Abstract
More than 80% of all cases of deafness are related to the death or degeneration of cochlear hair cells and the associated spiral ganglion neurons, and a lack of regeneration of these cells leads to permanent hearing loss. Therefore, the regeneration of lost hair cells is an important goal for the treatment of deafness. Atoh1 is a basic helix-loop-helix (bHLH) transcription factor that is critical in both the development and regeneration of cochlear hair cells. Atoh1 is transcriptionally regulated by several signaling pathways, including Notch and Wnt signalings. At the post-translational level, it is regulated through the ubiquitin-proteasome pathway. In vitro and in vivo studies have revealed that manipulation of these signaling pathways not only controls development, but also leads to the regeneration of cochlear hair cells after damage. Recent progress toward understanding the signaling networks involved in hair cell development and regeneration has led to the development of new strategies to replace lost hair cells. This review focuses on our current understanding of the signaling pathways that regulate Atoh1 in the cochlea.
Keywords: Atoh1, Huwe1, Cochlea, Hair cells, Regeneration, Post-translational regulation
1. Introduction
Hair cell formation is dependent on the basic helix-loop-helix (bHLH) transcription factor Atoh1, which is both necessary and sufficient for hair cell differentiation during development (Bermingham et al., 1999). Atoh1 overexpression in the inner ear via genetic or pharmacological manipulation has been shown to result in new hair cell formation in the cochlea and to restore hearing in experimental animal models (Zheng and Gao, 2000; Izumikawa et al., 2005; Gubbels et al., 2008). The transcriptional and post-translational regulations of Atoh1 have been shown to involve several overlapping signaling pathways. Transcriptional regulation of Atoh1 by Wnt or Notch signaling or post-translational regulation by the Huwe1-ubiquitin-proteasome pathway has been shown to sucessfully convert inner ear progenitor cells into hair cells (Shi et al., 2010; 2013; 2014; Mizutari et al., 2013; Bramhall et al., 2014; Cheng et al., 2016). These studies have led to novel approaches for the regeneration of hair cells in the inner ear. This review focuses on recent advances in our understanding of the signaling pathways that regulate Atoh1 in the cochlea.
2. Hearing impairment
Hearing impairment is one of the most prevalent disabilities in industrialized countries, and it is estimated that 360 million people have moderate to profound hearing deficiency globally (WHO, 2017). Most cases of hearing impairment involve sensorineural hearing loss caused by the degeneration or loss of two specific inner ear cell types: cochlear hair cells, which act as the primary mechanoreceptors for sound transduction, and the connected auditory nerve neurons, which transmit electrical signals from the synapsed inner hair cells to the brain (Davis, 1983).
Loss of cochlear hair cells caused by genetic mutations, ototoxic medications, noise overexposure, autoimmune disease, or aging is irreversible in mammals because they have very limited ability to spontaneously regenerate hair cells after loss, and there are currently very few methods to regenerate hearing function after hair cell damage, especially in adult animals (Izumikawa et al., 2005; Mizutari et al., 2013). Vestibular hair cells have a limited regenerative capacity (Forge et al., 1993; Warchol et al., 1993; Wang et al., 2015), but spontaneous hair cell regeneration after damage or hair cell loss has been reported only in the newborn murine cochlea (Bramhall et al., 2014; Cox et al., 2014). For deafness caused by loss of cochlear hair cells, cochlear implants are surgically implanted into the inner ear to stimulate the spiral ganglion. However, cochlear implants depend on the remaining functional spiral ganglion neurons, the loss of which can severely compromise their efficacy (Incesulu and Nadol, 1998). The regeneration or replacement of damaged cochlear hair cells is thus the ultimate goal for the treatment of hearing loss.
Several different approaches for hair cell regeneration have been attempted, including cell therapy, gene therapy, and pharmacological therapy. A few attempts at cell transplantation into the damaged cochlea have been reported, but most have shown extremely limited survival of transplanted cells and the inability of these cells to differentiate into mature hair cell types (Hu et al., 2005; Hu and Ulfendahl, 2006). Gene therapy involving the overexpression of Atoh1 has been shown to be a successful strategy for converting supporting cells into hair cells, thereby restoring hearing loss after damage (Izumikawa et al., 2005; Richardson and Atkinson, 2015). Gene augmentation (Akil et al., 2012; Askew et al., 2015; Pan et al., 2017), as well as emerging CRISPR-Cas9 genome editing methods (Zou et al., 2015; Zuris et al., 2015; Mianné et al., 2016), also provides promising experimental paradigms for treating genetic deafness. However, the complexity of the inner ear anatomy, as well as a lack of ideal vector systems and appropriate delivery methods, has made the delivery of foreign genetic material into the inner ear challenging (Husseman and Raphael, 2009; Shu et al., 2016). Pharmacotherapy using local delivery of small molecule compounds to convert inner ear progenitors into hair cells seems to be a more physiologically relevant approach to regenerating lost hair cells after damage (Mizutari et al., 2013). However, a successful therapy will require a better understanding of the underlying mechanisms controlling hair cell formation and regeneration.
