Plasma cell myeloma (PCM) is a clonal PC neoplasm that represents the second most common and mostly incurable hematopoietic malignancy comprising ~20% of all hematologic-related cancer deaths in the United States1,2. The diagnosis of PCM requires evidence of clonal expansion of PCs representing ≥10% of the bone marrow, elevated monoclonal protein concentration in the serum and/or urine, lytic bone lesions, and/or end organ damage3–5. Multiple risk stratification systems and prognostic-based therapeutic approaches have been devised incorporating a variety of clinical metrics including host factors such as age, performance status, comorbidities, serum beta-2 microglobulin, albumin, lactate dehydrogenase, and proliferation indices.
Cytogenetic analysis plays a critical role in risk stratification of patients with newly diagnosed PCM and subsequent disease progression3–5. Recurrent structural and numeric chromosomal abnormalities identified by fluorescence in situ hybridization (FISH) stratifies patients into standard- or high-risk groups that mainly estimate overall survival (OS)3–5. High-risk cytogenetic abnormalities include immunoglobulin heavy chain (IGH) translocations; t(4;14) (IGH/FGFR3), t(14;16) (IGH/MAF), t(14;20) (IGH/MAFB),17p (TP53) deletions, 1p deletions, and 1q duplications. Standard-risk abnormalities include hyperdiploidy (47–57 chromosomes) often involving gains of odd-numbered chromosomes and IGH translocations including t(11;14) (IGH/CCND1) and t(6;14) (IGH/CCND3)3,4. Beyond cytogenetic studies, next-generation sequencing (NGS) technologies have significantly expanded the ability to characterize the mutational landscape of PCM, thus providing additional genetic information of clinical value3,6.
Hyperhaploidy (24–34 chromosomes), a subtype of low hypodiploidy, is a unique cytogenetic subgroup in PCM that has rarely been described in the literature7–12. A recent retrospective clinical series of 33 patients with hyperhaploid PCM demonstrated a poor prognosis with a 5-year survival rate of 23%10. Curiously, the same set of odd-numbered disomies typically observed in hyperhaploid clones (chromosomes 3, 5, 7, 9, 11, 15, 18, 19, and 21) are the same set of odd-numbered trisomies typically observed in hyperdiploid PCM, with gains of chromosome 18 observed less than the odd-numbered trisomies7–13. Considering that certain monosomies in hyperhaploid clones are associated with high-risk abnormalities, including monosomies 1 (loss of 1p) and 17 (TP53 deletion), this may contribute to the unfavorable prognosis associated with this cytogenetic subgroup. However, additional likely contributing elements including mutation evaluation and morphologic description of hyperhaploid PCM have not been analyzed.
To further characterize this rare cytogenetic subgroup, we describe 22 cases of hyperhaploid PCM utilizing conventional chromosome, FISH and NGS studies along with morphologic and survivorship data (See supplementary materials and methods). To achieve this goal, we conducted a 10-year retrospective review of the Mayo Clinic cytogenetic database (2005–2015) following Institutional Review Board approval to identify hyperhaploid PCM karyotypes with chromosome modal numbers between 24 and 34 chromosomes, in addition to karyotypes with chromosome modal numbers between 48 and 68 that may represent a doubled hyperhaploid clone. Doubled hyperhaploid clones (48–68 chromosomes) in the absence of a hyperhaploid clone (24–34 chromosomes) were required to have supporting FISH evidence of hyperhaploidy. Karyotypes that included more than two marker chromosomes of unknown origin described in the stemline of the karyotype were excluded.
We identified 18 hyperhaploid cases analyzed by the Mayo Clinic Genomics Laboratory plus four cases from Hennepin Healthcare (HHC) Cytogenetics Laboratory, yielding a total of 22 hyperhaploid PCM cases (Table 1) comprised of 11 male and 11 female patients (M:F ratio, 1:1). Age at the time of first abnormal cytogenetic analysis (median age = 54 years; range: 39–80 years) was significantly lower compared to the age of a non-hyperhaploid cohort of patients with first abnormal chromosome and PCM FISH results collected during the same timeframe (2005–2015) (median age = 65 years; range: 28–87 years) (p-value = 0.001; Supplementary Table 1 and materials and methods). All patients were deceased at time of censorship with only one patient in our data survived beyond 4 years. Bone marrow aspirate and core biopsy morphology, light-chain status, and plasma cell labeling index (PCLI) or S-phase characteristics from eight patients are presented in Supplementary Table 2.
