Abstract
The 22q11.2 deletion syndrome (22q11DS) is a recurrent copy number variant with high penetrance for developmental neuropsychiatric disorders. Study of individuals with 22q11DS therefore may offer key insights into neural mechanisms underlying such complex illnesses. Resting-state functional connectivity MRI studies in idiopathic schizophrenia have consistently revealed disruption of thalamic and hippocampal circuitry. Here, we sought to test whether this circuitry is similarly disrupted in the context of this genetic high-risk condition. To this end, resting-state functional connectivity patterns were assessed in a sample of human youth with 22q11DS (n = 42; 59.5% female) and demographically matched healthy controls (n = 39; 53.8% female). Neuroimaging data were acquired via single-band protocols and analyzed in line with methods provided by the Human Connectome Project. We computed functional relationships between individual-specific anatomically defined thalamic and hippocampal seeds and all gray matter voxels in the brain. Whole-brain Type I error protection was achieved through nonparametric permutation-based methods. The 22q11DS patients displayed dissociable disruptions in thalamic and hippocampal functional connectivity relative to control subjects. Thalamocortical coupling was increased in somatomotor regions and reduced across associative networks. The opposite effect was observed for the hippocampus in regards to somatomotor and associative network connectivity. The thalamic and hippocampal dysconnectivity observed in 22q11DS suggests that high genetic risk for psychiatric illness is linked with disruptions in large-scale corticosubcortical networks underlying higher-order cognitive functions. These effects highlight the translational importance of large-effect copy number variants for informing mechanisms underlying neural disruptions observed in idiopathic developmental neuropsychiatric disorders.
SIGNIFICANCE STATEMENT Investigation of neuroimaging biomarkers in highly penetrant genetic syndromes represents a more biologically tractable approach to identify neural circuit disruptions underlying developmental neuropsychiatric conditions. The 22q11.2 deletion syndrome confers particularly high risk for psychotic disorders and is thus an important translational model in which to investigate systems-level mechanisms implicated in idiopathic illness. Here, we show resting-state fMRI evidence of large-scale sensory and executive network disruptions in youth with 22q11DS. In particular, this study provides the first evidence that these networks are disrupted in a dissociable fashion with regard to the functional connectivity of the thalamus and hippocampus, suggesting circuit-level dysfunction.
Keywords: 22q11, connectivity, fMRI, hippocampus, networks, thalamus
Introduction
Remarkable genetic and clinical heterogeneity presents a challenge for mapping pathological processes underlying neuropsychiatric disorders, such as schizophrenia and autism spectrum disorder (ASD). These disorders are increasingly viewed as developmental disruptions of neural circuitry with major genetic contributions (Insel, 2010; Geschwind and Flint, 2015). Thus, genetically defined syndromes with strong predisposition for neuropsychiatric illness provide powerful models to elucidate neural mechanisms underlying these complex disorders.
The 22q11.2 deletion syndrome (22q11DS), also known as DiGeorge or Velocardiofacial syndrome (OMIM #188400, #192430), occurs in ∼1 in 4000 live births (McDonald-McGinn et al., 2015). It represents one of the greatest known genetic risk factors for psychosis, ∼25 times population base rates (Bassett and Chow, 2008; Green et al., 2009), while additionally conferring elevated risk for multiple childhood disorders, including attention deficit hyperactivity disorder (ADHD), anxiety disorder, and ASD (Schneider et al., 2014).
Genes within the 22q11.2 locus are implicated in cortical circuit formation and functioning (Meechan et al., 2015; Paronett et al., 2015). Disrupted cortical interneuron migration has been observed in 22q11.2 mouse models (Meechan et al., 2012; Toritsuka et al., 2013). Correspondingly, deletion carriers present with a range of structural and functional brain abnormalities, including cortical surface area reductions, altered white-matter microstructure (Kates et al., 2001; Jalbrzikowski et al., 2014; Schmitt et al., 2015), and, importantly, disrupted large-scale network connectivity (Debbané et al., 2012; Padula et al., 2015). Recently, an independent components analysis revealed significant hypoconnectivity relative to controls within the anterior cingulate/precuneus and default mode networks, which reliably predicted 22q11DS case-control status in an independent cohort (Schreiner et al., 2017). Critically, due to its well-characterized genetic etiology, circuit-level abnormalities associated with 22q11DS can be experimentally manipulated in animals to generate causal links with circuit dysfunction. In humans, 22q11DS presents a compelling genetic high-risk model where anomalous circuitry can be investigated before overt illness development.
Specifically, aberrant connectivity of two key anatomically interconnected structures, the thalamus and hippocampus, has been implicated in neuropsychiatric disorders (Brown et al., 2017) and schizophrenia in particular (Samudra et al., 2015). The thalamus is a critical hub for flow of sensory and higher-order information, facilitating information integration across networks (Guo et al., 2017; Hwang et al., 2017). Consistent alterations of thalamocortical circuitry, involving a pattern of prefrontal-thalamic hypoconnectivity, concomitant with somatomotor (SOM)-thalamic hyperconnectivity, have been identified in schizophrenia patients and at-risk youth (Welsh et al., 2010; Woodward et al., 2012; Anticevic et al., 2014). Similarly, the hippocampus features prominently in schizophrenia neurobiology (Weinberger, 1987). Postmortem schizophrenia studies have demonstrated hippocampal alterations in excitatory pyramidal cells and local inhibitory interneurons. Hippocampal-prefrontal dysconnectivity during cognitive processing has been proposed as a translational phenotype for schizophrenia, as evidenced by a 22q11DS mouse model (Mukai et al., 2015) and by findings of altered connectivity in those at familial high-risk for schizophrenia (Meyer-Lindenberg, 2010). Critically, the thalamus and hippocampus are structurally and functionally interconnected (Jankowski et al., 2013; Nishio et al., 2014; Zakowski et al., 2017) and have been shown to exhibit opposing resting-state connectivity patterns in healthy adults (Stein et al., 2000). This may predict distinct or even interactive alterations along these circuits in a genetic risk model based on a copy number variant (CNV) that disrupts neural function broadly, thus providing a sensitive neuromarker. Yet, the pattern of thalamic and hippocampal neural alterations has not been systematically investigated in a genetic risk model, such as 22q11DS.
Here we take a hypothesis-based approach to study large-scale network alterations in 22q11DS by leveraging findings from animal models of the disorder and neuroimaging in humans. Using the Human Connectome Project (HCP) analytical pipeline, which yields exceptional cortical spatial alignment (Glasser et al., 2013), we computed functional relationships between subject-specific anatomically defined thalamic and hippocampal seeds in 22q11DS youth and matched controls. Relative to controls, 22q11DS youth exhibited thalamocortical hyperconnectivity with SOM cortex but hypoconnectivity with associative networks. A dissociable (i.e., interactive) pattern was found for hippocampal-cortical circuitry, suggesting that 22q11DS is associated with alterations in thalamic and hippocampal circuits.
Materials and Methods
Participants.
The total sample consisted of 81 participants (7–26 years of age; n = 42 22q11DS, 59.5% female; n = 39 demographically matched healthy controls [HCs], 53.8% female), recruited from an ongoing longitudinal study at the University of California, Los Angeles. The 22q11DS participants all had a molecularly confirmed 22q11.2 deletion (for demographic details, see Table 1). HCs and 22q11DS groups were statistically group-matched based on age, sex, handedness, paternal and maternal education level (in years), and the subjects' own education levels (Table 1). Exclusion criteria for all study participants were as follows: neurological or medical condition that might affect performance, insufficient fluency in English, and/or substance or alcohol abuse and/or dependence within the past 6 months. HCs additionally could not meet diagnostic criteria for any major mental disorder, based on information gathered during administration of the Structured Clinical Interview for DSM-IV Axis I Disorders (First et al., 1996). After study procedures had been fully explained, adult participants provided written consent, while participants under the age of 18 years provided written assent with the written consent of their parent or guardian. The University of California, Los Angeles Institutional Review Board approved all study procedures and informed consent documents.
Table 1.
