Abstract
Background and Objective
CYP2C9 catalyzes the biotransformation of indomethacin to its inactive metabolite O-desmethylindomethacin (DMI). The aim of this work was to determine the effect of CYP2C9 polymorphisms on indomethacin metabolism in pregnant women.
Methods
Plasma concentrations of indomethacin and DMI at steady state were analyzed with a validated LC-MS/MS method. DNA was isolated from subject blood and buccal smear samples. Subjects were grouped by genotype for comparisons of pharmacokinetic parameters.
Results
For subjects with the *1/*2 genotype, the mean CL/Fss of indomethacin was 13.5 ± 7.7 L/h (n=4) and the mean metabolic ratio (AUCDMI/AUCindomethacin) was 0.291 ± 0.133. For subjects with the *1/*1 genotype, these values were 12.4 ± 2.7 L/h and 0.221 ± 0.078, respectively (n=14). Of note, we identified one subject who was a carrier of both the *3 and *4 alleles, resulting in an amino acid change (I359P) which has not been reported previously. This subject had a metabolic ratio of 0.390 and a steady-state apparent oral clearance of indomethacin (24.3 L/h) that was nearly double the wild-type clearance.
Conclusion
Although our results are limited by sample size and are not statistically significant, these data suggest that certain genetic polymorphisms of CYP2C9 may lead to an increased metabolic ratio and an increase in the clearance of indomethacin. More data are needed to assess the impact of CYP2C9 genotype on the effectiveness of indomethacin as a tocolytic agent.
1. Introduction
Preterm delivery is a major cause of neonatal morbidity and mortality. Indomethacin, a prostaglandin synthetase inhibitor, is often prescribed to patients with spontaneous preterm labor in an effort to delay delivery. It is not used past 32 weeks of gestation in order to avoid the risk of constriction of the ductus arteriosus, but when used appropriately, it is deemed a relatively safe and effective tocolytic agent [1–3]. Gaining a better understanding of the factors affecting the pharmacokinetics of indomethacin tocolytic therapy is important because treatment failure may lead to preterm birth and its associated short- and long-term neonatal complications.
We previously reported an increase in the steady-state apparent oral clearance of indomethacin in pregnant subjects as compared to data from non-pregnant subjects [4]. Among the factors that may affect the pharmacokinetics of drugs during pregnancy (e.g., increased urinary excretion, decreased albumin concentration, or changes in hepatic metabolism) [5], increased biotransformation appears to be the most substantial factor affecting the clearance of indomethacin, likely due to an increase in the activity of the metabolizing enzyme CYP2C9 [4,6,7].
The biotransformation of indomethacin to its major metabolite O-desmethylindomethacin (DMI), which is inactive, occurs predominantly via the CYP2C9 enzyme [8]. Several studies have shown the effect of genetic polymorphisms of CYP2C9 on the pharmacokinetics of drugs [9–13], including indomethacin [14]. A major goal of this work is to determine the effect of such polymorphisms on the biotransformation of indomethacin in individual pregnant subjects. This is essential in order to select appropriate dosing to optimize individual pharmacotherapy and minimize adverse effects. In this study, plasma samples from our previous report [4] were further investigated for metabolite (DMI) concentrations. In addition, CYP2C9 genotype was analyzed in these subjects to investigate potential associations between CYP2C9 genetic variants and pharmacokinetics. Accounting for genetic differences in the activity of this enzyme may be crucial in determining individualized doses of indomethacin that will maximize the safety and efficacy of tocolytic therapy.
2. Materials and methods
2.1. Chemicals and biological reagent
Indomethacin was obtained from Sigma Aldrich (St. Louis, MO). DMI, d4-DMI and d4indomethacin were purchased from Toronto Research Chemicals Inc. (North York, Canada). LC/MS-grade acetonitrile and formic acid, and HPLC-grade chloroform and hydrochloric acid were purchased from Fisher Scientific (Fair Lawn, NJ). Pooled blank human plasma was purchased from Innovative Research, Inc. (Novi, MI).
