Abstract
Background:
Regulatory elements may be involved in the mechanisms by which 52 loci influence myocardial mass, reflected by abnormal amplitude and duration of the QRS complex on the electrocardiogram (ECG). Functional annotation thus far did not take into account how these elements are affected in disease context.
Methods:
We generated maps of regulatory elements on hypertrophic cardiomyopathy (HCM) patients (ChIP-seq N=14, RNA-seq N=11) and non-diseased hearts (ChIP-seq N=4, RNA-seq N=11). We tested enrichment of QRS-associated loci on elements differentially acetylated and/or directly regulating differentially expressed genes between HCM patients and controls. We further performed functional annotation on QRS-associated loci using these maps of differentially active regulatory elements.
Results:
Regions differentially affected in disease showed a stronger enrichment (p=8.6×10−5) for QRS-associated variants than those not showing differential activity (p=0.01). Promoters of genes differentially regulated between HCM patients and controls showed more enrichment (p=0.001) than differentially acetylated enhancers (p=0.8) and super-enhancers (p=0.025). We also identified 74 potential causal variants overlapping these differential regulatory elements. Eighteen of the genes mapped confirmed previous findings, now also pinpointing the potentially affected regulatory elements and candidate causal variants. Fourteen new genes were also mapped.
Conclusions:
Our results suggest differentially active regulatory elements between HCM patients and controls can offer more insights into the mechanisms of QRS-associated loci than elements not affected by disease.
Journal Subject Terms: Electrocardiology (ECG), Epigenetics, Gene Expression and Regulation, Heart Failure, Translational Studies
Keywords: ECG, heart failure, epigenetics, gene regulation, genetics, bioinformatics, functional annotation, OMICS data integration
Introduction
The QRS complex on the electrocardiogram (ECG) represents cardiac depolarization and conduction of the electrical signal through the ventricular muscle. Duration and amplitude of the QRS complex is used as a proxy for left ventricular mass1, 2. Abnormalities of the QRS complex are associated with an increased risk of cardiovascular (CV) mortality and morbidity3, 4.
A recent large-scale genome-wide association study (GWAS) meta-analysis of four correlated and clinically used QRS traits (Sokolow-Lyon, Cornell, 12-lead-voltage duration products (12-leadsum), and QRS duration) identified 52 independent loci at p<1×10−8 5. However, the identification of causal variants, their target genes and disturbed mechanisms remain an important challenge. In a given GWAS locus, the SNP with the most significant association with the disease (lowest p-value) is usually reported as the ‘lead’ SNP. This lead SNP is not necessarily the causal variant, and a SNP in high linkage disequilibrium (LD) with the lead SNP may be the causal one6. Only three of the 52 SNPs reported in the original study are located in coding regions of the genome5. A non-synonymous variant in high LD with a high deleteriousness score, such as CADD score > 12.377, can also be considered candidate for causality, and that is the case of 14 variants in nine QRS-associated loci (Supplemental Table 1). Potential mechanisms for causality in the remaining loci include disturbance of regulatory elements, genomic regions that play a crucial role in transcriptional regulation. Indeed, increasing evidence shows multiple GWAS variants regulate transcription8, 9. Non-coding variants often affect gene expression in a cell-type specific manner by altering the function of enhancer and promoter elements10, 11. Results from in silico analyses have suggested strong enrichment of QRS-associated variants in specific chromatin states associated with active enhancers, promoters, and transcription in the human heart12. By contrast, no enrichment was observed for transcriptionally repressive histone marks5, 13. Although using relevant cell-types, these previous studies did not include regulatory information obtained from tissue in disease state. Highlighting possible differences in transcription from control to diseased myocardial tissue may help understand the underlying mechanisms of changes in left ventricular mass. To gain insight into these mechanisms, we integrate genome, regulome and transcriptome information to test whether regions differentially affected in disease can be more informative than the regulatory landscape of non-diseased tissue. We use regulatory information obtained from non-diseased myocardium and diseased tissue from patients with hypertrophic cardiomyopathy (HCM). We also co-localize candidate causative QRS-associated variants and regulatory regions that show differential activity (proxied by H3K27ac levels or by gene expression) in HCM tissues compared to controls, in order to identify regulatory elements potentially altered by the variation. Taken together, our results may enhance our understanding on the regulatory mechanisms underlying increased myocardial mass.