Demographic and symptom measures for 22q11DS (n = 42) and HC subject (n = 39) groupsa
| HCS (n = 39) |
22q11DS (n = 42) |
Between-groups |
||||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | T or χ2 | p | |
| Age (yr) | 14.1 | 4.7 | 15.7 | 5.3 | 1.5 | 0.140 |
| Sex, N (% male)b | 46.2 | — | 40.5 | — | 0.7 | 0.403 |
| Handedness, N (% right-handed)b | 91.0 | — | 95.2 | — | 1.1 | 0.289 |
| Paternal education (yr) | 13.7 | 3.9 | 15.0 | 3.8 | 1.5 | 0.141 |
| Maternal education (yr) | 14.5 | 3.8 | 15.7 | 2.2 | 1.6 | 0.115 |
| Subject education (yr) | 7.9 | 4.7 | 8.5 | 3.8 | 0.7 | 0.499 |
| WASI full-scale IQ | 108.0 | 20.2 | 77.6 | 14.5 | 7.3 | 1.8 × 10−10 |
| Verbal IQ | 57.8 | 13.9 | 36.2 | 9.6 | 7.6 | 5.5 × 10−11 |
| Nonverbal IQ | 50.0 | 11.4 | 34.4 | 13.4 | 5.6 | 3.3 × 10−7 |
| Anxiety disorder, N (%) | 15.4 | — | 50.0 | — | — | — |
| ADHD, N (%) | 5.1 | — | 45.2 | — | — | — |
| ASD, N (%) | 0.0 | — | 54.8 | — | — | — |
| Mood disorder, N (%) | 12.8 | — | 23.8 | — | — | — |
| Psychotic disorder, N (%) | 0.0 | — | 7.1 | — | — | — |
| Prodromal syndrome (N% meeting COPS) | 0.0 | — | 35.7 | — | — | — |
| Antipsychotic medication, N (%) | 0.0 | — | 11.9 | — | — | — |
| Psychostimulant medication, N (%) | 0.0 | — | 14.3 | — | — | — |
| Congenital cardiac defect, N (%) | 0.0 | — | 31.0 | — | — | — |
| BOLD movement (% frames scrubbed per subject) | 5.0 | 9.8 | 4.4 | 6.8 | 0.3 | 0.755 |
| BOLD temporal SNR | 89.0 | 14.6 | 92.1 | 15.4 | 0.9 | 0.369 |
aVerbal IQ, WASI Vocabulary T score; Nonverbal IQ, WASI Matrix Reasoning T score; COPS, Criteria of Prodromal Syndromes, as part of the SIPS. Psychiatric diagnostic categories were based on SCID-IV criteria: n = 12 subjects had two comorbid diagnoses; n = 18 had three or more diagnoses.
bSex and handedness are reported as percentages for each group, but for these categorical variables (male/female; left/right) between-group differences were evaluated via appropriate χ2 tests, rather than t test. n = 1 HCS was ambidextrous and thus excluded from this calculation.
Neuroimaging acquisition.
All subjects were imaged on a 3 tesla Siemens TimTrio scanner with a 32-channel phased array head coil at the University of California, Los Angeles Center for Cognitive Neuroscience. Resting BOLD images were acquired in 34 interleaved axial slices parallel to the anterior–posterior commissure using a fast gradient-echo, echo-planar sequence (voxel size = 3 × 3 × 4 mm, TR = 2000 ms, TE = 30 ms, flip angle = 90°, matrix = 64 × 64, FOV = 192 × 192 mm). Acquisition lasted 5.1 min and produced 152 volumes. High-resolution T1w images were collected in 160 sagittal slices via an MP-RAGE (voxel size = 1 × 1 × 1 mm, TR = 2300 ms, TE = 2.91 ms, flip angle = 90°, matrix = 240 × 256, FOV = 240 × 256 mm).
Clinical assessment.
On the same day as the scan, demographic information and clinical measures were collected for each participant by trained master's level clinicians (Table 1). Verbal IQ was assessed via the Wechsler Abbreviated Scale of Intelligence (WASI) Vocabulary subtest, and nonverbal IQ was assessed via the WASI Matrix Reasoning subtest. Psychiatric and dimensional psychotic-like symptoms were assessed via the Structured Interview for Prodromal Syndromes (SIPS) (Miller et al., 2002). For more details on study ascertainment and recruitment procedures, see Jalbrzikowski et al. (2012, 2013) .
Neuroimaging data preprocessing.
Structural and fMRI data were first preprocessed according the methods provided by the HCP, outlined below, and described in detail by the WU-Minn HCP consortium (Glasser et al., 2013). These open-source HCP algorithms, which we further optimized for compatibility with legacy single-band data in this study, represent the current state-of-the-art approaches in spatial distortion correction, registration, and maximization of high-resolution signal-to-noise (SNR) (Glasser et al., 2016). All processing methods closely followed the minimal processing pipelines as outlined by Glasser et al. (2013), with a few key modifications.
The adapted HCP pipeline included the following steps: (1) The T1-weighted images were corrected for bias-field distortions and warped to the standard MNI-152 brain template through a combination of linear and nonlinear transformations using the FMRIB Software Library (FSL) linear image registration tool (FLIRT) and nonlinear image registration tool (FNIRT) (Jenkinson et al., 2002). (2) FreeSurfer's recon-all pipeline was used to compute brainwide segmentation of gray and white matter to produce individual cortical and subcortical anatomical segmentation (Reuter et al., 2012). (3) Next, cortical surface models were generated for pial and white matter boundaries as well as segmentation masks for each subcortical gray matter voxel. Using the pial and white matter surface boundaries, a “cortical ribbon” was defined along with corresponding subcortical voxels, which were combined to generate the Connectivity Informatics Technology Initiative (CIFTI) volume/surface “gray-ordinate” space for each individual subject, which drastically reduces file management for combined surface and volume analyses and visualization and establishes a combined cortical surface and subcortical volume coordinate system (Glasser et al., 2013). (4) the cortical surfaces were then registered to the group average HCP atlas using surface-based registration based on cortical landmark features, whereas the subcortical “volume” component of the image was brought into group atlas alignment via nonlinear registration (Glasser et al., 2013). (5) The BOLD data were motion corrected and aligned to the middle frame of every run via FLIRT. In turn, a liberal brain mask was applied to exclude signal from nonbrain tissue. After initial processing in Neuroimaging Informatics Technology Initiative (NIFTI) volume space, BOLD data were converted to the CIFTI gray matter matrix by sampling from the anatomically defined gray matter cortical ribbon, whereas the subcortical voxels were isolated using subject-specific FreeSurfer segmentation. The subcortical volume component of the BOLD data was then aligned to the group atlas as part of the NIFTI processing in a single transform step that concatenates all of the transform matrixes for each prior processing step (i.e., motion correction, registration, distortion correction). This produced a single nonlinear transformation to minimize interpolation cost. In turn, the cortical surface component of the CIFTI file was aligned to the HCP atlas using surface-based nonlinear deformation based on sulcal features.
Following these “minimal” HCP preprocessing steps, a high-pass filter (>0.008 Hz) was applied to the BOLD time series to remove low temporal frequencies and scanner drift. In-house MATLAB tools were then used to compute the signal in the ventricles, deep white matter, and across all gray matter voxels as proxy of global signal regression (GSR) to address spatially pervasive sources of artifacts (Power et al., 2017). These time series were modeled as nuisance variables and were regressed out of the gray matter voxels. Subsequent analyses used the residual BOLD time series following these denoising steps.
Of note, we calculated SNR for each participant by obtaining the mean signal and SD for a given slice across the BOLD run, while excluding all nonbrain voxels across all frames. In addition, we implemented “movement scrubbing,” as recommended by Power et al. (2012). Movement scrubbing refers to the practice of removing BOLD volumes that have been flagged for high motion to minimize movement artifacts. Specifically, all frames with possible movement-induced artifactual fluctuations in intensity were identified via two criteria: (1) frames in which the sum of the displacement across all 6 rigid body movement correction parameters exceeded 0.5 mm (assuming 50 mm cortical sphere radius) were identified; and (2) root mean square of differences in intensity between the current and preceding frame were computed across all voxels divided by mean intensity and normalized to time series median. Frames in which normalized root mean square exceeded the value of 3 were identified. The frames flagged by either criterion were marked for exclusion (logical or), as well as the one preceding and two frames following the flagged frame. Subjects with >50% frames flagged by either criterion were excluded from further analyses. Importantly, for the final sample, levels of motion and SNR did not relate to reported effects (see Results). Notably, we also entered percentage scrubbed frames as formal covariate into the whole-brain statistical models described below, which did not affect results.
Whole-brain seed-based functional connectivity analyses.
Thalamic and hippocampal seeds were first defined individually for each subject through automatic anatomical segmentation of high-resolution structural images via FreeSurfer software as part of the HCP minimal preprocessing pipelines. These structures were then used as “seeds,” as in our prior work (Anticevic et al., 2014). Specifically, Pearson correlations were computed between the mean BOLD signal time series in each seed and the BOLD time series at every other cortical and subcortical vertex in CIFTI gray-ordinate space. These correlation maps were then standardized for statistical analyses via Fisher r-to-Z transformation.