3. Atoh1, a bHLH transcription factor, is required for hair cell differentiation and regeneration after damage
bHLH transcription factors orchestrate cell fate commitment and specification during development (Lo et al., 1991; Ross et al., 2003), and a homologous group of “proneural” bHLH transcription factors plays a central role in neurogenesis. Several proneural bHLH transcription factors, including Atoh1, Neurog1, Neurod1, and Ascl1, contribute to the development of inner ear neurons and hair cells (Bertrand et al., 2002; Fritzsch, 2003; Tiveron et al., 2003; Jahan et al., 2013).
Atoh1, the mammalian homolog of the Drosophila gene atonal (also known as Math1 for “mouse atoh1” and HATH1 for “human atoh1”) (Ben-Arie et al., 1996), is a bHLH transcription factor that is both necessary and sufficient for the formation and differentiation of inner ear hair cells. Animal studies have shown that Atoh1-knockout mice fail to form cochlear and vestibular hair cells (Bermingham et al., 1999) as a result of progenitor cell apoptosis (Chen et al., 2002). In gain-of-function studies, overexpression of Atoh1 in mice by viral delivery led to ectopic hair cell formation in organ of Corti explants (Zheng and Gao, 2000). In vivo and in utero experiments in mice confirmed that forced upregulation of Atoh1 leads to the formation of extra hair cells (Woods et al., 2004; Izumikawa et al., 2005; Staecker et al., 2007; Gubbels et al., 2008; Liu et al., 2012; 2014; Atkinson et al., 2014), but very little is known about how cells regulate the level of Atoh1, especially in the cochlea.
4. Atoh1 level is critical for hair cell development and regeneration in mice
Proper spatiotemporal expression of Atoh1 is key for the differentiation and viability of hair cells. Atoh1 is expressed in mice in the inner ear progenitors before they have committed to the hair cell fate. It is first detected in the prosensory cells of the basal cochlear turn of the embryonic cochlea at embryonic day (E) 13.5. The expression of Atoh1 surges at the base of the cochlea by E14.5 and starts to appear in the apical turn by E17.5. Atoh1 expression begins to decline immediately after birth in the basal turn and by postnatal day 4 in hair cells in the apical turn (Yang et al., 2010; Pan et al., 2012).
Atoh1 affects not only hair cell differentiation, but also hair cell viability. Embryonic reduction of Atoh1 led to significant cochlear hair cell loss and abnormal hair cell bundle formation in a transgenic mouse model (Pan et al., 2012). Temporally, Atoh1 is critical for the survival of hair cell progenitors at the base of the cochlea in a 72-h window from E13.5 to E16.5, and deletion of Atoh1 outside this developmental window does not affect survival (Cai et al., 2013). Although Atoh1 overexpression in the supporting cells leads to ectopic hair cell formation, transgenic mouse models have shown that persistent Atoh1 overexpression in the cochlear hair cells leads to hair cell death and eventual hearing loss (Liu et al., 2012). A similar phenotype was also seen when Atoh1 protein degradation mechanisms were disrupted (Cheng et al., 2016).
5. Atoh1 is widely expressed
In addition to hair cell formation, Atoh1 plays important roles in the differentiation and formation of several other cell types. Atoh1 knockout induces cell loss in several tissues, including cerebellar granule cells (Ben-Arie et al., 1997; Flora et al., 2007; Forget et al., 2014), retrotrapezoid nucleus (RTN) neurons in the medulla (Wang et al., 2005; Huang et al., 2012; Ruffault et al., 2015), dorsal commissural interneurons in the spine (Miesegaes et al., 2009), intestinal goblet cells (Yang et al., 2001; VanDussen and Samuelson, 2010), and Merkel cells in the skin (Ben-Arie et al., 2000; Fröhlich et al., 2009; Morrison et al., 2009; Maksimovic et al., 2014; Wright et al., 2015). The distribution of Atoh1 in cells other than inner ear hair cells and the associated phenotypes in loss-of-function assays are listed in Table 1.
Table 1.
