Abstract
In the Alzheimer’s disease (AD) continuum, the prodromal state of mild cognitive impairment (MCI) precedes AD dementia and identifying MCI individuals at risk of progression is important for clinical management. Our goal was to develop generalizable multivariate models that integrate high-dimensional data (multimodal neuroimaging and cerebrospinal fluid biomarkers, genetic factors, and measures of cognitive resilience) for identification of MCI individuals who progress to AD within 3 years. Our main findings were i) we were able to build generalizable models with clinically relevant accuracy (~93%) for identifying MCI individuals who progress to AD within 3 years; ii) markers of AD pathophysiology (amyloid, tau, neuronal injury) accounted for large shares of the variance in predicting progression; iii) our methodology allowed us to discover that expression of CR1 (complement receptor 1), an AD susceptibility gene involved in immune pathways, uniquely added independent predictive value. This work highlights the value of optimized machine learning approaches for analyzing multimodal patient information for making predictive assessments.
Introduction
Alzheimer’s disease (AD) affects approximately 5.5 million people in the United States and more than 30 million people around the world, and imposes substantial personal and societal burdens1. Typically, AD progresses through a preclinical phase with underlying biomarker abnormalities, then a prodromal state of mild cognitive impairment (MCI), and finally frank AD dementia2. Annually, 10–15% of patients diagnosed with MCI progress to AD dementia3. Identification of factors contributing to progression from MCI to AD is crucial for clinical prognostication and risk stratification to guide counseling and selection of potential treatments.
In the last decade, biomarkers from cerebrospinal fluid (CSF), positron emission tomography (PET), and magnetic resonance imaging (MRI) have been increasingly used in AD clinical and research studies to assess the degree of AD related pathology. Increased amyloid pathology measured by decreased CSF Aβ42 and increased cerebral amyloid on PET, as well as increased neuronal injury assessed by increased CSF tau, hypometabolism on FDG-PET, and atrophy on structural MRI, are important factors in assessing the degree of brain changes due to AD pathology and as a surrogate for prediction of progression in individuals with MCI2. In addition4–6, clinical measures such as Mini-Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale-Cognition (ADAS-Cog) which reflect the current level of impairment in individuals have been shown to be useful for prediction of MCI progression4–6.
Two additional important factors in the context of MCI progression to AD are genetic factors and cognitive resilience. AD is a genetically complex disorder, with susceptibility thought to reflect the collective influences of multiple genetic risk and protective factors7. Other than the apolipoprotein E (APOE) ε4 allele, individual genetic variants associated with AD have shown modest population-level effect sizes, in keeping with current hypotheses about the genetic architecture of the disease8. Although the heritability of AD is thought to be 60–80%, beyond the well-studied effect of the APOE ε4 allele9, relatively little is known about the genetic factors specifically related to MCI-to-AD progression10, particularly regarding their added value above known biomarker profiles. Cognitive resilience represents the ability of an individual to delay the deleterious effects of neurodegenerative pathologies on onset of cognitive symptoms11. While cognitive resilience has been widely used to explain the pathology-cognition disconnect in cognitively unimpaired individuals with AD pathology (and normal cognition), its relative influence specifically on MCI-to-AD progression has not been fully evaluated12. Furthermore, the complex relationships among the well-known AD biomarkers, cognitive resilience, and genetic factors remain largely unknown and estimating the independent predictive values provided by each of these factors may open the door to alternative strategies to delay or prevent the onset of dementia.
Several machine learning (ML) based approaches have been proposed for predicting MCI-to-AD progression4,5,13,14 and clinical stage classification15 utilizing high-dimensional clinical and biomarker data among the potential predictors. A limitation of the ML-based approaches that utilize high-dimensional data is the potential for overfitting, such that the classifiers are so optimally trained to fit the primary dataset that they perform poorly on previously unseen test data, ultimately limiting the generalizability and wider interpretability15 of the predictive model. This is particularly important in this study because both a) an unbiased estimation of the contributions of cognitive resilience and genetic factors in predicting MCI-to-AD progression and b) the successful clinical translation of this technique, would require a generalizable model. However, prior studies on the predictability of MCI-to-AD progression have not performed satisfactory assessments of generalizability4,5,13,14. These approaches utilize non-linear classifications methods such as multi-kernel learning and artificial neural networks without establishing the need for non-linear models. Non-linear classification models are more susceptible to overfitting compared to linear classification models in problems involving high-dimensional data since they optimize relatively higher number of model parameters16. Whereas, a pitfall associated with linear models is that they provide suboptimal prediction performance when the data is not linearly separable. Therefore, our goals in this study with respect to the development of a predictive model are a) to understand whether linear classifiers are sufficient to provide clinically relevant accuracies in predicting MCI-to-AD progression based on a set of features derived from multiple modalities and b) to develop an analytical method that evaluates the generalizability of different classifiers and to aid model selection in clinical settings.
In this study, we used a machine learning-based approach and data from a well-characterized clinical cohort to identify individuals with MCI who rapidly progress to AD versus those with a more protracted course. Specifically, using a set of features derived from the ADNI multimodal biomarker and clinical dataset along with genetic factors and cognitive resilience measures, a linear model-based ML framework was developed for predicting MCI-to-AD progression. Based on a test of linear-separability17, we show that linear classification models perform just as good as non-linear models in this setting and a support vector machine classifier with a linear kernel provides the most generalizable performance with a test set area under ROC curve (AUC) value of 0.93. Using this framework, we evaluated the predictive values of previously underexplored factors of cognitive resilience and genetic factors and gauged their relative importance compared to commonly used CSF and imaging predictors. We found that, while the markers of AD pathophysiology (amyloid, tau, and neuronal injury) provided very high predictive values, the genetic factors and brain regions associated with cognitive resilience also displayed independent predictive values.
Materials and Methods
Study participants
This report utilized data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), a multisite longitudinal study of older adults representing clinical stages along the continuum from normal aging to AD18. All study participants provided written informed consent, and study protocols were approved by each local site’s institutional review board. Further information about ADNI, including full study protocols, complete inclusion and exclusion criteria, and data collection and availability can be found at http://www.adni-info.org/. All methods as stated on the website were performed with the relevant guidelines and regulations. Since all the analyses were performed on de-identified ADNI data which is publically available for download, IRB Review was not required. In addition, all methods were carried out in accordance with the approved guidelines.
In choosing ADNI participants to study, we used these inclusion criteria: i) the person had at least three years of follow-up; ii) the person had all the data modalities of interest (as specified below); and iii) the person was diagnosed with MCI at the baseline evaluation. We identified 135 participants who met those criteria. A total of 39 of the 135 progressed to AD within three years of the MCI diagnosis (and are referred to as the MCI-P group); the remaining 96 progressed to AD after three years or remained in MCI until their last follow-up after three years (and are referred to as the MCI-NP group).
Predictive factors
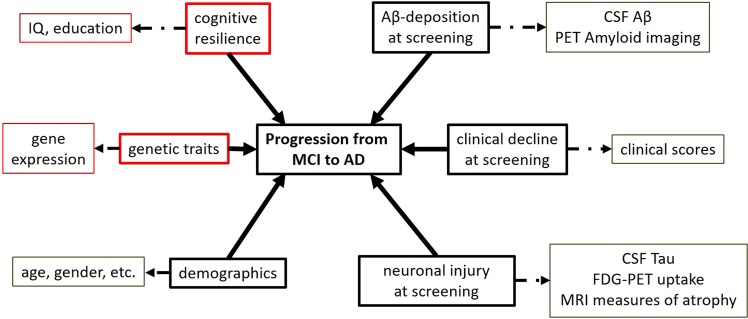
Figure 1 illustrates potential factors that may impact progression from MCI to AD. Below we describe the specifics of the individual biomarkers utilized in this work.