As noted, the thalamus and hippocampus in humans exhibit distinct resting-state connectivity profiles (Stein et al., 2000). Indeed, this would predict distinct alterations in a genetic risk model based on a de novo CNV that uniformly affects neural circuits. In turn, combining two “seed” regions, both of which may be affected, but with opposing predicted directions of alterations, constitutes a more powered neural marker. Put differently, we hypothesized a Group × Seed interaction whereby 22q11DS may exhibit distinct bidirectional alterations across the hippocampal and thalamic systems. To confirm the viability of this logic, we computed seed-based thalamic and hippocampal functional connectivity in a sample of 339 unrelated healthy adults derived from the HCP, comparing the whole-brain connectivity profiles of the two seeds via a dependent-samples t test as well as Pearson correlation. This provided the basis for the expected interactive effects between thalamic and hippocampal seeds in the core between-group analysis. In other words, the purpose of the HCP dataset here was to serve as a large normative sample to provide an empirical independent basis for the proposed hypotheses.
Next, to test the Group main effect and the Group × Seed interaction effect at the whole-brain level, we computed a two-way repeated-measures ANOVA with a factor of Group (22q11DS vs HCs) and Seed (thalamus vs hippocampus) with the BOLD resting-state functional connectivity MRI (rs-fcMRI) as the dependent measure. Whole-brain Type I error protection was applied via nonparametric permutation testing with FSL's Permutation Analysis of Linear Models (PALM) algorithm (Winkler et al., 2014) with 10,000 permutations. Of note, the main effect of Group (i.e., collapsing across seeds) did not reveal significant effects. In the results, we report the threshold-free main effect map for completeness along with simple main effects. We also computed additional PALM GLMs that included covariates for Age, Sex and Head Motion (expressed as percentage flagged frames by the aforementioned scrubbing criteria), which did not alter the results. Whole-brain familywise Type I error correction was implemented using the Threshold-Free Cluster Enhancement method to obtain clusters of spatially contiguous voxels that are above a given statistical criterion (Smith and Nichols, 2009). Collectively, this approach circumvents the distributional assumptions (e.g., normality) that may result in Type I error inflation (Eklund et al., 2016). Here we were specifically interested in the possibility that thalamic and hippocampal circuits may show distinct patterns of dysconnectivity across groups, namely, the Group × Seed interaction.
A priori functional network-based analyses.
We also independently repeated the seed-based analyses across seven a priori functional networks described by Yeo and colleagues to test for network specificity of the hypothesized effects (Buckner et al., 2011; Yeo et al., 2011; Choi et al., 2012). For the a priori network-based analysis of thalamic and hippocampal functional connectivity, BOLD time series were averaged across all voxels within each of the seven a priori defined networks before calculating functional connectivity. The logic of this approach was to test whether the observed effects were indeed network-specific, based on an a priori functional selection. Consequently, the dimensionality reduction (averaging across seven large-scale networks) reduces Type I error rate, compared with the initial 91282 CIFTI gray-ordinate vertices, under the assumption that the effects are occurring within functional networks. As described above for the whole-brain results, we computed an ANOVA, only here focused on the network effects, with factors of Seed (thalamus vs hippocampus), Group (22q11DS vs HCs), and Network (seven functionally defined a priori networks). We explicitly evaluated the interaction of Group × Network × Seed, which was designed to test whether between-group effects for the thalamus and hippocampus differ across the seven functional networks.
Clustering of thalamic and hippocampal dysconnectivity.
To quantify the contribution of distinct thalamic and hippocampal subregions to overall group differences, we used multiple clustering algorithms to partition voxels within each region based on the whole-brain group-difference rs-fcMRI effect at each voxel. Specifically, we used k-means and hierarchical clustering to provide two complementary solutions. (1) For the thalamus and hippocampus, k-means clustering was computed based on the group-difference rs-fcMRI correlation distance matrix, with k = 1 through k = 7 solutions. To evaluate the optimal number of clusters across these solutions, the within-cluster sum of squared distances was first heuristically compared via the “Elbow Method” (Thorndike, 1953). In turn, the Gap Criterion (Tibshirani et al., 2001) was computed to explicitly test the within-cluster error term for each k-means solution against a null reference distribution. This null distribution was generated by sampling n = 100 Monte Carlo replicates of a uniform distribution aligned with the principal component of the original data. (2) For the hierarchical clustering analysis within the thalamus and hippocampus, we used the Ward's minimum variance agglomeration method (Ward, 1963) to hierarchically cluster voxels based on the group-difference rs-fcMRI Euclidean distance matrix. Multiple cluster solutions emerged for each approach, which we submitted to additional analyses to determine the most parsimonious cluster solution across structures. For secondary analyses, we elected to focus on the two-cluster solutions for the thalamus and hippocampus, which was consistent across methods. Seed-based functional connectivity was subsequently computed for each of the four resultant k-means clusters (two per seed). Each cluster's whole-brain connectivity matrix was then correlated with the whole-brain connectivity matrix previously computed for the entire seed. Within the thalamus and hippocampus, the two resulting Pearson coefficients were compared using Steiger's Z test to determine which cluster's connectivity profile most resembled that of the whole seed (Steiger, 1980). For the thalamus, the connectivity profiles of the whole seed, as well as the anterior and posterior clusters derived from the k-means analysis, were also compared with the functional connectivity of seven a priori anatomical seeds derived from the FSL thalamic atlas.
Diagnostic classification analyses.
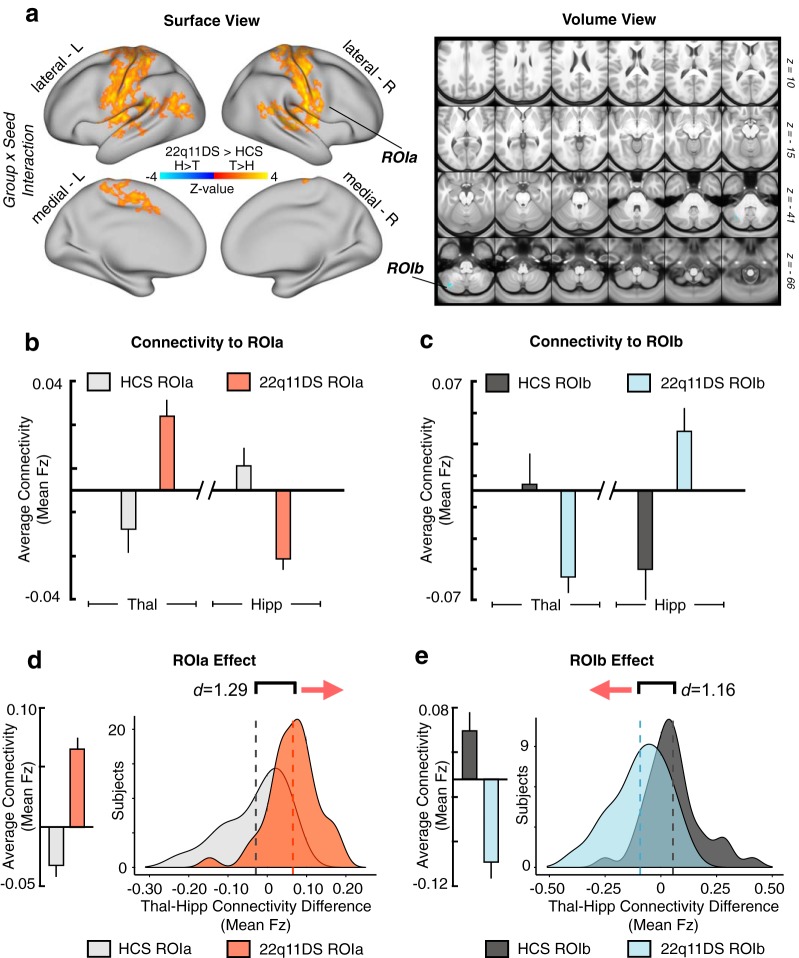
The utility of the observed rs-fcMRI effects for individual classification accuracy was assessed via a supervised binary classification algorithm. A total n = 1000 iterations of a support vector machine (SVM) were computed, each randomly splitting the n = 81 pooled subjects and training on n = 41, then using split-half cross-validation with the remaining n = 40 to build a distribution of receiver operator curves. The SVMs were trained and tested on a single feature, which consisted of the linear combination of thalamic and hippocampal connectivity to each of the interaction-derived ROIs ([thalamus-to-ROIa + hippocampus-to-ROIb] − [thalamus-to-ROIb + hippocampus-to-ROIa]). This was repeated for the network-derived results ([thalamus-to-SOM + hippocampus-to-frontoparietal (FPN)] − [thalamus-to-FPN + hippocampus-to-SOM]).
Post hoc confound analyses.
