Abstract
The current influenza vaccine has the inevitable risk of antigenic discordance between the vaccine and the circulating strains, which diminishes vaccine efficacy. This necessitates new approaches that provide broader protection against influenza. Here, we design a vaccine utilizing the hypervariable receptor-binding domain (RBD) of virus hemagglutinin displayed on a nanoparticle (np) able to elicit antibody responses that neutralize H1N1 viruses spanning over 90 years. Co-displaying RBDs from multiple strains across time, so that the adjacent RBDs are heterotypic, provides an avidity advantage to cross-reactive B cells. Immunization with the mosaic RBD-np elicited broader antibody responses than those induced by an admixture of nps encompassing the same set of RBDs as separate homotypic arrays. Furthermore, we identified a broadly neutralizing monoclonal antibody in a mouse immunized with mosaic RBD-np. The mosaic antigen array signifies a unique approach that subverts monotypic immunodominance and allows otherwise subdominant cross-reactive B cell responses to emerge.
Influenza A viruses have circulated among avian and mammalian species including humans for over 500 years1. As a result of prolonged circulation, the viruses evolve unique antigenic traits that diversify under selective pressure2, 3, 4, 5. Humoral immune responses to the viral hemagglutinin (HA) are the primary driving force of the selective evolution of influenza (i.e., antigenic drift), illustrating the importance of HA as a vulnerable target for protective antibody responses. The existence of broadly neutralizing antibodies (bnAbs) against influenza viruses6 demonstrates the possibility of generating universal influenza immunity in humans either through natural infections7 and/or by active immunizations8, 9, 10, 11, 12. There are two structurally defined antigenic supersites on the HA molecule being targeted by bnAbs. One is the receptor-binding site (RBS) within the globular head region which binds to sialic acid moieties on host cell surface glycoproteins and glycolipids13, 14, 15, 16. The other is a site on the HA stem centered at the hydrophobic groove surrounding the Trp21HA2ref 17, 18, 19, 20. There are other neutralization-sensitive antigenic sites on HA that are conserved within subtype, but not between subtypes21, 22, 23. These sites may represent alternative vaccine targets as the antibodies targeting those sites are often less strain-dependent than RBS-directed antibodies and have higher potency than stem-directed antibodies. Despite immense efforts to develop candidate universal influenza vaccines that elicit bnAb responses to any of the aforementioned viral sites of vulnerability, this goal has not been achieved. Although induction of antibody-mediated heterosubtypic protective immunity against lethal influenza virus challenge in animal models has been shown, it has not been associated with significant neutralizing activity24, 25, 26, 27.
During the course of antigen exposure from infection or vaccination antigen-specific B cells are stimulated and undergone a process called somatic hypermutation (SHM) to fine-tune the affinity and specificity of their B cell receptors (BCRs) through germinal center (GC) reactions28. This specialization process of B cells is essential to develop high affinity BCR and eventually generating highly neutralizing antibody responses. However, overspecialization of the immunodominant B cells with limited breadth against influenza viruses may impair or delay the emergence of B cells targeting conserved antigenic supersites. Since elicitation of cross-reactive B cell responses to antigenically hypervariable targets is of great interest for developing vaccines against rapidly evolving viruses such as influenza, hepatitis C virus, or human immunodeficiency virus type 1ref 29, 30, reshaping the intrinsic hierarchy of immunodominance is of critical importance for vaccine design.
Here, we developed a novel mosaic array by colocalizing heterotypic influenza HA antigens on a single np to diminish or avoid activation of strain-specific B cells, and allow selective engagement of B cells that tolerate antigenic variability. This would promote cross-reactive antibody responses by adaptively targeting conserved antigenic surfaces. The heterotypic mosaic np immunogen elicits quantitatively and qualitatively superior B cell responses compared to those elicited by antigenically homotypic immunogens even when multiple specificities are admixed together. A monoclonal antibody (mAb) isolated from a mouse immunized with the heterotypic mosaic np possesses exceptional neutralization capacity to H1N1 viruses spanning over 90 years. Finally, the structural studies of this antibody define a site of vulnerability that should inform pan-subtypic influenza vaccine designs.
Results
Design and characterization of heterotypic influenza HA receptor-binding domain (RBD) mosaic nanoparticles
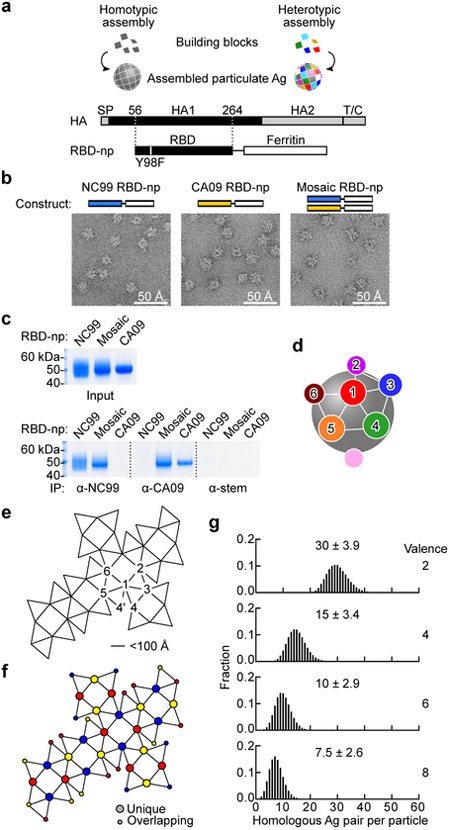
We theorized that the mosaic arrays of heterotypic antigens would reduce the likelihood of activating high avidity B cells expressing a BCR with narrow-specificity, and increase opportunities for B cells expressing a cross-reactive BCR to be activated, hence altering the hierarchy of B cell response frequencies to favor the epitopes of interest. To empirically test the hypothesis, a modular self-assembling np system based on the ferritin np scaffold was developed25, 31, 32. This system allows us to manipulate homogeneity and heterogeneity of antigens displayed as an array on the assembled np. H1N1 influenza HA was chosen as a model antigen to evaluate the impact of antigenic heterogeneity on induction of cross-reactive antibodies because of the extensive genetic history2, 33 and availability of reagents. Monomeric RBDs were expressed from a fusion construct linked to an engineered ferritin sequence31 (Fig. 1a). RBD-np expressed in transfected cells spontaneously self-assembles to form particles that are secreted into the culture supernatant. This system enables to make homogeneously assembled RBD-np (building blocks with a single RBD sequence) as well as heterogeneously co-assembled mosaic RBD-np (building blocks with multiple RBD sequences) (Fig. 1a). To prove co-assembly of mosaic RBD-np, we used an antigenically distinct pair of HA RBDs from two H1N1 strains, A/New Caledonia/20/99 (NC99) and A/California/04/09 (CA09), and generated three RBD-np preparations (i.e., homotypic NC99 or CA09, or co-assembled heterotypic mosaic particles) (Fig. 1b). Particle formation and antigenicity of these RBD-nps were confirmed by electron microscopy (EM) and immunoprecipitation (IP) using monoclonal antibodies (mAbs) specific to either NC99 or CA09 HA34. We found that the co-assembled mosaic RBD-np displayed analogous morphology to homotypically assembled NC99 or CA09 RBD-np by negative stain EM and possessed antigenicity of both NC99 and CA09 by IP as expected (Fig. 1b,c). In addition, the co-assembled mosaic RBD-np had distinct biochemical characteristics as evidenced by a unique profile on anion-exchange chromatography, indicating each individual particle contained both NC99 and CA09 RBD components (Supplementary Fig. 1 a,b). Spacing of model antigens between 50 and 100 Å apart is known to be optimal for B cell activation35. Optimal distance between antigenic sites displayed in an orderly, symmetrical array strengthens the recognition of the vaccine antigen as a foreign pathogen-associated molecular pattern (PAMP)36. Therefore, placing antigens in a repetitive, symmetrical array with 50–100 Å spacing was intended to maximize activation of cognate B cells via BCR crosslinking and/or BCR microclustering. To estimate the likelihood of identical antigens co-localizing adjacent to each other on a mosaic np composed of multiple distinct specificities, we modeled the location of randomly distributed subunits graphically. A 24-meric RBD-np has 24 vertices, each of which are within 50–100 Å of 5 neighboring vertices (Fig. 1d,e). The RBD antigen placement on ferritin is a snub cubical geometry (Fig. 1e) with 60 edges, 38 faces, and has a chromatic number of 3, therefore, in theory, it is possible to create a perfectly heterogeneous mosaic particle with no adjacent homologous antigen pairs at the 24 vertices by co-assembling 3 different antigens (valency of 3) (Fig. 1f). However, the random assembly of antigens that comprise the particle allows adjacent homologous antigen pairs even at valencies of 3 or greater. Therefore, we calculated the likelihood of adjacent homologous antigen pairs forming at different valencies, making the assumption of random assortment. When two different RBDs co-assemble in a single particle, 30 ± 3.9 out of the 60 possible adjacent pairs are made of 2 identical RBDs (i.e. A-A or B-B) and the remaining pairs are heterologous (i.e. A-B) (Fig. 1g). By increasing the valency to 8, the likelihood of homologous antigen pairs is reduced to 7.5 ± 2.6 per particle or about one eighth of RBD pairs (Fig. 1g), thus minimizing the chance for a single BCR to bivalently bind two neighboring homologous antigens or to form BCR microclusters leading to B cell activation.
Fig. 1. Design and characterization of a mosaic array of heterotypic antigens on self-assembling nps.