Figure 1.
Factors that can predict progression from MCI to AD. The extent of Aβ-deposition, clinical decline, and neuronal injury at baseline represent the clinical severity of the disease in the MCI subjects. Cognitive resilience, genetic traits, and demographic factors are measures of heterogeneity within the cohort. Aβ-deposition is generally measured using CSF-Aβ and PET amyloid imaging. Neuronal injury is measured using CSF-Tau, FDG-PET, glucose uptake, and MRI atrophy measures. Clinical cognitive decline is measured via clinical scores such as Mini Mental State Examinations (MMSE). Cognitive resilience in a subject can be measured using IQ and level of education. The genetic traits of an individual can be measured using gene expression (RNA) measures. Demographic factors like age, gender, and disease risk factors can also influence the progression. Indicated using solid arrows are factors that influence MCI-to-AD progression and broken lines indicate measurements that were used to measure those factors. Factors and measurements highlighted in red are those that have not been studied in previous MCI-to-AD progression studies in conjunction with the rest.
All the biomarkers utilized in this work were downloaded from the LONI data archive (https://ida.loni.usc.edu/) where the pre-processed ADNI data are hosted.
Cerebrospinal fluid (CSF) biomarkers
CSF levels of amyloid beta (Aβ) and total (T-tau) and phosphorylated (P-tau) tau proteins were assayed by the ADNI Biomarker Core as previously described19.
Magnetic resonance imaging biomarkers
Structural MRI (SMRI) scans at baseline were downloaded and processed as described previously20. FreeSurfer v5.1 was used to obtain volume and thickness measures for standard regions of interest (ROIs) as surrogates for cerebral atrophy. We scaled the volumes by total intracranial volume. In addition, we also included volumetric measurements of hippocampal subfields.
Positron emission tomography (PET) biomarkers
Fluorodeoxyglucose (FDG) and F-florbetapir PET imaging from the baseline visit were analyzed as surrogates for neuronal injury and amyloid pathology, respectively18. For FDG-PET, AD-specific ROIs representing the temporal, angular, and posterior cingulate gyri were utilized21. For F-florbetapir PET, we included regional amyloid deposition assessed by standardized uptake value ratio (SUVR) in the temporal, parietal, and cingulate cortex, as well as a composite global measure of multiple regions22.
Cognition
Scores on the Mini-Mental State Examination (MMSE)23 at baseline were utilized as measures of cognitive performance.
Cognitive resilience
The number of errors on the American National Adult Reading Test (ANART) (which is an estimate of pre-morbid verbal IQ) and years of education were utilized as surrogate measures of cognitive resilience.
Genetic factors
Genotype and gene expression data from peripheral blood samples in ADNI were obtained as previously described24. In this study, we specifically analyzed APOE ε4 allele status (carrier vs. non-carrier) as well as expression data for the top genes with validated associations to AD (APOE, BIN1, CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, and CD2AP) as listed in the AlzGene database25.
Demographics
Gender and age at the baseline visit were utilized in the predictive model.
Data aggregation and ML preprocessing
A total of 94 potential predictive factors were included for analysis. A matrix was generated with 135 rows (representing study participants) and 94 columns (representing the potential predictive factors for MCI-to-AD progression). Prior to further analysis, we centered and standardized all data on a feature-by-feature basis by subtracting the mean and then dividing by the standard deviation.
ML-based prediction framework
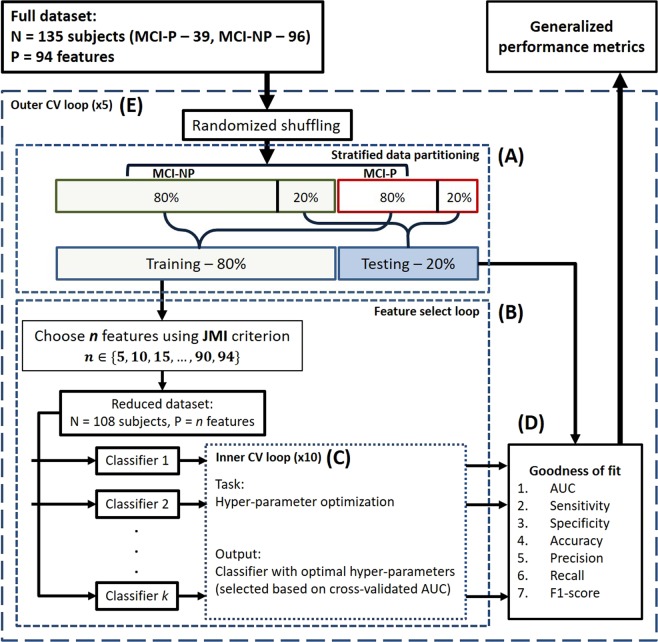
Figure 2 shows a flow diagram of the prediction framework developed for this study. The framework consists of four major steps: information-theoretic feature selection, classifiers and hyper-parameter optimization, goodness-of-fit evaluation, and generalized performance evaluation. They are explained in the following paragraphs. A unique aspect of this workflow is the ability to specify the number of parameters optimized in the classification model by using an information-theoretic feature selection method. This is particularly useful in assessing the generalizability of a classifier as a function of the number of model parameters.
Figure 2.
A flow diagram illustrating the prediction framework. The framework uses a machine learning-based approach to learn a classifier using 80% of the full dataset and to test its performance on the remaining 20% of the data. Specific details of each step in the framework are as follows. (A) Stratified data partitioning: After the order of the subjects is randomized, the MCI-NP and MCI-P groups are separately partitioned with an 80%-training/20%-testing split. The respective training and testing sets from MCI-NP and MCI-P groups are combined to form the overall training and testing data for a single cross-validation. (B) Feature select loop: The top n out of 94 features that best jointly correlate with the class labels (P-MCI or MCI-NP) are selected using the joint mutual information (JMI) criterion. (C) Inner CV loop: A combination of hyper-parameters is selected for each classifier based on a tenfold cross-validation. (D) Goodness-of-fit metrics: The classifier learned in the previous step is tested on the testing dataset and measured on its performance. (E) Outer CV loop: A fivefold cross-validation is utilized to produce generalized performance metrics accounting for non-uniformly distributed data.
Feature reduction using joint mutual information (JMI)
Mutual information between two random variables quantifies the amount of information shared between them. Mutual information is a more comprehensive measure of the relationships between random variables than statistical correlation-based approaches, which measure linear relationships only. Mathematically, mutual information is denoted by and defined for discrete random variables Q and R as shown in Eq. 1, where and denote the alphabets of Q and R, respectively.
| 1 |
When Fk is one of the attributes in a set of attributes and Y is an outcome or class that can be predicted by the attributes, mutual-information-based approaches can be used to select the most predictive attributes. One such approach is to treat the attributes as independent random variables, rank them in descending order based on their mutual information with respect to the outcome Y, and select the top n number of attributes. One limitation of that approach is that useful and parsimonious sets of features should be both (i) individually relevant, and (ii) not highly correlated with each other. Joint mutual information shared between and Y is defined as shown in Eq. 2, where Fk and Y denote the alphabets of Fk and Y, respectively.
| 2 |
A JMI-based feature selection method starts with an empty set of attributes and iteratively adds Fis that, when added, provide the maximum increase in the joint mutual information shared between the set of attributes and the outcome26. It has been shown to be the most stable and flexible feature selection method27 among all the information-theoretic feature selection methods developed to date.