Several analyses were performed to address confounds potentially introduced during data acquisition or processing. For BOLD images, frames with significant head movement were flagged based on algorithms and intensity thresholds recommended for single-band data, as described above (Power et al., 2012). Temporal SNR (calculated for each subject as the ratio of mean BOLD signal to its SD over time) and head motion (% of flagged frames per subject) were tested for correlation with obtained rs-fcMRI effects. In turn, to assess medication as a potential confound, two-sample t tests were computed between rs-fcMRI effects in medicated versus unmedicated 22q11DS patients for the subsets taking antipsychotic medications and dopaminergic stimulants. Additionally, a series of post hoc linear model analyses were computed to evaluate the main thalamic-hippocampal results with regards to Age, Sex, IQ, and the presence of psychiatric comorbidities (anxiety, ADHD, ASD, lifetime mood disorder, and psychotic disorder) and congenital cardiac defects. Mean gray matter signal regression was included as a preprocessing step for the main analyses, but functional connectivity was also recomputed for the data without GSR to ensure that the effects were comparable at the whole-brain level and within the specific ROIs derived from permutation testing.
Overview of Type I error correction strategies across analyses.
There are several analyses in this paper that require somewhat distinct Type I error protection considerations, which we summarize here. (1) The core analysis examined the Group (22q11DS vs HCs) × Seed (thalamus vs hippocampus) interaction at the whole-brain level. As noted, here significance was evaluated using nonparametric permutation testing via FSL's PALM software, which implements Threshold-Free Cluster Enhancement (Smith and Nichols, 2009). The same methods were used to evaluate the main effect of Group for the thalamus and hippocampus seeds separately and together. (2) For the a priori network-based analysis of thalamic and hippocampal functional connectivity, we used a factorial ANOVA design to examine the significance of the Group × Network × Seed interaction. Post hoc Cohen's d calculations were computed to provide a qualitative sense of how the observed effects manifested. (3) Subsequent analyses (e.g., the SVM classification) rely on data extracted from the whole-brain permutation-corrected and a priori network-based analyses, and thus benefit from the original Type I error protection. This is noted explicitly in each figure legend when applicable. (4) The significance of our multiple k-means results was formally evaluated compared with expectations under a null distribution via the Gap Criterion (Tibshirani et al., 2001). (5) Of note, the initial comparison of thalamic and hippocampal seed-based resting fcMRI in heathy adults (using HCP data) is shown unthresholded, as well as with a qualitatively chosen T value of >5 given the robustness of this general population effect. This, and any other threshold-free brain maps presented in the paper, is not intended to provide any statistical inference, which we note explicitly in each figure legend when applicable.
Results
22q11DS is associated with distinct functional dysconnectivity for thalamus and hippocampus
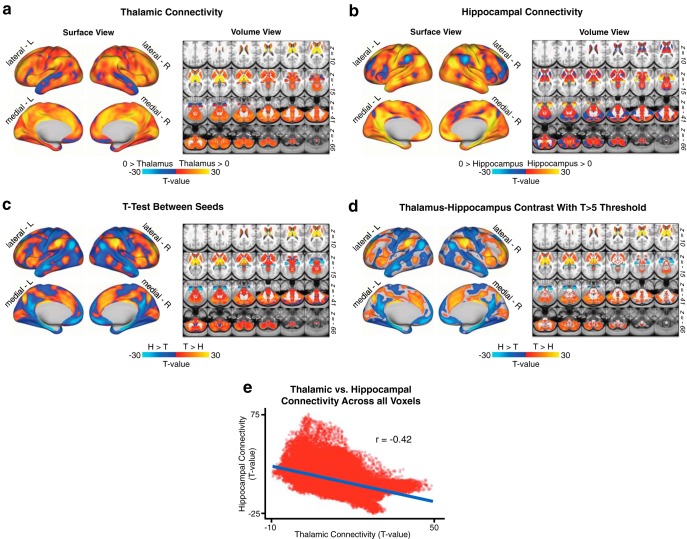
As noted, we sought to test whether 22q11DS is characterized by disruptions in thalamic as well as hippocampal resting-state functional connectivity (rs-fcMRI). We hypothesized dissociable effects across thalamic and hippocampal seeds, given known differences in the functional connectivity patterns of these structures. To further establish support for this hypothesized effect, we first conducted a “control” analysis in the n = 339 healthy adult subjects collected by the HCP (Fig. 1). Results showed that the rs-fcMRI profiles of the thalamus and hippocampus are intrinsically anticorrelated at the whole-brain level, with bidirectional differences observed in a broad set of regions overlapping with sensory and executive networks. Next, we tested whether these rs-fcMRI disruptions exhibit common or interactive effects for 22q11DS versus HCs. To this end, we computed a whole-brain GLM with factors of Group (22q11DS vs HCs) and Seed (thalamus vs hippocampus). Of note, results did not reveal any significant main effects of Group across seeds.
Figure 1.
Hippocampal versus thalamic seed connectivity. N = 339 healthy adults (HCP dataset). Comparison of thalamic and hippocampal resting-state functional connectivity in the HCP dataset. a, Surface and volume maps showing the connectivity profile of the thalamus as a t test against zero in the n = 339 healthy subjects. b, Same as in a, showing hippocampal functional connectivity. c, Threshold-free dependent-samples t test between thalamic and hippocampal functional connectivity in the HCP sample. d, The same contrast, masked at a threshold of T > 5 for illustrative purposes. e, Scatterplot of the voxelwise relationship between thalamic and hippocampal scores from a and b, showing the anticorrelation between whole-brain thalamic and hippocampal functional connectivity in a normative sample of healthy adults (Pearson r = −0.42, p < 2.2 × 10−6).
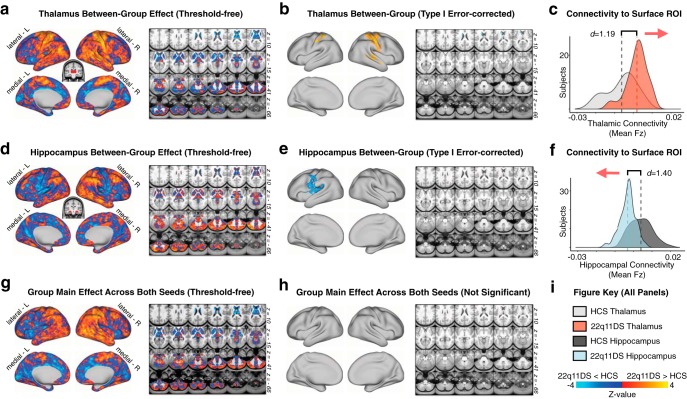
As predicted, results revealed a significant 2 × 2 Group × Seed interaction across two sets of regions: (1) sensory-motor regions, marked by hyperconnectivity for the thalamus but hypoconnectivity with the hippocampus; (2) a cerebellar region marked by hypoconnectivity for the thalamus but hyperconnectivity with the hippocampus (for a table of all regions surviving Type I error correction, see Figure 2-1; Fig. 2). Put differently, the interaction effect reflects the 22q11DS group exhibiting significantly increased thalamic connectivity (but decreased hippocampal connectivity) with bilateral SOM regions, including the precentral and postcentral gyri and superior temporal gyrus, whereas the opposite effect (decreased thalamic and increased hippocampal connectivity) was observed for a region in the left cerebellum. While this effect was localized to the cerebellum following Type I error correction, the threshold-free maps show a broader set of prefrontal and parietal regions that trend toward significance (for threshold-free, and Type I error-corrected post hoc individual seed effects and the main effect of Group, see Fig. 3). Throughout the subsequent text and figures, ROIa refers to the interaction-derived Type I error-corrected region that includes the precentral, postcentral, and superior temporal gyri (i.e., the regions around the SOM cortex). In turn, ROIb refers to the cerebellar region that was identified via the same whole-brain analysis.
Figure 2.
Results of the whole-brain Group × Seed interaction. Resting-state functional connectivity of the thalamus and hippocampus in 22q11DS and HCs. a, Surface and volume maps showing Type I error-protected* group-level contrast for the 2 × 2 interaction between Group (22q11DS vs HCs) and Seed (thalamus vs hippocampus). Yellow-orange (ROIa) represents an effect whereby 22q11DS showed thalamic hyperconnectivity but hippocampal hypoconnectivity relative to HCs. Blue contrast (ROIb) represents an effect whereby 22q11DS showed thalamic hypoconnectivity but hippocampal hyperconnectivity relative to HCs. b, Group mean connectivity to ROIa (mean Fz) for the thalamic and hippocampal seeds. c, Same as b for ROIb, showing effects in the opposite direction as ROIa. d, Difference scores between thalamic and hippocampal connectivity to ROIa (mean Fz) across subjects in each group. Group means (left) and distributions (right) shown to illustrate the direction of the effect. e, Same as d for ROIb. Histograms are based on the data extracted from the maps presented in a. *Type I error is controlled for through nonparametric permutation-based testing implemented in PALM (Winkler et al., 2014) using Threshold-Free Cluster Enhancement (Smith and Nichols, 2009) at the whole-brain level (for a detailed table of all regions surviving Type I error correction, see Figure 2-1).