(a) Design of HA RBD-np. Alteration in the residue 98 (Y98F) was made to abrogate sialic acid-binding property of HA. SP, signal peptide; T/C, transmembrane/cytoplasmic domains. (b) Negative-stain EM images of self-assembled HA RBD-nps. RBD-np were made using either single building blocks (left and middle) or two different building blocks (right). Shown are representative images from one experiment. (c) Antigenic characterization of RBD-np by immunoprecipitation (IP). The mAbs 3u-u (anti-NC99), 2D1 (anti-CA09) and C179 (anti-HA stem) were used to pull-down NC99, CA09 RBDs, and HA stem (control), respectively. Similar results were obtained from two independent experiments. (d) Schematic model of ferritin-based np. Twenty-four spatially dispersed antigens (colored individually) are displayed on the surface. Positions 2–6 are localized within a 100 Å distance from position 1. (e) Ferritin snub cube net with positions numbered similarly to panel d. Connected lines indicate adjacent positions located within 50–100 Å. (f) Twenty-four positions are colored according with the chromatic number (three) to avoid the same color from being located within 100 Å radius of each other. (g) Simulated likelihood of homologous antigen pairs made within 100 Å radius on a single particle by using 2, 4, 6 or 8 different building blocks (valence).
Immune induction with heterotypic co-assembled mosaic RBD-np
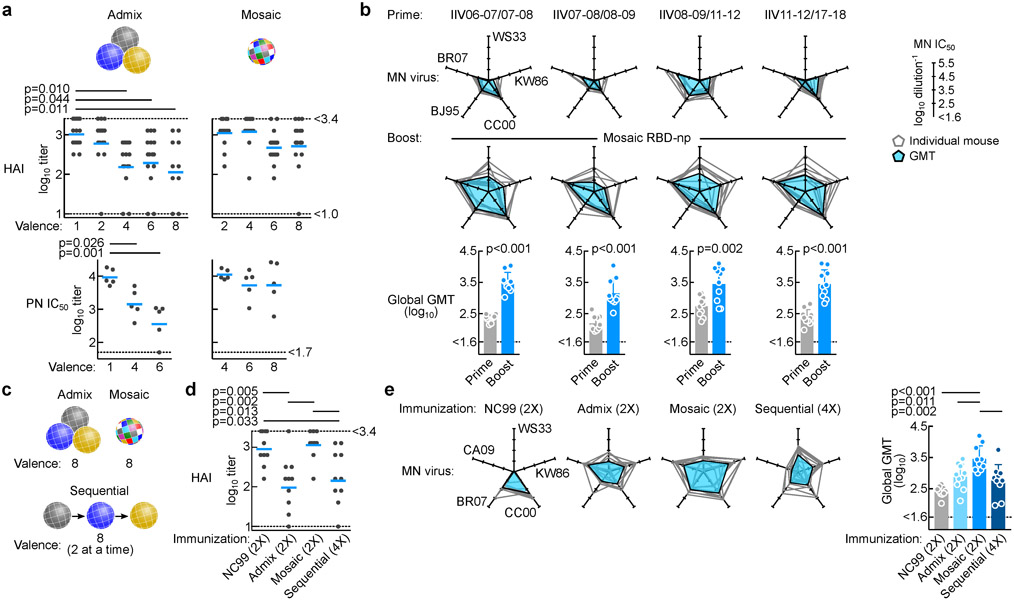
To assess the impact of per-particle antigen heterogeneity on its immunogenicity, we created RBD-nps from a selection of H1N1 strains with a range of valencies (Supplementary Fig. 1c–f). Mice were immunized with RBD-nps presenting between 1 and 8 different RBDs derived from various H1N1 strains either individually (i.e., admixture of up to 8 homotypic particles) or altogether as a single mosaic particle (co-assembly using up to 8 different RBDs) (Fig. 2a). Immunization dose was kept constant across different groups, thus decreasing the amount of each component as the valency increased. After two immunizations with adjuvant, hemagglutination inhibition (HAI) and neutralizing activity against homologous H1N1 NC99 virus were detected in nearly all mice, as NC99 RBD was a component of all immunogens (Fig. 2a). As expected, responses to NC99 progressively diminished as the NC99 antigen content was reduced in the admixed np groups (Fig. 2a, left). The reduced antibody response could be explained by antigen dilution and/or antigenic competition between different RBDs. The effect of antigen dilution on antibody responses was not observed in groups immunized with mosaic nps regardless of antigen valency (Fig. 2a, right). Dose de-escalation of NC99 RBD in the mosaic nps did not diminish response as observed in admixed np-immunized groups. Although total NC99 antigen content decreases as valency increases, the number of particles containing NC99 RBDs should be relatively constant in the mosaic np preparations. In contrast, the number of particles possessing NC99 RBDs decreases proportionally to the total antigen content in admixed preparations. The dilution effect on antibody response in mice immunized with admixed nps was also observed for other H1N1 strains (A/Brisbane/59/2007 (BR07) and CA09) (Supplementary Fig. 2a,b). We next assessed serum microneutralization titers generated in mice immunized with the Northern hemisphere formulation of either 2006–7, 2007–8, or 2008–9 trivalent inactivated influenza vaccine (TIV) and compared the levels to those of mice immunized with mosaic RBD-np (8-valent). All three TIV groups were able to elicit similar levels of neutralizing activity against A/Christchurch/1/2003 (CC03) virus as the antigenic properties of this virus are similar to vaccine viruses. In contrast, responses to antigenically dissimilar viruses were much lower compared to mice vaccinated with mosaic RBD-np (Supplementary Fig. 2c). These results indicate that the mosaic RBD-np is capable of eliciting qualitatively different responses with greater breadth than conventional influenza vaccines. To ask if prior influenza exposure would impact on the subsequent vaccine-induced response, mice were immunized with two distinct commercial vaccines derived from different seasons to establish background influenza immunity, and subsequently immunized with mosaic RBD-np (8-valent). Prior to the mosaic RBD-np immunization, serum neutralizing antibody was detectable, but had low magnitude activity against antigenically mismatched viruses (e.g. A/Wilson-Smith/1933 (WS33), A/Kawasaki/6/1986 (KW86) or A/Beijing/262/1995 (BJ95)). Neutralization activity against those viruses was substantially boosted after the mosaic RBD-np immunization, suggesting that the mosaic RBD-np was capable of eliciting broad neutralizing antibody responses in the presence of pre-existing influenza immunity (Fig. 2b). Importantly, the enhancement was observed in differentially conditioned mice primed with different combinations of commercial vaccines, indicating that the individualized memory B cell repertoires established by prior influenza exposures did not adversely impact on the subsequent response induced by the mosaic RBD-np. Collectively, the mosaic antigen array avoids antigenic competition, induces comparable immune responses with much lower antigen load, and also effectively broadens the breadth of neutralizing antibody responses in mice with preexisting immunity.
Fig. 2. Immune induction by HA RBD-nps displaying homogeneous- and mosaic antigen arrays.
(a) Antibody responses to homologous NC99 virus. Serum HAI and pseudotype neutralization (PN) IC50 titers are measured after 2 immunizations of indicated valences of either admixed or mosaic RBD-nps with Sigma adjuvant system. For HAI, cumulative data of 3 independent experiments (n = 15) except for 8-valent admix group (2 independent experiments, n = 10) are shown. For PN, shown are representative data from one experiment (n = 5). Experiments are independently performed 3 times (2 times for 8-valent admix group) with similar results. Lines indicate means. Statistical analyses are done with one-way ANOVA with Tukey’s multiple comparisons post-hoc test. Non-statistical significance between groups is not shown for clarity. (b) Microneutralization (MN) IC50 titers against 5 different viruses in mice after priming with TIV and after boosting with mosaic RBD-np. Immune sera were collected at week 5 (prime) and 12 (boost). MN IC50 titers against each virus is plotted on five axes and connected by a line (n = 10). MN IC50 across 5 H1N1 viruses is shown as GMT ± geometric s.d. Statistical analyses are done with two-tailed paired t-test. Data are from one experiment. (c) Schematic representation of different immunization modalities. (d) Serum HAI titers to NC99 virus after 2 (NC99, admix and mosaic groups) or 4 (sequential) immunizations with adjuvant. Shown are representative data from one experiment (n = 10). Experiments are independently performed two times with similar results. Statistical analysis is done with one-way ANOVA with Tukey’s multiple comparison post-hoc test. (e) MN IC50 titers against 5 H1N1 viruses. MN IC50 titers are plotted as b. Shown are representative data from one experiment (n = 10). Experiments are independently performed two times with similar results. Statistical analysis is done with one-way ANOVA with Tukey’s multiple comparisons post-hoc test.
Differences in immune induction by immunization regimens
We next compared immune responses elicited by different immunization regimens, namely, repeated immunizations (2 immunizations of NC99 RBD-np alone, admix of 8 RBD-nps, or mosaic nps made of 8 different RBDs) or sequential immunizations (4 immunizations with 2 unique homotypic RBD-nps per immunization (total of 8 different RBD-nps)) (Fig. 2c). We determined HAI antibody titers to NC99 virus after immunization and again observed that the group immunized with admixed RBD-nps had significantly reduced neutralization activity against NC99 relative to mosaic RBD-np and homogeneous NC99 RBD-np groups, which had comparable titers to each other (Fig. 2d). Microneutralization using a diverse set of H1N1 viruses, CA09, BR07, CC03, KW86 and WS33 revealed superior serum neutralization breadth and potency in animals immunized with mosaic np compared to responses in other groups (Fig. 2e). Neutralization geometric mean titers across 5 viruses were significantly higher in mosaic np-immunized group than other groups (Fig. 2e, right), indicating that the mosaic RBD-np elicited greater neutralizing antibody breadth than groups immunized with regimens of repeated admixed nps or sequential immunizations of homotypic RBD-nps.