Classification methods
We evaluated a number of classifiers in this study to understand the concepts of linear separability and generalizability in the context of predicting MCI-to-AD progression. Support vector machine (SVM), multiple kernel learning (MKL), and generalized linear models (GLM) with elastic-net regularization are the classifiers used in this study. SVM classifier, because it allows the transformation of features using linear and non-linear kernels, provides us the ability to evaluate the suitability of linear and non-linear classifiers for this problem. On the other hand, MKL allows the application of different kernel transformations for features from different modalities while optimizing more model-parameters than an SVM classifier. Although MKL and SVM are similar classification paradigms, MKL facilitates the integration of multiple modalities at the expense of potential overfitting. Finally, GLM classification with elastic-net regularization is an extension of the commonly known logistic regression classifier with additional regularization to minimize overfitting. In this study, we evaluated the overall generalizability of these classifiers that have distinct optimization objectives.
A support vector machine is a binary classifier that finds the maximum margin hyperplane that separates the two classes in the data28. Suppose that the data being classified are denoted by (N subjects and P features), and that the data from subject i are denoted by . Let us also use to denote the class labels for all the subjects (where and are numerical labels for the two classes), and to denote the class label for subject i. The optimization problem to find the optimal hyperplane (described by weights and intercept term ) is shown in Eq. 3.
| 3 |
Once the optimal hyperplane has been found, the predicted class label for subject i is obtained as the sign of . This formulation assumes that the data are fully linearly separable between the two classes. When that is not the case (but there is still a linear classification task), slack variables and a tolerance parameter (box-constraint) can be introduced to obtain separating hyperplanes that tolerate small misclassification errors.
Dual formulation of SVM has received considerable interest due to its ability to use different kernel transformations of the original feature space without altering the optimization task, and due to its advantages in complexity when the data are high-dimensional29,30. The dual form of the above optimization problem can be written as shown in Eq. 4, where the operation denotes an inner product between two vectors.
| 4 |
Notably, the dual formulation can be expressed simply in terms of the inner product between and (which is a scalar value). Therefore, any transformation of the features can replace the original features, i.e., can be replaced by , where is a feature mapping. Furthermore, for any such feature transformation , we can define a kernel function K such that . With the kernel transformation, the cases where the original data are not linearly separable, may be solved as transforming the data to higher dimensions may introduce linear separation in the transformed domain. Linear, radial basis function (RBF), and polynomial kernels are widely used kernels in this context.
Multiple kernel learning is a classification technique that builds upon the above property of SVM and introduces additional variables in order to weight the kernel transformation of31 individual features. It does so by replacing the kernel function with , where is the kernel function of the mth feature and is its weight. This technique is particularly useful when the data contain different modalities of features that need to be weighted differently. In a standard kernel-SVM, either the kernel transformation is uniformly weighted, or the weights are determined manually. It has been shown that the same dual formulation of SVM can be utilized to find the optimal weightings of the kernel transformation under the constraint that .
Logistic regression is a subclass of generalized linear models (GLM) and is well-suited for binary classification tasks32. It models the response variable as a binomially distributed random variable whose parameters are described by predictor variables and model parameters. Regularization of the model parameters is utilized to avoid effects related to overfitting. The objective function of a regularized GLM model is
| 5 |
where is the logistic function defined as and is the regularization term with the elasticity parameter . The regularization term in general might contain both -norm terms and -norm terms, a situation that is commonly referred to as elastic net regularization with 33. The elasticity parameter ranges in with values of α → 0 approaching ridge regression and α → 1 approaching LASSO regression.
Hyper-parameter optimization
The hyper-parameters of the three classifiers are SVM’s kernel, kernel scale σ, and box-constraint , MKL’s kernel, and GLM’s penalty term and elastic-net parameter . The optimal values of hyper-parameters are chosen by performing a grid-search of parameter values with a k-fold cross-validation within the training dataset.
Goodness-of-fit evaluation and generalized performance metrics
Goodness of fit of a classifier is evaluated by i) predicting the classes of the test dataset by using the classifier that was trained on the training dataset, and ii) comparing the predictions against the true class labels of the test dataset. The comparison is performed using standard performance metrics such as receiver operating characteristics (ROC) curve analysis, area under ROC curve (AUC), sensitivity, specificity, accuracy, precision, recall, and F1-score.
Because of heterogeneity in the data, choosing one partition of training and test datasets is not sufficient to credibly evaluate the performance of a classifier. A common practice to obviate the effect of heterogeneity in the data is to perform an k-fold training-testing cross-validation of the dataset. One run of this procedure is carried out by randomly choosing a subset of the dataset as training data, and testing on the rest of the dataset. That is repeated different times, and the performance metrics of all evaluations are averaged. In addition to this, the proportion of the different classes in the randomly chosen training dataset was kept constant in our approach via a stratified data-partitioning to eliminate any variability in the performance of the classifiers due to class imbalances in the data16. We performed those extra steps to obtain a generalizable set of performance metrics for the analyzed classifier.
Implementation of the prediction framework
The prediction framework was implemented in MATLAB version R2017b and the code is publicly made available at https://gitlab.engr.illinois.edu/varatha2/adni_mci2ad_prediction. We utilized a standard fivefold cross-validation with 80% training data and 20% testing data to evaluate the performance of the classifiers. Training and testing data selection was performed using the stratified data-partitioning approach explained previously. The JMI criterion was used to identify features based on the training dataset and corresponding labels. Using the reduced dataset, we trained SVM with a linear kernel, SVM with an RBF kernel, MKL with a linear kernel, LR with elastic net regularization, and RF. The highest average AUC obtained with a tenfold cross-validation within the training set was used as the selection criterion for the optimal hyper-parameters. We generated performance metrics for each classifier with the best identified hyper-parameters by predicting the labels of the test set and comparing them against the true class labels of the test set. The performance metrics of all five of the (outer) cross-validation runs were averaged to generate generalized performance metrics.
Test for linear separability
Linear separability of a dataset with two classes signifies that a linear classifier (or a hyperplane) can separate the two classes in the data as well as nonlinear classifiers can. We show that by using a slight modification of the histogram of projections17 method, which is considered a test for class separability using SVM classification. The histogram of projections method17, is as follows. An SVM classifier is trained using the training data and a chosen kernel. Then, the test samples are projected on the 1-dimensional line perpendicular to the maximum margin hyperplane learned by the SVM model. Histograms of these projections for the two different classes are compared to evaluate the degree of separation achieved by the classifier. In our approach, we applied a sigmoid transformation to the projections of the test samples to obtain likelihood probabilities, and then, based on the likelihood probabilities, we created histograms of the two classes. This modification was performed in order to eliminate the effects of scaling when different kernel transformations are applied in SVM classification and to maintain the range of histograms within a closed set .
Results
Our results are organized as follows. Table 1 represents the overall summary statistics of the dataset analyzed in this study with some important demographic factors. First, we present the results of our evaluation of whether linear models are sufficient to classify this dataset. Second, we report the analytic method we developed to evaluate generalizability and the results on the generalizability of the analyzed linear classifiers. Third, we report the relative importance of the biomarkers and clinical variables utilized in this study evaluated using the most generalizable classifier selected based on the previous result. Finally, we report the analysis based on LASSO regression34 and a set of identified features that provide independent information in predicting MCI-to-AD progression.
Table 1.
