The intestinal microbiota contains trillions of commensal microorganisms that shape multiple aspects of host physiology and disease. In contrast to the host’s genome, the microbiome is amenable to change over the course of an organism’s lifetime, providing an opportunity to therapeutically modulate the microbiome’s impact on human pathophysiology.
KEYWORDS: microbiome, short-term and long-term interventions, temporal dynamics
ABSTRACT
The intestinal microbiota contains trillions of commensal microorganisms that shape multiple aspects of host physiology and disease. In contrast to the host’s genome, the microbiome is amenable to change over the course of an organism’s lifetime, providing an opportunity to therapeutically modulate the microbiome’s impact on human pathophysiology. In this Perspective, we highlight environmental factors that regulate the temporal dynamics of the intestinal microbiome, with a particular focus on the different time scales at which they act. We propose that the identification of transient and intermediate states of microbiome responses to perturbations is essential for understanding the rules that govern the behavior of this ecosystem. The delineation of microbiome dynamics is also helpful for distinguishing cause and effect in microbiome responses to environmental stimuli. Understanding the dimension of time in host-microbiome interactions is therefore critical for therapeutic strategies that aim at short-term or long-term engineering of the intestinal microbial community.
PERSPECTIVE
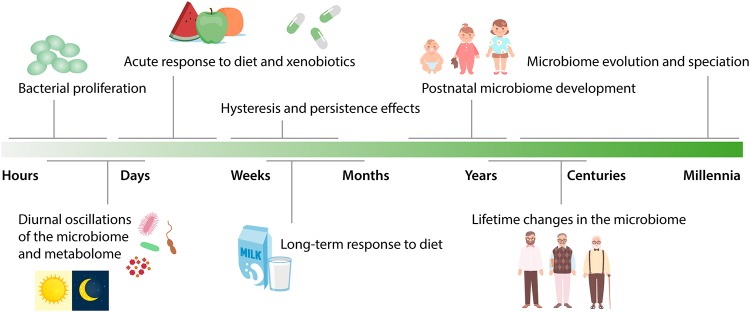
The intestinal microbiome has been recognized as a major component of the human endocrine system whose digestive and secretory activities profoundly influence most other organs of the human body (1, 2). As such, the microbiome’s activity is critical for human health, and aberrations in the function of the microbiome are involved in the pathophysiology of multiple diseases, ranging from inflammatory and metabolic to neoplastic and neurological diseases (3–7). Consequently, intensive research efforts are currently focused on modulating the intestinal microbiota to promote health and to prevent or counteract disease (8). A property of the intestinal microbial ecosystem that needs to be well understood for therapeutic modulations to be effective is its dynamic behavior over time. In the following sections, we will provide a broad overview of some of the knowns and unknowns regarding the dimension of time in microbiome research. The temporal dynamics of the microbiome important for host physiology range from the scale of hours, reflecting the shortest bacterial generation times, to the scale of centuries and millennia, during which intestinal microorganisms have coevolved with their host (Fig. 1).
FIG 1.
Time scales of microbiome dynamics. Schematic overview of major factors impacting the microbiome and the time scales at which they act.
HOURS: BACTERIAL PROLIFERATION AND DIURNAL RHYTHMS
The fastest processes studied in the context of the gut microbial ecosystem have been bacterial proliferation and infection by bacteriophages, some of which occur at the subhour scale (9). Microbiome-wide proliferation rates have been assessed by metagenomic inference, implying widespread hour-scale changes in genomic abundances (10, 11).
A particular organizing principle of such short-term dynamics is diurnal oscillations. The relative and absolute abundances of intestinal bacteria, their biogeography in the gastrointestinal tract, and the intestinal metabolite profile undergo hour-scale oscillations over the course of a day (12–19), indicating that the microbiome is highly dynamic and contains autocorrelative features (self-similarity over a time course) even at short-term intervals. These daily microbial fluctuations are strongly influenced by the time and type of food intake (13, 15, 16, 20). The fact that the intestinal microbiota is fluctuating in composition and function at such short time scales may affect numerous short-term responses of the intestinal community to foreign elements entering the gastrointestinal system, including pathogens, xenobiotics (substances that are foreign to the body, such as drugs and environmental toxins), and dietary nutrients (18, 21).