Atoh1 expression and its function in cell types other than hair cells
| Cell type | Function of Atoh1 | Phenotype in loss-of-function assays | Reference |
| Cerebellar granule cells | Maturation and proliferation | Missing external germinal layer (EGL) of the developing cerebellum | Ben-Arie et al., 1997; Wang et al., 2005; Flora et al., 2007; Forget et al., 2014 |
| Retrotrapezoid nucleus (RTN) neurons of the medulla | Migration of neurons and establishment of essential connections with the pre-Bötzinger complex | RTN neurons fail to develop and establish connections with the respiratory rhythm-generating center in mammals | Rose et al., 2009; Huang et al., 2012; Ruffault et al., 2015 |
| D1 commissural interneurons of the spinal cord | Ventral migration and fate determination of D1 interneurons | Progenitors adopting roof plate or D2 interneuron fates | Miesegaes et al., 2009 |
| Intestinal goblet cells | Secretory lineage fate determination | Failure to form intestinal secretory cells | Yang et al., 2001; Shroyer et al., 2007; VanDussen and Samuelson, 2010 |
| Merkel cells | Specification of Merkel cells | Absence of Merkel cells | Ben-Arie et al., 2000; Morrison et al., 2009; van Keymeulen et al., 2009; Maksimovic et al., 2014 |
Atoh1 expression has also been reported to be associated with tumorigenesis. Atoh1 acts as a tumor suppressor gene in the colon and skin, as it antagonizes tumor formation and growth, and many colorectal cancer and Merkel cell carcinoma patients show genetic mutations of Atoh1 (Bossuyt et al., 2009). However, Atoh1 acts as an oncogene in medulloblastoma (Zhao H. et al., 2008; Flora et al., 2009; Ayrault et al., 2010), indicating its multiple roles in differentiation and proliferation in different cell types.
6. Signaling pathways that regulate Atoh1
6.1. Notch signaling pathway
Notch signaling plays a key role in Atoh1 regulation and inner ear development (Jarriault et al., 1995; Lanford et al., 1999; Brooker et al., 2006). The major role of Notch signaling is to regulate cell fate decisions through lateral inhibition during development. It acts as a mediator between prosensory cells during development, leading these cells to differentiate into hair cells and supporting cells. Thus, Notch signaling is utilized to establish the structural pattern of hair cells and supporting cells in the cochlea (Kelley, 2003).
The inner ear is derived from the otic placode, which is recognized initially as a thickening near the hindbrain at E8 in mice. The otic placode invaginates by E10.5 to form an otocyst that contains prosensory cells and ganglion neuroblasts. The determination of hair cell or supporting cell fate is influenced by Notch signaling, and developing hair cells express Atoh1 along with the Notch ligands Jag2 and Delta1. These ligands bind to Notch1 in adjacent cells and induce the release of Notch intracellular domain (NICD) from their membranes. Upregulation of inhibitory bHLH transcription factors such as Hes1 and Hes5 follows after the increase of NICD, and expression of Atoh1 is blocked, leading to inhibition of hair cell fate and development of supporting cells (Kelley, 2003; 2006).
Pharmacological inhibition of Notch signaling by γ-secretase inhibitors increases the number of cochlear hair cells in neonatal cochlear explants (Yamamoto et al., 2006; Takebayashi et al., 2007; Bramhall et al., 2014; Li et al., 2015; Maass et al., 2015) and increases the differentiation of inner ear stem cells into hair cells (Jeon et al., 2011). In an in vitro lineage-tracing study involving genetically marking supporting cells with a fluorescent reporter, treatment with γ-secretase inhibitors after aminoglycoside-induced hair cell damage has been shown to promote hair cell regeneration through the direct transdifferentiation of supporting cells in the cochlea (Bramhall et al., 2014). An in vivo study also showed that direct round-window application of γ-secretase inhibitors resulted in recovery of cochlear function after noise-induced hair cell damage. This was evidenced by increased Atoh1 expression and direct conversion of supporting cells into hair cells, as measured by a similar genetic tagging model (Mizutari et al., 2013).
The responsiveness of the cochlea to γ-secretase inhibitors has been shown to be position-dependent and age-dependent. The apical region of the cochlea exhibits a stronger response in terms of transdifferentiation of supporting cells into hair cells compared to the basal region, and the response declines rapidly with age (Mizutari et al., 2013; Bramhall et al., 2014; Maass et al., 2015).
6.2. Wnt/β-catenin signaling pathway
The Wnt pathway plays a critical role in patterning and cell fate specification in the early development of the inner ear (Stevens et al., 2003; Gregorieff and Clevers, 2005; Riccomagno et al., 2005; Ohyama et al., 2006). In canonical Wnt signaling, Wnt binds to receptors on the cell membrane surface to disrupt the destruction complex consisting of axin, adenomatosis polyposis coli (APC), glycogen synthase kinase 3β (GSK-3β), and Disheveled (Dvl). This prevents β-catenin from being degraded in the cytoplasm. β-Catenin then translocates into the nucleus and subsequently forms nuclear β-catenin/Tcf complexes that drive the expression of Wnt target genes. In non-canonical Wnt signaling, Wnt proteins can work in a β-catenin-independent manner via the planar cell polarity (PCP) and Wnt/calcium pathways (Jansson et al., 2015).
Wnt/β-catenin signaling induces patches of hair cells and supporting cells in the auditory sensory epithelium and is involved in establishing or maintaining the distinction between sensory domains that contain hair cells and non-sensory domains in the inner ear (Stevens et al., 2003). Overexpression of β-catenin increases Atoh1 expression in neuroblastoma cells and neural progenitor cells. This upregulation is due to the interaction of β-catenin with the 3'-enhancer of the Atoh1 gene (Shi et al., 2010). Conditional knockout of β-catenin inhibits hair cell formation from sensory progenitors, while constitutive upregulation of β-catenin expands sensory progenitors and results in the formation of extra hair cells (Shi et al., 2012; 2014; McLean et al., 2017).