Methods
The procedures for obtaining human samples were approved by the scientific advisory board of the biobank of the University Medical Center Utrecht (protocol number 12/387), the Washington University School of Medicine Ethics Committee (Institutional Review Board) and the local ethics committee of the Erasmus MC, and written consent was obtained. Biopsies on HCM patients are septal myomectomy specimens. Control samples were obtained from donor hearts not used for transplantation. Mutation and clinical data of patients are given in Table 1.
Table 1.
Sample characteristics.
| Sample name | ChIP-seq | RNA-seq | Sex | Caridac region |
Age (at operation) |
Mutation | Type of mutation |
BMI | Ejection Fraction (%, via Echo) |
Cause of death |
|---|---|---|---|---|---|---|---|---|---|---|
| HCM_1 | x | x | M | Septum | 32 | MYBPC3 | truncation | |||
| HCM_2 | x | M | Septum | 60 | MYBPC3 | truncation | ||||
| HCM_3 | x | x | M | Septum | 17 | MYBPC3 | truncation | |||
| HCM_4 | x | M | Septum | 26 | MYBPC3 | truncation | ||||
| HCM_5 | x | x | M | Septum | 33 | MYBPC3 | truncation | |||
| HCM_6 | x | x | M | Septum | 27 | MYBPC3 | truncation | |||
| HCM_7 | x | x | F | Septum | 24 | MYBPC3 | truncation | |||
| HCM_8 | x | x | M | Septum | 33 | MYBPC3 | truncation | |||
| HCM_9 | x | x | M | Septum | 49 | MYBPC3 | truncation | |||
| HCM_10 | x | x | F | Septum | 21 | MYBPC3 | truncation | |||
| HCM_11 | x | x | F | Septum | 53 | MYBPC3 | truncation | |||
| HCM_12 | x | x | M | Septum | 53 | MYBPC3 | truncation | |||
| HCM_13 | x | M | Septum | 48 | MYBPC3 | truncation | ||||
| HCM_14 | x | x | M | Septum | 22 | MYBPC3 | truncation | |||
| CONTROL_ChIP_1 | x | F | Septum | NA | NA | NA | ||||
| CONTROL_ChIP_2 | x | M | LV | NA | NA | NA | ||||
| CONTROL_ChIP_3 | x | M | Septum | NA | NA | NA | ||||
| CONTROL_ChIP_4 | x | F | Septum | NA | NA | NA | ||||
| CONTROL_RNA_1 | x | M | RV | 46 | 32.9 | 60-65 | Intracranial hemorrhage/Stroke | |||
| CONTROL_RNA_2 | x | M | LV | 46 | 32.9 | 60-65 | Intracranial hemorrhage/Stroke | |||
| CONTROL_RNA_3 | x | M | RV | 50 | 27.4 | 65 | Intracranial hemorrhage/Stroke | |||
| CONTROL_RNA_4 | x | F | RV | 56 | 31.4 | 60-65 | Intracranial hemorrhage/Stroke | |||
| CONTROL_RNA_5 | x | F | LV | 56 | 31.4 | 60-65 | Intracranial hemorrhage/Stroke | |||
| CONTROL_RNA_6 | x | F | LV | 59 | 22.8 | 45-50 | Head trauma | |||
| CONTROL_RNA_7 | x | M | RV | 46 | 25.4 | 55 | Head trauma | |||
| CONTROL_RNA_8 | x | M | LV | 46 | 25.4 | 55 | Head trauma | |||
| CONTROL_RNA_9 | x | M | RV | NA | NA | NA | NA | |||
| CONTROL_RNA_10 | x | F | RV | 61 | 40.9 | NA | Cerebrovascular/stroke | |||
| CONTROL_RNA_11 | x | F | LV | 61 | 40.9 | NA | Cerebrovascular/stroke |
Because of the sensitive nature of the data collected for this study, requests to access the dataset from qualified researchers trained in human subject confidentiality protocols may be sent to the corresponding author at F.W.Asselbergs@umcutrecht.nl. The methods of our study are detailed in the supplemental data.