It should be noted that these conclusions about daily oscillations of the gut microbiome are mostly based on mouse studies, and that the majority of human data is so far only available from a limited number of stool samples and only on the level of relative taxonomical composition. Recent studies have assessed the concept of diurnal microbiome oscillations in human saliva, a tissue more accessible for repeated sampling over the course of a day. These studies have likewise documented oscillatory behavior in taxonomic composition and the dependency of this diurnal property on the timing of food intake (22–24).
The realization that microbiome taxonomy and metabolic function are distinct during day and night at various body sites provides an example for how a better understanding of temporal microbiome behavior can improve the design of analytical studies and therapeutic interventions that harness the optimal time of day for microbiome modulation. For instance, delivery of drugs or probiotics at the ideal time of day in terms of microbiome biotransformation reactions or colonization resistance harbors great optimization potential for already existing therapies.
DAYS: ACUTE RESPONSES TO INGESTED FOOD AND XENOBIOTICS
Diet and ingested xenobiotics strongly shape the intestinal microbial community and offer opportunities for noninvasive strategies for microbiome modulation. The preponderance of evidence suggests that marked alterations in diet can perturb the taxonomical configuration of the microbiota within a few days.
One of the first studies that longitudinally measured daily responses of the microbiome to environmental perturbations found rapid adaptations of the intestinal microbial community to changes in diet (25). A controlled study of healthy volunteers consuming defined diets showed that microbiota composition shifts caused by dietary intervention were effective already within 24 h (26). Short-term consumption (4 days) of diets based entirely on either animal or plant products was sufficient to introduce distinct community-wide alterations of the intestinal microbiota, coupled to rapid changes in the concentration of intestinal short-chain fatty acids (26).
Microbiome responses to other ingested substances can be similarly rapid. In particular, it has been shown in humans and animal models that antibiotics dramatically alter the gut microbial composition within a few days. Within 4 days of broad-spectrum antibiotic treatment, microbiome diversity plummets, resulting in long-lasting deviations of community structure from the initial composition even several months later (27, 28).
It is important to note that these findings have primarily been based on sampling of the stool microbiome, which is distinct from the microbial communities in upper areas of the small and large intestines (29). The time scales at which different regions of the gastrointestinal tract respond to dietary and xenobiotic perturbations, and the degree of synchronization or codependency between different anatomic regions, remain to be investigated.
WEEKS AND MONTHS: DIETARY PATTERNS
Diet also influences the composition and function of the microbiota on longer time scales. In particular, long-term dietary patterns kept over several months are a strong factor influencing the overall composition of the microbiota (30). As such, stable exposure to external factors is a critical determinant of long-term microbiota behavior—a concept that is important for using microbiome features as biomarkers for clinical outcome and prognostic or diagnostic disease indicators.
In addition, several recent studies have uncovered additional properties of microbiome temporal dynamics: hysteresis (dependence on past stimuli) and persistence (prolonged effect after disappearance of the initial stimulus). Repeated exposures to alternating diets in mice lead to a hysteresis effect, whereby the microbiota retains characteristics from the previous cycle of exposure in each subsequent cycle (31); hence, the return to the original configuration is impeded with each cycle. Similarly, episodes of low-fiber or high-fat diet lead to long-term persistence of specific microbial features despite the return to normal dietary conditions (32, 33).
Several studies have revealed an annual cyclic reconfiguration of the microbiome that reflects the seasonal availability of different types of food. Evidence for this phenomenon has been found in squirrels (34), wild great apes (35), and human indigenous hunter-gatherer populations (36).