The Notch and Wnt/β-catenin signaling pathways interact with each other. As mentioned earlier, β-catenin increases Atoh1 expression through interaction with the 3' enhancer of Atoh1, and this accounts for the effect of Notch inhibition on Atoh1. Shi et al. (2010) found that inhibition of Notch signaling increases β-catenin expression, while simultaneous inhibition of β-catenin abolishes the increase in Atoh1 expression caused by Notch inhibition. Thus, β-catenin expression is required not only for increased expression of Atoh1, but also for the mitotic generation of hair cells in the cochlea after Notch inhibition (Li et al., 2015). Interestingly, a combination of Notch inhibition and Wnt signaling activation seems to have synergistic effects on the proliferation and transdifferentiation of progenitors or supporting cells in the neonatal cochlea, especially in the basal turn that usually shows only limited ability to form new hair cells after birth (Ni et al., 2016; McLean et al., 2017).
6.3. Post-translational regulation of Atoh1: the CK1-Huwe1-ubiquitin-proteasome pathway
Increasing evidence has shown that Atoh1 gene expression is tightly regulated by overlapping pathways, some of which have been described above. Post-translational regulation of Atoh1, however, is only now beginning to be understood. The ubiquitin-proteasome pathway (UPP) is the major post-translational regulatory system in eukaryotes, and it plays an important role in the development and physiology of eukaryotic cells (Tai and Schuman, 2008). The UPP has been implicated in the regulation of stem cell differentiation and lineage commitment via proteolytic degradation of key regulatory proteins. Specific protein substrates are conjugated by polyubiquitylation in an adenosine triphosphate (ATP)-dependent process and then targeted by the proteasome, the machinery by which proteins are degraded. The system is highly selective and tightly regulated. It not only degrades misfolded or damaged proteins, but is also essential for regulating cell-signaling pathways (Naujokat and Šarić, 2007).
The UPP consists of three key enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). E3 catalyzes polyubiquitin (and sometimes monoubiquitin) chain formation by transferring ubiquitin that has been activated by E1 and E2 onto the lysine residues of specific substrates. Huwe1 (HECT, UBA, and WWE domain containing 1) is a large 4371-amino acid HECT-domain E3 ubiquitin ligase that is involved in proteasomal degradation of several protein substrates, including Atoh1 in the cerebellum and in the cochlea (Forget et al., 2014; Cheng et al., 2016). Huwe1 binds to Atoh1 and forms a lysine 48-cojugated polyubiquitin chain on Atoh1 as a signal for proteasomal degradation (Cheng et al., 2016). As a result, overexpression of Huwe1 decreases Atoh1 levels, while inhibiting Huwe1 increases the half-life of the Atoh1 protein.
A “degron” is a sequence element or modification that is sufficient to be recognized or targeted by an E3 ligase to promote ubiquitylation (Varshavsky, 1991). Atoh1 is found to be enriched in serine residues in the C-terminus, and many of the serines are conserved among different species. Casein kinase 1 (CK1), a serine/threonine selective protein kinase, phosphorylates Atoh1 at its serine-enriched C-terminal region, specifically serine 334, to form a phosphodegron that enhances the Atoh1-Huwe1 interaction and the subsequent ubiquitylation of Atoh1 (Cheng et al., 2016). The overexpression of CK1 increases the degradation of Atoh1, while inhibition of CK1 stabilizes Atoh1.
Interestingly, other sites, including other conserved serines at the C-terminus, have been reported to contribute to Atoh1 stability. Forget et al. (2014) found that sonic hedgehog (SHH) controls the phosphorylation of serine 328 and serine 339 of Math1 through protein phosphatase 2A (PP2A) activity to protect Atoh1 from degradation. Tsuchiya et al. (2007) reported that Atoh1 protein is expressed in normal colon tissue, but it is degraded by GSK3β in cancer tissue through phosphorylation of serine 54 and serine 58 of Hath1 (the equivalent of serine 52 and serine 56 of Math1).