Qualitative differences in HA-specific B cells between homogeneous- and heterogeneous RBD-np immunizations
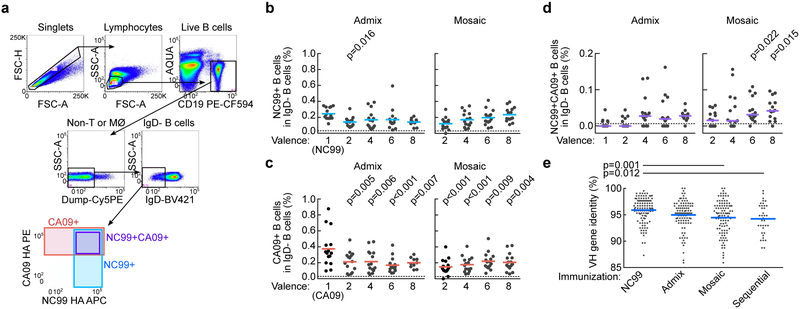
We next asked whether there were qualitative differences in immune responses by assessing cross-reactivity of HA-specific B cells in immunized mice. Peripheral blood cells were collected after three immunizations with either NC99 alone, admix or mosaic RBD-np and probed with two distinct H1N1 HAs (NC99 and CA09) to identify HA-specific B cells by flow cytometry9 (Fig. 3a). Frequency of IgD– B cells reactive to NC99 HA in NC99 RBD-np immunized mice were the highest (0.245 ± 0.074%) among all groups, but the frequency was not statistically different from that in 8-valent admix or mosaic RBD-np immunized mice (0.140 ± 0.049% or 0.235 ± 0.087%, respectively)(Fig. 3b). We observed lower frequencies of B cells reactive to CA09 HA in all the admix- or mosaic RBD-np groups (ranging from 0.151% to 0.221%) compared to CA09 RBD-np immunized group (0.374 ± 0.226%)(Fig. 3c). Interestingly, B cells stained with both NC99 and CA09 HAs were rare but detected in the majority of mice immunized with nps with 2 or more RBDs delivered admixed or as mosaic nps (Fig. 3d). However, these cells were detected more consistently and frequently in mice receiving the mosaic nps with RBD valency of 6 or 8 compared to other immunization groups (Fig. 3d). This suggests that increased valency promotes induction of B cells that bind conserved regions on adjacent heterologous antigens (i.e., cross-reactivity to multiple HAs) leading to broad antibody responses. To determine how antigen recognition patterns impact maturation of HA-specific B cells we next characterized the variable region of immunoglobulin (Ig) heavy chain (VH) gene of sorted NC99 HA+ B cells from different immunization groups. The Ig genes were recovered by single-cell RT-PCR37, sequenced and analyzed. As expected, VH sequences from mice receiving NC99 RBD-np had diverged from germline sequences by 4.1 ± 0.2% (n = 108) as a result of SHM. Mean mutation rate in groups immunized with admixed RBD-np was slightly higher at 5.0 ± 0.3% (n = 98). Mosaic RBD-nps and sequential immunization groups yielded the highest mutation rates of 5.5% ± 0.3% (n = 101) and 5.8 ± 0.5% (n = 36), respectively. The differences between groups receiving NC99 RBD-np and mosaic RBD-np or sequential RBD-np were statistically significant (Fig. 3e), although the frequency of HA-specific B cells and serum antibody responses in the sequential immunization group were relatively low. These data suggest HA-specific B cells from mosaic np and sequential immunization groups acquired more SHMs compared to those from repeated homotypic np immunizations.
Fig. 3. Induction of cross-reactive HA-specific B cells by RBD-nps.
(a) Gating strategy for identifying HA-specific B cells by flow cytometry. Anti-CD3, CD8, CD14, and F4/80 were combined and used to separate T cells, monocytes, and macrophages (MØ). A disparate pair of HAs (NC99 and CA09) was used to define cross-reactivity of HA-specific B cells. Frequencies of IgD− B cells specific to NC99 HA (b), CA09 (c), or cross-reactive to NC99 and CA09 (d) in PBMCs of mice immunized with different RBD-nps (n = 15, cumulative data of 3 independent experiments except for 8-valent admix group (n = 10, cumulative of 2 independent experiments). Statistical analyses are done with one-way ANOVA with Tukey’s multiple comparisons post-hoc test by using valence = 1 as a comparator. (e) Mutation rate in IGHV genes of individually sorted NC99 HA+ B cells isolated from immunized mice (n = 3, NC99, admix and mosaic; n = 2, sequential). Total productive IGHV genes obtained are 108, 98, 101 and 36 for NC99, admix, mosaic and sequential RBD-np groups, respectively. Statistical analysis is done with one-way ANOVA with Tukey’s multiple comparisons post-hoc test.
Isolation and characterization of broadly neutralizing H1N1 antibody 441D6
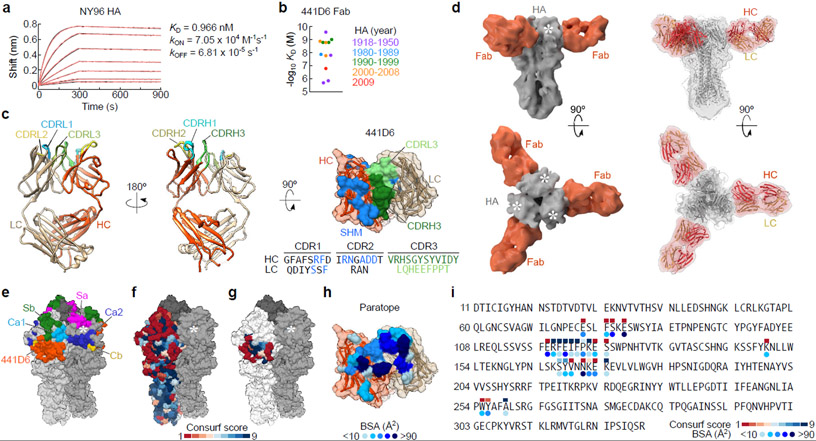
As a proof of concept, we sought to isolate mAbs to determine whether examples of cross-reactive B cells induced by mosaic np contributing to broad serum neutralization could be detected. We sorted B cells co-stained by NC99 and CA09 HAs and cloned Ig genes recovered from individual cells into expression vectors containing constant regions of corresponding Ig heavy and light chains. Out of 17 paired Ig genes, we identified 4 recombinant mAbs that bound to NC99 HA as well as other H1N1 HAs. Three out of four mAbs also bound to H5N1 (A/Indonesia/05/2005) HA (Supplementary Fig. 3a). The remaining mAb termed 441D6 is specific to H1N1 yet it had the highest neutralization potency of the four antibodies against homologous NC99 and a heterologous H1N1 virus BJ95 (Supplementary Fig. 3b). None of the mAbs had detectable HAI activity. We next assessed binding kinetics of the antigen-binding fragment (Fab) of 441D6 to various H1N1 HAs by biolayer interferometry (Fig. 4a,b, Supplementary Fig. 4). Affinities of 441D6 Fab to HAs from a diverse set of 12 H1N1 viruses varied from sub-nanomolar to micromolar interactions with no obvious association between apparent KD and collection year of viruses (Fig. 4b). To better understand how 441D6 targets diverse HAs we solved the structure of unliganded 441D6 Fab by X-ray crystallography to 2.0 Å and determined the structure of the HA–Fab complex by cryo-EM at a resolution of 8.0 Å (Fig. 4c,d, Supplementary Table 1 and Supplementary Fig. 5). Stoichiometry of HA-binding by 441D6 was three Fabs per trimer, resulting in a propeller-like topology when viewed along the longitudinal axis of HA trimer (Fig. 4d). Atypical for HA head-directed antibodies, 441D6 Fab approaches HA nearly perpendicular to the HA trimer vertical axis (79º relative to HA longitudinal axis) (Fig. 4d). Coordinates corresponding to A/New York/653/1996 (H1N1) HA and 441D6 Fab fit well into the cryo-EM density map with only 260 Fab atoms (8% total, 3% backbone), and 994 HA atoms (9% total, 2% backbone) outside the cryo-EM density (Fig. 4d). The epitope of 441D6 on HA is located on the head proximal and opposite to the sialic acid-binding pocket (i.e., RBS), and distinct from the known classical antigenic sites of H1N1 HA38, 39 (Fig. 4e–g). The HA footprint on the 441D6 Fab clusters mostly on the complementarity determining region (CDR) loops and overlaps with residues that had undergone SHM (Fig. 4c,h). Despite the high variability of the HA head region even within subtype, the epitope is centered on a relatively conserved patch shared by H1N1 subtype viruses (Fig. 4f,g,i). In order to determine whether the 441D6 epitope on mosaic RBD-np is accessible for antibody and/or B cell recognition we carried out EM to visualize the nanoparticle–Fab complex. Unlike the mosaic RBD-np alone which yields homogeneous spherical particles with protruding short spikes with an average radius of 13.0 nm, the mosaic RBD-np in complex with 441D6 Fab resulted in larger particles with conformationally heterogeneous densities surrounding the mosaic RBD-np structure (average radius of 15.5 nm)(Supplementary Fig. 6), indicating that the 441D6 Fab was able to recognize the RBD molecules displayed on the mosaic RBD-np. These results demonstrate that the 441D6-binding sites on the mosaic RBD-np are readily accessible for immune recognition.
Fig. 4. Biophysical and structural characterization of broadly-neutralizing antibody 441D6.