Interestingly, succession of microbial community assembly in natural environments likewise spans time periods of weeks to months, as has been documented after resolution of enteric infection (37) and during mammalian corpse decomposition (38).
YEARS: LIFETIME MICROBIOME CHANGES
While the microbiome remains relatively stable throughout adult life (39), two periods of life are particularly unstable with regard to the overall taxonomy of the intestinal microbial community: early life and old age. In the early years of life, there are different factors influencing the development of the microbiome, including the delivery mode, breastfeeding, and the introduction of solid food (40, 41). After around 3 years, the relative proportions of microbial taxa remain mostly stable, but the microbiome composition can be altered over time by changes in diet as well as by antibiotics (39, 42), which may even have an intergenerational effect (32, 43). This early susceptibility of the microbiome to community perturbations is physiologically meaningful, since long-lasting detrimental effects on host health have been documented in cases of early-life microbiome disruption by antibiotics or caesarian section (44–46).
In the elderly, the microbiome composition was found to feature a distinct taxonomy compared to the average microbiome of healthy adults (47, 48) This difference has been associated with several age-related processes, such as weakened gut barrier integrity, intestinal pro- versus anti-inflammatory balance, immune and cardiometabolic health, reduced mobility, hospitalizations, and use of medications (47–50). However, the direction of causality is unclear, and dissimilar results have been reported for elderly populations at different geographical locations, highlighting the necessity for strictly controlled studies to discern age-related from confounding environmental factors (51).
CENTURIES AND BEYOND: MICROBIOME-HOST COEVOLUTION
The essential role of the microbiome for human health makes it particularly interesting to consider long-term microbiome changes throughout human evolution. Given the technical challenges associated with archeological human microbiome samples and their sparsity, studies probing the evolution of the microbiota have so far relied on comparisons of humans with phylogenetically related species. A major question regarding the long-term evolution of the human microbiome is whether its ability to synthesize key components of human metabolism, such as vitamins, and to digest complex carbohydrates have undergone major alterations following the agricultural and later the industrial revolution.
Comparative analysis of microbiomes from African apes demonstrated that host phylogenetic divergence scales with microbiome divergence. Interestingly, compositional changes have accelerated during human evolution and have deviated from the formerly clock-like pace during African ape diversification (52). This analysis further revealed that major taxa of the commensal microbiota have cospeciated with the hominid host, i.e., they have been maintained exclusively within an evolutionary lineage for hundreds of thousands of generations and have diversified alongside host evolution (53).
Experiments in mice have indicated that the microbiota is primarily vertically transmitted, but that there are certain taxa that undergo horizontal transmission, particularly those that potentially cause pathogenicity to the host (54).
Recent studies have shown that within-host evolution of the microbiome can also occur at rapid time scales, even within the life span of a host. For instance, bacterial phenotypes are rapidly varied by invertible promoters upon selective pressure (55). Other facilitators of such rapid within-host evolution of commensal bacteria are homologous recombination and de novo mutations (56, 57).
OUTLOOK
The last decade of microbiome research has brought numerous surprising insights into the diverse impact of the intestinal bacterial community on seemingly unrelated aspects of host physiology, from hepatic metabolism to blood-brain barrier function (58–60). Consequently, microbiome modulatory strategies are of vast academic and medical interest. Surprisingly little is known about the requirements for timing and duration of such interventions, although these factors are probably fundamental for the success of any treatment strategy. While the major principles underlying the temporal dynamics of the intestinal microbiome still need to be unraveled, a number of paradigms have emerged from recent studies.
First, the microbiome is exquisitely susceptible to change early in life. Thus, the ecosystem is particularly vulnerable to perturbation during early childhood, resulting from skin contacts, mode of delivery, and neonatal diet. On the other hand, this period also offers unique opportunities for therapeutic interventions with long-lasting effects. Likewise, the microbiome might be more susceptible to taxonomic alterations in the elderly, although more experimental and empirical data are required to support this notion.