The biological roles of Huwe1 in development and neurogenesis have recently been described (Zhao X. et al., 2008; 2009; D'Arca et al., 2010; Dominguez-Brauer et al., 2016; Urbán et al., 2016). Atoh1 degradation by Huwe1 is required for proper neuronal migration and differentiation in the cerebellum (Forget et al., 2014) because uncontrolled Atoh1 expression in the cerebellum leads to early postnatal lethality and interferes with cerebellar development (Helms et al., 2001). Spatiotemporal control of the Atoh1 protein level by Huwe1 is essential for cochlear hair cell fate determination and survival (Cheng et al., 2016), and conditional knockout of Huwe1 in the cochlear supporting cells at the embryonic or early postnatal stage leads to extra hair cells in the cochlea. However, hair cell death is observed when Huwe1 is conditionally knocked out in the cochlear hair cells, and this phenotype appears to be correlated with an increase in Atoh1 protein expression. This is in line with a previous study showing that Atoh1 overexpression in the hair cell causes hair cell death (Liu et al., 2012). Thus, the termination of Atoh1 activity after hair cell differentiation and maturation seems to be critical for normal cell patterning in the cochlea. Although several substrates of Huwe1 have been found to be associated with cell apoptosis, including Mcl-1, protein phosphatase 5 (PP5), p53, Myc, and cdc6 (Chen et al., 2005; Hall et al., 2007; D'Arca et al., 2010; Kurokawa et al., 2013), further studies are required to unravel the mechanisms of hair cell death underlying Huwe1 deletion and Atoh1 overexpression.
7. Concluding remarks
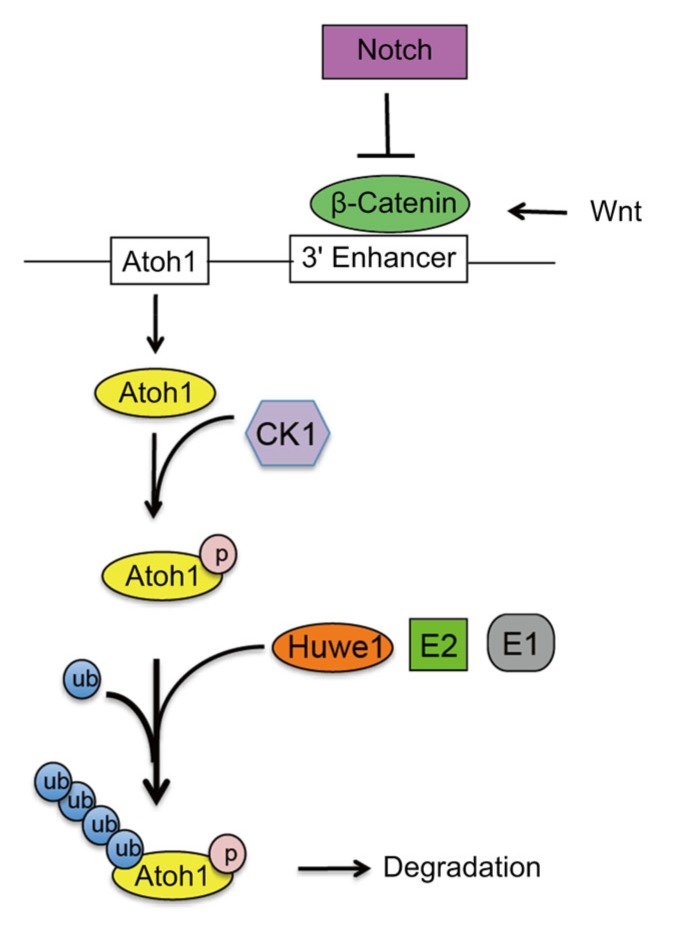
Atoh1 is a key transcription factor in the development and regeneration processes of the inner ear. Several signaling pathways have been shown to be involved in the regulation of its expression in the cochlea, including the Notch, Wnt, and ubiquitin-proteasome pathways. The studies reviewed in this article have shown that proper spatiotemporal control of Atoh1 expression is essential for inner ear development, not only at the transcriptional level, but also at the post-translational level (concluded in Fig. 1). Forcing Atoh1 overexpression or manipulating Atoh1-enhancing signaling pathways is one of most efficient strategies known for regenerating damaged hair cells. Incorporating knowledge of how Atoh1 is regulated will lead to a better understanding of cochlear development and to more insights into therapeutic strategies for hair cell regeneration.
Fig. 1.
Summary of Atoh1 regulation
Wnt signaling activates Atoh1 expression through interactions between β-catenin and the 3' enhancer of Atoh1. Overexpression of Notch signaling decreases β-catenin and decreases Atoh1 expression, while inhibition of Notch leads to upregulation of β-catenin and increased Atoh1 expression. At the post-translational level, CK1 binds to and phosphorylates Atoh1 to facilitate interaction with the E3 ubiquitin ligase Huwe1, which orchestrates ubiquitin transfer events from E1 (ubiquitin-activating enzyme) and E2 (ubiquitin-conjugating enzyme). This in turn leads to the polyubiquitylation and proteasomal degradation of Atoh1. Figure adapted and modified from Shi et al. (2010) and Cheng et al. (2016)
Footnotes
Compliance with ethics guidelines: Yen-Fu CHENG declares that he has no conflict of interest.
This article does not contain any studies with human or animal subjects performed by the author.
