilization in ice-cold methanol for 20 min. Samples were blocked for 1 hr in 10% goat serum and stained for 4 hr in primary antibodies (rabbit-anti-mouse cytokeratin 19, rat-anti-mouse F4/80). After washing, secondary antibody (488 goat-anti-rabbit, 568 goat-anti-rat) was applied for 1 hr. Samples were washed, and images were captured using an Olympus IX83 microscope. Background reduction was performed in CellSens software (Olympus) and images were subsequently processed using EBImage software analysis in R to perform color deconvolution, adaptive thresholding of F4/80 signal, gaussian blur, and binarization of signal. Size exclusion was applied to remove particles with a radius less than 10 pixels or greater than 200 pixels. The green (CK) signal was normalized intrinsically for each image to minimize variability in staining intensity. The phagocytic index was determined by calculating the normalized green (CK) intensity within each F4/80+ cells.
Anti-tumor activity assay.
Luciferase-labeled PDAC cells were harvested as a single cell suspension and seeded into a 96-well tissue culture plates at 5×104 cells/well. Pretreated BMDMs were harvested by mechanical detachment and seeded at 0:1, 1:1, 2:1, 5:1 macrophage-to-tumor ratios. After 48-hour co-culture, tumor cell survival was determined using Luciferase assay system (Promega). Luminescence measurements were performed using a SpectraMax M3 reader (Molecular Devices). Tumor cell survival was determined by normalizing luminescence to tumor-only controls.
Immunohistochemistry.
Cryosectioned murine PDAC tumors were fixed in 4% PFA and treated with 0.1% hydrogen peroxide (Sigma). Samples were blocked in 10% goat serum (Vector Laboratories) in PBS and treated overnight with primary antibodies (Supplementary Table 2). Secondary antibody was applied for 1 hour, and samples were stained with ABC HRP kit (Vectastain) per manufacturer instructions. Samples were then treated with DAB (3, 3 -diaminobenzidine) HRP substrate (Vector Labs) prior to counterstaining with hematoxylin (Sigma) and dehydration. Images were captured using the Olympus BX-43 microscope.
Immunofluorescence microscopy.
Treated BMDMs were harvested by mechanical detachment and seeded into glass chamber slides (Nunc). Samples were then fixed in 4% PFA, permeablized in 0.1% Triton-X for 15 min, and blocked with 10% goat serum (Vector Laboratories) in PBS. Primary antibodies and isotype control were applied overnight and washed; secondary antibody was applied for 1 hour and washed. Samples were stained with DAPI for 15 min and mounted prior to image acquisition. Images were captured using the Olympus IX83 microscope.
Membrane Fluidity.
BMDMs were seeded at 2.5×105 cells/well in 24-well plates, and stimulated under mock, CpG, CpG + ACLY-inhibitor (BMS 303141), or CpG + etomoxir conditions for 72 hr. Samples were then assessed using the membrane fluidity kit (Abcam). After stimulation, plates were washed and labeled with 10 uM pyrenedecanoic acid + 0.08% pluronic F127 for 30min at 37oC. Cells were harvested by gentle scraping, washed, and analyzed for fluorescence of monomer (400nm) and excimers (470nm) using a FACS Canto. Per manufacturer recommendations, membrane fluidity was quantified using the ratio of excimer:monomer mean fluorescence intensity.
Tumor growth studies.
PDAC cells were harvested from culture, washed, and prepared as a single cell suspension for implantation into the subcutis of mice. Rat-anti-mouse anti-CD47 (MIAP301, BioXcell) and isotype control antibodies (2A3, BioXcell) were diluted in saline, and 50 μg of each antibody was delivered intratumorally on days 10 and 14 after tumor implantation, as previously described (Suppplementary Table 3). Tumor-bearing mice were treated with vehicle (PBS) or 50 μg CpG by intraperitoneal injection every other day, beginning on day 10. For macrophage depletion, 200 μl of clodronate-encapsulated liposomes (clodronateliposomes.org) was delivered by retroorbital injection into mice, 400 μg of anti-CSF1R Ig (AFS98, BioXcell) diluted in saline was delivered by intraperitoneal injection on days 8, 10, 13, 16, or GW2580 (AdooQ) was delivered daily at 160 mg/kg by oral gavage52. For natural killer cell depletion, 200 μg anti-NK1.1 Ig (PK136, BioXcell) diluted in saline was delivered by intraperitoneal injection on days 8, 10, 13, and 16. For dendritic cell depletion in CD11c-DTR/eGFP, diphtheria toxin (Sigma) was administered by intraperitoneal injection at 8 ng/gram body weight. Etomoxir (Sigma) was resuspended in PBS and delivered daily at 40 mg/kg by intraperitoneal injection as previously described (Supplementary Table 3). Tumor volume was monitored by caliper measurement and calculated using a formula for an ellipsoid: Volume = 1/2 × (Length) × (Width)2. For survival studies, mice were euthanized when tumors reached 1,000 mm3.