Results
In this study, we functionally annotated and fine-mapped 52 QRS-associated loci. We used regulatory information (promoters from RNA-seq experiments, H3K27ac regions highlighting active promoters and enhancers, and super-enhancers from ChIP-seq experiments) obtained from non-diseased myocardium and diseased tissue from HCM patients (Table 1, Supplemental Table 2). Categorizing as expressed genes with average RPKM > 0.5 across samples of each group, 12,136 genes were expressed on HCM group and 12,540 on control group (Supplemental Tables 2 and 3). Differential expression analysis between the 11 HCM patients and 11 controls resulted in 1557 up-regulated and 1202 down-regulated genes (Supplemental Table 4). Differential acetylation analysis between 14 HCM patients and 4 controls resulted in 4068 up-acetylated and 2983 down-acetylated regions (Supplemental Table 5). From the total set of super-enhancers identified, 1048 were unique to the HCM group, not overlapping with the control group (Supplemental Table 6). Promoter, enhancer and super-enhancer regions were further narrowed down to those overlapping regions of open chromatin, in order to retain only sequences accessible to TF binding. We used these regulatory features to test the enrichment of QRS-associated variants and vicinity in LD. In order to identify an optimal LD cutoff, we identified variants in LD with the 52 QRS-associated SNPs, and divided them into bins, as described in the Methods section. We overlapped each bin with regulatory elements differentially regulated/acetylated in HCM, and performed 10k permutation tests to calculate enrichment (Supplemental Figure 1). Although LD thresholds showed fluctuations on enrichment, we observed a continuous decrease in enrichment as the LD threshold becomes more lenient. We defined LD r2>0.5 as cutoff to expand the set of candidate causal QRS-associated variants, from 52 lead SNPs to 4620 SNPs. We co-localized these candidate causal QRS-associated variants and regulatory regions that show differential activity in disease, in order to identify regulatory elements potentially altered by the variation. We also investigated which set of regulatory features showed more enrichment for QRS-associated variants, and thus has the potential to be more informative for fine-mapping efforts.
Disease-affected regulatory regions show more enrichment for QRS-associated variants than all tissue-specific regulatory regions identified in control and disease patients
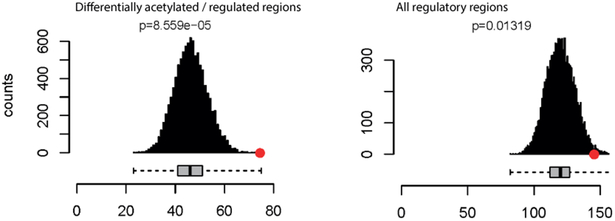
We performed enrichment tests aiming to identify which set of regulatory regions is more informative for fine-mapping efforts. Regulatory features differentially regulated/acetylated in HCM were more enriched (p=8.6×10−5) than those not showing differential activity (p=0.01), suggesting these can offer more insight into mechanisms altered by genetic variation than general tissue-specific regulatory elements (Figure 1A).
Figure 1.