References
- 1.Akil O, Seal , R.P. , Burke K, et al. Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron. 2012;75(2):283–293. doi: 10.1016/j.neuron.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askew C, Rochat C, Pan B, et al. Tmc gene therapy restores auditory function in deaf mice. Sci Transl Med. 2015;7(295):295ra108. doi: 10.1126/scitranslmed.aab1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson PJ, Wise AK, Flynn BO, et al. Hair cell regeneration after ATOH1 gene therapy in the cochlea of profoundly deaf adult guinea pigs. PLoS ONE. 2014;9(7):e102077. doi: 10.1371/journal.pone.0102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayrault O, Zhao H, Zindy F, et al. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res. 2010;70(13):5618–5627. doi: 10.1158/0008-5472.CAN-09-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Arie N, McCall AE, Berkman S, et al. Evolutionary conservation of sequence and expression of the bHLH protein atonal suggests a conserved role in neurogenesis. Hum Mol Genet. 1996;5(9):1207–1216. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Arie N, Bellen HJ, Armstrong DL, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390(6656):169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Arie N, Hassan BA, Bermingham NA, et al. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127(5):1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- 8.Bermingham NA, Hassan BA, Price SD, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284(5421):1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3(7):517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 10.Bossuyt W, Kazanjian A, de Geest N, et al. Atonal homolog 1 is a tumor suppressor gene. PLoS Biol. 2009;7(2):e1000039. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bramhall NF, Shi F, Arnold K, et al. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2014;2(3):311–322. doi: 10.1016/j.stemcr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133(7):1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- 13.Cai T, Seymour ML, Zhang H, et al. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J Neurosci. 2013;33(24):10110–10122. doi: 10.1523/JNEUROSCI.5606-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Kon N, Li M, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121(7):1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Johnson JE, Zoghbi HY, et al. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129(10):2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 16.Cheng YF, Tong M, Edge ASB. Destabilization of Atoh1 by E3 ubiquitin ligase Huwe1 and casein kinase 1 is essential for normal sensory hair cell development. J Biol Chem. 2016;291(40):21096–21109. doi: 10.1074/jbc.M116.722124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox BC, Chai R, Lenoir A, et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo . Development. 2014;141(4):816–829. doi: 10.1242/dev.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Arca D, Zhao X, Xu W, et al. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. USA. 2010;107(13):5875–5880. doi: 10.1073/pnas.0912874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis AC. Hearing disorders in the population: first phase findings of the MRC national study of hearing. In: Lutman ME, Haggard MP, editors. Hearing Science and Hearing Disorders. Academic Press Inc. (London) Ltd., London; 1983. pp. 35–60. [DOI] [Google Scholar]
- 20.Dominguez-Brauer C, Hao Z, Elia AJ, et al. Mule regulates the intestinal stem cell niche via the Wnt pathway and targets EphB3 for proteasomal and lysosomal degradation. Cell Stem Cell. 2016;19(2):205–216. doi: 10.1016/j.stem.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flora A, Garcia J, Thaller C, et al. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc. Natl. Acad. Sci. USA. 2007;104(39):15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flora A, Klisch TJ, Schuster G, et al. Deletion of Atoh1 disrupts sonic hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326(5958):1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forge A, Li L, Corwin JT, et al. Ultrastructural evidence for hair cell regeneration in the mammalian inner ear. Science. 1993;259(5101):1616–1619. doi: 10.1126/science.8456284. [DOI] [PubMed] [Google Scholar]
- 24.Forget A, Bihannic L, Cigna SM, et al. SHH signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev Cell. 2014;29(6):649–661. doi: 10.1016/j.devcel.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Fritzsch B. Development of inner ear afferent connections: forming primary neurons and connecting them to the developing sensory epithelia. Brain Res Bull. 2003;60(5-6):423–433. doi: 10.1016/S0361-9230(03)00048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fröhlich A, Kisielow , J. , Schmitz , I. , et al. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science. 2009;324(5934):1576–1580. doi: 10.1126/science.1172815. [DOI] [PubMed] [Google Scholar]
- 27.Gregorieff A, Clevers , H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19(8):877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 28.Gubbels SP, Woessner DW, Mitchell JC, et al. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455(7212):537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall JR, Kow E, Nevis KR, et al. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18(9):3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helms AW, Gowan K, Abney A, et al. Overexpression of MATH1 disrupts the coordination of neural differentiation in cerebellum development. Mol Cell Neurosci. 2001;17(4):671–682. doi: 10.1006/mcne.2000.0969. [DOI] [PubMed] [Google Scholar]