Summary characteristics of participants at baseline.
| MCI-P (n = 39) | MCI-NP (n = 96) | |
|---|---|---|
| Average age (and range of ages) | 72 (55–84) | 71 (55–88) |
| Number of females (and percent female) | 19 (49%) | 48 (50%) |
| Average years of education (and range) | 16 (9–20) | 16 (12–20) |
| Number of APOE carriers (and percent) | 6 (15%) | 3 (3%) |
Here we list the summary statistics of the participants in this study.
Linear separability
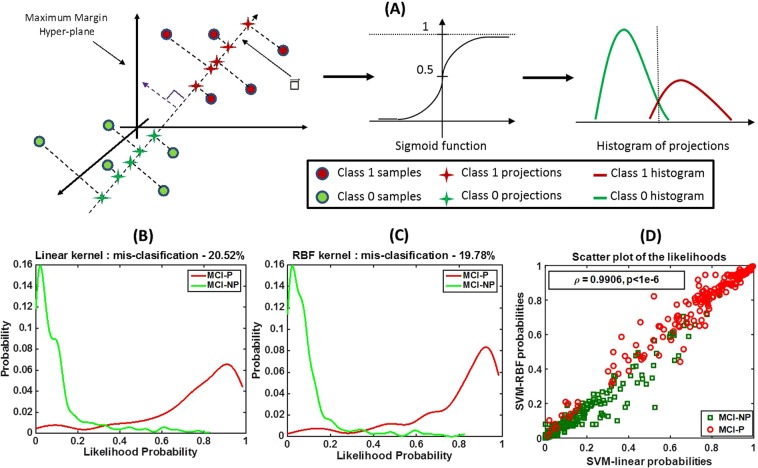
First, we evaluated the linear separability of the data in order to understand whether linear classifiers, which are less susceptible to overfitting, were sufficient to perform prediction of MCI-to-AD progression. We applied the modified histogram of projections method to our dataset by training two SVM classifiers with linear and RBF kernels and by plotting histograms of sigmoid transformations of the projections of test samples.
Figure 3A shows a generic illustration of our approach based on the histogram of projections method. Figure 3B,C show the histograms obtained using our approach for linear and RBF kernels, respectively. The two histograms have similar shapes, and the misclassification errors obtained by choosing an appropriate threshold are also close (20.52% for the linear kernel and 19.78% for the RBF kernel). Figure 3D shows a grouped scatter plot of the probabilities obtained using linear versus RBF kernels for the MCI-P and MCI-NP classes. There is a significantly high correlation between the probabilities obtained using the two kernels ( = 0.99, p < 1e-6), indicating that the classification boundaries learned using the linear and nonlinear kernels are similar. This experiment suggests that the dataset being analyzed is linearly separable. Hence, we limited our analyses to linear classification techniques for the rest of this paper.
Figure 3.
An illustration of the method that evaluates linear separability of the data. We utilize a slightly modified version of the histogram of projections method to evaluate linear separability of the data. (A) A maximum margin hyperplane is learned using SVM with a choice of kernel. All samples are projected onto the line perpendicular to the hyperplane to obtain the projections. The projection lengths are transformed to a probability value via the sigmoid function. Histograms of the probabilities for the two classes are plotted separately. (B) Histograms of the probabilities of the MCI-P and MCI-NP samples in our dataset obtained using a linear kernel. (C) Histograms of the probabilities of the MCI-P and MCI-NP samples in our dataset obtained using an RBF kernel. (D) A grouped scatter plot of the probabilities obtained using linear and RBF kernels for MCI-P and MCI-NP classes. The similar histogram shapes and similar misclassification errors in (B,C), and the high correlation ( = 0.99, p < 1e-6) between the probabilities obtained using the two kernels, indicate that linear and nonlinear kernels result in similar boundaries for classification; hence, this dataset is linearly separable.
Generalizability of classifiers
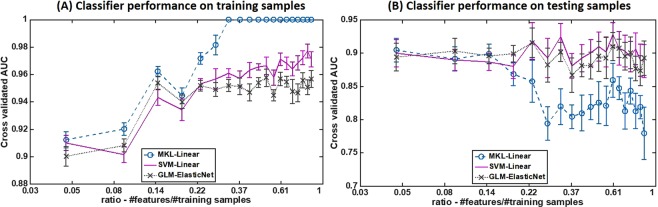
Second, we evaluated the overfitting characteristics of the linear classifiers in order to identify models that were likely to provide highly generalizable performance on this dataset. Three linear classifiers—multiple kernel learning (MKL) with linear kernels, support vector machine (SVM) with linear kernel, and generalized linear model (GLM) with elastic-net regularization—were trained using 80% of the whole dataset as the training set but using only a subset of all the features. We selected the subset (out of a total of 94 features) using the joint mutual information (JMI) criterion, by varying the number of features used each time as {5, 10, 15, …, 85, 90, 94}. We utilized tenfold stratified cross-validation to obtain average training and testing area under ROC curve (AUC) metrics and their respective standard deviations on each occasion. Figure 4 shows plots of the cross-validated AUCs with their standard deviations against the ratio for both training (4A) and test (4B) sets. On the training set, the AUCs of all the classifiers showed better performance with increasing numbers of features used in training. But on the test set, only SVM and GLM showed a relatively steady trend in the AUC. MKL, on the other hand, showed a decreasing testing AUC trend with increasing numbers of features used in training. Those data suggest that the MKL classifier may be overfitting the training data when the ratio is not small (it is >0.14 in this case).
Figure 4.
An evaluation of generalizability of linear classifiers. Three linear classifiers—multiple kernel learning (MKL) with linear kernels, support vector machine (SVM) with a linear kernel, and generalized linear model (GLM) with elastic-net regularization—were trained multiple times using 80% of the data as the training set but with a variable number of features each time. We plotted the cross-validated AUCs with their standard deviations against the ratio for both training (A) and testing (B) sets. While all the classifiers show an increasing AUC trend on the training set with increased numbers of features used in training, only SVM and GLM show a relatively steady trend on the test set. MKL on the other hand, shows a decreasing testing AUC trend with increased numbers of features used in training.
Those results suggest that both SVM with a linear kernel and GLM with elastic-net regularization have good generalizability (with a consistent AUC value of approximately 0.9) regardless of the ratio of the number of features to the number of training samples, and MKL had good generalizable performance only when this ratio was small. Further, particularly for this dataset, all linear classifiers showed a reasonable test set predictability when the number of features was appropriately chosen during the training phase.
Prediction performance of linear classifiers
Based on the previous experiment, we selected the subset of features that provided the most generalizable predictive performance for each linear classifier. Table 2 lists all the cross-validated goodness-of-fit metrics (AUC, sensitivity, specificity, accuracy, precision, recall, and F1-score) obtained using MKL with linear kernels, SVM with a linear kernel and GLM with elastic-net regularization, with their respective subsets of features. All linear classifiers produced comparable results, while SVM with a linear kernel provided the best result.
Table 2.
Cross-validated goodness-of-fit metrics for linear classifiers.
| AUC | Sensitivity | Specificity | Accuracy | Precision | Recall | F1-score | Training size | #Features | |
|---|---|---|---|---|---|---|---|---|---|
| MKL-Linear | 0.90(0.02) | 0.86(0.03) | 0.78(0.03) | 0.80(0.02) | 0.63(0.03) | 0.86(0.03) | 0.72(0.03) | 80% | 5 |
| SVM-Linear | 0.93(0.02) | 0.93(0.03) | 0.77(0.03) | 0.81(0.02) | 0.64(0.03) | 0.93(0.02) | 0.75(0.03) | 80% | 65 |
| GLM-Elastic Net | 0.92(0.02) | 0.91(0.02) | 0.76(0.02) | 0.81(0.02) | 0.62(0.03) | 0.91(0.04) | 0.74(0.03) | 80% | 25 |
Here we list the goodness-of-fit metrics (AUC, sensitivity, specificity, accuracy, precision, recall, and F1-score) obtained for the test dataset (20% of the whole dataset), using the subset of features that provided the most generalizable result, as shown in Fig. 4. Their average values and standard deviations were computed using a tenfold stratified cross-validation.