Table 1.
Patient characteristics of hyperhaploid PCM cohort
| Case | Site | Sex | Age (years) | Karyotype | FISH results | TP53 mutation | Time to death (months) |
|---|---|---|---|---|---|---|---|
| 1 | Mayo | F | 39 | 33,X,add(1)(p11), +3, +del(5)(q13q35), +add(6)(p23), +add(7)(p22),add(8)(q22), +9, +11, +14, +15, +16, +18,add(20)(q11.2)[1]/46,XX[29] | Monosomies 13, 17; three copies of CCND1 | NT | 41.3 |
| 2 | Mayo | M | 54 | 34,X,dup(1)(q21q32), +del(3)(q12), +5, +7, +8, +9, +11, +14, +15, +18, +19, +21[1]/54~69,idemx2[cp18]/46,XY[1] | Monosomies 13, 17; 5’IGH deletion; near-tetraploid | p.Arg196* | 4 |
| 3 | Mayo | M | 71 | 32,X, +3, +5, +7, +9, +add(10)(q24), +15, +18, +22, +mar[1]/46,XY[19] | Monosomies 13, 14, 17 | NT | 12.9 |
| 4 | HHC | F | 61 | 60~62,XX, +3, +3, +6, +7, +7, +8, +9, +9, +11, +11, +11, +15, +15, +16,−18,−18, +19, +19, +20, +2mar[cp2]/46,XX[18] | Monosomies 1, 8, 13, 14, 17 | NT | 1.4 |
| 5 | Mayo | M | 49 | 31~32,del(X)(q13), +add(3)(p21), +add(7)(p22), +add(9)(q11), +11, +15,−18, +21, +2mar[cp14]/ 61~62,idemx2, +2mar[cp6] | Not performed | NT | 0.9 |
| 6 | Mayo | F | 52 | 30,X, +3, +7, +9, +11, +18, +add(19)(p13.3), +21[3]/51~54,idemx2,−3,−4,−7,−18,−add(19)(p13.3),−22, +2~3mar[cp8] | Not performed | None | 30.1 |
| 7 | Mayo | F | 55 | 32~34,X, +3, +5, +7, +9, +add(11)(p15),add(14)(p11.2), +18, +19, +21, +r, +1~2mar[cp7]/66,idemx2[1]/46,XX[12] | Monosomies 13, 14, 15, 17 | None | 25.8 |
| 8 | Mayo | F | 64 | 31,X, +3, +7, +9, +11, +add(15)(p11.1),add(16)(q12.1), +18, +19, +21[10]/46,XX[10] | Monosomies 13, 14, 15, 17; near-tetraploid | p.Phe270Ser | 11.2 |
| 9 | Mayo | F | 49 | 33,X, +3, +7, +9, +10, +11, +15, +18, +19, +21, +mar[3]/XX[15] | Not performed | NT | 9.9 |
| 10 | Mayo | M | 56 | 31,X,add(1)(q32), +add(3)(q21), +add(5)(q11.2), +7, +9, +11, +15, +18, +del(22)(q13)[19]/46,XY[1] | Not performed | NT | 4.2 |
| 11 | Mayo | M | 50 | 32,X, +3, +5, +7, +9, +11, +15,der(16)t(16;17)(q13;q11.2), +18, +19, +21[5]/46,XY[15] | Monosomies 13, 14, 17 | None | 4.4 |
| 12 | Mayo | F | 72 | 33,X, +1, +2, +6, +7, +11, +14, +18, +19, +21, +22[6]/46,XX[7] | Not performed | p.Arg156Pro | 75.5 |
| 13 | Mayo | M | 50 | 31~33,XY, +3, +5, +7, +add(9)(p13), +11, +15, +18, +19, +mar[cp7]/46,XY[3] | Monosomies 13, 14, 17; trisomy 9 | NT | 6.8 |