2.2. Subjects
Inclusion and exclusion criteria for the enrollment of pregnant subjects were reported previously [4]. All women were enrolled with written informed consent under a protocol that was approved by the Institutional Review Boards at the University of Texas Medical Branch and the University of Pittsburgh. The blood sample collection protocol is described in detail in our previous report [4]. Briefly, blood samples were collected at 0, 0.1, 1, 1.5, 2, 3, 4 and 6 hours post steady-state dosing and were centrifuged immediately. Plasma was stored at −70°C until further analysis.
2.3. Analytical methods (LC-MS/MS)
Indomethacin and DMI were analyzed on an Agilent 1200 HPLC system coupled with an API 4000 triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA). The API 4000 triple quadrupole mass spectrometer was equipped with a Turbo (V) ion source (ESI) and was operated in a positive mode. Multiple reactions monitoring (MRM) mode was applied for the quantification of DMI and indomethacin. MRM of indomethacin, indomethacin-d4, DMI and DMI-d4 was m/z 358→139, m/z 362→143, m/z 344→139 and m/z 348→143, respectively. The source/gas dependent MS parameters are provided in supplementary Table S1. Chromatographic separation was performed on a Waters Symmetry C18 column (2.1 mm × 100 mm, 3.5 μm) connected to a Phenomenex C18 guard column. The mobile phase was composed of (A) acetonitrile with 0.05% formic acid (v/v) and (B) 0.05% formic acid aqueous solution (v/v); the gradient elution was as follows: 0–3 min 60% A to 80% A and 3–4 min 80% A to 90% A, followed by washing the HPLC column for 1 min with 90% A and then equilibrated with 60% A for the next 5 min. The flow rate of the mobile phase was 0.3 mL/min. For the preparation of calibration standards and quality control (QC) samples (low, medium and high concentrations), stock solutions for indomethacin, DMI and their internal standards (ISs: d4-indomethacin and d4-DMI) were dissolved in 30% methanol followed by the addition of 10 μL of working standard solution at the appropriate concentration into 150 μL of pooled blank plasma samples.
The retention times of indomethacin and DMI were 3.2 min and 1.7 min, respectively. The calibration curves were fitted using weighted (1/x) least-squares linear regression of peak area ratio versus concentration. Good linearity was obtained for the concentration range of indomethacin from 6.22 ng/mL to 1.59×103 ng/mL, and for DMI from 1.18 ng/mL to 303 ng/mL. The lower limits of quantification for indomethacin and DMI were 6.22 ng/mL and 1.18 ng/mL, respectively. The accuracy for indomethacin and DMI ranged between 83–106% and between 91–105%, respectively. Precision of less than 6% was obtained for indomethacin, and less than 11% for DMI in three days’ validation.
2.3.1. Sample treatment
For subject plasma samples from our previous study [4], 10 μL of IS working solution and 20 μL of 1 M hydrochloric acid were added into 150 μL of patient plasma samples, followed by 30 sec of vortexing. To this, a 1 mL mixture of chloroform and ethyl acetate (1:1) was added and samples were vortexed for 3 min, followed by centrifugation for 3 min at 16,000×g. At the end of the centrifugation, the organic layer was carefully collected to a clean tube and dried under a stream of nitrogen at 40°C. The residues were then reconstituted with 100 μL of the initial mobile phase. After centrifugation at 16,000×g for 3 minutes, an aliquot of 5 μL of each sample was analyzed by LC-MS/MS for indomethacin and DMI concentrations.