Figure 3.
Separate seed-based connectivity maps. Group contrasts for functional connectivity of separate thalamic and hippocampal seeds. a, Surface and volume maps showing the threshold-free effect (22q11DS vs HCs) for the thalamic seed. Yellow-orange contrast represents 22q11DS > HCs. Blue represents HCs > 22q11DS. b, Same contrast as in a, restricted to regions remaining significant after Type I error protection (applying the same nonparametric permutations methods as in Fig. 2). c, Distributions of group mean thalamic connectivity to the yellow-orange surface ROI in b. d–f, Same as in a–c, showing the main effect of Group for the hippocampal seed. g, Threshold-free main effect of Group (i.e., regions that exhibit a consistent pattern of alterations regardless of seed) is shown for completeness and visual inspection. h, No main effects of Group survived Type I error correction. i, Color key for brain maps and distribution plots.
All Regions Showing Significant Interaction-Derived Effects. List of all region coordinates (x,y,z), locations (left/right hemisphere or subcortex), anatomical names, sizes, and 22q11DS vs. HCS comparative statistics (Cohen's d, t-value, and related p-value) for every brain region found to exhibit a significant Group x Seed interaction. Download Figure 2-1, DOCX file (48.7KB, docx)
Ruling out motion, SNR, and medication effects
To ensure that the observed effects were not attributable to differential motion between groups, or to differential SNR profiles, we correlated both measures with the functional connectivity values for both seeds to both interaction-derived ROIs, as well as with the linear combination of these four connectivity values. No significant relationships were observed between functional connectivity and head motion or SNR for the 22q11DS or HC groups (Table 2). Within the 22q11DS group, mean rs-fcMRI effects were also compared between cohorts of medicated and unmedicated patients (with regards to antipsychotic and stimulant medication). No significant effects of either medication type were observed (Table 3).
Table 2.
Movement and SNR relationshipsa
| HCS |
22q11DS |
|||
|---|---|---|---|---|
| Pearson r | p | Pearson r | p | |
| Movement | ||||
| Combined rs-fcMRI effects | 0.09 | 0.60 | 0.26 | 0.09 |
| Thalamus-to-ROIa | 0.17 | 0.31 | 0.26 | 0.10 |
| Thalamus-to-ROIb | −0.18 | 0.27 | −0.11 | 0.47 |
| Hippocampus-to-ROIa | −0.13 | 0.44 | −0.15 | 0.34 |
| Hippocampus-to-ROIb | −0.20 | 0.22 | 0.15 | 0.35 |
| SNR | ||||
| Combined rs-fcMRI effects | −0.05 | 0.77 | −0.18 | 0.26 |
| Thalamus-to-ROIa | −0.16 | 0.33 | −0.17 | 0.29 |
| Thalamus-to-ROIb | 0.02 | 0.89 | −0.05 | 0.75 |
| Hippocampus-to-ROIa | 0.01 | 0.95 | 0.09 | 0.59 |
| Hippocampus-to-ROIb | 0.04 | 0.81 | −0.23 | 0.15 |
aPearson correlations showing no significant relationship between functional connectivity effects (mean Fz connectivity values) and measures of head movement (% BOLD frames scrubbed per subject) and SNR. Combined rs-fcMRI effects, linear combination of connectivity values from the thalamus and hippocampus to the two ROIs (ROIa and ROIb) derived from the Type I error-protected Group × Seed interaction analysis (see Whole-brain seed-based functional connectivity analyses). The linear combination was obtained as follows: [thalamus-to-ROIa + hippocampus-to-ROIb] − [thalamus-to-ROIb + hippocampus-to-ROIa]. Connectivity between each seed and ROI (e.g. thalamus-to-ROIa) was also individually tested for correlation with motion and SNR.
Table 3.
Comparison of medicated versus unmedicated 22q11DSa
| Antipsychotic |
Stimulant |
|||
|---|---|---|---|---|
| t | p | t | p | |
| Combined rs-fcMRI effects | −1.64 | 0.144 | 0.20 | 0.849 |
| Thalamus-to-ROIa | −0.34 | 0.735 | −0.58 | 0.581 |
| Thalamus-to-ROIb | 1.00 | 0.361 | 0.39 | 0.710 |
| Hippocampus-to-ROIa | −0.56 | 0.603 | −1.00 | 0.346 |
| Hippocampus-to-ROIb | −1.13 | 0.302 | 0.60 | 0.565 |
aFunctional connectivity scores for the subset of patients medicated with antipsychotics were compared with the rest of the 22q11DS patients (and similarly for 22q1DS with and without dopaminergic stimulants). Results are presented from two-sample t test, with the dependent variable considered as the linear combination of functional connectivity values from both seeds and ROIs. Combined rs-fcMRI effects, linear combination of connectivity values from the thalamus and hippocampus to the two ROIs (ROIa and ROIb) derived from the Type I error-protected Group × Seed interaction analysis (see Whole-brain seed-based functional connectivity analyses). The linear combination was obtained as follows: [thalamus-to-ROIa + hippocampus-to-ROIb] − [thalamus-to-ROIb + hippocampus-to-ROIa].
Characterizing 22q11DS dysconnectivity across thalamic and hippocampal subregions
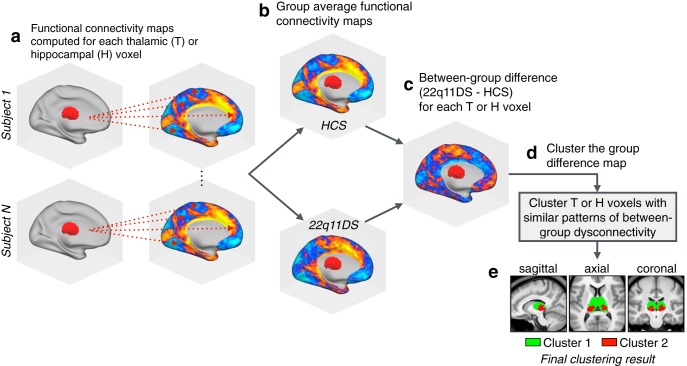
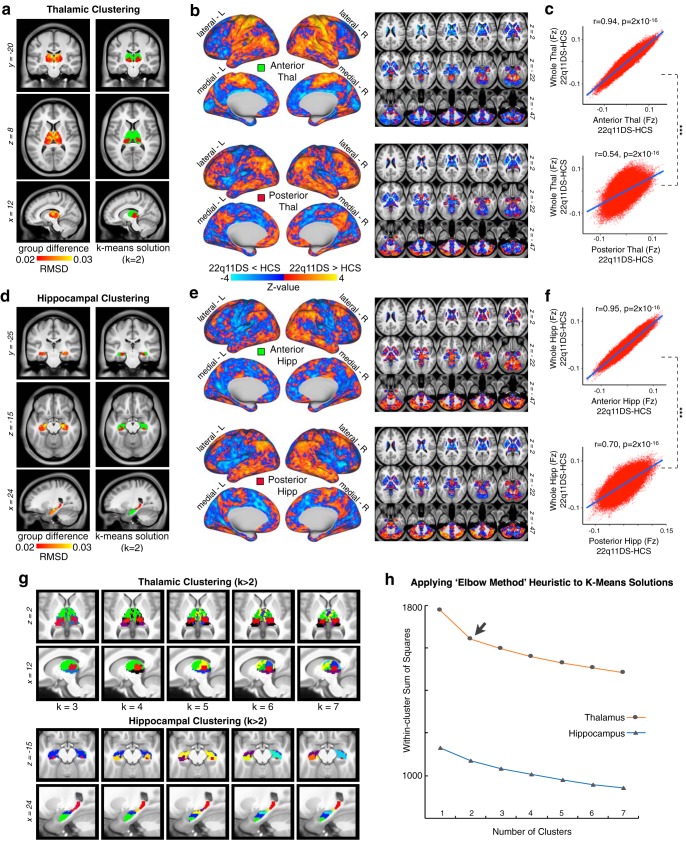
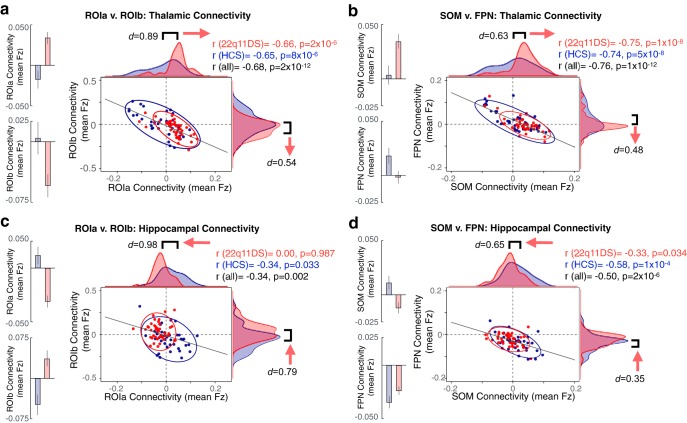
The thalamus and hippocampus are both heterogeneous structures which can be divided into multiple nuclei with distinct physiologies and connectivity profiles (Haber and McFarland, 2001). To assess differential functional connectivity disruptions across thalamic and hippocampal subregions, we used a k-means algorithm to cluster thalamic and hippocampal voxels based on unique between-group connectivity differences. The implementation of this algorithm is outlined in Figure 4. Figure 5 shows the k-means solutions for the thalamus and the hippocampus, both of which reveal distinct anterior and posterior clusters. The anterior thalamic cluster encompasses “associative” thalamic nuclei (e.g., the mediodorsal nucleus), whereas the posterior cluster is centered on visual lateral geniculate and pulvinar nuclei. The hippocampus was similarly divided along an anterior–posterior axis. Seed-based rs-fcMRI was subsequently computed for each thalamic and hippocampal cluster (group contrasts shown in Fig. 5b). For both the thalamus and hippocampus, the whole-brain connectivity matrices for the anterior cluster were quantitatively more similar to the whole-seed effect (Fig. 5c; whole-seed effects are shown above in Fig. 3).