| 14 | Mayo | M | 44 | 34,X, +3, +5, +add(7p22), +9, +10, +11, +15, +18, +19, +add(21)(q22), +mar[6]/46,XY[14] | Monosomies 13, 14, 17 | None | 45.8 |
| 15 | Mayo | M | 43 | 34,X, +3, +5, +7, +8, +9, +9, +11, +15, +18, +19, +21[2]/46,XY[28] | Monosomies 13, 14, 17 | p.Asp48Alafs*5 | NA |
| 16 | Mayo | F | 80 | 32,X, +3, +5, +7, +9, +11, +15, +add(15)(p11.2), +19, +21[5]/46,XX[12] | Monosomies 13, 14, 17 | p.Pro250Thr | 18 |
| 17 | Mayo | F | 53 | 33,X,der(1;21)(q10;q10), +3, +5, +7, +9, +11, +add(14)(q32), +15, +18, +19, +21[4]/46,XX[16] | Monosomies 4, 6, 13, 16, 17, 20; IGH/MYC fusion | p.Thr253Pro | 12.4 |
| 18 | Mayo | F | 71 | 30,X, +3, +5, +7, +9, +add(11)(q25), +15, +add(19)(q13.3),−21, +1~2mar[cp3]/46,XX[20] | Monosomies 8, 13, 17, 20 | p.Tyr220Metfs*27 | 22.4 |
| 19 | HHC | F | 54 | 32,X, +der(3)t(2;3)(q33;q29),der(4)inv(4)(p16q13)ins(4;?)(p16;?), +5, +7, +8, +9, +11, +15, +19, +21[3]/46,XX[17] | Monosomies 1, 4, 13, 14, 16, 17 | NT | NA |
| 20 | HHC | M | 53 | 34,X,del(1)(q11),ins(1;?)(q21;?),add(1)(q32), +der(3)add(3)(p23)add(3)(q22), +add(3)(q22), +5, +7, +8, +9, +11, +add(11)(q23), +14, +add(14)(p11.1), +15, +18, +19, +21[cp20]/46,XY[1] | Not performed | NT | NA |
| 21 | HHC | M | 58 | 30,X, +2, +3, +8, +11, +15, +18, +19,−22, +mar[4]/46,XY[3] | Monosomies 1, 4, 13, 14, 16, 17 | NT | NA |
| 22 | HHC | M | 50 | 33,X, +3, +5, +7, +9, +10, +11, +15, +18, +19, +21[5]/46,XY[15] | Monosomies 1, 4, 13, 14, 16, 17 | NT | NA |
Mayo, cases from Mayo Clinic Cohort; HHC, cases from Hennepin Health Care
NT not tested; NA Time to death information not available for HHC cases
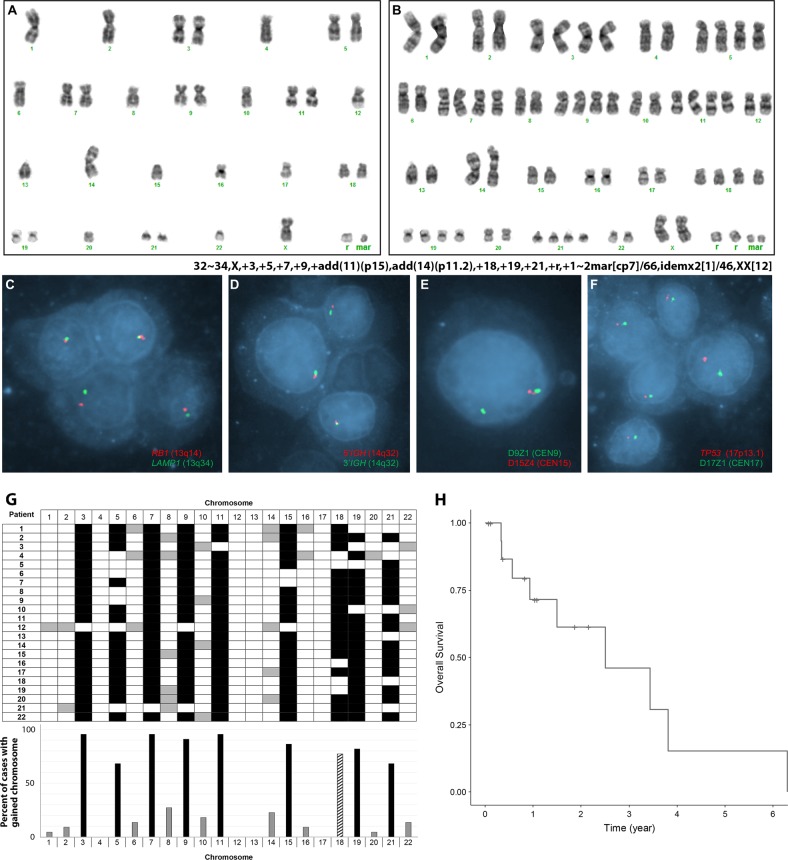
Eighteen cases had at least two hyperhaploid metaphases and met criteria for an abnormal clone for this study (Table 1). Of four cases that did not meet these criteria, three cases (patients 1, 2, and 3) had a single hyperhaploid metaphase cell accompanied by supporting FISH results, and 1 case (patient 4, Supplementary Figure 1) had two metaphases (60–62 chromosomes) representing an apparent doubled hyperhaploid clone supported by FISH results. Six cases (patients 2, 4, 5, 6, 7, and 8) had chromosomal or FISH evidence of both a hyperhaploid and doubled hyperhaploid clone (Fig. 1a–f).