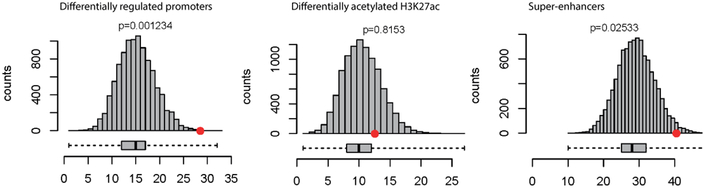
A) Mean number of regulatory elements overlapping with QRS-associated loci (red circle) compared with 10,000 matched control sets (gray bars). Differentially acetylated / regulated elements in HCM show more enrichment (p=8.6e-5) for QRS-associated candidate causal variants than those not showing differential activity (p=0.01). B) Promoters of genes differentially regulated between HCM patients and controls showed more enrichment (p=0.001) for QRS-associated variants than differentially acetylated enhancers (p=0.8) and super-enhancers (p=0.025).
Promoters of differentially expressed genes show more enrichment than regions highlighted by differential acetylation
Given the strong enrichment of QRS-associated variants in differentially regulated promoters and differentially acetylated regulatory elements in HCM (Figure 1A), we overlapped candidate causative SNPs and enriched regulatory features to identify variants that might be altering the function of these elements. Of the 4620 candidate QRS-associated SNPs, 74 co-localized with differential regulatory features (Figures 1 and 2, Supplemental Table 1), more than expected by chance (p=8.6e-5). These variants show more enrichment in differentially expressed promoters from RNA-seq experiments (p=0.001) than differentially acetylated regulatory elements highlighted by H3K27 from ChIP-seq experiments (enhancers p=0.8, super-enhancers p=0.02) (Figure 1B). These results highlight the potential of integrating regulatory elements that show differential behavior in disease, especially promoter regions.
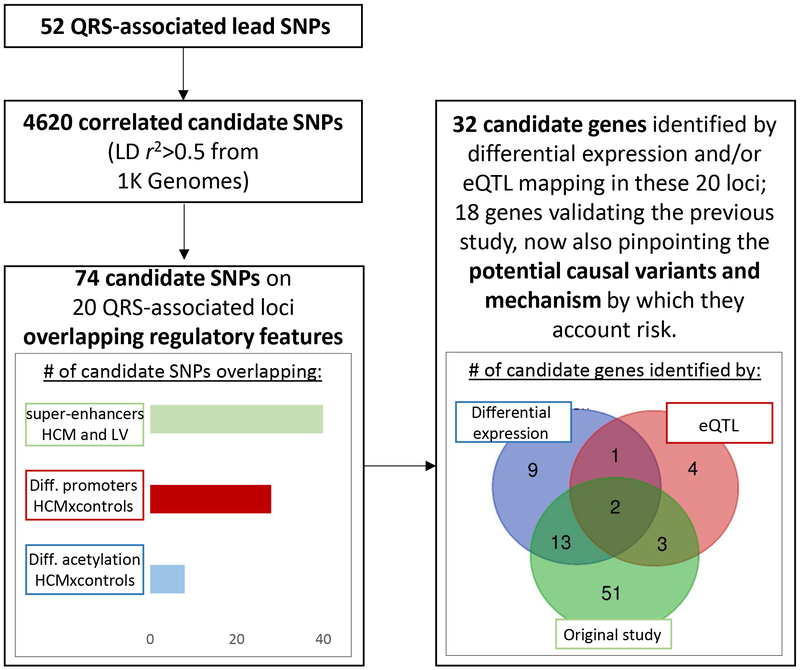
Figure 2.
Overview of functional annotation pipeline. We expanded the search from 52 lead SNPs to include variants in LD (r2>0.5), leading to 4620 candidate variants. Of these, 74 overlapped one or more regulatory feature of interest: differentially acetylated (H3K27ac) regions between HCM and control patients, differentially expressed promoter regions between HCM and control patients, and super-enhancers specific to HCM or LV. In the 20 loci where these overlaps were observed, differential expression analysis on HCM patients and controls and or/eQTL mapping in LV identified 32 candidate genes. Eighteen of these validate the findings of the previous study 5, now also pinpointing causal variants and the potential mechanisms by which they account risk. In addition, 14 new candidate genes were identified, some with cardiac function and previously implicated in other studies on ECG traits (Table 2, Supplemental Tables 1 and 7).