(a) Binding kinetics of 441D6 Fab to NY96 HA determined by biolayer interferometry (BLI). Measured sensorgram and calculated curve fit (1:1 binding model) are shown in black and red, respectively. Experiments were independently performed 2 times with similar results. (b) Summary of binding affinities of 441D6 Fab to a diverse set of 12 H1N1 HAs. Each HA–Fab interaction was plotted and color-coded based on year of virus identification. Binding kinetics details are found in Supplementary Fig. 4. (c) 2.0 Å crystal structure of unliganded 441D6 Fab. Somatic mutations of 441D6 Fab (right). Residues that underwent SHM are colored in blue. Amino acid sequence of CDRH1–3 and CDRL1–3 loops are shown with SHM residues in blue. (d) Cryo-EM structure of NY96 HA trimer in complex with 441D6 Fab. Side view along the longitudinal axis of an HA trimer (top) and top view looking down on the 3-fold axis of an HA trimer from the membrane distal end (bottom) are shown. Superimposition of NY96 HA (homology model, grey) and 441D6 Fab (orange-red) coordinates into the cryo-EM density map (right). White asterisks indicate sialic acid-binding pocket on each HA protomer. (e) Updated antigenic sites of H1N1 HA. Known antigenic sites Sa, Sb, Ca1, Ca2, and Cb are shown along with the site recognized by 441D6. (f) Surface conservation of 1,368 non-overlapping H1N1 HAs. Conservation scores were calculated by the ConSurf server (http://consurf.tau.ac.il) and colored on one NY96 HA protomer with dark blue equating to highest conservation. Predicted 441D6 epitope (colored) mapped on NY96 HA (g) and paratope of 441D6 (h). Each paratope residue is colored according to buried surface area (BSA) contribution. (i) Amino acid sequence of HA1 subunit of NY96 HA. 441D6 epitope residues are indicated by conservation score and BSA.
Neutralization profile of 441D6 and immune sera elicited by mosaic RBD-np
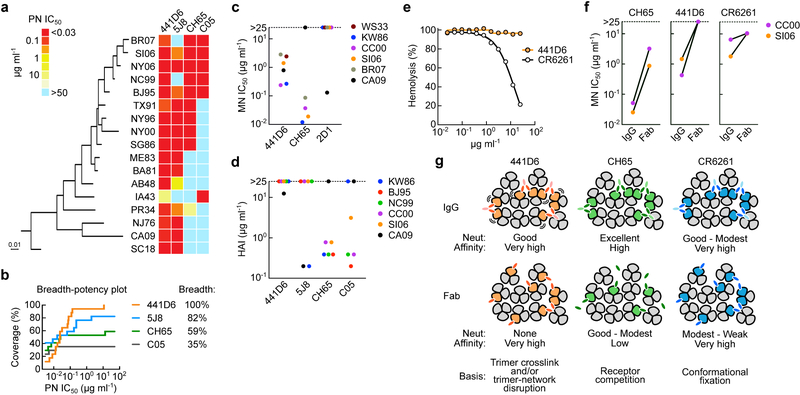
To determine the breadth of neutralization of 441D6 we employed an extended panel of 17 H1N1 pseudoviruses (see a full list in Online Methods). 441D6 neutralized all the H1N1 viruses tested in the assay that represent ~90 years of antigenic evolution (Fig. 5a), and all except A/Iowa/1943 (IA43) were potently neutralized. This neutralization breadth of 441D6 is noteworthy as it exceeds the breadth of well-characterized bnAbs directed to the RBS and vicinity (5J8, CH65 and C05)13, 15, 40, and 441D6 requires ~0.1 µg ml−1 to achieve a half maximal neutralizing activity against most viruses (Fig. 5a,b). Importantly, 441D6 also neutralized viruses in microneutralization assays as demonstrated by using 6 authentic influenza viruses (WS33, KW86, CC03, A/Solomon Islands/3/2006 (SI06), BR07, and CA09) with IC50 values between 0.2 and 2.8 µg ml−1 (Fig. 5c) and provided protection against lethal H1N1 infections (both CA09 and A/Puerto Rico/8/1934) in murine models when given therapeutically at 24 hours post infection (Supplementary Fig. 7a,b). To dissect the basis for virus neutralization of 441D6, we tested its HAI activity against multiple different H1N1 viruses. As we predicted from HA–Fab structure, there was no HAI activity against any virus with the exception of CA09 for which we detected weak activity (Fig. 5d). Additionally, we assessed the pH-dependent hemolysis inhibition activity41 of 441D6, and found no inhibitory activity (Fig. 5e). These results suggest that 441D6 neutralizes viruses without competing with receptor-binding or inhibiting the viral fusion machinery. We next tested virus neutralization of 441D6 in its Fab form, and observed that 441D6 Fab completely lost virus neutralizing activity compared to its IgG form (Fig. 5f). Therefore, we propose that the basis for 441D6 virus neutralization is virus aggregation which effectively reduces inoculum size. Alternatively, cross-linking neighboring HA trimers with two Fab arms of IgG, or binding HA perpendicular to its longitudinal axis could disrupt the geometric arrangement of HA trimers on the virion, and cause interfere with the formation of fusion pores on the host endosomal membrane (Fig. 5g).
Fig. 5. Breadth, potency, and potential mechanism of neutralization of 441D6.
(a) Neutralization profile of bnAbs determined by a panel of 17 H1N1 pseudoviruses. Potency was displayed as a heatmap along with associated HA phylogenetic tree (maximum-likelihood method with full-length HA sequences). Antibodies 5J8 and CH65 (pan-H1N1 bnAbs targeting RBS) and C05 (cross-subtypic bnAb targeting RBS) were used as controls. Experiments are independently performed two times with similar results. (b) Neutralization breadth-potency plot generated from data displayed in a. Breadth denotes the neutralization coverage of a panel of 17 H1N1 strains representing > 90 years of antigenic evolution. MN IC50 (c) and HAI (d) of 441D6 against 6 different H1N1 viruses. Experiments are independently performed two times with similar results. (e) Hemolysis inhibitory activity of 441D6. Hemolysis inhibition assay was performed by using PR34 virus. The hemolysis is calculated with a formula: hemolysis (%) = {[Absexperimental - AbsRBC-only] / [Absno antibody - AbsRBC-only]} × 100. Experiments are independently performed two times. (f) MN activity of 441D6 in its IgG and Fab forms. MN IC50 concentrations of CH65, 441D6, and CR6261 were determined by MN assays with 2 H1N1 viruses. Experiments are independently performed two times with similar result. (g) Proposed model for neutralization by 441D6, CH65, and CR6261.
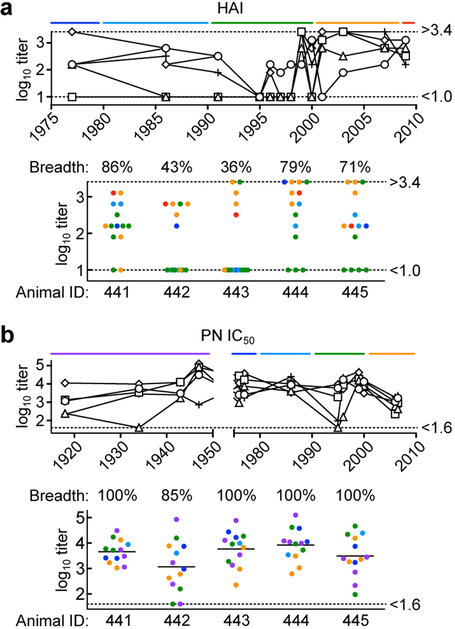
We next asked if immunization with the mosaic RBD-np elicits cross-neutralizing antibody responses to diverse H1N1 viruses based on 441D6-like antibodies. The breadth of serum HAI activity was measured using a panel of 14 H1N1 viruses representing ~30 years of antigenic drift. We observed variation in HAI titers to different viruses, and breadth of response ranged between 36–86% in a group of 5 animals (Fig. 6a). Strikingly, when immune sera were tested for neutralizing activity using a panel of 13 H1N1 pseudoviruses (~90 years of coverage) we observed broad neutralizing activity against 85–100% (97.0 ± 6.7%, n = 5) strains with geometric mean reciprocal IC50 titers of 4,643 across all viruses (Fig. 6b). The neutralization profile of immune sera resembles the breadth of 441D6, and is independent of HAI activity. These data suggest co-displaying heterotypic HA RBDs on nanoparticles can elicit potent and broadly neutralizing antibodies that are not necessarily focused on the hypervariable sequences around the sialic acid-binding pocket that tend to be immunodominant and strain-specific.
Fig. 6. Neutralization activity in immune sera elicited by mosaic RBD-np.
(a) Serum HAI titers against a panel of 14 H1N1 viruses (> 30 years of coverage). HAI titers of immune sera elicited by mosaic RBD-np (8-valent) are plotted against the year of virus isolation (top) or plotted individually with breadth (bottom). (b) Serum PN IC50 titers against a panel of 13 H1N1 pseudoviruses (~90 years of coverage). Data are similarly represented as panel c. Shown are representative data from one experiment (n = 5). Experiments are independently performed two times with similar results.