Fig. 1.
a Karyograms from patient 7 with a hyperhaploid and b doubled hyperhaploid clones with supporting FISH results in c–f. g Depiction of the gained chromosomes (in reference to a haploid clone) from the cohort of 22 patients with hyperhaploid PCM in both grid format per patient (top) and overall percentage of cases with the gained chromosome (bottom). The most commonly gained chromosomes are indicated by a black bar, less commonly gained chromosomes by a gray bar and chromosome 18 indicated by a pattern bar in graph. Overall survival (OS) was calculated from time of diagnosis to date of death for patients seen at Mayo Clinic (h)
The chromosome complement in 21 of 22 cases (excluding patient 4 with only a doubled hyperhaploid clone) ranged from 30 to 34 (mean 32, median 32). The most frequently observed chromosomal gains compared to a haploid chromosome complement (23,X) were chromosomes 3 (n = 21; 95%), 11, (n = 21; 95%), 7 (n = 21; 95%), 9 (n = 20; 91%), 15 (n = 19; 86%), 19 (n = 18; 82%), 18 (n = 17; 77%), 21 (n = 15; 68%), 5 (n = 15; 68%), 8 (n = 6; 27%), 14 (n = 5; 23%), 10 (n = 4; 18%), 22 (n = 3; 14%), 6 (n = 3; 14%), 16 (n = 2; 9%), 2 (n = 2; 9%), 1 (n = 1; 5%), and 20 (n = 1; 5%) (Fig. 1g). No gains of chromosomes 4, 12, 13, or 17 were observed in any of the 22 cases.
Sixteen of 22 cases had concurrent plasma cell FISH studies performed, all of which demonstrated monosomies 13 and 17. In addition to FISH signal patterns suggestive of hyperhaploidy, patient 17 had an IGH/MYC fusion, and patient 2 had a 5’IGH deletion without an IGH translocation to the common PCM translocation partners CCND1, CCND3, MAF, MAFB, and FGFR3. Eleven of 22 cases had subsequent NGS testing performed and revealed a mutation within the TP53 gene in seven (64%) (patients 2, 8, 12, 15, 16, 17, and 18) (Table 1 and supplementary table 3). Patient 18 also exhibited pathogenic or likely pathogenic mutations in CDKN1B and RB1. Several patients had co-existing TP53 mutations and mutations in TGFBR2 gene (patient 15), KRAS (patient 17), or NRAS (patient 2). For 17 cases with survivorship data, the median survival was 2.51 years, with an estimated 3-year survival of 46% (95% CI, 22–95%) (Fig. 1h).