Fine-mapping pinpoints potential causal variants and candidate genes
The 74 variants overlapping regulatory elements affected in HCM are spread through 20 QRS-associated loci (Supplemental Figures 2–21). We investigated which genes are the potential targets of the 74 QRS-associated variants. We retrieved the three genes nearest to each of the 74 variants. We found 13 of the nearest genes are down-regulated and 12 are up-regulated in HCM versus controls (Supplemental Table 1). eQTL mapping with LV tissue from GTEx portal14 confirmed the involvement of these differentially regulated genes with QRS-associated candidate regulatory variants in three loci (ACP2/MADD, FADS1/FADS2, PROCR/EDEM2). Eighteen of the 68 candidate genes that were identified by the original study were confirmed with this new approach 5. The functional annotation performed in this study pinpoints the likely regulatory elements affected by the variation that in turn affect expression of these genes, as well as candidate causative SNPs (Supplemental Tables 1 and 7). The function of the new fourteen candidate genes potentially involved in myocardial mass is described in Table 2. Visualization and further description of each of the 20 QRS-associated loci overlapping regulatory regions of interest can be found on the Supplemental Material.
Table 2.
Description of new candidate genes identified mapped to QRS-associated loci overlapping regulatory regions with abnormal expression / acetylation between HCM and control patients.
| LIPH - Lipase H | Lipase H is a phosphatidic acid-selective phospholipase A1 (PLA1) that produces 2-acyl lysophosphatidic acid (LPA). LPA is a lipid mediator with diverse biologic properties, including induction of platelet aggregation, smooth muscle contraction, and stimulation of cell proliferation15. |
| CPNE5 - Copine 5 | This gene is one of several genes that encode a calcium-dependent protein containing two N-terminal type II C2 domains and an integrin A domain-like sequence in the C-terminus. Has been implicated in previous GWAS in heart rate16. |
| DPY19L1 - Dpy-19 Like C-Mannosyltransferase 1 | Function unknown. Automated annotations supported by experiments on knockout mouse models associate this gene to abnormal heart morphology and enlarged heart17. |
| TTC39A - Tetratricopeptide repeat domain 39A | Function unknown. Automated annotations supported by experiments on knockout mouse models associate this gene to increased heart weight and increased circulating phosphate level18. |
| PROCR - Endothelial protein C receptor | The protein encoded by this gene is a receptor for activated protein C, a serine protease activated by and involved in the blood coagulation pathway. The encoded protein is an N-glycosylated type I membrane protein that enhances the activation of protein C. Mutations in this gene have been associated with venous thromboembolism, myocardial infarction and coronary artery disease19, 20. |
| PRKCD - Protein kinase C delta | Protein kinase C (PKC) is a family of serine- and threonine-specific protein kinases that can be activated by calcium and the second messenger diacylglycerol. Studies both in human and mice demonstrate that PRKCD kinase is involved in B cell signaling and in the regulation of growth, apoptosis, and differentiation of a variety of cell types21. |
| CORO6 - Coronin 6 | Involved in actin filament binding. Overexpressed in heart and skeletal muscle22. Has been implicated in previous GWAS in coronary artery disease19. |
| TCEA3 - Transcription Elongation Factor A3 | This gene is associated to other measure of amplitude of the electrocardiogram, ST-T wave23 and QT-interval24. Overexpressed in heart22. |
| FUT11 - Fucosyltransferase 11 | Function unknown. |
| CENPA - Centromere protein A | Histone H3-like variant which exclusively replaces conventional H3 in the nucleosome core of centromeric chromatin at the inner plate of the kinetochore25. Required for recruitment and assembly of kinetochore proteins, mitotic progression and chromosome segregation. May serve as an epigenetic mark that propagates centromere identity through replication and cell division26, 25, 27, 28. |
| AGBL5 - ATP/GTP binding protein-like 5 | Function unknown. |
| KCNK3 - Potassium channel, subfamily K, member 3 | pH-dependent, voltage-insensitive, background potassium channel protein. Rectification direction results from potassium ion concentration on either side of the membrane. Acts as an outward rectifier when external potassium concentration is low29. |