Predictive ability of individual modalities
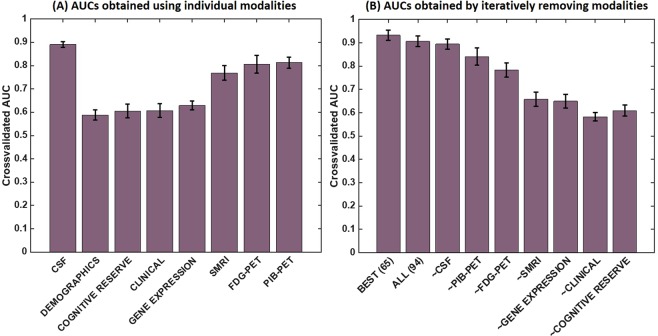
To understand the predictive ability of individual modalities in a statistically impartial manner, we restricted our analysis in this subsection to only one linear classifier. Based on Table 2, we chose the SVM classifier with a linear kernel, as it provided the best performance in terms of the AUC metric. Then, we repeated our classification procedure using a stratified tenfold cross-validation for each of the modalities (using all the variables in the respective modality each time). Figure 5A shows a bar-chart of the average AUCs obtained by individual modalities. CSF proteomic markers (including Aβ, total-Tau, and phosphorylated-Tau) provided the best individual prediction capability, followed by the imaging markers in the order amyloid PET, FDG-PET, and SMRI. Gene expression, clinical, cognitive resilience, and demographic markers showed lower predictive ability than did CSF and imaging-based markers. Figure 5B shows a different bar-chart of the average AUCs obtained by iteratively removing the respective features of the individual modalities in a descending order based on their individual predictive abilities seen in Fig. 5A. A gradual decline in the AUC is observed, with the largest drop occurring when SMRI features were removed.
Figure 5.
An evaluation of the relative predictive abilities of modalities. (A) The cross-validated AUCs obtained using an SVM classifier with a linear kernel separately for each of the modalities. (B) The AUCs obtained by iteratively removing the modalities in a descending order based on AUCs obtained in (A). (Modalities with high AUC values per (A) were removed first.) In (B), “~X” indicates that modality X was removed while the modalities that are less predictive than X were kept, to obtain the respective AUC.
Best predictors of MCI-to-AD progression
Next, we sought to identify a minimal set of predictors required to model MCI-to-AD progression. This was motivated by an observation about Fig. 5: although CSF features provided the best individual AUC (Fig. 5A), the removal of CSF features from the data (Fig. 5B) did not result in a substantial drop in the AUC (the reduction in AUC is within the error limits). That suggests that other modalities provide overlapping information that may be correlated with the CSF markers, and also indicates that all the modalities might have a notable level of correlation with each other. We used a generalized linear model (GLM) classification method with L1 regularization (known as LASSO regression) to identify a sparse set of features with minimal within-correlation and maximal prediction potential of clinical progression34. We chose to use LASSO regression instead of SVM classification for this task because the feature weights assigned by the SVM classifier with a linear kernel do not represent the independent predictive values of individual features when the features themselves are correlated with each other. However, the L1-norm-based regularization of LASSO regression may enable identification of the smallest possible set of predictors, since it penalizes the classifiers that utilize a large number of features in the resulting predictive function. Table 3 shows the results from this approach, identifying the factors that provided the best spread of independent information to predict MCI-to-AD progression. This model included PET, MRI, and CSF variables in addition to age and expression of CR1 (complement receptor 1) and was able to predict MCI-to-AD progression with an AUC of 0.92.
Table 3.
A minimal set of markers that are predictive of MCI-to-AD progression. A generalized linear model (GLM) classifier with L1 regularization was utilized to identify a small set of minimally correlated features that can predict MCI-to-AD progression.
| CSF | PIB-PET | FDG-PET | SMRI | Genetic | Demographics |
|---|---|---|---|---|---|
| Total-Tau | COMPOSITE REFNORM | Angular-Left | Cortical Thickness Average of Insula | CR1 | AGE |
| Amyloid Beta | TEMPORAL | Temporal-Left | Cortical Thickness Average of SuperiorFrontal | ||
| Phosphorylated-Tau | Cingulum-Bilateral | Cortical Thickness Average of InferiorParietal | |||
| Temporal-Right | Cortical Thickness Average of Parahippocampal | ||||
| Cortical Thickness Average of MedialOrbitofrontal | |||||
| Cortical Thickness Average of CaudalAnteriorCingulate | |||||
| Cortical Thickness Average of SuperiorParietal | |||||
| Cortical Thickness Average of IsthmusCingulate | |||||
| Cortical Thickness Average of ParsTriangularis | |||||
| Cortical Thickness Average of PosteriorCingulate | |||||
| Hippocampal Subfield Volume of Subiculum |
Those features are categorized here based on their feature modality.
Discussion
In this work, we developed an accurate and generalizable machine learning-based methodology for predicting short-term progression of MCI to AD dementia in a well-characterized clinical cohort. A unique aspect of this work was our evaluation of important genetic markers and cognitive resilience markers in addition to the neuroimaging biomarkers and clinical examination. The results suggest that a combination of selected neuroimaging, blood and CSF biomarkers, and demographic traits reflect the underlying pathophysiology and factors that drive clinical progression in the AD spectrum. The methodology allowed us to discover that expression of CR1 and AD-pattern neuropathology and neurodegeneration (MRI measures in the frontal lobes) in brain regions associated with cognitive resilience added independent predictive values in predicting MCI-to-AD progression. We also evaluated the relative importance of individual predictors and identified a minimal set of predictors that are important for predictive modeling of MCI progression to AD.
Predictors of MCI progression to AD
The broad pathogenesis of AD has been well-described conceptually, from initial alterations in molecular and cellular pathology to neurodegeneration and eventually to clinical impairment sufficient to cause dementia2. However, it is not yet fully understood what specific factors “shift the curve” to either promote or inhibit the development of dementia. As a result, we initially approached our study in a relatively unbiased fashion, casting a broad net for potential predictive factors out of multidimensional clinical, neuroimaging, and other biomarker data. Through our ML approach, we found that measures of AD neuropathology (CSF amyloid and tau and cerebral amyloid assessed by PET) and neuronal injury (assessed by FDG-PET and structural MRI) explained the most variance in separating fast versus slow progression from MCI to AD dementia. The importance of biomarkers in predicting progression has been studied in the past, but the approach we took allowed us to rank various predictors (including the biomarkers) as well as identify the minimal predictor set that are key to the overall models. When the biomarkers were excluded, predictors such as cognitive performance, cognitive resilience, and expression of selected genes individually explained less of the outcome variance and collectively appeared to offer less new information to the predictive model. Our results support the hypothesis that CSF biomarkers and imaging can be used as surrogates for neuropathology and brain health and can serve as key indicators of future MCI prognosis.