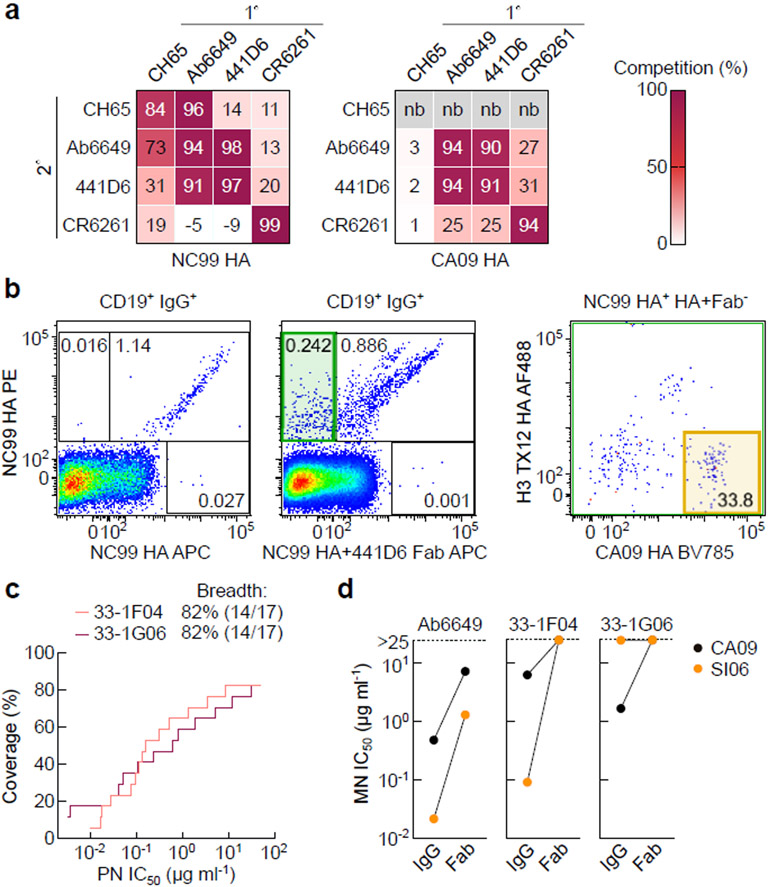
We next identified the two residues on HA that were critically involved in 441D6-binding and made an HA variant possessing two point mutations that specifically knock out binding of 441D6 (Supplementary Fig. 8a). By using this HA variant, termed 441D6KO, we detected a subtle but statistically significant difference between binding of serum antibody to wild-type HA and the 441D6KO HA in mice immunized with the mosaic RBD-np and with the admix RBD-np, while the difference was not observed in mice immunized with NC99 RBD-np or sequential RBD-np (Supplementary Fig. 8b). The results align well with the cross-reactive B cell data and suggest that the mosaic RBD-np and the admix RBD-np elicit detectable serum antibody responses specific to the site recognized by 441D6. Further, to see if humans are capable of generating 441D6-like antibodies through infection and/or vaccination, we tested 26 serum samples collected from healthy volunteers. Significant differences were detected between antibody binding to wild-type HA and the 441D6KO HA (Supplementary Fig. 8c), suggesting that serum antibodies directed to the 441D6-binding site can also be induced in humans through natural exposure to influenza virus and/or conventional vaccines. Indeed, recently, Raymond et al. have identified a family of human antibodies42 (e.g. Ab6649) targeting the overlapping site of vulnerability on HA that we have determined as the 441D6 epitope (Fig. 4e and Fig. 7a). This lineage of antibodies broadly neutralizes H1N1 virus isolates from 1977 to 2009, suggesting that humans are capable of targeting the 441D6 epitope and can generate bnAb responses targeting this site. To directly prove the human anamnestic B cell repertoire to the site we sorted human PBMCs with NC99 HA probe labeled with one fluorochrome and NC99 HA probe in complex with 441D6 Fab labeled with another fluorochrome to selectively enrich B cells with specificity similar to 441D6 (Fig. 7b). More than one third of B cells binding to NC99 HA but not to NC99 HA–Fab also bound to CA09 HA. By this targeted sorting strategy, we identified two lineages of antibodies and confirmed that the representative antibody from each lineage was capable of neutralizing 14 out of 17 (82%) H1N1 subtype viruses spanning >90 years of antigenic evolution (Fig. 7c). Interestingly, the neutralization activity was severely impaired when these antibodies were used as Fabs, suggesting that these antibodies have the similar neutralization mechanism as 441D6 (Fig. 7d). Taken together, both this and previously published work42 support the existence of antibodies with 441D6-like specificity exist in the human B cell repertoire, and based on our results in mice we foresee these B cells with this specificity would represent cross-reactive lineages preferentially induced by the mosaic RBD-np.
Fig. 7. Isolation and characterization of 441D6-like antibodies from human PBMCs.
(a) Antibody cross-competition profile. Antibody binding to NC99 HA (left) and CA09 HA (right) was measured by BLI in the presence or absence of competing (1º) antibody. Antibody competition was calculated by a formula: competition (%) = 100 - {[Bmaxexperimental / Bmaxnon-competed] × 100}. Experiments are independently performed two times with similar results. (b) HA staining of human B cells. The sample was collected with informed consent of volunteer, and approval was obtained under protocol number VRC 310 (Clinicaltrials.gov NCT01086657). CD19+IgG+ B cells were probed with NC99 HA with 2 different fluorochromes (left) or probed with NC99 HA PE and NC99 HA–441D6 Fab complex APC (middle). CD19+IgG+ B cells stained with NC99 HA but not stained with HA–Fab complex (middle, green gate) were collected by single-cell sorting. Reactivity of NC99 HA+ HA–Fab complex− cells to CA09 HA BV785 and H3 A/Texas/50/2012 (TX12) HA AF488 was shown (right). Experiments are independently performed two times with similar results. (c) Neutralization breadth-potency plot of two representative 441D6-like human antibodies, 33–1F04 and 33–1G06. Neutralization breadth and potency were determined by PN assays using the same 17 virus panel as shown in Fig. 5. Experiments are independently performed two times with similar results.
Model of immune induction by heterotypic mosaic antigen arrays
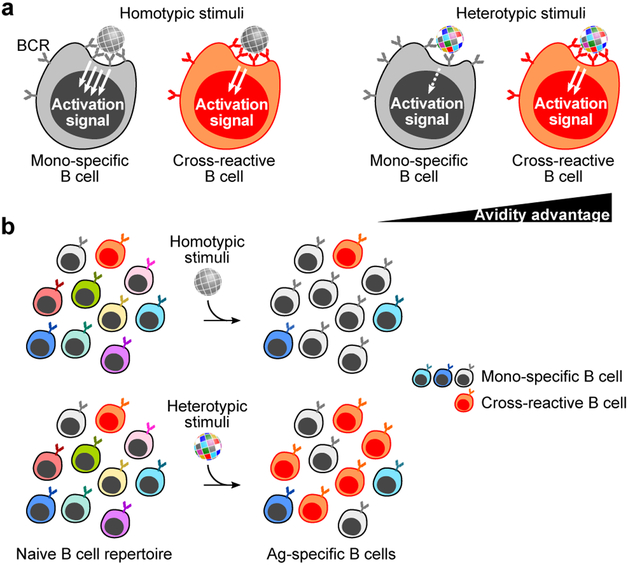
When the immune system encounters particulate substances that are not comprised of homotypic antigens the outcome is less certain since single-particle heterotypic arrays of the same protein do not exist in nature. In the scenario, B cells with cross-reactive BCRs would be favored for bivalent binding to neighboring heterologous antigen pairs, providing avidity advantage over strain-specific BCR interactions which are incapable of facilitating bivalent binding (Fig. 8a). Even though the initial affinity may be lower than for strain-specific BCRs, there would be less competition with high affinity BCRs, allowing the expansion of broadly cross-reactive B cells (Fig. 8b).
Fig. 8. Model of immunosubversion approach with mosaic antigen array.
(a) B cell activation by homotypic- or heterotypic antigen arrays. B cells possessing BCR specific to ‘gray’ antigen or cross-reactive to multiple antigenic variants are colored in gray or orange, respectively. Situations with particulate stimuli (e.g., virus, vaccine, etc.) made of homotypic (left) or heterotypic (right) antigens are shown. (b) Predicted immune outcome induced by homotypic- (top) and heterotypic (bottom) antigen arrays.
Discussion
The ultimate goal of universal influenza immunity is to achieve broad and durable protection against future drifted and pandemic strains of influenza viruses by active immunization43. However, antigenic variability of influenza viruses poses the greatest challenge for achieving effective vaccine-induced immunity beyond a single influenza season. Finding ways to induce bnAbs could provide an avenue for developing broadly protective vaccines that would be useful for multiple seasons. We envision three major approaches to achieve these aims. One would be to target B cell recognition to conserved antigenic supersites like the RBS or the Trp21HA2 groove in the HA stem (immunofocusing approach). Another approach would be to accumulate breadth by eliciting an ensemble of neutralizing antibodies to multiple sites on multiple strains including seasonal H1N1, H3N2 and influenza B viruses, and supplemented by including antigens from exotic HA subtypes, such as H5 and H7 (additive approach). This approach is inherently limited because it is dependent on and influenced by prior antigenic exposures, and tends to protect looking backwards rather than forward. Immunological imprinting from prior influenza infection and vaccination results in B cell repertoires that prevent recognition or induction of antibody specificities that would be more cross-reactive with newly emerging strains. Immunodominance of strain-specific regions of HA is a key obstacle to achieving broad cross-reactivity in the setting of an antigenically diverse and evolving target, and is addressed by what we have termed an immunosubversion approach. Allowing lower affinity cross-reactive B cells from the naïve repertoire to emerge by subverting the existing strain-specific responses could potentially provide protection against future drifted seasonal strains as well as pandemic strains, and may eliminate the need for annual reformulation and manufacturing that create a significant bottleneck in the current influenza vaccine approach44. In the antigen-naïve population (i.e. infants), the mosaic RBD-np would preferentially stimulate cross-reactive B cells by providing an avidity advantage when the antigen-binding sites of the BCR can recognize more than one HA specificity. Importantly, antigen-experienced humans appear to mount memory B cells specific to the 441D6 epitope through natural influenza exposures and/or vaccinations. These preexisting memory B cells might be selectively stimulated and differentiated into antibody producing cells upon receiving the mosaic RBD-np.