| KHK - ketohexokinase | Catalyzes the first step of metabolism of dietary fructose, conversion of fructose to fructose-1-phosphate. It has been shown that myocardial hypoxia influences fructose metabolism in human and mouse models of pathologic cardiac hypertrophy through hypoxia-inducible factor 1-alpha activation of SF3B1 and SF3B1-mediated splice switching of KHK-A to KHK-C30, 31. In mice, heart-specific depletion of SF3B1 or genetic ablation of KHK suppressed pathologic stress-induced fructose metabolism, growth, and contractile dysfunction31. |
| FADS1 - Fatty Acid Desaturase 1 | A member of the fatty acid desaturase (FADS) gene family. Component of a lipid metabolic pathway that catalyzes the biosynthesis of highly unsaturated fatty acids from precursor essential polyunsaturated fatty acids, linoleic acid, and alpha-linolenic acid. SNPs of the FADS gene cluster have been associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease32. |
Discussion
We performed functional annotation of 52 loci influencing myocardial mass, considering regulatory elements with differential activity between diseased (HCM) heart samples and control donors may act as mediators between genetic variation and gene expression. The 52 QRS-associated variants showed enrichment on the set of differential regulatory elements in comparison to non-differential regulatory elements. In addition, differentially expressed promoters identified by RNA-seq in HCM and controls showed more enrichment of QRS-associated variants, than the other differential regulatory elements analyzed. These results can aid the study design of future fine-mapping studies.
Our functional annotation showed twenty loci showed overlapping with the enriched differential regulatory regions. We identified the variants in LD with the lead QRS-associated SNP overlapping regulatory features, thus pinpointing the potential causative variants and mechanisms through which these variants account risk for disease. We observed that some loci showed enrichment of more than one candidate SNP in the same regulatory feature. Studies indeed show that several variants can be causal inside a disease-associated locus33, 34. In addition, more than one gene can be causal inside the same loci35, and we indeed identified more than one candidate gene per locus in some cases. Further assays, such as chromatin conformation capture36–40, can add evidence on the interaction between QRS-candidate regulatory SNPs and each candidate gene, preferably also taking into account the disease context41. These experiments can also reveal whether regions identified as promoters also function as enhancers42–44. Moreover, genes potentially influenced by the regulatory elements affected by QRS-associated variants that are not in the immediate vicinity can also be detected45.
We observed that 18 genes mapped by the previous study were confirmed by differential expression and/or eQTL mapping, adding evidence for the role of these genes in natural variance in myocardial mass. We also identified 14 genes not previously mapped to the QRS-associated variants, some with known cardiac function and previously implicated in studies on ECG parameters and/or heart phenotypes. Definitive assignment of function(s) to putative cis-regulatory elements requires perturbation of these elements46. A promising approach is deletion and manipulation of nucleases through CRISPR/Cas9, which was successful in recent studies47, 10.
Future efforts can unravel the mechanisms of the remaining QRS-associated loci by also taking into account other regulatory elements apart from enhancers and promoters, such as silencers, highlighted by different histone modifications. In addition, other histone modifications can also help identify additional enhancer elements. A subset of active enhancers has been identified in mouse embryonic stem cells that lack H3K27ac but are marked by H3K122ac and/or H3K64ac, H3K4me1 and enhancer RNAs (eRNAs)48. This evidence shows that although H3K27ac highlights enhancer activity, enhancers that lack H3K27ac are not necessarily inactive, and other histone marks can carry out the activating role of H3K27ac49. Moreover, as chromatin states are spatially and temporally dynamic along disease progression, regulatory information is needed in more different conditions including different stages of disease progression. This will allow a more precise and complete fine-mapping and functional annotation. It can also help answer whether QRS-associated SNPs promote aberrant function of regulatory elements, or the SNPs alter the activity of regulatory elements following disease establishment. Finally, the low number of control heart samples included in our ChIP-seq experiments might partly explain the lower degree of enrichment for enhancer/super-enhancer regions as compared to the promoter regions identified from RNA-seq experiments. Increasing sample sizes may help unravel more regulatory regions and their enrichment for QRS-associated variants.