ECAR and OCR measurements.
BMDMs were pretreated with TLR agonists or inhibitors and seeded into XF96-well plates (Agilent) at 1×105 cells/well. Prior to measurements, samples were washed and incubated in Seahorse media (Agilent) supplemented with 0.5 mM D-glucose (Sigma). The mitostress kit (Agilent) was prepared per manufacturer instructions by loading 1.5 μM oligomycin 1.5 μM FCCP, and 1 μM rotenone/antimycin A into injection ports. Measurements were made using an XF96 Extracellular Flux Analyzer (Agilent) and results processed with Wave v2.2.0 software.
Cytotoxicity studies.
BMDMs and tumor cells were seeded at 1×105 cells/well into a 96-well plate and treated with inhibitors diluted in Dulbecco’s Modified Eagle’s Media supplemented with 10% v/v fetal calf serum, 2 mM glutamax, and 10 ng/ml gentamicin. After 4 hours, the media was removed, and samples were stained with 0.05% w/v crystal violet (Sigma) in 20% ethanol/80% H2O. Samples were then washed and optical density measured at 570 nm using a Spectramax M3 spectrophotometer. Cell viability was determined by normalizing optical density of treated conditions to mock-treated controls.
13C metabolite tracing.
After pretreatment with CpG or vehicle for 96 hours, macrophages were counted and seeded at 4×106 cells per sample. For glucose and glutamine labeling, U-13C-glucose and U-13C-glutamine (Cambridge Isotope Laboratories) were substituted into DMEM supplemented with 10% v/v dialyzed FCS (Life Technologies), 4 mM L-glutamine (Sigma), and 25 mM glucose (Sigma). Cells were incubated at 37 °C in labeled media for 2 hours prior to harvest. For palmitate labeling, cells were cultured in DMEM supplemented with 100 μM U-13C-palmitate (Cambridge Isotope Laboratories),10% v/v charcoal-stripped FCS (Life Technologies), 4 mM L-glutamine (Sigma), and 25mM glucose (Sigma) for 4 hours. Both tracer experiments included unlabeled control samples not exposed to 13C-labeled substrates. After incubation, cells harvested and placed on ice. Samples were then pelleted, washed in cold PBS, and harvested in 750 μl of ice-cold 10% trichloroacetic acid (Sigma-Aldrich, St. Louis, MO cat. #T6399) and internal standard containing 13C315N1-labeled acyl-CoAs generated in pan6-deficient yeast culture were added to each sample in equal amounts51. Acyl-CoA thioester were analyzed by LC-MS/HRMS using an Ultimate 3000 autosampler coupled to a Thermo Q Exactive Plus instrument in positive ESI mode using the settings described previously52. Isotopologue enrichment in cells exposed to 13C labeled substrates was calculated using unlabeled control samples not exposed to 13C substrate as previously described53. For relative total pool quantitation, the total AUC for each acyl-CoA species (sum of all relevant isotopologues) was normalized to the AUC for the 13C315N1-labeled internal standard specific for that species. As previously described (Supplementary Table 3), samples were centrifuged at 1,200×g for 10 min at 4 °C and pulse-sonicated with a probe sonicator (Fisher Scientific). Lysates were centrifuged at 15,000×g for 15 min, and the supernatants were further purified by solid-phase extraction using (OASIS HLB) columns. Supernatants were then applied, and columns were washed with 1 ml H2O. Analytes were eluted in 25 mM ammonium acetate in methanol and evaporated to dryness overnight by N2 gas. Samples were resuspended in 50 μl of 5% 5-sulfosalicylic acid and 10 μl injections were applied in LC/ESI/MS/MS analysis. Isotopologues were designated as unlabeled (M+0), containing one 13C isotope (M+1), two 13C isotopes (M+2), etc following tracer labeling.