There are a few other vaccine approaches that might elicit cross-reactive responses including chimeric HAs which utilize engineered HA with an exotic HA head paired with an invariant stem to promote elicitation of stem-focused immune response in individuals with preexisting immune repertoires27, 45. Another approach is the COBRA HA which utilizes computationally optimized consensus HA sequences to help broaden the B cell reactivity46, 47. Both the chimeric and COBRA HA approaches have shown promise in preclinical models, however, there is no constraint on how the immune system responds to these immunogens, i.e. providing an equal opportunity to all antigen-specific B cells regardless of specificity. One unique feature of our heterotypic mosaic antigen array is that it gives an avidity advantage to cross-reactive B cells over strain-specific B cells, hence adds an immunological constraint for the immune system. This avidity advantage may compensate for the inherently low-affinity of cross-reactive BCRs and allow cross-reactive B cells to emerge.
Conventional influenza vaccines chase circulating viruses from year to year. This is effective as long as the antigenicity of vaccine strains match the circulating viruses because the current vaccines provide protection with a relatively narrow range. Using the “additive approach” by either serial or combined vaccination with multiple antigenic types might accumulate responses with non-overlapping specificities. Successful examples of this strategy include pneumococcal conjugate vaccines with 13 different antigens, or human papillomavirus vaccines with up to 9 different antigens in a single vaccine. Although these multivalent vaccines are highly efficacious in humans, the admixture could result in antigenic competition and eventually might lead to loss of efficacy for one or more components in the influenza vaccine, and the accumulation of breadth by serial immunization with conventional vaccine has not been effective for generating broad immunity48. Importantly, for influenza, there is substantial antigenic variation even within subtypes, so even a subtype-specific vaccine will need to induce considerable breadth to avoid escape through antigenic drift or reassortment from zoonotic reservoirs which leads to new viruses with pandemic potential. In contrast, the “Immunofocusing approach” employs engineered immunogens presenting a domain or a subdomain of a protein, attempting to make only the selected antigenic sites accessible to B cells so that a response is made to a surface that is immunologically subdominant or silent in its native configuration. An example of immunofocusing for influenza would be the “head-less” HA that focuses antibody responses on the conserved stem supersite and has shown some promise24, 25. Our “immunosubversion approach” described herein adds an immunological constraint –single-particle antigenic variability– to avoid dominant strain-specific antibody responses and create an opportunity for cross-reactive antibody responses to conserved sites with shared surfaces between diverse HA antigens. Together, we have shown that the mosaic antigen array of HAs from diverse influenza strains allows antibody responses to unique sites that are generally not targeted otherwise. This provides a proof-of-concept for vaccine-induced immunosubversion with a heterotypic mosaic antigen array to give a selective avidity advantage for cross-reactive B cells.
The viral site of vulnerability defined by 441D6 is an attractive pan-subtype vaccine target. Determining structural and molecular mechanism for virus neutralization by 441D6 informs immunogen designs that could target this site of vulnerability. Although precise modes of virus neutralization are different, other pan-subtypic sites of vulnerability have been discovered through the isolation and characterization of mAbs including F005–126 for H3N2ref 21, FLD194 for H5N1ref 23, and CR8033, CR8071, 46B8, and C12G6 for influenza B HAs17, 49, 50. These pan-subtypic mAbs except for C12G6 which targets RBS, target the vestigial esterase domain of HA that partially overlaps with the 441D6 epitope. It is likely that presentation of heterotypic mosaic antigen arrays would preferentially stimulate B cells that tolerate antigenic variability for other HA subtypes and influenza B lineages, and allow B cells targeting conserved antigenic sites to emerge.
We propose that the juxtaposition of heterotypic influenza HA antigens in a mosaic array will represent a novel immunological pattern, that preferentially engages B cells with cross-reactive potential and changes the hierarchy of immunodominance. This immunosubversion approach may be applicable to other pathogens with extreme antigenic variability, especially in cases where strain-specific B cells dominate the response and prevent the emergence of cross-reactive B cells through competition or other forms of immunodominance. Advances in protein engineering and manufacturing technologies make this approach achievable and providing a new tool to combat pathogens that exploit genetic plasticity and antigenic variation as a primary immunoevasion mechanism.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
Online Methods
Study design.
The aim of this study was to elucidate and characterize the immune response elicited by the non-natural mosaic arrays of antigenically heterotypic influenza antigens in mice. The experimental design involved in vivo and in vitro experiments, including design, purification, biochemical, biophysical, and antigenic characterizations of nanoparticle immunogens, mathematical simulation, ELISA, virus neutralization, FACS, single-cell reverse transcription PCR, immunogenetics analysis, recombinant antibody production, biolayer interferometry, X-ray crystallography, and cryo-EM. The animal experiments were not randomized. The investigators were not blinded to the allocation during experiments and analyses unless otherwise indicated. Experimental replication is indicated in the figure legends. Animal experiments were carried out in accordance with all federal regulations and NIH guidelines and approved by the Institutional Animal Care and Use Committee. All human serum samples for this study were collected with informed consent of volunteers, and approval for this study was obtained under protocol number VRC 700 (Clinicaltrials.gov NCT01262079)51. All volunteers were at least 6 years of age, healthy, and ranged in age from 7 to 93 years. Human PBMC sample was obtained from VRC 310, a single-site, phase 1, open-label, randomized clinical trial conducted at the National Institutes of Health (NIH) Clinical Center by the NIAID VRC (Clinicaltrial.gov NCT01086657)52. These studies were approved by the NIAID Intramural Institutional Review Board. US Department of Health and Human Services guidelines for conducting clinical research were followed.
Gene synthesis and vector construction.
All genes used in the study were human codon optimized (GenScript). The gene encoding ferritin (Helicobacter pylori-bullfrog hybrid) was designed and characterized previously31. Briefly, the ferritin was composed of residues 2–9 of bullfrog (Rana catesbeiana) ferritin lower subunit (UniProt: P07797 with a point mutation at residue 8 (N8Q) to abolish a potential N-glycosylation site) to H. pylori nonheme ferritin (UniProt: Q9ZLI1, residues 3–167) with mutations at residues 7 (I7E) and 19 (N19Q) to maintain the salt bridge found in bullfrog ferritin and to abolish a potential N-glycosylation site, respectively. All the HA RBD sequences used in the study correspond to residues 56–264 (H3 numbering) and possess a mutation which knocks out sialic acid-binding property (Y98F)9 and are derived from various H1N1 strains listed below. To construct RBD-np, a modified bovine prolactin signal sequence31 was attached upstream of the RBD and fused to the ferritin with a SG linker. All genes were then cloned into the CMV/R (VRC 8400) or CMV/R 8κb (VRC 8405) mammalian expression vector for protein production.
Biosynthesis of recombinant proteins and purification.
The expression vectors were transiently transfected (1.0 mg l−1 of 2.5 × 106 cells) into Expi293F cells (Thermo Fisher Scientific) using ExpiFectamine 293 transfection reagents (Thermo Fisher Scientific). For coassembly, cells were cotransfected with equal amount of multiple RBD-np plasmids (total of 1.0 mg l−1 of 2.5 × 106 cells). Four days after transfection, culture supernatants were harvested and cleared. The RBD-np were purified by ion-exchange chromatography using HiTrap Q HP columns at pH 8.0 (GE Healthcare) followed by size exclusion chromatography with a Superose 6 XK 16/70 PG column (GE Healthcare) in PBS. Recombinant HA ectodomain trimers constructed with T4 fibritin foldon and hexa-histidine tag were produced and purified as described elsewhere32, 53. Recombinant HA probes (Y98F) for FACS sorting were made as previously described9, 54.
Immunization.
Eight- to 10-week old female BALB/cAnNTac mice (Jackson Laboratories) were immunized (n = 5–10) intramuscularly with 2 μg of purified np in 100 μl of 50% (v/v) mixture of SAS adjuvant (Sigma) in PBS. For the experiments shown in Fig. 2a, mice were immunized at weeks 0 and 4. For the experiments shown in Fig. 2b, mice were immunized with 1.5 µg of TIV from indicated season at weeks 0 and 3 to generate preexisting influenza immunity, and then immunized with 2 µg of mosaic RBD-np (8-valent) formulated with SAS and given at weeks 6 and 10. For the experiments shown in Fig. 2 d and e, and Fig. 3, mice were immunized at weeks 0, 4 and 12 (NC99, admix, and mosaic) or weeks 0, 4, 8 and 12 (sequential). For the experiments shown in Fig. 6 a and b, mice were immunized at weeks 0, 4 and 8. Sera were collected prior to the first dose and periodically 2 to 3 weeks after each immunization.
H1N1 viruses and recombinant HAs.
Influenza virus strains used for microneutralization and HAI assays are: A/USSR/90/1977; A/Kawasaki/6/1986; A/Chile/4795/2000; A/Texas/36/1991; A/Shanghai/8/1996; A/Auckland/65/2001; A/Ostrava/801/1998; A/Johannesburg/159/1997; A/Beijing/262/1995; A/Brisbane/59/2007; A/Christchurch/1/2003; A/New Caledonia/20/1999; A/California/07/2009; A/Singapore/6/1986; and A/WSN/1933 (made by reverse genetics). Influenza virus strains used for pseudotype neutralization assays are: A/South Carolina/1/1918 (GenBank AF117241, SC18); A/Puerto Rico/8/1934 (GenBank CY009444, PR34); A/New Jersey/1976 (GenBank CY021957, NJ76); A/Singapore/6/1986 (GenBank CY020477, SG86); A/Beijing/262/1995 (GenBank CY033614, BJ95); A/New Caledonia/20/1999 (GenBank CY033622, NC99); A/Solomon Islands/3/2006 (GenBank CY031340, SI06); A/Brisbane/59/2007 (GenBank CY058487, BR07); A/California/04/2009 (GenBank GQ117044, CA09); A/Albany/4835/1948 (GenBank CY019947, AB48); A/Baylor/4052/1981 (GenBank CY021029, BA81); A/Memphis/39/1983 (GenBank ABO37988, ME83); A/New York/146/2000 (GenBank AAX56530, NY00); A/New York/653/1996 (GenBank ABF47649, NY96); A/New York/8/2006 (GenBank ABK79959, NY06); A/Texas/36/1991 (GenBank CY033655, TX91); A/Hong Kong/117/1977 (GenBank ABD60944, HK77); A/Iowa/1943 (GenBank ABO38373, IA43); and A/Fort Monmouth/1/1947 (GenBank CY009612, FM47). Recombinant HA proteins were made by fusing HA ectodomain with T4 fibritin trimerization motif (foldon) and purification tag as described previously53 for NY96, FM47, SI06, NC99, BJ95, BR07, SG86, SC18, NY00, CA09, IA43 and PR34.