2.4. Pharmacokinetic analysis
After the determination of concentrations of indomethacin and DMI from subject plasma samples, pharmacokinetic parameters including maximum plasma drug concentration (Cmax), minimum plasma drug concentration (Cmin), time to attain Cmax (tmax), area under the plasma concentration versus time curve at steady state (AUCss), apparent steady-state oral clearance (CL/Fss), and mean steady-state drug concentration (Cave) for indomethacin and DMI were calculated using Kinetica software version 5.0 (Thermo Scientific, Waltham, MA) as described previously [4]. Since DMI is a metabolite of the administered drug, indomethacin, the oral clearance for DMI was not calculated. Since the plasma concentrations of indomethacin from our previous subject study set were re-analyzed using a newly developed analytical method, the newly obtained indomethacin pharmacokinetic data were also compared with our previously reported values [4]. Supplementary Fig. S1 shows good agreement between the previously reported pharmacokinetic parameters for indomethacin and the data determined from the reanalyzed samples. To understand the metabolic activity in individual subjects, the metabolic ratio (AUCDMI/AUCindomethacin) was assessed on a molar basis using the most recently determined concentration data.
2.5. CYP2C9 genotyping
Whole blood (n=6) and buccal swabs (n=15) were collected from study subjects and stored at −80°C until DNA isolation for determination of CYP2C9 genotypes. DNA samples were not available for three subjects. DNA was isolated from blood samples using the Puregene Blood Core Kit (Qiagen Inc., Valencia, CA) and from buccal swabs using the MasterAmp™ Buccal Swab DNA Extraction Kit (Epicentre, Madison, WI,). DNA quality and concentration were determined using the DeNovix DS-11 FX spectrophotometer (Wilmington, DE). All subjects were genotyped for rs1799853 (430C>T), rs1057910 (1075A>C), and rs7900194 (449G>A) SNPs, which denote the CYP2C9 *2, *3 and *8 alleles, respectively. In addition, we also genotyped for the rs56165452 (1076T>C) SNP which denotes the rare CYP2C9*4 allele, since it is adjacent to the 1075A>C SNP (rs1057910) and thus may confound the CYP2C9*3 genotyping results. Since the rs7900194 SNP is triallelic, resulting in a 449G>A substitution (denoting the *8 allele) or a 449G>T substitution (denoting the CYP2C9*27 allele), for added accuracy in identifying the *8 allele we also genotyped all subjects for the 449G>T SNP. Allele discrimination assays using the TaqMan probes (Thermo Fisher Scientific Inc., Waltham, MA) C__25625805_10, C__27104892_10, C__30634131_20, C__25625804_10 and C_25625804D_20 were used for the determination of CYP2C9*2, CYP2C9*3, CYP2C9*4, CYP2C9*8 and CYP2C9*27 alleles, respectively. Typical PCR reactions contained 5 μl of TaqMan GTXpress master mix, 10 ng of DNA and 0.5 μl of 20× TaqMan probes. Genotyping was performed using a Roche LightCycler 96 (Roche, Indianapolis, IN).
2.6. Statistical Analysis
All the pharmacokinetic data are presented as mean values ± SD. Two-tailed Student’s t-test was performed to compare pharmacokinetic parameters with respect to genotype. P-values <0.05 were considered statistically significant.
3. Results
3.1. Pharmacokinetic analysis
The obtained plasma concentrations of indomethacin determined by the newly developed analytical method were consistent with our previously reported values [4] and there was no statistically significant difference observed between the calculated pharmacokinetic parameters (Student’s t-test). This observed sample stability for the plasma samples stored at –80°C suggested that we could feasibly determine DMI concentrations from those same samples.