Figure 4.
Clustering algorithm flow-chart. A graphical illustration of the procedure for k-means clustering of thalamic voxels. This same procedure was repeated for the hippocampal k-means analyses, as well as both hierarchical clustering models. a, First, functional connectivity was computed at the single-subject level for each voxel in the thalamus or hippocampus (with regards to all other voxels in the brain). b, The resulting single-subject connectivity maps were then averaged within the HCS and 22q11DS groups (separately for thalamus and hippocampus connectivity). c, For each seed, a single between-group difference map was computed between the HCS and 22q11DS group average connectivity maps. d, The group-difference maps generated in c serve as the input for the clustering algorithms (k-means and hierarchical clustering). e, Example clustering result showing a k-means (k = 2) solution for the thalamus (see Figure 5).
Figure 5.
Data-driven clustering of thalamic and hippocampal voxels. k-means clustering of thalamus and hippocampus by group. a, Left, Group difference map (root mean square) for the thalamus, highlighting the anterior and medial dorsal nuclei. Right. The k = 2 solution splits the thalamus into an anterior and posterior cluster. b, Surface and volume maps show the group difference (22q11DS vs HCs) for the brainwide functional connectivity of the anterior thalamic cluster (top) and posterior cluster (bottom). Yellow-orange contrast represents 22q11DS > HCs. Blue represents HCs > 22q11DS. c, Voxelwise relationship between the brainwide functional connectivity maps for each thalamic cluster versus the whole-thalamus seed (see Fig. 3). The correlation with the whole-seed effect is significantly larger for the anterior cluster (top) compared with the posterior cluster (bottom) (Steiger's z = 263, p = 2 × 10−16). d–f, Replication of a–c for the hippocampus, showing a similar anterior–posterior distinction (z = 240, p = 2 × 10−16). g, Distinct k-means solutions ranging from k = 2 to k = 7 always reveal an anterior solution (green). h, Elbow plot illustrating percent variance explained by each progressive cluster solution.
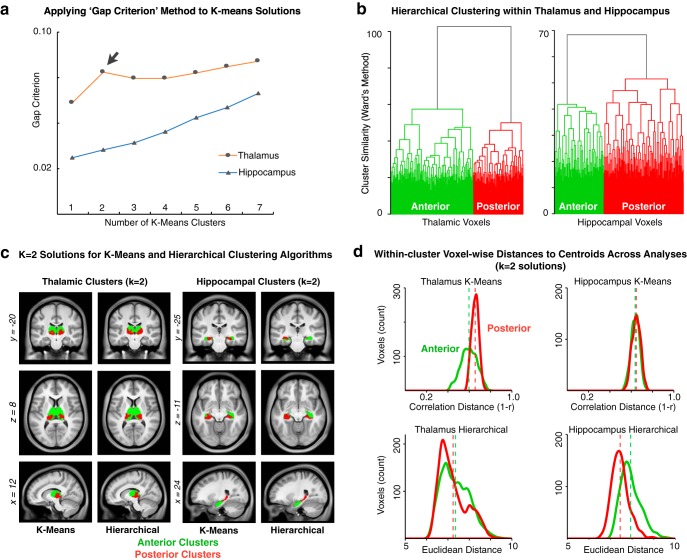
In turn, to provide a more rigorous statistical evaluation of the cluster solutions, we used the Gap Criterion, as described by Tibshirani et al. (2001), which formally tests the within-cluster error terms for each k-means solution compared with an appropriately generated null reference distribution (for details, see see Materials and Methods). Notably, the thalamus k-means solutions (Fig. 6a, orange line) show a clear local maximum for the Gap Criterion at k = 2, which suggests that this solution may represent a particularly good fit for the data. This result is also in line with the qualitative “Elbow” observed at k = 2 (Fig. 5h). For the hippocampus (Figs. 5h, 6a, blue line), neither the “Elbow” nor the Gap Criterion methods reveal local extrema implicating any particular “optimal” clustering solution. Next, using an independent clustering approach, we generated full hierarchical clustering models for the thalamus and hippocampus, using the method of Ward (1963) to build a bottom-up set of linkages minimizing within-cluster variances at each step (Fig. 6b). For both the thalamus (Fig. 6b, left) and hippocampus (Fig. 6b, right), the full dendrograms show a complex set of hierarchical clusters that ultimately converge into well-separated anterior and posterior groups (i.e., k = 2) for both structures. Importantly, both the k-means and hierarchical clustering solutions yielded highly comparable results in both seeds (Fig. 6c). To evaluate the robustness of either k-means or hierarchical cluster solutions for outliers, we computed distributions of distances between cluster centroids and each voxel assigned to that cluster (Fig. 6d). These qualitative analyses show consistent distance distributions across solutions, indicating similarly robust results for each cluster in both structures.
Figure 6.
Additional clustering analyses. a, The “Gap Criterion,” as described by Tibshirani et al. (2001), formally tests the within-cluster error terms for each k-means solution compared with an appropriately generated null reference distribution (for details, see Materials and Methods). b, Hierarchical clustering approach for the thalamus and hippocampus, using the method of Ward (1963), is with full dendrograms that converge into well-separated anterior and posterior groups for both structures (i.e., k = 2). c, Side-by-side presentation of the k = 2 solutions derived via k-means and hierarchical clustering in both seeds, highlighting the overall similarity between results. d, For each thalamic and hippocampal k-means and hierarchical k = 2 solutions, distributions are plotted for the distances between cluster centroids and each voxel assigned to that cluster. Green represents anterior clusters. Red represents posterior clusters. Vertical dashed lines indicate the average within-cluster distance-to-centroid for each cluster.
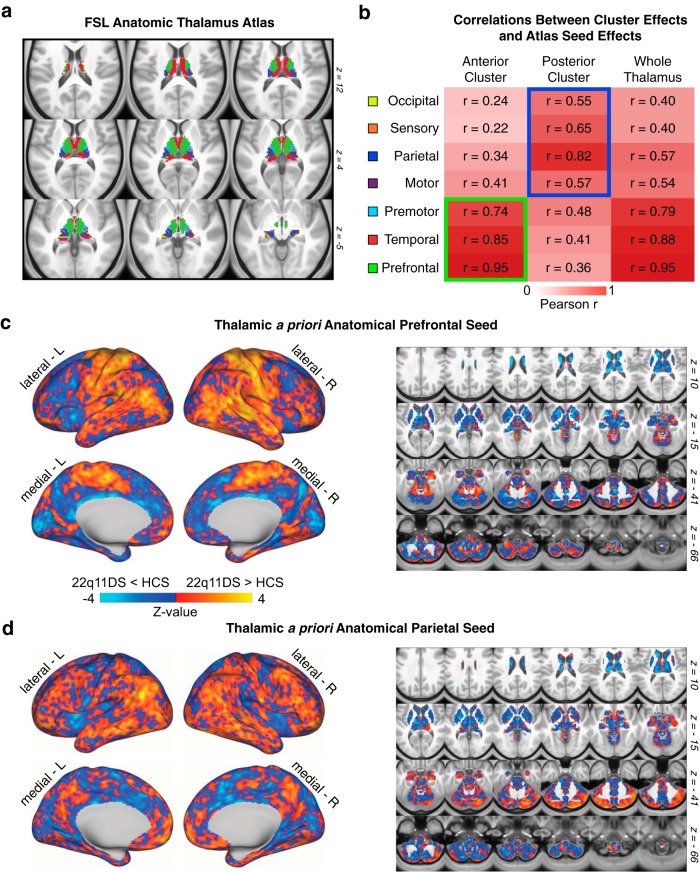
For the thalamus specifically, due to its well-defined neuroanatomical subdivisions in humans, we also investigated how the k-means two-cluster solutions compare to the functional connectivity profiles of seven a priori anatomically defined seeds derived from an FSL diffusion-weighted imaging thalamic atlas (Fig. 7) (Behrens et al., 2003). As expected, both the anterior thalamic cluster and the whole-thalamus effects were most similar to a set of “associative” thalamic seeds (prefrontal, temporal, premotor). In contrast, the posterior cluster effect was most similar to a set of “sensory” thalamic seeds (occipital, sensory, parietal, and motor) (Fig. 7b).