Hemagglutination inhibition assay.
Seed stocks of the influenza viruses were obtained from the Division of Viral Products, U.S. Food and Drug Administration (FDA) and the viruses were propagated in Madin-Darby canine kidney (MDCK) cells. Immune sera were pretreated with receptor-destroying enzyme (RDE II; Denka Seiken) and HAI assays were performed using 4 hemagglutinating units well−1 and 0.5% turkey red blood cells (Lampire Biological Laboratories). Viruses used in the assays are listed above.
Flow cytometry.
Mouse peripheral white blood cells were prepared from heparinized whole blood by lysing red blood cells with ACK lysing buffer (Thermo Fisher Scientific). After hemolysis, white blood cells were washed and stained with fluorochrome-conjugated mAbs to mouse CD3-Cy5PE (BD Bioshiences, 553065, clone 145–2C11, 1:300 dilution), CD4-Cy5PE (BD Biosciences, 553654, clone H129.19, 1:300 dilution), CD8-Cy5PE (BD Biosciences, 553034, clone 53–6.7, 1:300 dilution), F4/80-Cy5PE (Biolegend, 1231111, clone BM8, 1:300 dilution), CD19-PE-CF594 (BD Biosciences, 562329, clone 1D3, 1:300 dilution), IgD-BV421 (BD Biosciences, 744291, clone 11–26c.2a, 1:600 dilution). PE- and APC-labeled HA probes were prepared as described previously9. AQUA dead cell stain was added for live or dead discrimination (Thermo Fisher Scientific). Samples were analyzed on a LSR II (BD Biosciences) and data analysis was done in FlowJo 9 (TreeStar). To single-cell sort, HA-specific B cells were stained as above and HA+ IgD- B cells were sorted as single cells into 96-well plates using a FACS Aria II (BD Biosciences). Addition of two HA probes and index sorting was used to determine the binding of each sorted B cell to HA of two strains simultaneously.
Single-cell immunoglobulin amplification and cloning.
Reverse transcription was performed on sorted cells and multiplexed PCR was used to amplify immunoglobulin heavy and light chain genes as described elsewhere37. PCR products were sequenced by Beckman Coulter or ACGT and analyzed using IMGT55. Heavy and light chain sequences were synthesized and cloned by Genscript into murine IgG2a and kappa expression vectors. To produce antibodies recombinantly, 293F cells were transfected with plasmids encoding Ig heavy and light chain pairs with 293fectin (Thermo Fisher Scientific). Monoclonal antibodies were purified from the cell supernatant using Protein A Sepharose Fast Flow (GE Healthcare).
Virus neutralization assay.
All influenza viruses except for A/WSN/1933 were obtained from Division of Viral Products, FDA. A/WSN/1933 virus was made by reverse genetics with 8 plasmids encoding each influenza A segment under control of biscistronic polI/polII promoters obtained from St. Jude Children’s Research Hospital. All viruses were propagated in MDCK cells in the presence of TPCK-treated trypsin (1 µg ml−1, Sigma) and titrated in MDCK cells as described elsewhere (http://www.who.int/influenza/gisrs_laboratory/2010_12_06_serological_diagnosis_of_influenza_by_microneutralization_assay.pdf). Briefly, DMEM-TPCK (DMEM with 1 µg ml−1 TPCK-trypsin and penicillin/streptomycin) was used to make four-fold serial dilutions of RDE-treated sera or mAbs and to dilute influenza viruses to a final concentration of 100 TCID50 per well. In a 96-well plate, serum or antibody and virus were mixed and incubated 1 h at 37°C prior to adding to substrate cells. Control wells of virus alone (VC) and diluent alone (CC) were included on each plate. Cells were seeded at 1.5 × 104 cells per well 24 h prior to the assays and washed once with PBS before use. Fifty microliters of antibody/virus mixture were then added to wells of pre-washed cells in duplicate and the plates were incubated for 18 h at 37°C and 5% CO2 humidified atmosphere. The cells were then fixed with 80% cold acetone and allowed to air dry. The presence of viral nucleoprotein (NP) was detected by ELISA with biotin-conjugated anti-influenza NP antibodies (MAB8257B and MAB8258B, EMD Millipore), and visualized with HRP-conjugated streptavidin and SureBlue TMB Microwell Peroxidase Substrate (KPL). Absorbance was read at 450 nm (A450) and 650 nm (A650) with the SpectraMax Paradigm microplate reader (Molecular Devices). The A650 was used to subtract plate background. The percent neutralization was calculated by constraining the VC control as 0% and the CC control as 100%, and plotted against serum/antibody concentration. A curve fit was generated by a four-parameter nonlinear fit model in Prism 7 (GraphPad). The half-maximal inhibitory concentration (IC50) was obtained from the curve fit for each serum sample or mAb, respectively. The pseudotype neutralization assay was performed as previously described56, 57. Briefly, pseudovirus was produced by transfection of 293T cells of HA and corresponding NA along with the lentiviral packaging and reporter plasmids. A plasmid encoding human type II transmembrane serine protease TMPRSS2 gene was also cotransfected for efficient proteolytic activation of HA. Forty-eight hours after transfection supernatants were harvested, filtered, and frozen until use.
Electron microscopy.
For negative stain EM analysis of mosaic RBD-np alone and in complex with 441D6 Fab, 50 µg ml−1 of samples were adsorbed to freshly glow-discharged carbon-coated grids, rinsed with distilled water, and stained with 2% ammonium molybdate, 0.7% uranyl formate or phosphotungstic acid (PTA). Images were recorded on a T12 microscope operating at 100 kV (FEI) with a OneView camera (Gatan), using SerialEM for semi-automated data collection58. 2D class averages were made with RELION, rotationally averaged images were created from RELION class averages using EMAN2 image processors. Rotationally averaged images were analyzed with FIJI59 to yield a profile of image intensity as a function of radius.
Immunoprecipitation.
Antibodies (5 µg) specific to NC99 (3u-u)53 and CA09 (2D1)34, 60 or HA stem (C179)19 were incubated with purified nanoparticles (5 µg) at ambient temperature for 30 min. Protein G Dynabeads (Thermo Fisher Scientific) were then added and incubated for another 30 min. Immune complexes were separated by magnetic force, washed and eluted in Laemmli buffer containing reducing agent. Half of each reaction was analyzed by SDS-PAGE followed by InstantBlue staining (Expedeon).
Simulation of particle assembly with multiple antigens.
The ferritin particle was represented as a snub cube graph with 24 vertices and 60 edges. For k different antigens (colors), a graph coloring was generated by assigning one of the k possible colors to each of the 24 vertices in the graph. A total of 106 random graphs were generated for each k. In each of the 106 graphs, the number of homologous edges (where an edge exists between two vertices of the same color) was counted, and the fraction of 106 graphs with 0, 1, 2, …, 60 homologous edges was computed. For a given k, the number of homologous edges was reported as mean ± s.d.
Biolayer interferometry.
All biosensors were hydrated in PBS prior to use. Recombinant HA proteins were immobilized either on SA or HIS1K biosensors through conjugated biotin or hexa-histidine tag, respectively. After briefly dipping in assay buffer (1% BSA in PBS), the biosensors were dipped in a 2-fold dilution series of Fab for 5 min. Biosensors were then dipped in assay buffer to allow Fab to dissociate from HA for 10 min. All assay steps were performed at 30ºC with agitation set at 1,000 rpm in the Octet HTX instrument (fortéBio). Correction to subtract non-specific baseline drift was carried out by subtracting the measurements recorded for a sensor loaded with Epstein-Barr virus gp350 incubated with the Fabs as previously described61. Data analysis and curve fitting were carried out using Octet analysis software (version 9.0). Experimental data were fitted with the binding equations describing a 1:1 interaction. Global analyses of the complete data sets assuming binding was reversible (full dissociation) were carried out using nonlinear least-squares fitting allowing a single set of binding parameters to be obtained simultaneously for all concentrations used in each experiment.
X-ray crystallography.
The 441D6 antigen-binding fragment (Fab) was generated by digestion of the engineered antibody which has a HRV 3C cleavage site introduced in the hinge region, using HRV 3C protease (EMD Millipore) at 10 units mg−1 of Fab for 3–4 h at ambient temperature. The digestion reaction was then passed over a Protein A affinity column (GE Healthcare) to remove the Fc fragments. The flow-through and PBS wash was pooled and concentrated to ~ 8 mg ml−1 and 768 crystallization conditions were screened in 192 well plates (Corning) using a Mosquito crystallization robot. Initial crystal conditions were observed in a condition containing 12% PEG 3350, 0.1 M HEPES pH7.5, 5 mM cobalt chloride, 5 mM calcium chloride, 5 mM magnesium chloride, 5 mM nickel chloride, with the final optimized crystals grown in 12 % PEG 3350, 0.1 M HEPES pH7.5, 5 mM magnesium chloride, 5 mM nickel chloride. Crystals were cryoprotected using 15% 2R, 3R, butanediol and diffraction data were collected at the Advanced Photon Source (Argonne National Laboratory) SER-CAT ID-22 beamline, at a wavelength of 1.00 Å, temperature of 100 K, and processed with HKL2000ref 62. Iterative cycles of model building and refinement were carried out using COOT63 and PHENIX64 software packages respectively, with 5% of the data acting as an R-free cross validation test set. All structural images were generated in UCSF Chimera65.