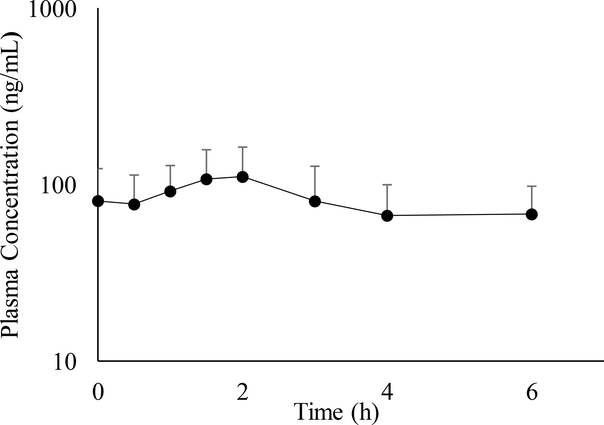
Plasma concentrations of DMI could be determined for 24 of the subjects from our prior study [4], but the remaining plasma volume of Subject #1 was insufficient for analysis of DMI. The mean plasma concentration versus time profile is presented in Fig.1. Maximum plasma DMI concentration (Cmax), minimum plasma DMI concentration (Cmin), time to DMI Cmax (tmax) and total area under the plasma DMI concentration versus time curve at steady state over a 6-h dosing interval (AUCss) are shown in Table 1. The average Cmax and Cmin of DMI were found to be 129.8 ± 56.0 ng/mL and 43.5 ± 17.0 ng/mL, respectively. The average time to reach the Cmax for DMI was 2.2 ± 1.4 hours and the average AUCss was 466 ± 183 ng/mL*h. Individual pharmacokinetic data and demographic information can be found in the supplementary data section (Tables S2 and S3). No significant differences between smokers (n=7) and non-smokers (n=17) were observed. In addition, differences in maternal age, indication, and adverse events (e.g., oligohydramnios) did not reveal any salient trend with pharmacokinetic parameters.
Fig. 1.
Mean steady-state plasma concentration of DMI (O-desmethylindomethacin) versus time profile for 24 pregnant subjects receiving 25-mg oral doses of indomethacin every 6 h. Error bars represent the standard deviation.
Table 1.
Steady-state pharmacokinetic parameters of DMI (O-desmethylindomethacin) for 24 pregnant subjects receiving 25-mg oral doses of indomethacin every 6 h.
| Parameter | Median | Mean ± SD |
|---|---|---|
| Cmax (ng/mL) | 115.9 | 129.8 ± 56.0 |
| Cmin (ng/mL) | 39.2 | 43.5 ± 17.0 |
| AUCss (ng/mL*h) | 458 | 466 ± 183 |
| tmax (h) | 2.0 | 2.2 ± 1.4 |
Abbreviations: Cmax = maximum plasma drug concentration; Cmin = minimum plasma drug concentration; tmax = time to Cmax; AUCss = total area under the plasma concentration versus time curve at steady state over a 6-h dosing interval.
3.2. Genotyping
The detailed results for each subject are shown in the supplementary data section (Table S4). As shown in Table 2, four subjects were heterozygous for the *2 allele (CYP2C9*1/*2), one sample was homozygous for the *3 allele (CYP2C9*3/*3), one sample was heterozygous for the *8 allele (CYP2C9*1/*8), and one sample was homozygous for the *3 allele and also heterozygous for the *4 allele (CYP2C9*3/*4). The remaining 14 samples were homozygous wild-type (CYP2C9*1/*1). Details pertaining to each subject are provided in supplementary Table S3. We have also compared the CYP2C9 allele nucleotide sequences causing amino acid changes at position 359 of the CYP2C9 protein (Table 3). Supplementary Fig. S2 shows the breakdown of genotype by race/ethnicity, presented as overall frequency.
Table 2.
Pharmacokinetic data grouped by CYP2C9 genotype.†
| CYP2C9 Genotype | n | AUCDMI/AUCindomethacin | Indomethacin CL/Fss (L/h) | Indomethacin AUCss (μg·h/mL) |
|---|---|---|---|---|
| *1/*1 | 14 | 0.221 ± 0.078 | 12.4 ± 2.7 | 2.10 ± 0.43 |
| *1/*2 | 4 | 0.291 ± 0.133 | 13.5 ± 7.7 | 2.22 ± 0.89 |
| *1/*8 | 1 | 0.242 | 14.9 | 1.68 |
| *3/*3 | 1 | 0.259 | 11.0 | 2.28 |
| *3/*4‡ | 1 | 0.390 | 24.3 | 1.03 |
Abbreviations: CL/Fss = apparent steady-state oral clearance CL/Fss, AUCss = total area under the plasma concentration versus time curve at steady state over a 6-h dosing interval.
This table includes only subjects for whom genotype data were available.
This subject was homozygous for the *3 allele and a heterozygous carrier of the *4 allele.