Second, the long-term evolution of the human microbiome seems to be accompanied by loss of species diversity, at least in the last century (61). This loss of species diversity might be disadvantageous for human health, and strategies should be considered of how to rediversify the human microbiome.
Third, the microbiome is amenable to long-term changes. These changes are provoked by persistent lifestyle changes, including long-term dietary patterns and geographical localization. Such long-term approaches might ultimately turn out to be the most optimally suited interventions to engineer the human microbiome for health improvements.
Fourth, on intermediate time scales, the intestinal microbiome features hysteresis and “memory-like” properties. As such, prior exposures to environmental stimuli leave long-lasting signatures in the taxonomic composition and functional properties of the microbiome which influence the response to subsequent stimuli.
An extension of this list of insights into the temporal dynamics of the human microbiome is an essential goal of microbiome research in the years to come. To achieve this, the microbiome research community needs to embrace strategies to incorporate the factor of time in study designs and sampling frequencies. To resolve the spectrum of time scales discussed here, ideal protocols would include sampling frequencies of hours to days (62). The power of such protocols is exemplified by the identification of decaying autocorrelation, i.e., loss of self-similarity over time, in highly time-resolved data sets of the commensal microbiome (28, 63). Such sampling strategies would also enable deeper insights into the phenomenon of rare anaerobe blooms that seem to occur independently of dietary patterns (63).
A higher sampling frequency would facilitate yet another goal of the microbiome research community: the quest to distinguish correlation from causation in human studies. Enhanced temporal resolution in microbiome intervention studies will highlight transient and intermediate stages in microbial community responses to a given perturbation. Identifying and characterizing these intermediate stages will be essential not only to better understand ecological dynamics in the gut but also to distinguish cause and effect in the transition of the microbiome from one state to another. These temporal chains of cause and effect, in turn, are exactly the type of insight that we need in order to design rational therapeutic interventions targeted at the microbiome. As the tools now exist, microbiome research is ready for a venture into the dimension of time.
ACKNOWLEDGMENTS
We thank the members of the Thaiss lab for valuable input and apologize to those colleagues whose relevant work could not be cited owing to space constraints.
C.A.T. is supported by the Edward Mallinckrodt, Jr. Foundation, the Global Probiotics Council, and grants by the Penn Institute for Immunology, the PennCHOP Microbiome Program, the Penn Center for Molecular Studies in Digestive and Liver Diseases, and the Penn Skin Biology and Diseases Resource-based Center.
REFERENCES
- 1.Schroeder BO, Backhed F. 2016. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med 22:1079–1089. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]
- 2.Levy M, Thaiss CA, Elinav E. 2015. Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med 7:120. doi: 10.1186/s13073-015-0249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. 2016. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 167:1469.e12–1480.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar J-P, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 8.Vieira AT, Fukumori C, Ferreira CM. 2016. New insights into therapeutic strategies for gut microbiota modulation in inflammatory diseases. Clin Transl Immunology 5:e87. doi: 10.1038/cti.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkova VV, Lu Z, Besser T, Grohn YT. 2014. Modeling the infection dynamics of bacteriophages in enteric Escherichia coli: estimating the contribution of transduction to antimicrobial gene spread. Appl Environ Microbiol 80:4350–4362. doi: 10.1128/AEM.00446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N, Sirota-Madi A, Thaiss CA, Pevsner-Fischer M, Sorek R, Xavier RJ, Elinav E, Segal E. 2015. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349:1101–1106. doi: 10.1126/science.aac4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CT, Olm MR, Thomas BC, Banfield JF. 2016. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 34:1256–1263. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang X, Bushman FD, FitzGerald GA. 2015. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci U S A 112:10479–10484. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. 2014. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 14.Thaiss CA, Zeevi D, Levy M, Segal E, Elinav E. 2015. A day in the life of the meta-organism: diurnal rhythms of the intestinal microbiome and its host. Gut Microbes 6:137–142. doi: 10.1080/19490976.2015.1016690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarrinpar A, Chaix A, Yooseph S, Panda S. 2014. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. 2015. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weger BD, Gobet C, Yeung J, Martin E, Jimenez S, Betrisey B, Foata F, Berger B, Balvay A, Foussier A, Charpagne A, Boizet-Bonhoure B, Chou CJ, Naef F, Gachon F. 2019. The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab 29:362.E8–382.E8. doi: 10.1016/j.cmet.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, Federici S, Zmora N, Zeevi D, Dori-Bachash M, Pevsner-Fischer M, Kartvelishvily E, Brandis A, Harmelin A, Shibolet O, Halpern Z, Honda K, Amit I, Segal E, Elinav E. 2016. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167:1495.E12–1510.E12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, Haraguchi A, Ikeda Y, Fukuda S, Shibata S. 2018. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep 8:1395. doi: 10.1038/s41598-018-19836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaiss CA, Levy M, Elinav E. 2015. Chronobiomics: the biological clock as a new principle in host-microbial interactions. PLoS Pathog 11:e1005113. doi: 10.1371/journal.ppat.1005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tognini P, Thaiss CA, Elinav E, Sassone-Corsi P. 2017. Circadian coordination of antimicrobial responses. Cell Host Microbe 22:185–192. doi: 10.1016/j.chom.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Skarke C, Lahens NF, Rhoades SD, Campbell A, Bittinger K, Bailey A, Hoffmann C, Olson RS, Chen L, Yang G, Price TS, Moore JH, Bushman FD, Greene CS, Grant GR, Weljie AM, FitzGerald GA. 2017. A pilot characterization of the human chronobiome. Sci Rep 7:17141. doi: 10.1038/s41598-017-17362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takayasu L, Suda W, Takanashi K, Iioka E, Kurokawa R, Shindo C, Hattori Y, Yamashita N, Nishijima S, Oshima K, Hattori M. 2017. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res 24:261–270. doi: 10.1093/dnares/dsx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collado MC, Engen PA, Bandin C, Cabrera-Rubio R, Voigt RM, Green SJ, Naqib A, Keshavarzian A, Scheer F, Garaulet M. 2018. Timing of food intake impacts daily rhythms of human salivary microbiota: a randomized, crossover study. FASEB J 32:2060–2072. doi: 10.1096/fj.201700697RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108 Suppl 1:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB, Federici S, Cohen Y, Linevsky R, Rothschild D, Moor AE, Ben-Moshe S, Harmelin A, Itzkovitz S, Maharshak N, Shibolet O, Shapiro H, Pevsner-Fischer M, Sharon I, Halpern Z, Segal E, Elinav E. 2018. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174:1388.E21–1405.E21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 30.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. 2015. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, Dohnalova L, Braverman S, Rozin S, Malitsky S, Dori-Bachash M, Kuperman Y, Biton I, Gertler A, Harmelin A, Shapiro H, Halpern Z, Aharoni A, Segal E, Elinav E. 2016. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 540:544–551. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 34.Carey HV, Walters WA, Knight R. 2013. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am J Physiol Regul Integr Comp Physiol 304:R33–R42. doi: 10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hicks AL, Lee KJ, Couto-Rodriguez M, Patel J, Sinha R, Guo C, Olson SH, Seimon A, Seimon TA, Ondzie AU, Karesh WB, Reed P, Cameron KN, Lipkin WI, Williams BL. 2018. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat Commun 9:1786. doi: 10.1038/s41467-018-04204-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG, Sonnenburg JL. 2017. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David LA, Weil A, Ryan ET, Calderwood SB, Harris JB, Chowdhury F, Begum Y, Qadri F, LaRocque RC, Turnbaugh PJ. 2015. Gut microbial succession follows acute secretory diarrhea in humans. mBio 6:e00381-15. doi: 10.1128/mBio.00381-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metcalf JL, Xu ZZ, Weiss S, Lax S, Van Treuren W, Hyde ER, Song SJ, Amir A, Larsen P, Sangwan N, Haarmann D, Humphrey GC, Ackermann G, Thompson LR, Lauber C, Bibat A, Nicholas C, Gebert MJ, Petrosino JF, Reed SC, Gilbert JA, Lynne AM, Bucheli SR, Carter DO, Knight R. 2016. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 351:158–162. doi: 10.1126/science.aad2646. [DOI] [PubMed] [Google Scholar]