Figure 7.
Quantifying convergence between k-means solution and independent anatomically defined thalamic seeds. a, FSL anatomic atlas of the thalamus derived from diffusion-weighted imaging. b, Relationship between whole-brain maps of group differences derived from FSL's atlas and the k = 2 k-means solution, indicating a strong correspondence between the “anterior” cluster and executive thalamic nuclei. c, Thalamic a priori anatomically defined prefrontal-projecting thalamic seed used to compute between-group differences, which matches the anterior k = 2 effect. d, Thalamic a priori anatomically defined parietal-projecting thalamic seed used to compute between-group differences, which matches the posterior k = 2 effect.
Effects of GSR on 22q11DS dysconnectivity profiles
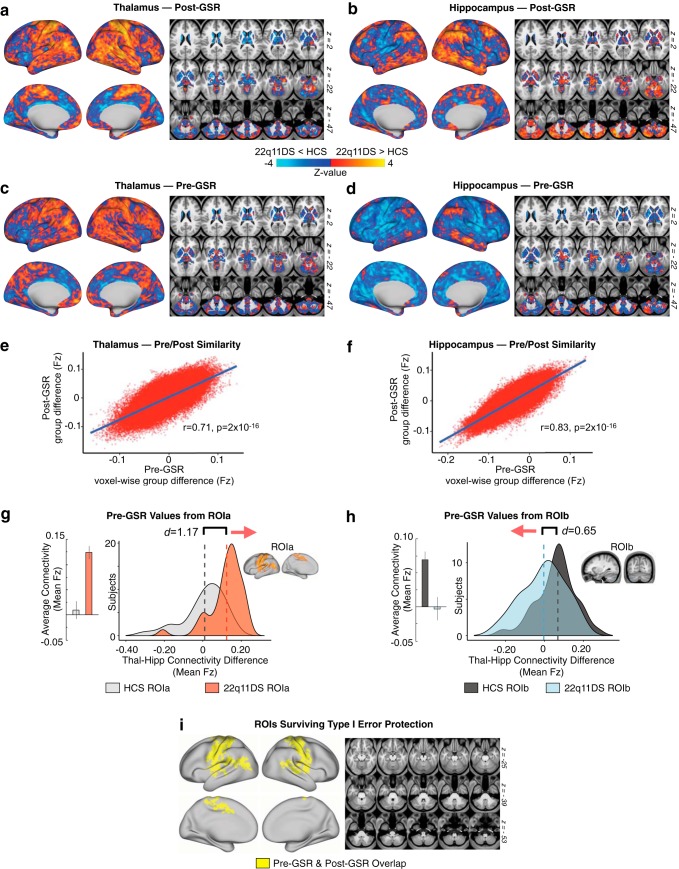
As noted, before the main rs-fcMRI analyses, BOLD data were “denoised” via mean GSR, to attenuate the contribution from spatially pervasive sources of artifact, such as fluctuations in the magnetic field; and non-neural physiological processes, such as respiration (Power et al., 2017). Nevertheless, there is ongoing development regarding the best practices for GSR in situations involving clinical populations (Glasser et al., 2018). To test whether core-observed rs-fcMRI effects are robust to GSR, we recomputed the main analyses without applying GSR. Notably, whole-brain thalamic and hippocampal functional connectivity maps were highly correlated before and after GSR. Furthermore, the pre-GSR data extracted from the original interaction-derived ROIs (ROIa and ROIb, first shown in Fig. 2) showed the same interactive thalamic and hippocampal effects between groups. Finally, the Type I error-corrected map for the pre-GSR results (as shown in Fig. 8) fully overlapped with the original Figure 2 mask, but with somewhat greater spatial extent (for a detailed list of regions, see Fig. 8-1). As such, results appear robust to GSR.
Figure 8.
Stability of effects before and after GSR. Here we show a comparison of functional connectivity effects computed with and without GSR as a preprocessing step. a, b, Post-GSR threshold-free connectivity for thalamus and hippocampus (same as Fig. 3). c, d, Thalamic and hippocampal connectivity before GSR. e, f, Pearson correlation between pre- and post-GSR matrices. g, h, Pre-GSR data extracted from ROIa and ROIb (see Fig. 2). i, Overlapping regions (logical AND) for Type I error-corrected interaction effect pre-GSR and post-GSR (for a detailed list of all regions found to exhibit a significant Group × Seed effect pre-GSR, but not post-GSR, see Figure 8-1). The cerebellar effect observed in the original analysis (see Fig. 2 ROIb) was not found to be significant in the case where GSR was not applied, but the distributions presented here in h show a qualitatively similar (though weaker) functional connectivity effect for that ROI without GSR.
Regions Only Showing Significant Interaction-Derived Effects Before GSR. Region coordinates (x,y,z), locations (left/right hemisphere or subcortex), anatomical names, sizes, and 22q11DS vs. HCS comparative statistics (Cohen's d, t-value, and related p-value) for every brain region found to exhibit a significant Group x Seed interaction only when the main whole-brain permuted type I error-protected results were re-computed as a follow-up analysis excluding global signal regression (GSR) as a preprocessing step. In other words, the whole-brain type I error protected effect was not significant for these ROIs after GSR was applied (see Figure 2). Download Figure 8-1, DOCX file (60.9KB, docx)
Dissociable 22q11DS disruptions across sensory and executive networks
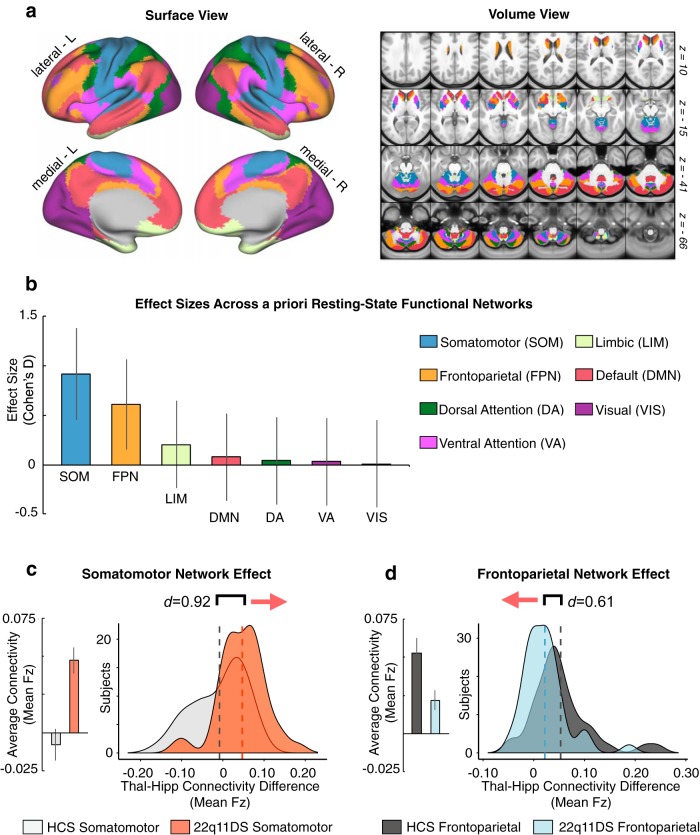
As noted, while we observed a focused Type I error corrected effect in the cerebellum, the interactive results appeared substantially more widespread (as presented threshold-free in Fig. 3). Therefore, we tested whether 22q11DS patients indeed exhibit a network-level dissociation for thalamic versus hippocampal connectivity. To this end, we repeated the seed-based analyses focusing on thalamic and hippocampal connectivity to a priori networks derived from a data-driven parcellation of the human cortex, cerebellum, and striatum (Buckner et al., 2011; Yeo et al., 2011; Choi et al., 2012) (Fig. 9). Here, functional connectivity was computed between the thalamic and hippocampal seeds and each of the seven a priori networks. In other words, we examined the connectivity between the thalamus or hippocampus with the entire brainwide average of each functional network, yielding 14 values (i.e., seven thalamus-to-each-network and seven hippocampus-to-each-network rs-fcMRI values). As predicted, the 22q11DS group exhibited increased thalamic but decreased hippocampal connectivity to brainwide SOM network regions, whereas the opposite effect was observed for the brainwide FPN network. A full factorial ANOVA was computed to test this Group × Network × Seed interaction, which yielded a significant effect (F(6,474) = 4.30, p = 3.15 × 10−4), indicating that the between-group effects indeed differ across a priori networks as a function of seed. Critically, across subjects, for both the interaction-derived and a priori network-derived effects, the magnitude of the rs-fcMRI effect in the sensory ROI/network was inversely related to the magnitude of the effect in the associative ROI/network (Figs. 10, 11). We quantified this relationship via Pearson correlations between the effects defined in the data-driven interaction-derived ROIs and the a priori networks (SOM and FPN).