- 31.Hu , Z. Andäng M, Ni D, et al. Neural cograft stimulates the survival and differentiation of embryonic stem cells in the adult mammalian auditory system. Brain Res. 2005;1051(1):137–144. doi: 10.1016/j.brainres.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Huang W.H., Tupal S., Huang TW, et al. Atoh1 governs the migration of postmitotic neurons that shape respiratory effectiveness at birth and chemoresponsiveness in adulthood. Neuron. 2012;75(5):799–809. doi: 10.1016/j.neuron.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Husseman J, Raphael Y. Gene therapy in the inner ear using adenovirus vectors. In: Ryan AF, editor. Gene Therapy of Cochlear Deafness. Advances in Oto-Rhino-Laryngology. Karger, Basel, Vol. 66; 2009. pp. 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Incesulu A, Nadol JB. Correlation of acoustic threshold measures and spiral ganglion cell survival in severe to profound sensorineural hearing loss: implications for cochlear implantation. Ann Otol Rhinol Laryngol. 1998;107(11):906–911. doi: 10.1177/000348949810701102. [DOI] [PubMed] [Google Scholar]
- 35.Izumikawa M, Minoda R, Kawamoto K, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11(3):271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- 36.Jahan I, Pan N, Kersigo J, et al. Beyond generalized hair cells: molecular cues for hair cell types. Hear Res. 2013;297:30–41. doi: 10.1016/j.heares.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansson L, Kim GS, Cheng AG. Making sense of Wnt signaling-linking hair cell regeneration to development. Front Cell Neurosci, 9:66. 2015 doi: 10.3389/fncel.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarriault S, Brou C, Logeat F, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 39.Jeon SJ, Fujioka M, Kim SC, et al. Notch signaling alters sensory or neuronal cell fate specification of inner ear stem cells. J Neurosci. 2011;31(23):8351–8358. doi: 10.1523/JNEUROSCI.6366-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelley MW. Cell adhesion molecules during inner ear and hair cell development, including notch and its ligands. Curr Top Dev Biol. 2003;57:321–356. doi: 10.1016/S0070-2153(03)57011-9. [DOI] [PubMed] [Google Scholar]
- 41.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 42.Kurokawa M, Kim J, Geradts J, et al. A network of substrates of the E3 ubiquitin ligases MDM2 and HUWE1 control apoptosis independently of p53. Sci Signal. 2013;6(274):ra32. doi: 10.1126/scisignal.2003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanford PJ, Lan Y, Jiang R, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21(3):289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Wu J, Yang J, et al. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc. Natl. Acad. Sci. USA. 2015;112(1):166–171. doi: 10.1073/pnas.1415901112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Dearman JA, Cox BC, et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J Neurosci. 2012;32(19):6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, Fang J, Dearman J, et al. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic Atoh1 expression. PLoS ONE. 2014;9(2):e89377. doi: 10.1371/journal.pone.0089377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo LC, Johnson JE, Wuenschell CW, et al. Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5(9):1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- 48.Maass JC, Gu R, Basch ML, et al. Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front Cell Neurosci, 9:110. 2015 doi: 10.3389/fncel.2015.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maksimovic S, Nakatani M, Baba Y, et al. Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature. 2014;509(7502):617–621. doi: 10.1038/nature13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLean WJ, Yin X, Lu L, et al. Clonal expansion of Lgr5-positive cells from mammalian cochlea and high-purity generation of sensory hair cells. Cell Rep. 2017;18(8):1917–1929. doi: 10.1016/j.celrep.2017.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mianné J, Chessum L, Kumar S, et al. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome Med. 2016;8(1):16. doi: 10.1186/s13073-016-0273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miesegaes GR, Klisch TJ, Thaller C, et al. Identification and subclassification of new Atoh1 derived cell populations during mouse spinal cord development. Dev Biol. 2009;327(2):339–351. doi: 10.1016/j.ydbio.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizutari K, Fujioka M, Hosoya M, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77(1):58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrison KM, Miesegaes GR, Lumpkin EA, et al. Mammalian Merkel cells are descended from the epidermal lineage. Dev Biol. 2009;336(1):76–83. doi: 10.1016/j.ydbio.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naujokat C, Šarić T. Concise review: role and function of the ubiquitin-proteasome system in mammalian stem and progenitor cells. Stem Cells. 2007;25(10):2408–2418. doi: 10.1634/stemcells.2007-0255. [DOI] [PubMed] [Google Scholar]
- 56.Ni W, Lin C, Guo L, et al. Extensive supporting cell proliferation and mitotic hair cell generation by in vivo genetic reprogramming in the neonatal mouse cochlea. J Neurosci. 2016;36(33):8734–8745. doi: 10.1523/JNEUROSCI.0060-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohyama T, Mohamed OA, Taketo MM, et al. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133(5):865–875. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- 58.Pan B, Askew C, Galvin A, et al. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat Biotechnol. 2017;35(3):264–272. doi: 10.1038/nbt.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan N, Jahan I, Kersigo J, et al. A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS ONE. 2012;7(1):e30358. doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riccomagno M, Takada S, Epstein D. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19(13):1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richardson RT, Atkinson PJ. Atoh1 gene therapy in the cochlea for hair cell regeneration. Expert Opin Biol Ther. 2015;15(3):417–430. doi: 10.1517/14712598.2015.1009889. [DOI] [PubMed] [Google Scholar]