Information added by genetic factors and cognitive resilience
An important aspect of this work is the incorporation of genetic factors and cognitive resilience into our predictive model. Most cases of AD are thought to be genetically complex, with multiple factors presumed to contribute to susceptibility and protection, with the largest known factor being the APOE ε4 allele35. Understanding of the genetic architecture of AD has greatly expanded over the last decade as a result of genome-wide and rare-variant association studies, among other approaches7. However, the specific genetic factors that influence progression at various clinical stages of AD are still not well-characterized. Our ML approach identified expression of CR1 as a key factor in predicting fast versus slow progression to AD dementia, which is a novel finding of this study. Interestingly, our final minimal set model included CR1 expression but did not include APOE ε4 allele status, suggesting that the former provided unique information for clinical course prediction while the latter was already represented by surrogate biomarkers, specifically amyloid deposition36. Polymorphisms (genotype variations) in CR1 have been associated with AD status and endophenotypes in numerous large-scale studies37–42. CR1 encodes a receptor involved in complement activation, a major immune mechanism with a wide array of functions; it has been proposed that it impacts the clearance of amyloid in AD43. Previous findings on CR1 have been illuminated by a more recent and extensive body of work highlighting immune system pathways as potential cruxes in AD pathophysiology44–48. Our new findings relating CR1 expression to progression from MCI to AD dementia provide further validation of that previous work and argue for greater focus on CR1—and on genetic variation in MCI-to-AD progression more broadly49—to enable better understanding of the mechanisms underlying AD and its clinical trajectories.
It is not unexpected that in our ML-based prediction model for MCI to AD progression, CR1 gene expression contributed relatively less to explaining variance than biomarkers of neuropathology and neuronal injury. Surrogates of pathology (fluid biomarker and neuroimaging) reflect the extent of disease related changes and are likely more proximal to clinical manifestations of disease compared to gene expression which may be upstream of these changes and thus possibly modifiable by concomitant forces over time. An example is the relationship between APOE ɛ4 and amyloid: while APOE ɛ4 is a key driver of amyloidosis, the measured effect of amyloid load on cognition is significantly stronger than the impact of APOE ɛ4 on cognition36,50. Complementary methods to incorporate collective effects of multiple genes, including pathway analyses and polygenic risk scoring51–54, could be incorporated into our ML approach in future. In addition, for this study we analyzed microarray gene expression (rather than genotype) data, which offers the conceptual advantage of being a dynamic (rather than static) marker of late-life conditions but which has the disadvantage of representing a downstream effect of influences earlier in life with the potential to be modified by interactions with other heritable and non-heritable factors. Finally, for this study we limited the focus to the AlzGene Top 10 list, but other genes with less well-known population-level effects on case-control status may have larger contributions to late-life clinical progression which could be discovered with an unbiased genome-wide approach. Despite these constraints, our approach serves as a proof of concept that incorporating genetic data can add value to ML-based clinical prediction models and highlights CR1 for further study on its potential effects at the inflection of MCI to AD progression.
Although cognitive resilience has been shown to contribute significantly to delaying the onset of clinical impairment, neither education nor verbal IQ was identified as a key predictor of MCI progression. There are two possible explanations for those results. (i) Preservation of structure or lack of atrophy, as observed via MRI, may be a better surrogate of resilience than are estimates of pre-morbid IQ or educational attainment. It has been shown that cognitive resilience is captured well by greater volume and metabolism, especially in the frontal and cingulate regions55. (ii) Cognitive resilience may be more relevant before the onset of cognitive symptoms or impairment12, and thus may be of less importance in cognitively impaired individuals, who are the focus of this study.
Strengths of the computational approach
We presented a machine learning-based prediction framework for predicting MCI-to-AD progression using state-of-the-art classification techniques under a widely accepted cross-validation setup that accounts for sample bias and unbiased hyper-parameter optimization. Our results indicate that linear classifiers are sufficient to identify patients who have the potential to progress from MCI to AD within 3 years, via the use of multimodal measurements including imaging, CSF, genetic, and clinical data. The reason for this might be that all the features were engineered prior to application of machine learning in such a way that a dichotomy within a feature correlates with disease progression. Linear classifiers are less susceptible to overfitting and thus can be appropriate for clinical translation. In addition, we developed an analytic method for evaluating the generalizability of three different linear classification approaches, namely GLM-ElasticNet, multi-kernel SVM with linear kernels, and standard SVM with a linear kernel. Multi-kernel SVM showed a slightly greater tendency to overfit than the other linear classifiers in our experiments. Because multi-kernel SVM optimizes twice as many model parameters as standard SVM or GLM, it seems that the number of samples used in this study was not enough to train multi-kernel SVM in a generalizable manner. However, as indicated in the experiments, when the number of features used in the classification was small, multi-kernel SVM seemed to provide a more generalizable performance compared to when more features were used in the classification, since there were fewer optimized model parameters in the former scenario. Even so, standard SVM and GLM provided better generalizability and achieved higher classification accuracies with more features. In comparison with previously published predictors of MCI-to-AD progression, our approach achieves superior predictive performance, with a 0.93 area under ROC curve providing a 6% improvement over the current best predictor4, which utilized only CSF and imaging modalities for the prediction of short-term MCI-to-AD progression. It is our firm belief that AUCs of test sets are the most reliable measure of a classifier’s general performance, and hence that our evaluation presents a fair assessment of the proposed ML-based predictor.
Limitations and future directions
Our study has several limitations, which may be addressed by future work. We used publicly available data from the ADNI and ultimately analyzed only a modest sample, because we could only use data from patients for whom we had complete data for all the assessed predictive variables. Replication of our methods in similar and larger cohorts would help validate our approach. In addition, application of our final model to an independent dataset could extend our findings by assessing their success in predicting clinical progression from known baseline data. Further, use of alternative surrogate variables—such as verbal episodic memory performance (rather than MMSE scores) as a reflection of cognitive impairment, and lifetime intellectual enrichment (rather than, or in addition to, the AMNART or educational attainment) as a reflection of cognitive resilience—as well as newer biomarkers such as Tau-PET might produce a more optimal model. We also chose to focus on differentiation of faster versus slower progression from MCI. However, it is not yet known whether the factors that are important at that clinical stage are more broadly generalizable to earlier stages of AD at which clinical intervention might have greater impact. Finally, a broader investigation of genetic factors, including genome-wide genotype and expression data, might increase the accuracy of our model and raise additional loci for follow-up.
In conclusion, we used a machine learning approach to integrate data from multiple sources to predict progression from MCI to AD dementia. By analyzing the linear-separability of the dataset, we established that linear classifiers are sufficient for predicting MCI-to-AD progression in the proposed setting. Furthermore, we also developed an analytical method to assess the generalizability of classifiers based on the number of model parameters. We also showed that, the markers of AD pathophysiology (amyloid, tau, and neuronal injury) provided very high predictive values, the genetic factors and brain regions associated with cognitive resilience also displayed independent predictive values. Our findings, particularly in relation to the impact of genetic factors and cognitive resilience in the setting of biomarkers and clinical tests, warrant further investigation using larger datasets and may have downstream benefits leading to improved prognostication and risk stratification in AD.
Acknowledgements
This work was partly supported by National Institute of Health grants (R01 NS097495 and R01 AG056366), National Science Foundation grants (CNS-1337732 and CNS-1624790), Mayo Clinic and Illinois Alliance Fellowship for Technology-based Healthcare Research and an IBM faculty award. We would like to thank Dr. Duygu Tosun for providing clarification regarding the ADNI Freesurfer data. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author Contributions
Y.V. and P.V. designed the study. Y.V. developed the methods, conducted the analyses, and drafted the paper. Y.V., V.R., R.I. and P.V. participated in the interpretation of the results and critical revision of the manuscript. All the ADNI consortium authors contributed to the acquisition, processing, and public availability of ADNI data.