Table 3.
Comparison of CYP2C9 allele nucleotide sequences at positions 1074–1077 and the resulting amino acid change at position 359 of the CYP2C9 protein.
| Allele | Nucleotide sequence (1074–1077) | Amino acid change |
|---|---|---|
| CYP2C9*1 (wild-type) | CATT | I359 (wild-type) |
| CYP2C9*3 | CCTT | I359L |
| CYP2C9*4 | CACT | I359T |
| Both CYP2C9*3 and CYP2C9*4 | CCCT | I359P |
4. Discussion
The effects of biotransformation on the pharmacokinetics of drugs are important to consider when administering pharmacologic therapy during pregnancy. Pregnancy-induced changes in the activity of metabolizing enzymes may alter the pharmacokinetic profiles of certain drugs as compared to the non-pregnant state. Inter-individual differences in drug disposition may be observed due to genetic polymorphisms in enzymes and transporters [15]. The activity of the CYP2C9 enzyme is reported to be increased during pregnancy and genetic polymorphisms in the CYP2C9 gene are well established [2,16]. There are about 60 SNPs reported in the CYP2C9 gene which may possess functional differences [17,18].
When the pregnant subjects in our study were grouped by genotype, the mean metabolic ratio (AUCDMI/AUCindomethacin) was 0.291 ± 0.133 and the mean CL/Fss of indomethacin was 13.5 ± 7.7 L/h (n=4) for carriers of the *2 allele. For individuals with the CYP2C9*1 (wild-type) genotype, these values were 0.221 ± 0.078 and 12.4 ± 2.7 L/h, respectively (n=14). This may suggest increased metabolism of indomethacin for carriers of the *2 allele. Perhaps due to the limited number of subjects with each genotype, no statistically significant differences were observed. On the other hand, reduced metabolism of warfarin has been observed for carriers of CYP2C9*2 and CYP2C9*3 as compared to the *1/*1 (wild-type) genotype. Accordingly, the FDA has recommended a lower daily dose of warfarin for carriers of the CYP2C9*2 and CYP2C9*3 alleles [19]. Similar results were observed for celecoxib [20]. It should be noted, however, that for both studies with warfarin and celecoxib, the patient population was non-pregnant. Although decreased activity of CYP2C9*2 has been suggested in the literature for non-pregnant subjects [21,22], Table 2 shows that the mean indomethacin AUCss for our pregnant CYP2C9*1/*2 subjects (n=4) was approximately the same as the average AUCss for our CYP2C9*1/*1 subjects. Although limited by sample size, the observed differences in the activity of the same enzyme in our study compared to previous studies of warfarin and celecoxib in non-pregnant subjects may be due to pregnancy-induced changes, the effect of genotype, and/or drug specificity.
Rodrigues predicted an AUC ratio of 1.8 for CYP2C9*3/*3 versus CYP2C9*1/*1 subjects following an oral dose of indomethacin [23], and in vitro studies by Tracy et al. also suggest reduced CYP2C9*3-mediated drug demethylation [24]. Our findings are limited due to having only one CYP2C9*3/*3 subject, whose indomethacin AUCss was only 8.6% higher than the average AUCss for CYP2C9*1/*1 subjects (n=14).