Hyperhaploidy in PCM is a rare, high-risk cytogenetic subgroup that exhibits the same set of odd-numbered chromosome gains (in reference to a haploid clone) as in hyperdiploid PCM with the exception of chromosome 187–13. The poor prognosis of hyperhaploid PCM underscores the importance of correctly identifying this cytogenetic subgroup.
Of the 61 total cases of hyperhaploid PCM reported in the literature (including the current study), 39 (64%) had evidence of only a hyperhaploid clone, 19 (31%) had evidence of both hyperhaploid and a doubled hyperhaploid clone, and 3 cases (5%) only had evidence of a doubled hyperhaploid clone7–12. However, this latter subgroup may be under-recognized and erroneously misclassified as hyperdiploid without careful evaluation of the apparent chromosome gains and correlative FISH studies. Detection of recurrent structural chromosomal abnormalities in PCM currently relies heavily upon locus- and centromere-specific FISH probes and according to the 2017 revised WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, the minimal PCM FISH panel should include probes targeting t(4;14) (IGH/FGFR3), t(14;16) (IGH/MAF), and TP5314. While using this minimal FISH panel could detect commonly observed monosomies in hyperhaploidy including 4, 14, 16, and 17, doubling of a hyperhaploid clone in the absence of a hyperhaploid clone could be incorrectly interpreted as a “normal” result without a more thorough FISH evaluation. Considering that hyperhaploidy shares the same chromosome gains as hyperdiploidy, centromere-specific probes for chromosomes 3, 7, 9, and 15 should be included in the standard PCM FISH panel in order to not miss the opportunity to discriminate this high-risk cytogenetic subtype. However, newer methodologies to comprehensively evaluate PCM clones, including SNP arrays or NGS assays, will likely eliminate the need for large FISH panels.
Mutations in TP53 are rarely detected at disease presentation and have been reported in ~3% of newly diagnosed PCMs often in the context of 17p deletions. TP53 mutations have been reported to increase in frequency with disease progression and are associated with inferior clinical outcome15,16. Given 64% of our hyperhaploid cohort (n = 11) had pathogenic or likely pathogenic mutations in TP53 suggests that TP53 mutation may at least in part account for the poor overall survival of patients with hyperhaploid PCM.
As a large reference genomics laboratory that is unable to routinely obtain complete patient histories, a limitation in this study is our inability to ascertain the exact date of PCM diagnosis for patients not treated at our institution. Therefore, patient survival (as established from the date of the abnormal chromosome study) may be underestimated. The limited sample size of this study (22 patients) also limits the power of the survival estimates. Nevertheless, given that the 4-year survival for PCM exceeds 80% for those patients eligible for ASCT17, the survival of patients with hyperhaploid PCM in contrast to all PCM remains poor.
The frequency of hyperhaploidy in various neoplasms is low (~0.2–0.3% of tumors) and is often hallmarked by a non-random pattern of disomic chromosomes distributed among different malignancies8. The biological incentive and mechanism for a clone to undergo a massive loss of genomic material with retention of specific chromosomes is currently unclear, but a selective advantage resulting from altered gene expression contributing from the retained chromosomes is likely contributory. In addition, abnormalities in the spindle apparatus necessary for separation of sister chromatids during cell division resulting in specific non-disjunction patterns may possibly contribute to this phenomenon and also to the generation of hyperdiploidy8,18. While hyperhaploid and hyperdiploid PCM share a similar pattern of gained chromosomes, how these two entities are related to one another is unclear. While it is possible that hyperhaploidy represents a subclone of hyperdiploidy, we do not see evidence of both hyperdiploidy and hyperhaploidy together within the same specimen by chromosome or FISH studies (Table 1). Although hyperdiploidy and hyperhaploidy share similarly gained chromosomes, IGH gene rearrangements are rarely observed and retention of chromosome 18 along with TP53 mutations are commonly observed in hyperhaploidy and likely provide a unique selective advantage to this high-risk genetic subgroup. Taken together, hyperhaploid PCM is associated with multiple monosomies including monosomy 17, frequent TP53 mutations, younger age at the time of abnormal cytogenetic analysis, and decreased overall survival.