- 39.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, A DL, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:343ra382. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R. 2011. Moving pictures of the human microbiome. Genome Biol 12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulfer AF, Battaglia T, Alvarez Y, Bijnens L, Ruiz VE, Ho M, Robinson S, Ward T, Cox LM, Rogers AB, Knights D, Sartor RB, Blaser MJ. 2018. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol 3:234–242. doi: 10.1038/s41564-017-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. 2019. The human microbiome and child growth - first 1000 days and beyond. Trends Microbiol 27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. 2012. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez KA II, Devlin JC, Lacher CR, Yin Y, Cai Y, Wang J, Dominguez-Bello MG. 2017. Increased weight gain by C-section: functional significance of the primordial microbiome. Sci Adv 3:eaao1874. doi: 10.1126/sciadv.aao1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Toole PW, Jeffery IB. 2015. Gut microbiota and aging. Science 350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 49.Zapata HJ, Quagliarello VJ. 2015. The microbiota and microbiome in aging: potential implications in health and age-related diseases. J Am Geriatr Soc 63:776–781. doi: 10.1111/jgs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 51.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC, Clavel T, Koebnick C, Zunft HJ, Dore J, Blaut M. 2006. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, Ochman H. 2014. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci U S A 111:16431–16435. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, Pusey AE, Peeters M, Hahn BH, Ochman H. 2016. Cospeciation of gut microbiota with hominids. Science 353:380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moeller AH, Suzuki TA, Phifer-Rixey M, Nachman MW. 2018. Transmission modes of the mammalian gut microbiota. Science 362:453–457. doi: 10.1126/science.aat7164. [DOI] [PubMed] [Google Scholar]
- 55.Jiang X, Hall AB, Arthur TD, Plichta DR, Covington CT, Poyet M, Crothers J, Moses PL, Tolonen AC, Vlamakis H, Alm EJ, Xavier RJ. 2019. Invertible promoters mediate bacterial phase variation, antibiotic resistance, and host adaptation in the gut. Science 363:181–187. doi: 10.1126/science.aau5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao S, Lieberman TD, Poyet M, Groussin M, Gibbons SM, Xavier RJ, Alm EJ. 2018. Adaptive evolution within the gut microbiome of individual people. bioRxiv 10.1101/208009. [DOI] [PMC free article] [PubMed]
- 57.Garud NR, Good BH, Hallatschek O, Pollard KS. 2019. Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol 17:e3000102. doi: 10.1371/journal.pbio.3000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy M, Thaiss CA, Elinav E. 2016. Metabolites: messengers between the microbiota and the immune system. Genes Dev 30:1589–1597. doi: 10.1101/gad.284091.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, Korecka A, Bakocevic N, Ng LG, Guan NL, Kundu P, Gulyás B, Halldin C, Hultenby K, Nilsson H, Hebert H, Volpe BT, Diamond B, Pettersson S. 2014. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blaser MJ. 2018. The past and future biology of the human microbiome in an age of extinctions. Cell 172:1173–1177. doi: 10.1016/j.cell.2018.02.040. [DOI] [PubMed] [Google Scholar]
- 62.Gerber GK. 2014. The dynamic microbiome. FEBS Lett 588:4131–4139. doi: 10.1016/j.febslet.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 63.Gibbons SM, Kearney SM, Smillie CS, Alm EJ. 2017. Two dynamic regimes in the human gut microbiome. PLoS Comput Biol 13:e1005364. doi: 10.1371/journal.pcbi.1005364. [DOI] [PMC free article] [PubMed] [Google Scholar]