Cytokine analysis.
Peripheral blood was collected by tail vein bleed on day 14, and the serum fraction was isolated following centrifugation at 10,000×g for 15 min. For in vitro experiments, 1×106 macrophages were cultured in 1 ml of media. Supernatant was collected following 48 hr activation with cytokines and TLR agonists. Serum and supernatants were analyzed per manufacturer instructions using cytokine bead array kits (BD Biosciences) for IL-1β, IL-4, IL-6, IL-10, IL-12, IFN-γ, TNF, and CCL2. Beads were analyzed using a FACS Canto II.
Statistics and software analysis.
P-values were calculated using a two-tailed unpaired Student’s t-test and Hochberg correction for multiple comparison testing, unless stated otherwise. Linear mixed effects models (LMEM) were built to include random effects for each cell and image fields; statistical differences between treatment conditions were determined by ANOVA comparison of LMEM models. For survival analysis, significance was determined using the Kaplan-Meier log-rank test. P-values of 0.05 or less were considered significant. Error bars indicate standard deviation unless stated otherwise. Data analysis and graphical design was performed using R software (v3.4.3), R-studio, and additional R-packages: ggplot2, dplyr, reshape2, EBimages, lme4, kmsurv, and heatmap.2 (Supplementary Table 3). Flow cytometric analysis was completed using FlowJo (v10.3), and figure design utilized Adobe Illustrator CS6.
Data availability.
The data and code that support the findings of this study are available within the paper and its supplementary information files and are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
We’d like to thank all members of the Beatty lab for helpful suggestions, and Drs. B. Keith, A. Rustgi, A. Minn, T. Ridky, and K. Wellen for their scientific critiques. We thank Drs. J. Benci and O. Kawalekar for their assistance with CRISPR/Cas9 and Seahorse experiments; Dr. M Stone for her assistance with immunohistochemistry; Dr. A. Rech for manuscript review; Dr. K. Foskett for sharing his Seahorse bioanalyzer; John Scholler and Dr. A. Posey for assistance with lentivirus production; and the Molecular Biology and Molecular Pathology and Imaging Cores of the Penn Center supported by a Molecular Studies in Digestive and Liver Diseases grant (P30-DK050306) from the National Institutes of Health. This work was supported by grants from the NIH (R01 CA197916 to G.L.B., R03 HD092630 to N.W.S., and F30 CA196124 to M.L), and the Seed Grant Program from the American Medical Association Foundation (M. Liu).
Footnotes
Competing interests
The authors declare no competing interests
References
- 1.Mantovani A, Marchesi F, Malesci A, Laghi L & Allavena P Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas SK & Mantovani A Orchestration of metabolism by macrophages. Cell Metab 15, 432–437 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Colegio OR et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long KB et al. IFNgamma and CCL2 Cooperate to Redirect Tumor-Infiltrating Monocytes to Degrade Fibrosis and Enhance Chemotherapy Efficacy in Pancreatic Carcinoma. Cancer Discov 6, 400–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyonteck SM et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature medicine 19, 1264–1272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerriero JL et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature 543, 428–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majeti R et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiskopf K et al. Engineered SIRPalpha variants as immunotherapeutic adjuvants to anticancer antibodies. Science 341, 88–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldenborg PA, Gresham HD & Lindberg FP CD47-signal regulatory protein alpha (SIRPalpha) regulates Fcgamma and complement receptor-mediated phagocytosis. J Exp Med 193, 855–862 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willingham SB et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A 109, 6662–6667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cioffi M et al. Inhibition of CD47 Effectively Targets Pancreatic Cancer Stem Cells via Dual Mechanisms. Clin Cancer Res 21, 2325–2337 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Chen J et al. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature 544, 493–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass CK & Natoli G Molecular control of activation and priming in macrophages. Nature immunology 17, 26–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosser DM & Edwards JP Exploring the full spectrum of macrophage activation. Nat Rev Immunol 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue J et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jha AK et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Huang SC et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nature immunology 15, 846–855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu PS et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nature immunology 18, 985–994 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Vats D et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab 4, 13–24 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura M et al. Fatty acid oxidation in macrophage polarization. Nature immunology 17, 216–217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herber DL et al. Lipid accumulation and dendritic cell dysfunction in cancer. Nature medicine 16, 880–886 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hossain F et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol Res 3, 1236–1247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly B & O’Neill LA Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res 25, 771–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sastry PS & Hokin LE Studies on the role of phospholipids in phagocytosis. J Biol Chem 241, 3354–3361 (1966). [PubMed] [Google Scholar]