Data Availability
Data used in preparation of this article were obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). Thus, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in this analysis or the writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf/. For additional details and up-to-date information, see http://www.adni-info.org.
Competing Interests
The authors declare no competing interests.
Footnotes
A comprehensive list of consortium members appears at the end of the paper
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yogatheesan Varatharajah, Email: varatha2@illinois.edu.
Prashanthi Vemuri, Email: Vemuri.Prashanthi@mayo.edu.
For the Alzheimer’s Disease Neuroimaging Initiative:
Michael W. Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, Andrew J. Saykin, William Jagust, John Q. Trojanowki, Arthur W. Toga, Laurel Beckett, Robert C. Green, John Morris, Leslie M. Shaw, Zaven Khachaturian, Greg Sorensen, Maria Carrillo, Lew Kuller, Marc Raichle, Steven Paul, Peter Davies, Howard Fillit, Franz Hefti, David Holtzman, M. Marcel Mesulam, William Potter, Peter Snyder, Adam Schwartz, Tom Montine, Ronald G. Thomas, Michael Donohue, Sarah Walter, Devon Gessert, Tamie Sather, Gus Jiminez, Archana B. Balasubramanian, Jennifer Mason, Iris Sim, Danielle Harvey, Matthew Bernstein, Nick Fox, Paul Thompson, Norbert Schuff, Charles DeCArli, Bret Borowski, Jeff Gunter, Matt Senjem, David Jones, Kejal Kantarci, Chad Ward, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, Nigel J. Cairns, Erin Franklin, Lisa Taylor-Reinwald, Virginia Lee, Magdalena Korecka, Michal Figurski, Karen Crawford, Scott Neu, Tatiana M. Foroud, Steven Potkin, Kelley Faber, Sungeun Kim, Kwangsik Nho, Leon Thal, Neil Buckholtz, Marilyn Albert, Richard Frank, John Hsiao, Jeffrey Kaye, Joseph Quinn, Lisa Silbert, Betty Lind, Raina Carter, Sara Dolen, Lon S. Schneider, Sonia Pawluczyk, Mauricio Beccera, Liberty Teodoro, Bryan M. Spann, James Brewer, Helen Vanderswag, Adam Fleisher, Judith L. Heidebrink, Joanne L. Lord, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Rachelle S. Doody, Javier Villanueva-Meyer, Valory Pavlik, Victoria Shibley, Munir Chowdhury, Susan Rountree, Mimi Dang, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Beau Ances, Maria Carroll, Mary L. Creech, Erin Franklin, Mark A. Mintun, Stacy Schneider, Angela Oliver, Daniel Marson, David Geldmacher, Marissa Natelson Love, Randall Griffith, David Clark, John Brockington, Erik Roberson, Hillel Grossman, Effie Mitsis, Raj C. Shah, Leyla deToledo-Morrell, Ranjan Duara, Maria T. Greig-Custo, Warren Barker, Chiadi Onyike, Daniel D’Agostino, Stephanie Kielb, Martin Sadowski, Mohammed O. Sheikh, Anaztasia Ulysse, Mrunalini Gaikwad, P. Murali Doraiswamy, Jeffrey R. Petrella, Salvador Borges-Neto, Terence Z. Wong, Edward Coleman, Steven E. Arnold, Jason H. Karlawish, David A. Wolk, Christopher M. Clark, Charles D. Smith, Greg Jicha, Peter Hardy, Partha Sinha, Elizabeth Oates, Gary Conrad, Oscar L. Lopez, Mary Ann Oakley, Donna M. Simpson, Anton P. Porsteinsson, Bonnie S. Goldstein, Kim Martin, Kelly M. Makino, M. Saleem Ismail, Connie Brand, Adrian Preda, Dana Nguyen, Kyle Womack, Dana Mathews, Mary Quiceno, Allan I. Levey, James J. Lah, Janet S. Cellar, Jeffrey M. Burns, Russell H. Swerdlow, William M. Brooks, Liana Apostolova, Kathleen Tingus, Ellen Woo, Daniel H. S. Silverman, Po H. Lu, George Bartzokis, Neill R Graff-Radford, Francine Parfitt, Kim Poki-Walker, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Jared R. Brosch, Scott Herring, Christopher H. van Dyck, Richard E. Carson, Martha G. MacAvoy, Pradeep Varma, Howard Chertkow, Howard Bergman, Chris Hosein, Sandra Black, Bojana Stefanovic, Curtis Caldwell, Ging-Yuek Robin Hsiung, Benita Mudge, Vesna Sossi, Howard Feldman, Michele Assaly, Elizabeth Finger, Stephen Pasternack, Irina Rachisky, John Rogers, Dick Trost, Andrew Kertesz, Charles Bernick, Donna Munic, Emily Rogalski, Kristine Lipowski, Sandra Weintraub, Borna Bonakdarpour, Diana Kerwin, Chuang-Kuo Wu, Nancy Johnson, Carl Sadowsky, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Jerome Yesavage, Joy L. Taylor, Barton Lane, Allyson Rosen, Jared Tinklenberg, Marwan N. Sabbagh, Christine M. Belden, Sandra A. Jacobson, Sherye A. Sirrel, Neil Kowall, Ronald Killiany, Andrew E. Budson, Alexander Norbash, Patricia Lynn Johnson, Thomas O. Obisesan, Saba Wolday, Joanne Allard, Alan Lerner, Paula Ogrocki, Curtis Tatsuoka, Parianne Fatica, Evan Fletcher, Pauline Maillard, John Olichney, Charles DeCarli, Owen Carmichael, Smita Kittur, Michael Borrie, T.-Y. Lee, Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Pierre Tariot, Anna Burke, Ann Marie Milliken, Nadira Trncic, Adam Fleisher, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Douglas W. Scharre, Maria Kataki, Brendan Kelly, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Laura A. Flashman, Marc Seltzer, Mary L. Hynes, Robert B. Santulli, Kaycee M. Sink, Leslie Gordineer, Jeff D. Williamson, Pradeep Garg, Franklin Watkins, Brian R. Ott, Geoffrey Tremont, Lori A. Daiello, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, David Perry, Jacobo Mintzer, Kenneth Spicer, David Bachman, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Susan K. Schultz, Karen Ekstam Smith, Hristina Koleva, Ki Won Nam, Hyungsub Shim, Norman Relkin, Gloria Chaing, Michael Lin, Lisa Ravdin, Amanda Smith, Balebail Ashok Raj, and Kristin Fargher
References
- 1.Alzheimer’s Association. 2018 Alzheimer’s disease facts and figures. Alzheimer’s & Dementia14(3), 367–429, 10.1016/j.jalz.2018.02.001 (2018). [DOI] [PubMed]
- 2.Jack CR, Jr., et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. The Lancet. Neurology. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farias ST, Mungas D, Reed BR, Harvey D, DeCarli C. Progression of mild cognitive impairment to dementia in clinic- vs community-based cohorts. Arch Neurol. 2009;66:1151–1157. doi: 10.1001/archneurol.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korolev IO, Symonds LL, Bozoki AC. Predicting Progression from Mild Cognitive Impairment to Alzheimer’s Dementia Using Clinical, MRI, and Plasma Biomarkers via Probabilistic Pattern Classification. PloS one. 2016;11:e0138866. doi: 10.1371/journal.pone.0138866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts RO, et al. Higher risk of progression to dementia in mild cognitive impairment cases who revert to normal. Neurology. 2014;82:317–325. doi: 10.1212/wnl.0000000000000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tifratene K, Robert P, Metelkina A, Pradier C, Dartigues JF. Progression of mild cognitive impairment to dementia due to AD in clinical settings. Neurology. 2015;85:331–338. doi: 10.1212/wnl.0000000000001788. [DOI] [PubMed] [Google Scholar]
- 7.Naj AC, Schellenberg GD. & Alzheimer’s Disease Genetics, C. Genomic variants, genes, and pathways of Alzheimer’s disease: An overview. Am J Med Genet B Neuropsychiatr Genet. 2017;174:5–26. doi: 10.1002/ajmg.b.32499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Cauwenberghe C, Van Broeckhoven C, Sleegers K. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med. 2016;18:421–430. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elias-Sonnenschein LS, Viechtbauer W, Ramakers IH, Verhey FR, Visser PJ. Predictive value of APOE-epsilon4 allele for progression from MCI to AD-type dementia: a meta-analysis. Journal of neurology, neurosurgery, and psychiatry. 2011;82:1149–1156. doi: 10.1136/jnnp.2010.231555. [DOI] [PubMed] [Google Scholar]
- 10.Reitz C, Mayeux R. Use of genetic variation as biomarkers for mild cognitive impairment and progression of mild cognitive impairment to dementia. Journal of Alzheimer’s disease: JAD. 2010;19:229–251. doi: 10.3233/JAD-2010-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. The Lancet. Neurology. 2012;11:1006–1012. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vemuri P, et al. Association of lifetime intellectual enrichment with cognitive decline in the older population. JAMA neurology. 2014;71:1017–1024. doi: 10.1001/jamaneurol.2014.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landau SM, et al. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75:230–238. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D, Shen D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PloS one. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suk HI, Lee SW, Shen D. & Alzheimer’s Disease Neuroimaging, I. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage. 2014;101:569–582. doi: 10.1016/j.neuroimage.2014.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastie, T., Friedman, J. & Tibshirani, R. In The Elements of Statistical Learning: Data Mining, Inference, and Prediction 193–224 (Springer New York, 2001).