Supplementary information
Acknowledgements
This work was Supported by the Mayo Clinic Department of Laboratory Medicine and Pathology.
Authors’ contributions
K.E.P., C.M.W., and L.A.F. provided technical support and analyzed raw data; J.F.P. and L.B.B. wrote the manuscript; M.Y.P. was the study statistician; P.T.G., R.P.K., S.K., D.S.V., J.M.F., K.K.R., J.F.P., L.B.B., R.A..R., M.Y.P., C.A.M., and D.L.V. contributed samples, interpreted data, and reviewed and approved final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at (10.1038/s41408-019-0182-z).
References
- 1.Fonseca R, et al. Trends in overall survival and costs of multiple myeloma, 2000-2014. Leukemia. 2017;31:1915–1921. doi: 10.1038/leu.2016.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa LJ, et al. Recent trends in multiple myeloma incidence and survival by age, race, and ethnicity in the United States. Blood Adv. 2017;1:282–287. doi: 10.1182/bloodadvances.2016002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar SK, Rajkumar SV. The multiple myelomas—current concepts in cytogenetic classification and therapy. Nat. Rev. Clin. Oncol. 2018;15:409–421. doi: 10.1038/s41571-018-0018-y. [DOI] [PubMed] [Google Scholar]
- 4.Fonseca R, Monge J, Dimopoulos MA. Staging and prognostication of multiple myeloma. Expert Rev. Hematol. 2014;7:21–31. doi: 10.1586/17474086.2014.882224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palumbo A, et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J. Clin. Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker BA, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoctor VT, Campbell LJ. Hyperhaploid plasma cell myeloma. Cancer Genet. 2012;205:414–418. doi: 10.1016/j.cancergen.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Mandahl N, Johansson B, Mertens F, Mitelman F. Disease-associated patterns of disomic chromosomes in hyperhaploid neoplasms. Genes Chromosomes Cancer. 2012;51:536–544. doi: 10.1002/gcc.21947. [DOI] [PubMed] [Google Scholar]
- 9.Mohamed AN, Bentley G, Bonnett ML, Zonder J, Al-Katib A. Chromosome aberrations in a series of 120 multiple myeloma cases with abnormal karyotypes. Am. J. Hematol. 2007;82:1080–1087. doi: 10.1002/ajh.20998. [DOI] [PubMed] [Google Scholar]
- 10.Sawyer JR, et al. Hyperhaploidy is a novel high-risk cytogenetic subgroup in multiple myeloma. Leukemia. 2017;31:637–644. doi: 10.1038/leu.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smadja NV, et al. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98:2229–2238. doi: 10.1182/blood.V98.7.2229. [DOI] [PubMed] [Google Scholar]
- 12.Pantou D, et al. Cytogenetic manifestations of multiple myeloma heterogeneity. Genes Chromosomes Cancer. 2005;42:44–57. doi: 10.1002/gcc.20114. [DOI] [PubMed] [Google Scholar]
- 13.Chretien ML, et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015;126:2713–2719. doi: 10.1182/blood-2015-06-650242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swerdlow SH, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thanendrarajan S, et al. The level of deletion 17p and bi-allelic inactivation of TP53 has a significant impact on clinical outcome in multiple myeloma. Haematologica. 2017;102:e364–e367. doi: 10.3324/haematol.2017.168872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lionetti M, et al. Molecular spectrum of TP53 mutations in plasma cell dyscrasias by next generation sequencing: an Italian cohort study and overview of the literature. Oncotarget. 2016;7:21353–21361. doi: 10.18632/oncotarget.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajkumar SV. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2016;91:719–734. doi: 10.1002/ajh.24402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawyer JR, Morgan GJ. Hyperhaploid karyotypes in multiple myeloma. Oncotarget. 2017;8:78259–78260. doi: 10.18632/oncotarget.20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.