Cryo-EM.
Samples of complexes of HA NY96 and 441D6 Fab purified by gel-filtration chromatography were applied to glow discharged Quantifoil R2/2 300 mesh grids (Quantifoil) at a concentration of 1 mg ml−1 and plunge frozen in a Vitrobot Mark IV plunge freezer (FEI) under 100% humidity. The sample was blotted for 4 sec after a blot delay of 1 sec, and then the grid was rapidly plunged into liquid ethane to form vitreous ice. Plunge frozen grids were imaged using a Titan Krios electron microscope (FEI) operating at 300 kV at a magnification of 59,000 × using a Falcon 2 direct electron detector (FEI). The detector was operated in movie-mode to collect multiple frames per micrograph at a pixel size of 1.38 Å. Exposures were 1 sec total, dose fractionated into 7 frames. The accumulated dose per micrograph was 34 e-/Å2. A total of 464 micrographs were collected, with defocus values ranging between −2 µm and −7 µm. Micrographs were motion corrected by frame alignment using MotionCorr66. Defocus values were determined using CTFFIND467. 1,800 particles were manually picked, and these particles were used to create 2D class averages with RELION68. The best resolved of these 2D class averages were used as templates for auto-picking with RELION69 from all micrographs, which yielded 225,000 particles. Batches of particles were subjected to 2D classification using EMAN270 to separate out overlapping particles and false positive auto-picked particles, finding 195,314 particles judged to be HA-Fab complexes. Particles were transitioned back to RELION for further 2D classification. Three-fold symmetry was readily observable in the 2D class averages, thus we created an initial model from 2D class averages using EMAN2, and this 3D model was used for refinement and additional 3D classification in RELION. The project was transitioned to RELION271, and original micrographs were motion corrected using MotionCor272, before micrograph coordinates corresponding to the curated working set of particles from RELION were used to re-extract particles from dose-weighted motion corrected micrographs. A final set of 75,233 particles was refined using the “gold-standard” protocol as implemented in RELION2, post-processed with an ad hoc B-factor of −300, then the map resolution was determined as 8.0 Å using a FSC cutoff of 0.143. Molecular modelling of the HA-Fab complex and epitope footprint was performed by rigid docking using UCSF Chimera65. The coordinates of HA and the Fab were independently fit into the cryo-EM map of the complex, and the position of both molecules was well determined by the map. Images rendered with surfaces were created using vertices exported from UCSF Chimera, and rendered with Blender (Blender Foundation, www.blender.org), an open-source 3D rendering program.
Virus challenge in mice.
All virus challenge studies were conducted in an ABSL2 facility at Bioqual, Inc. in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee. Groups of 10 BALB/cAnNHsd mice (5–9 weeks old, female, Envigo) were anesthetized and infected intranasally with 10 LD50 A/California/07/2009 (H1N1) or A/Puerto Rico/8/1934 (H1N1) and given a purified monoclonal IgG antibody via intraperitoneal route 24 h later. The animals were monitored twice daily for development of clinical signs and weighed daily for 14 days. Any animals that had lost 20% or more of their initial body weight were humanely euthanized.
Statistical analysis.
Multi-group comparisons were done by one-way ANOVA with the Tukey’s post-hoc test performed in Prism 7 (GraphPad) unless mentioned otherwise. Differences were considered significant when P values were less than 0.05. Statistical methods and exact P values can be found in figure legends and figures, respectively.
Data and material availability.
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary information. Coordinates for 441D6 Fab structure and cryo-EM density map have been deposited to the Protein Data Bank (PDB) under PDB ID 5TR8 and EM Data Bank (EMDB) under EMD-7021. HA RBD-np and antibody sequences are deposited to the GenBank under accession no. MK273069–MK273076 and MK283705–MK283710. Requests for materials should be addressed to M.K. and B.S.G.
Supplementary Material
Acknowledgements:
We thank J. Weir (Food and Drug Administration) for providing influenza viruses; R. Webby (St. Jude Children’s Research Hospital) for influenza reverse genetics plasmids; M. Dillon, K. Wuddie, G. Sarbador, C. Chiedi, A. Taylor, H. Bao, D. Scorpio (VRC) for help with animal studies; A. Kumar (VRC) for help with protein production; V. Nair and E. Fischer (NIAID) for cryo-EM data collection; M. Chen (VRC), D. Angeletti and J. Yewdell (NIAID) for help with initial epitope mapping. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov), and also the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the NIAID, NIH. Use of insertion device 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences (under contract W-31–109-Eng-38). Support for this work was provided by the Intramural Research Programs at VRC and Division of Intramural Research, NIAID, NIH. This work was also supported in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH (under contract HHSN261200800001E).
Footnotes
Accession codes: PDB ID 5TR8; EMDB EMD-7021; GenBank Accession MK273069–MK273076 and MK283705–MK283710.
Competing interests:
Authors declare that the National Institutes of Health has filed a patent application based on the studies presented in this paper. M.K., H.M.Y., and B.S.G. are listed as inventors for the patent application.
Supplementary information
Supplementary information includes Supplementary Table 1, Supplementary Figs. 1–8.
References:
- 1.Morens DM & Taubenberger JK Historical thoughts on influenza viral ecosystems, or behold a pale horse, dead dogs, failing fowl, and sick swine. Influenza Other Respir Viruses 4, 327–337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford T et al. Integrating influenza antigenic dynamics with molecular evolution. Elife 3, e01914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koel BF et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Smith DJ et al. Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Webster RG, Laver WG, Air GM & Schild GC Molecular mechanisms of variation in influenza viruses. Nature 296, 115–121 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Air GM Influenza virus antigenicity and broadly neutralizing epitopes. Curr Opin Virol 11, 113–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrammert J et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208, 181–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce MG et al. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166, 609–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whittle JR et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 88, 4047–4057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li GM et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109, 9047–9052 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt AG et al. Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews SF et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Ekiert DC et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekiert DC et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittle JR et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108, 14216–14221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee PS et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5, 3614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfus C et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekiert DC et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuno Y, Isegawa Y, Sasao F & Ueda S A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67, 2552–2558 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corti D et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Iba Y et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol 88, 7130–7144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause JC et al. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85, 10905–10908 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong X et al. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc Natl Acad Sci U S A 112, 9430–9435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Impagliazzo A et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Yassine HM et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21, 1065–1070 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Mallajosyula VV et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A 111, E2514–2523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krammer F, Pica N, Hai R, Margine I & Palese P Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87, 6542–6550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Victora GD & Nussenzweig MC Germinal centers. Annu Rev Immunol 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Doria-Rose NA & Joyce MG Strategies to guide the antibody affinity maturation process. Curr Opin Virol 11, 137–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanzavecchia A, Fruhwirth A, Perez L & Corti D Antibody-guided vaccine design: identification of protective epitopes. Curr Opin Immunol 41, 62–67 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Kanekiyo M et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 162, 1090–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanekiyo M et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmer SM & Burke DS Historical perspective--Emergence of influenza A (H1N1) viruses. N Engl J Med 361, 279–285 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Xu R et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328, 357–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dintzis HM, Dintzis RZ & Vogelstein B Molecular determinants of immunogenicity: the immunon model of immune response. Proc Natl Acad Sci U S A 73, 3671–3675 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachmann MF et al. The influence of antigen organization on B cell responsiveness. Science 262, 1448–1451 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Tiller T, Busse CE & Wardemann H Cloning and expression of murine Ig genes from single B cells. J Immunol Methods 350, 183–193 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Gerhard W, Yewdell J, Frankel ME & Webster R Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290, 713–717 (1981). [DOI] [PubMed] [Google Scholar]
- 39.Caton AJ, Brownlee GG, Yewdell JW & Gerhard W The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–427 (1982). [DOI] [PubMed] [Google Scholar]
- 40.Hong M et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 87, 12471–12480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yewdell JW, Gerhard W & Bachi T Monoclonal anti-hemagglutinin antibodies detect irreversible antigenic alterations that coincide with the acid activation of influenza virus A/PR/834-mediated hemolysis. J Virol 48, 239–248 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raymond DD et al. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A 115, 168–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neu KE, Henry Dunand CJ & Wilson PC Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease? Curr Opin Immunol 42, 48–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambert LC & Fauci AS Influenza vaccines for the future. N Engl J Med 363, 2036–2044 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Hai R et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 86, 5774–5781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter DM et al. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets pre-immune to historical H1N1 influenza viruses. J Virol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Carter DM et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemagglutinin Vaccine for H1N1 Influenza Viruses. J Virol 90, 4720–4734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belongia EA et al. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 16, 1–14 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Chai N et al. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun 8, 14234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen C et al. A multimechanistic antibody targeting the receptor binding site potently cross-protects against influenza B viruses. Sci Transl Med 9 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Ngwuta JO et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7, 309ra162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ledgerwood JE et al. Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. J Infect Dis 208, 418–422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei CJ et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329, 1060–1064 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Wheatley AK et al. H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. J Immunol 195, 602–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brochet X, Lefranc MP & Giudicelli V IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong WP et al. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc Natl Acad Sci U S A 103, 15987–15991 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang ZY et al. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317, 825–828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu X et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455, 532–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Misasi J et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science 351, 1343–1346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Li X et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheres SH Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol 189, 114–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Kimanius D, Forsberg BO, Scheres SH & Lindahl E Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.