- 17.Cherkassky V, Dai WY. Empirical Study of the Universum SVM Learning for High-DimensionalData. Lect Notes Comput Sc. 2009;5768:932–941. doi: 10.1007/978-3-642-04274-4_96. [DOI] [Google Scholar]
- 18.Weiner MW, et al. Impact of the Alzheimer’s Disease Neuroimaging Initiative, 2004 to 2014. Alzheimer’s & Dementia. 2015;11:865–884. doi: 10.1016/j.jalz.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw LM, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jack CR, Jr., et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. Journal of magnetic resonance imaging: JMRI. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jagust WJ, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2010;6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landau SM, et al. Measurement of longitudinal beta-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J Nucl Med. 2015;56:567–574. doi: 10.2967/jnumed.114.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. Journal of psychiatric research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 24.Saykin, A. J. et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plan. Alzheimer’s & dementia: the journal of the Alzheimer’s Association (In Press) (2015). [DOI] [PMC free article] [PubMed]
- 25.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nature genetics. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 26.Yang H. H., M. J. Feature Selection Based on Joint Mutual Information. Proceedings of International ICSC Symposium on Advances in Intelligent Data Analysis, 22–25 (1999).
- 27.Brown G, Pocock A, Zhao MJ, Lujan M. Conditional Likelihood Maximisation: A Unifying Framework for Information Theoretic Feature Selection. J Mach Learn Res. 2012;13:27–66. [Google Scholar]
- 28.Cortes C, Vapnik V. Support-Vector Networks. Mach Learn. 1995;20:273–297. doi: 10.1007/Bf00994018. [DOI] [Google Scholar]
- 29.Scholkopf B, Smola AJ. A short introduction to learning with kernels. Lect Notes Artif Int. 2002;2600:41–64. [Google Scholar]
- 30.Scholkopf B, et al. Comparing support vector machines with Gaussian kernels to radial basis function classifiers. Ieee T Signal Proces. 1997;45:2758–2765. doi: 10.1109/78.650102. [DOI] [Google Scholar]
- 31.Gonen M, Alpaydin E. Multiple Kernel Learning Algorithms. J Mach Learn Res. 2011;12:2211–2268. [Google Scholar]
- 32.Hosmer, D. W., Lemeshow, S. & Sturdivant, R. X. Applied Logistic Regression, 3rd Edition. Wiley Ser Probab St, 1–500, 10.1002/9781118548387 (2013).
- 33.Zou H, Hastie T. Regularization and variable selection via the elastic net (vol. B 67, pg 301, 2005) J Roy Stat Soc B. 2005;67:768–768. doi: 10.1111/j.1467-9868.2005.00527.x. [DOI] [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the Lasso. J Roy Stat Soc B Met. 1996;58:267–288. [Google Scholar]
- 35.Ramanan VK, Saykin AJ. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. American journal of neurodegenerative disease. 2013;2:145–175. [PMC free article] [PubMed] [Google Scholar]
- 36.Vemuri P, et al. Age, vascular health, and Alzheimer disease biomarkers in an elderly sample. Ann Neurol. 2017;82:706–718. doi: 10.1002/ana.25071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert JC, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature genetics. 2009;41:1094–U1068. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 38.Keenan BT, et al. A coding variant in CR1 interacts with APOE-epsilon4 to influence cognitive decline. Human molecular genetics. 2012;21:2377–2388. doi: 10.1093/hmg/dds054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barral S, et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012;78:1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chibnik LB, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jun G, et al. Meta-analysis Confirms CR1, CLU, and PICALM as Alzheimer Disease Risk Loci and Reveals Interactions With APOE Genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thambisetty M, et al. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73:422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sims R, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nature genetics. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones L, et al. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PloS one. 2010;5:e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamelin L, et al. Early and protective microglial activation in Alzheimer’s disease: a prospective study using 18F-DPA-714 PET imaging. Brain. 2016;139:1252–1264. doi: 10.1093/brain/aww017. [DOI] [PubMed] [Google Scholar]
- 48.Ramanan VK, et al. GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain. 2015;138:3076–3088. doi: 10.1093/brain/awv231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lacour, A. et al. Genome-wide significant risk factors for Alzheimer’s disease: role in progression to dementia due to Alzheimer’s disease among subjects with mild cognitive impairment. Mol Psychiatry, 10.1038/mp.2016.18 (2016). [DOI] [PMC free article] [PubMed]
- 50.Lim YY, et al. Stronger effect of amyloid load than APOE genotype on cognitive decline in healthy older adults. Neurology. 2012;79:1645–1652. doi: 10.1212/WNL.0b013e31826e9ae6. [DOI] [PubMed] [Google Scholar]
- 51.Desikan RS, et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 2017;14:e1002258. doi: 10.1371/journal.pmed.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiology of aging. 2017;49:214 e217–214 e211. doi: 10.1016/j.neurobiolaging.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 53.Mormino EC, et al. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology. 2016;87:481–488. doi: 10.1212/WNL.0000000000002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arenaza-Urquijo EM, et al. Relationships between years of education and gray matter volume, metabolism and functional connectivity in healthy elders. NeuroImage. 2013;83:450–457. doi: 10.1016/j.neuroimage.2013.06.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used in preparation of this article were obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). Thus, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in this analysis or the writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf/. For additional details and up-to-date information, see http://www.adni-info.org.