The indomethacin AUCss for the one CYP2C9*3/*4 subject was 51% less than the wild-type mean. Of note, this particular subject was identified to have a genotype that has not been reported previously in the literature. This subject had a combination of both the *3 (homozygous) and *4 (heterozygous) alleles, resulting in an amino acid change that had not been reported previously in the CYP2C9 protein. Table 3 compares the wild-type (*1) allele nucleotide sequence to the nucleotide sequence of the *3 and *4 alleles, as well as the nucleotide sequence observed in this particular subject. This newly observed combination of the *3 and *4 alleles results in an amino acid change at position 359 from isoleucine to proline. Reports in the literature suggest that the *3 and *4 alleles individually—which change isoleucine-359 to leucine and threonine, respectively—cause reductions in the rates of CYP2C9-mediated drug metabolism [23]. This is in agreement with what we had observed in one subject carrying the *3 allele, in whom the apparent oral clearance of indomethacin (11.0 L/h) seemed slightly below the average clearance amongst carriers of the wild-type allele (12.4 ± 2.7 L/h). However, for the individual having both the *3 and *4 alleles, the clearance of indomethacin was observed to be 24.3 L/h, which is approximately double the mean wild-type clearance. This particular amino acid change (I359P) had not been reported previously and has not yet been characterized. It is possible that the presence of proline in this position could cause a conformational change in the CYP2C9 protein, resulting in an accelerated biotransformation of indomethacin. It has been suggested that substitutions in residue 359 may affect the active site of the CYP2C9 protein [23,25]. We seek to further investigate this allele combination and its implications, as it may help us to better understand how genomic differences affect the clearance and efficacy of indomethacin in our efforts to optimize individual dosing requirements to improve indomethacin tocolytic therapy.
78% of the African American subjects in our study had the wild-type genotype (*1/*1). This is in fair agreement with an earlier report which stated that 71.7% of African Americans had *1/*1 [26]. One African American was a carrier of the *8 allele, which is more commonly seen in African Americans as compared to other races [26]. The *1/*2 genotype is more common in Hispanics and non-Hispanic whites [18]; amongst our subjects, two carriers of the *2 allele were Hispanic, one was African American, and one was white/non-Hispanic. When stratified for race/ethnicity, the average metabolic ratio (AUCDMI/AUCindomethacin) was 0.219 ± 0.101 for white/non-Hispanic subjects (n=7), 0.247 ± 0.070 for African Americans (n=11), and 0.239 ± 0.119 for Hispanics (n=6). The apparent oral clearance of indomethacin was 11.6 ± 5.8 L/h for white/non-Hispanic subjects, 14.3 ± 4.5 L/h for African Americans, and 11.6 ± 1.6 L/h for Hispanics. None of these comparisons were statistically significant. However, it is interesting to note that if the carrier of both the *3 and *4 alleles were excluded from consideration as an extensive-metabolizing outlier, the clearance of indomethacin in the other six white/non-Hispanic subjects (9.5 ± 1.7 L/h) would be significantly lower than that of our African American subjects (14.3 ± 4.5 L/h, P < 0.05). One possible explanation is that estradiol—a major hormone of pregnancy—increases the activity of CYP2C9 in vitro [27], and estradiol levels during pregnancy are higher in African American women than in white/non-Hispanic women [28]. Accordingly, more detailed investigations are warranted regarding the influence of estradiol on the pharmacokinetics of indomethacin during pregnancy. Interestingly, we had previously observed a significant difference (P < 0.05) in BMI between those subjects treated with indomethacin for preterm labor who had delivered within 3 days of their last dose versus those for whom delivery was at least 11 days after their last dose of indomethacin [4].
Considering the gestational age at the time of delivery as a pharmacodynamic outcome for those prescribed indomethacin for preterm labor, subjects who delivered prior to 32 weeks of gestation had a mean metabolic ratio (AUCDMI/AUCindomethacin) of 0.251 ± 0.104 (n=8), whereas for those who delivered after 32 weeks of gestation, the mean metabolic ratio was 0.188 ± 0.045, (n=7, P=0.16). The data in Table 2 suggest that CYP2C9 genotype may affect the biotransformation of indomethacin to its metabolite DMI. Genetic polymorphisms may lead to an increased metabolic ratio and an increase in the clearance of indomethacin, which may affect the pharmacodynamic action of the drug and its utility as a tocolytic therapy. Nevertheless, we must point out a major limitation of this analysis, i.e., limited sample size. Carriers of some alleles determined in this study (*8, *3, and *3+*4) had only one representative each, which makes it difficult to draw any substantive conclusions at this time regarding the effect of genetic polymorphisms on the pharmacokinetics of indomethacin in pregnant women. We anticipate that expansion of this study to enroll additional subjects will strengthen the statistical power needed to assess dose optimization based on genotype or other factors (such as estradiol or BMI).