- 25.Lokesh BR & Wrann M Incorporation of palmitic acid or oleic acid into macrophage membrane lipids exerts differential effects on the function of normal mouse peritoneal macrophages. Biochim Biophys Acta 792, 141–148 (1984). [DOI] [PubMed] [Google Scholar]
- 26.Beatty GL et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science 331, 1612–1616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng CW et al. Pretreatment with Cisplatin Enhances E7-Specific CD8+ T-Cell-Mediated Antitumor Immunity Induced by DNA Vaccination. Clin Cancer Res 14, 3185–3192 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behrens EM et al. Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J Clin Invest 121, 2264–2277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beatty GL et al. Exclusion of T Cells From Pancreatic Carcinomas in Mice Is Regulated by Ly6C(low) F4/80(+) Extratumoral Macrophages. Gastroenterology 149, 201–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanford DE et al. Inflammatory Monocyte Mobilization Decreases Patient Survival in Pancreatic Cancer: A Role for Targeting the CCL2/CCR2 Axis. Clin Cancer Res 19, 3404–3415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatel C et al. Exposure to Bacterial CpG DNA Protects from Airway Allergic Inflammation by Expanding Regulatory Lung Interstitial Macrophages. Immunity 46, 457–473 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Feng M et al. Macrophages eat cancer cells using their own calreticulin as a guide: roles of TLR and Btk. Proc Natl Acad Sci U S A 112, 2145–2150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray PJ et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanjuan MA et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Wu D et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity 44, 1325–1336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ecker J et al. Induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes. Proc Natl Acad Sci U S A 107, 7817–7822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koberlin MS et al. A Conserved Circular Network of Coregulated Lipids Modulates Innate Immune Responses. Cell 162, 170–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagnon E et al. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110, 119–131 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Barkal AA et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nature immunology 19, 76–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Sullivan D et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 41, 75–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leidi M et al. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than m1 cells in vitro. J Immunol 182, 4415–4422 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Han CZ & Ravichandran KS Metabolic connections during apoptotic cell engulfment. Cell 147, 1442–1445 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha B et al. ‘Clustering’ SIRPalpha into the plasma membrane lipid microdomains is required for activated monocytes and macrophages to mediate effective cell surface interactions with CD47. PLoS One 8, e77615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sosale NG et al. Cell rigidity and shape override CD47’s “self”-signaling in phagocytosis by hyperactivating myosin-II. Blood 125, 542–552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz LX et al. The Lipid-Modifying Enzyme SMPDL3B Negatively Regulates Innate Immunity. Cell Rep 11, 1919–1928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian Y et al. Cytokine secretion requires phosphatidylcholine synthesis. J Cell Biol 181, 945–957 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krieg AM Development of TLR9 agonists for cancer therapy. J Clin Invest 117, 1184–1194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirsh V et al. Randomized phase III trial of paclitaxel/carboplatin with or without PF-3512676 (Toll-like receptor 9 agonist) as first-line treatment for advanced non-small-cell lung cancer. J Clin Oncol 29, 2667–2674 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Manegold C et al. A phase III randomized study of gemcitabine and cisplatin with or without PF-3512676 (TLR9 agonist) as first-line treatment of advanced non-small-cell lung cancer. Ann Oncol 23, 72–77 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Mitchem JB et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res 73, 1128–1141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder NW et al. Production of stable isotope-labeled acyl-coenzyme A thioesters by yeast stable isotope labeling by essential nutrients in cell culture. Anal Biochem 474, 59–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frey AJ et al. LC-quadrupole/Orbitrap high-resolution mass spectrometry enables stable isotope-resolved simultaneous quantification and (1)(3)C-isotopic labeling of acyl-coenzyme A thioesters. Anal Bioanal Chem 408, 3651–3658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trefely S, Ashwell P & Snyder NW FluxFix: automatic isotopologue normalization for metabolic tracer analysis. BMC Bioinformatics 17, 485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and code that support the findings of this study are available within the paper and its supplementary information files and are available from the corresponding author upon request.