Taken together, our results show the importance of using differential regulatory elements in disease tissue on fine-mapping studies, since influence from cell conditions are crucial for epigenetic modifications, chromatin accessibility, TF binding and consequently gene regulation50. We also expand the view of the epigenetic and regulatory aspects involved in myocardial mass by pinpointing potential causal variants, mechanisms potentially affected by genetic variation, and candidate genes. Validation of the identified candidate causal variants through methods such as genome-editing technologies will broaden our knowledge on how QRS-associated variants contribute to disease.
Supplementary Material
Acknowledgments:
This work was supported by the National Institute of health (NIH) grant LM010098. DH is supported by the National Council for the Improvement of Higher Education (CAPES) and Science without Borders Project, process nº 13259/13–0. M.H. is supported by Wilhelmina Children’s Hospital research funding OZF/14 and by personal research fund VENI ZonMW/NWO2016‐016.176.136. FWA is supported by UCL Hospitals NIHR Biomedical Research Centre. We acknowledge support from the Netherlands Cardiovascular Research Initiative—an initiative with support of the Dutch Heart Foundation, CVON: The Netherlands CardioVascular Research Committee, CVON2014–40 DOSIS. We gratefully acknowledge the contribution of Noortje van den Dungen and Nico Lansu from the Department of Paediatrics, UMCU, as well as Joyce van Kuik and Erica Sierra-de Koning from the Department of Pathology, UMCU, and the Utrecht Sequencing Facility. We also acknowledge Gregory Crawford Lab from Duke Center For Genomic And Computational Biology and the ENCODE consortium for the public dataset of open chromatin in heart used in this study.
Source of Funding: This work was supported by the National Institute of health (NIH) grant LM010098 (United States of America). DH is supported by the National Council for the Improvement of Higher Education (CAPES) and Science without Borders Project, process nº 13259/13–0 (Brazil). M.H. is supported by Wilhelmina Children’s Hospital research funding OZF/14 and by personal research fund VENI ZonMW/NWO2016‐016.176.136 (The Netherlands). FWA is supported by UCL Hospitals NIHR Biomedical Research Centre. UCL Hospitals NIHR Biomedical Research Centre (United Kingdom). We acknowledge support from the Netherlands Cardiovascular Research Initiative—an initiative with support of the Dutch Heart Foundation, CVON: The Netherlands CardioVascular Research Committee, CVON2014–40 DOSIS (The Netherlands).
Footnotes
Disclosures: None
References:
- 1.Levy D, et al. Determinants of sensitivity and specificity of electrocardiographic criteria for left ventricular hypertrophy. Circulation. 1990;81:815–820. [DOI] [PubMed] [Google Scholar]
- 2.Okin PM, et al. Time-voltage QRS area of the 12-lead electrocardiogram: detection of left ventricular hypertrophy. Hypertension. 1998;31:937–942. [DOI] [PubMed] [Google Scholar]
- 3.Kannel WB, et al. Left ventricular hypertrophy by electrocardiogram. Prevalence, incidence, and mortality in the Framingham study. Ann Intern Med. 1969;71:89–105. [DOI] [PubMed] [Google Scholar]
- 4.Usoro AO, et al. Risk of mortality in individuals with low QRS voltage and free of cardiovascular disease. Am J Cardiol. 2014;113:1514–1517. [DOI] [PubMed] [Google Scholar]
- 5.van der Harst P, et al. 52 Genetic Loci Influencing Myocardial Mass. J Am Coll Cardiol. 2016;68:1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corradin O, et al. Enhancer variants: evaluating functions in common disease. Genome Med. 2014;6:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaub MA, et al. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RM, et al. A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression. Cell. 2017;170:522–533 e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao L, et al. Functional annotation of colon cancer risk SNPs. Nat Commun. 2014;5:5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parmar PG, et al. International Genome-Wide Association Study Consortium Identifies Novel Loci Associated With Blood Pressure in Children and Adolescents. Circ Cardiovasc Genet. 2016;9:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium GT, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonoda H, et al. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J Biol Chem. 2002;277:34254–34263. [DOI] [PubMed] [Google Scholar]
- 16.den Hoed M, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.“Gene: Dpy19l1.”. Retrieved June 11, 2018, from http://www.mousephenotype.org/data/genes/MGI:1915685.