5. Conclusion
Although our results are limited by sample size, these data suggest that CYP2C9 genotype may affect the biotransformation of indomethacin to its metabolite DMI. Genetic polymorphisms may lead to an increased metabolic ratio and an increase in the clearance of indomethacin. More data are needed to understand the effects of CYP2C9 polymorphisms on the pharmacokinetics and pharmacodynamics of indomethacin as a tocolytic therapy. This should enable individualized dose optimization in order to enhance therapeutic outcomes and reduce adverse effects.
Supplementary Material
Keypoints.
The obtained data suggest that CYP2C9 genotype may affect the biotransformation of indomethacin to its metabolite DMI. Genetic polymorphisms which cause an increased metabolic ratio may increase the clearance of indomethacin, which may affect the drug’s therapeutic action.
We identified one subject who was a carrier of both the *3 and *4 alleles, resulting in an amino acid change (I359P) which has not been reported previously. Clearance of indomethacin for this newly identified subject was approximately double that of the mean for wild-type subjects.
Funding
The authors are grateful for research support from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (U10HD04789108 and R01HD083003).
Footnotes
Compliance with Ethical Standards
Conflict of Interest
Authors Mansi Shah, Meixiang Xu, Poonam Shah, Xiaoming Wang, Shannon M. Clark, Maged Costantine, Holly A. West, Tatiana N. Nanovskaya, Mahmoud S. Ahmed, Sherif Z. Abdel-Rahman, Raman Venkataramanan, Steve N. Caritis, Gary D.V. Hankins, and Erik Rytting declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its actual amended version, the International Conference on Harmonization-Good Clinical Practices (ICH-GCP) guidelines.
Informed Consent
Informed consent was obtained from all participants included in this study.
References
- 1.GOLDENBERG R The management of preterm labor*1. Obstet Gynecol. 2002. November;100(5):1020–37. [DOI] [PubMed] [Google Scholar]
- 2.Bivins HA, Newman RB, Fyfe DA, Campbell BA, Stramm SL. Randomized trial of oral indomethacin and terbutaline sulfate for the long-term suppression of preterm labor. Am J Obstet Gynecol. 1993. October;169(4):1065–70. [DOI] [PubMed] [Google Scholar]
- 3.Vermillion ST, Scardo JA, Lashus AG, Wiles HB. The effect of indomethacin tocolysis on fetal ductus arteriosus constriction with advancing gestational age. Am J Obstet Gynecol. 1997. August;177(2):256–9–61. [DOI] [PubMed] [Google Scholar]
- 4.Rytting E, Nanovskaya TN, Wang X, Vernikovskaya DI, Clark SM, Cochran M, et al. Pharmacokinetics of Indomethacin in Pregnancy. Clin Pharmacokinet. 2014. Jun 4; 53(6):545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson GD. Pregnancy-Induced Changes in Pharmacokinetics. Clin Pharmacokinet. 2005;44(10):989–1008. [DOI] [PubMed] [Google Scholar]
- 6.Alqahtani S, Kaddoumi A. Development of Physiologically Based Pharmacokinetic/Pharmacodynamic Model for Indomethacin Disposition in Pregnancy. Subbian S, editor. PLoS One. 2015. October 2;10(10):e0139762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isoherranen N, Thummel KE. Drug metabolism and transport during pregnancy: how does drug disposition change during pregnancy and what are the mechanisms that cause such changes? Drug Metab Dispos. 2013. February;41(2):256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyatt JE, Pettit WL, Harirforoosh S. Pharmacogenetics of nonsteroidal anti-inflammatory drugs. Pharmacogenomics J. 2012. December 9;12(6):462–7. [DOI] [PubMed] [Google Scholar]
- 9.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999. February 27;353(9154):717–9. [DOI] [PubMed] [Google Scholar]