- 62.Rose MF, Ahmad KA, Thaller C, et al. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for math1. Proc. Natl. Acad. Sci. USA. 2009;106(52):22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39(1):13–25. doi: 10.1016/S0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 64.Ruffault PL, D'Autréaux F, Hayes JA, et al. The retrotrapezoid nucleus neurons expressing Atoh1 and Phox2b are essential for the respiratory response to CO2 . eLife, 4:e07051. 2015 doi: 10.7554/eLife.07051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi F, Cheng YF, Wang XL, et al. β-Catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3' enhancer. J Biol Chem. 2010;285(1):392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi F, Kempfle JS, Edge ASB. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32(28):9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi F, Hu L, Edge ASB. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl. Acad. Sci. USA. 2013;110(34):13851–13856. doi: 10.1073/pnas.1219952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi F, Hu L, Jacques BE, et al. β-Catenin is required for hair-cell differentiation in the cochlea. J Neurosci. 2014;34(19):6470–6479. doi: 10.1523/JNEUROSCI.4305-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shroyer NF, Helmrath MA, Wang VYC, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132(7):2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 70.Shu Y, Tao Y, Wang Z, et al. Identification of adeno-associated viral vectors that target neonatal and adult mammalian inner ear cell subtypes. Hum Gene Ther. 2016;27(9):687–699. doi: 10.1089/hum.2016.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Staecker H, Praetorius M, Baker K, et al. Vestibular hair cell regeneration and restoration of balance function induced by Math1 gene transfer. Otol Neurotol. 2007;28(2):223–231. doi: 10.1097/MAO.0b013e31802b3225. [DOI] [PubMed] [Google Scholar]
- 72.Stevens CB, Davies AL, Battista S, et al. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Dev Biol. 2003;261(1):149–164. doi: 10.1016/S0012-1606(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 73.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9(11):826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 74.Takebayashi S, Yamamoto N, Yabe D, et al. Multiple roles of Notch signaling in cochlear development. Dev Biol. 2007;307(1):165–178. doi: 10.1016/j.ydbio.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 75.Tiveron MC, Pattyn A, Hirsch MR, et al. Role of Phox2b and Mash1 in the generation of the vestibular efferent nucleus. Dev Biol. 2003;260(1):46–57. doi: 10.1016/S0012-1606(03)00213-6. [DOI] [PubMed] [Google Scholar]
- 76.Tsuchiya K, Nakamura T, Okamoto R, et al. Reciprocal targeting of Hath1 and β-catenin by Wnt glycogen synthase kinase 3β in human colon cancer. Gastroenterology. 2007;132(1):208–220. doi: 10.1053/j.gastro.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 77.Urbán N, van den Berg DLC, Forget A, et al. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353(6296):292–295. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.VanDussen KL, Samuelson LC. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev Biol. 2010;346(2):215–223. doi: 10.1016/j.ydbio.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Keymeulen A, Mascre G, Youseff KK, et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J Cell Biol. 2009;187(1):91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Varshavsky A. Naming a targeting signal. Cell. 1991;64(1):13–15. doi: 10.1016/0092-8674(91)90202-A. [DOI] [PubMed] [Google Scholar]
- 81.Wang T, Chai R, Kim GS, et al. Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle. Nat Commun, 6:6613. 2015 doi: 10.1038/ncomms7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48(1):31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 83.Warchol ME, Lambert PR, Goldstein BJ, et al. Regenerative proliferation in inner ear sensory epithelia from adult guinea pigs and humans. Science. 1993;259(5101):1619–1623. doi: 10.1126/science.8456285. [DOI] [PubMed] [Google Scholar]
- 84.WHO (World Health Organization) Deafness and hearing loss. Fact sheet, WHO Media Centre.2017. [Google Scholar]
- 85.Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7(12):1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- 86.Wright MC, Reed-Geaghan EG, Bolock AM, et al. Unipotent, Atoh1 + progenitors maintain the Merkel cell population in embryonic and adult mice. J Cell Biol. 2015;208(3):367–379. doi: 10.1083/jcb.201407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto N, Tanigaki K, Tsuji M, et al. Inhibition of Notch/RBP-J signaling induces hair cell formation in neonate mouse cochleas. J Mol Med. 2006;84(1):37–45. doi: 10.1007/s00109-005-0706-9. [DOI] [PubMed] [Google Scholar]
- 88.Yang H, Xie X, Deng M, et al. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis. 2010;48(6):407–413. doi: 10.1002/dvg.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang Q, Bermingham NA, Finegold MJ, et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294(5549):2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 90.Zhao H, Ayrault O, Zindy F, et al. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22(6):722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao X, Heng JIT, Guardavaccaro D, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10(6):643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao X, D'Arca D, Lim WK, et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17(2):210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3(6):580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- 94.Zou B, Mittal R, Grati M, et al. The application of genome editing in studying hearing loss. Hear Res. 2015;327:102–108. doi: 10.1016/j.heares.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo . Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu Z, Andäng M, Ni D, et al. Cell replacement therapy in the inner ear . Stem Cells Dev. 2006;15(3):449–459. doi: 10.1089/scd.2006.15.449. [DOI] [PubMed] [Google Scholar]