- 18.“Gene: Ttc39a.” Retrieved June 11, 2018, from http://www.mousephenotype.org/data/genes/MGI:2444350.
- 19.van der Harst P, et al. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ Res. 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis J, et al. The endothelial protein C receptor (PROCR) Ser219Gly variant and risk of common thrombotic disorders: a HuGE review and meta-analysis of evidence from observational studies. Blood. 2012;119:2392–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verweij N, et al. Twenty-eight genetic loci associated with ST-T-wave amplitudes of the electrocardiogram. Hum Mol Genet. 2016;25:2093–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arking DE, et al. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orthaus S, et al. Assembly of the inner kinetochore proteins CENP-A and CENP-B in living human cells. Chembiochem. 2008;9:77–92. [DOI] [PubMed] [Google Scholar]
- 26.Shelby RD, et al. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan BA, et al. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. [DOI] [PubMed] [Google Scholar]
- 29.Plant LD, et al. SUMOylation silences heterodimeric TASK potassium channels containing K2P1 subunits in cerebellar granule neurons. Sci Signal. 2012;5:ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirtschink P, et al. Hypoxia-driven glycolytic and fructolytic metabolic programs: Pivotal to hypertrophic heart disease. Biochim Biophys Acta. 2016;1863:1822–1828. [DOI] [PubMed] [Google Scholar]
- 31.Mirtschink P, et al. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature. 2015;522:444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malerba G, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. [DOI] [PubMed] [Google Scholar]
- 33.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flister MJ, et al. Identifying multiple causative genes at a single GWAS locus. Genome Res. 2013;23:1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295:1306–1311. [DOI] [PubMed] [Google Scholar]
- 37.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet. 2006;38:1348–1354. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. [DOI] [PubMed] [Google Scholar]
- 39.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Javierre BM, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384 e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosa-Garrido M, et al. High-Resolution Mapping of Chromatin Conformation in Cardiac Myocytes Reveals Structural Remodeling of the Epigenome in Heart Failure. Circulation. 2017;136:1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Catarino RR, et al. Promoting transcription over long distances. Nat Genet. 2017;49:972–973. [DOI] [PubMed] [Google Scholar]
- 43.Dao LTM, et al. Genome-wide characterization of mammalian promoters with distal enhancer functions. Nat Genet. 2017;49:1073–1081. [DOI] [PubMed] [Google Scholar]
- 44.Diao Y, et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat Methods. 2017;14:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandhu KS, et al. Large-scale functional organization of long-range chromatin interaction networks. Cell Rep. 2012;2:1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elkon R, et al. Characterization of noncoding regulatory DNA in the human genome. Nat Biotechnol. 2017;35:732–746. [DOI] [PubMed] [Google Scholar]
- 47.Xia Q, et al. The type 2 diabetes presumed causal variant within TCF7L2 resides in an element that controls the expression of ACSL5. Diabetologia. 2016;59:2360–2368. [DOI] [PubMed] [Google Scholar]
- 48.Pradeepa MM, et al. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat Genet. 2016;48:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atlasi Y, et al. The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet. 2017;18:643–658. [DOI] [PubMed] [Google Scholar]
- 50.Liu L, et al. Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res. 2015;43:3873–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.