- 10.Ke AB, Nallani SC, Zhao P, Rostami-Hodjegan A, Unadkat JD. Expansion of a PBPK model to predict disposition in pregnant women of drugs cleared via multiple CYP enzymes, including CYP2B6, CYP2C9 and CYP2C19. Br J Clin Pharmacol. 2014. March;77(3):554–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin JH, Begg EJ, Kennedy MA, Roberts R, Barclay ML. Is cytochrome P450 2C9 genotype associated with NSAID gastric ulceration? Br J Clin Pharmacol. 2001. June 1;51(6):627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu WJ, Zhang S, Yang Y, Zhang N, Wang W, Liu SY, et al. Efficient inhibition of human colorectal carcinoma growth by RNA interference targeting polo-like kinase 1 in vitro and in vivo. Cancer Biother Radiopharm. 2011/07/30. 2011;26(4):427–36. [DOI] [PubMed] [Google Scholar]
- 13.Jaja C, Patel N, Scott SA, Gibson R, Kutlar A. CYP2C9 allelic variants and frequencies in a pediatric sickle cell disease cohort: implications for NSAIDs pharmacotherapy. Clin Transl Sci. 2014. October;7(5):396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CJ, Ryckman KK, Bahr TM, Dagle JM. Polymorphisms in CYP2C9 are associated with response to indomethacin among neonates with patent ductus arteriosus. Pediatr Res. 2017. July 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong H Altered drug metabolism during pregnancy: hormonal regulation of drug-metabolizing enzymes. Expert Opin Drug Metab Toxicol 2010. June;6(6):689–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinney SK, Patil AS, Flockhart DA. Is personalized medicine achievable in obstetrics? Semin Perinatol. 2014. December;38(8):534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. https://www.pharmvar.org/gene/CYP2C9. Accessed 7 Feb 2018.
- 18.Xie H-G, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev. 2002. November;54(10):1257–70. [DOI] [PubMed] [Google Scholar]
- 19.Dean L Warfarin Therapy and the Genotypes CYP2C9 and VKORC1 Medical Genetics Summaries. National Center for Biotechnology Information (US); 2012. [PubMed] [Google Scholar]
- 20.Dean L Celecoxib Therapy and CYP2C9 Genotype Medical Genetics Summaries. National Center for Biotechnology Information (US); 2012. [PubMed] [Google Scholar]
- 21.Maekawa K, Adachi M, Matsuzawa Y, Zhang Q, Kuroki R, Saito Y, et al. Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9. Biochemistry. 2017. October 17;56(41):5476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niinuma Y, Saito T, Takahashi M, Tsukada C, Ito M, Hirasawa N, et al. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 2014. April 11;14(2):107–14. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues AD. IMPACT OF CYP2C9 GENOTYPE ON PHARMACOKINETICS: ARE ALL CYCLOOXYGENASE INHIBITORS THE SAME? Drug Metab Dispos. 2005;33(11). [DOI] [PubMed] [Google Scholar]
- 24.Tracy TS, Hutzler JM, Haining RL, Rettie AE, Hummel MA, Dickmann LJ . Polymorphic variants (CYP2C9*3 and CYP2C9*5) and the F114L active site mutation of CYP2C9: effect on atypical kinetic metabolism profiles. Drug Metab Dispos. 2002. April;30(4):385–90. [DOI] [PubMed] [Google Scholar]
- 25.Imai J, Ieiri I, Mamiya K, Miyahara S, Furuumi H, Nanba E, et al. Polymorphism of the cytochrome P450 (CYP) 2C9 gene in Japanese epileptic patients: genetic analysis of the CYP2C9 locus. Pharmacogenetics. 2000. February;10(1):85–9. [DOI] [PubMed] [Google Scholar]
- 26.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009. August;10(8):1243–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi S-Y, Koh KH, Jeong H. Isoform-Specific Regulation of Cytochromes P450 Expression by Estradiol and Progesterone. Drug Metab Dispos. 2013. February 1;41(2):263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson BE, Bernstein L, Ross RK, Depue RH, Judd HL. The early in utero oestrogen and testosterone environment of blacks and whites: potential effects on male offspring. Br J Cancer. 1988. February;57(2):216–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.