Figure 9.
Examining effects across a priori functional networks. Replication of thalamic and hippocampal seed-based analysis (Fig. 2) using a priori functionally derived networks, mapped into CIFTI space. a, Surface and volume components of the map showing seven distinct a priori functional networks derived from prior work that parcellated the cortex, striatum, and cerebellum. Colors represent distinct functional networks, following the same labeling pattern as the original work. b, Effect sizes (Cohen's d) with 95% CIs comparing 22q11DS and HCs groups with regard to the difference scores between thalamic and hippocampal connectivity to each of the seven networks. c, Thalamus-hippocampus difference scores illustrated for the SOM network across subjects in each group. Group means (left) and distributions (right) illustrate the direction of the effect. d, Same as c for the FPN network, showing an effect in the opposite direction compared with the SOM network.
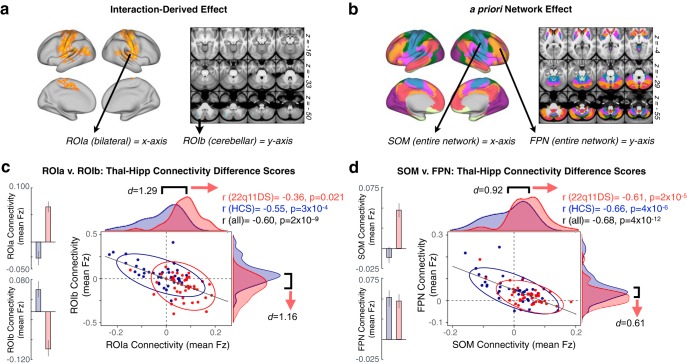
Figure 10.
Relationship between identified regions of disrupted thalamic and hippocampal connectivity. a, Surface and volume maps indicating the whole-brain interaction ROIs from which the functional connectivity data are plotted below in c. b, Surface and volume maps indicating the a priori functional networks from which the functional connectivity data are plotted below in d. For considerations regarding Type I error protection, see original Fig. 2 and Fig. 9 analyses. c, Across subjects, ROIa and ROIb are significantly negatively related in terms of the overall connectivity effect (thalamus-hippocampus Fz difference score). The distribution of 22q11DS subjects (red) is shifted relative to controls (blue), showing greater thalamic connectivity relative to hippocampal connectivity for ROIa, and the inverse for ROIb. d, As presented in c, connectivity to a priori SOM and FPN networks shows dissociable group effects for thalamus versus hippocampus with large-scale sensory and associative networks. The 22q11DS subjects show increased thalamic and decreased hippocampal connectivity to SOM networks, but the opposite effect in FPN networks. Expanded effects are presented for the thalamus and hippocampus seeds and ROIs in Figure 11 .
Figure 11.
Distinct effects across thalamic and hippocampal seeds. Expanding on Figure 10, showing distributions and relationships across subjects for whole thalamic and hippocampal seeds individually. The thalamic effect shifts in the opposite direction to the hippocampal effect. a, Thalamic seed-based connectivity to the ROIs derived from the whole-brain interaction analysis (i.e., ROIa and ROIb as shown in Fig. 2). Red represents 22q11DS distributions. Blue represents HCs. b, Thalamic connectivity to SOM and FPN a priori derived functional networks (Yeo et al., 2011). c, Hippocampal connectivity to ROIa and ROIb. d, Hippocampal connectivity to SOM and FPN networks. For considerations regarding Type I error protection, see original Figure 2 and Figure 9 analyses.
Prediction of 22q11 case-control status from data-driven and network-level dysconnectivity effects
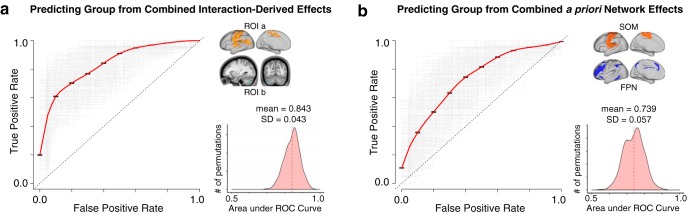
To test the hypothesis that the observed rs-fcMRI effects have potential utility as a neural biomarker, we conducted an SVM analysis (Fig. 12). One-dimensional SVMs, computed based on the unweighted linear combination of thalamic and hippocampal connectivity to ROIa and ROIb (interaction-derived ROIs), correctly predicted diagnosis at rates well above chance (for n = 1000 iterations, mean area under the curve [AUC] = 0.843, SD = 0.043). The unweighted combination of thalamic and hippocampal connectivity to entire a priori SOM and FPN networks was also able to provide moderate diagnostic accuracy (for n = 1000 iterations, mean AUC = 0.739, SD = 0.057).
Figure 12.
Diagnostic classification via SVM. SVM classification based on combined thalamic and hippocampal functional connectivity effects. a, Receiver operating characteristic (ROC) curves for a binary classifier predicting group membership (22q11DS or HCs) based on the linear combination of connectivity values (mean Fz) from the thalamus and hippocampus to ROIa and ROIb. Here ROIa and ROIb refer to the interaction-derived effects from the whole-brain analysis shown in Figure 2. The linear combination was obtained in the following way: ([thalamus-to-ROIa + hippocampus-to-ROIb] − [thalamus-to-ROIb + hippocampus-to-ROIa]). This is formally equivalent to an “interactive” effect of thalamic and hippocampal connectivity across the ROIs and produces a single value for classification. Gray represents ROC curves for each of the n = 1000 iterations. Red represents the vertical average. The distribution of areas under the n = 1000 ROC curves (AUC) is plotted on the right, where an AUC of 1 would represent a perfect classifier. b, As presented in a, connectivity to a priori SOM and FPN networks produced a linear combination for the ROC analyses, obtain in the following way: ([thalamus-to-SOM + hippocampus-to-FPN] − [thalamus-to-FPN + hippocampus-to-SOM]). For considerations regarding Type I error protection, see Figure 2 and Figure 9 analyses.
Effects of age and sex
We conducted several follow-up analyses to evaluate potential relationships between the main reported functional connectivity effects and demographic factors of age and sex. To investigate whether our effects manifest differentially across the age range in 22q11DS versus HCs, the combined thalamic and hippocampal functional connectivity to ROIa and ROIb was evaluated in a GLM with the predictors Group and Age. No significant interaction was observed (F(1,79) = 0.013, p = 0.910), indicating that the relationship between functional connectivity and Age was similar across groups. The effect of Sex (male or female) on the combined fcMRI score was evaluated in a similar linear model, with no significant interaction observed between Group and Sex (F(1,79) = 0.12, p = 0.728). Finally, we examined whether Age alone or Age and Sex combined alter the whole-brain permuted analyses when included formally as a covariate in the PALM whole-brain GLM. Here we also tested a model that included each subject's percentage of flagged high-motion frames as a covariate (along with Age and Sex), given the expectation that there may be differences in head motion across the age range that interact with Group. Notably, all three models with additional covariates showed the same general pattern as reported in the original results. We confirmed this by correlating the derived Z-scored interaction maps for each PALM model, which each yielded a highly comparable map compared with the original effect (r > 0.98 and p < 2 × 10−16).
Characterizing clinical and cognitive measures in relation to fcMRI effects
As indicated in Table 1, 22q11DS is accompanied by a range of psychiatric and medical comorbidities in a proportion of the sample. To test whether these comorbidities explain variance in the main effects beyond that explained by Group (22q11DS vs HCs), the combined hippocampal and thalamic functional connectivity scores to ROIa and ROIb were evaluated in several post hoc hierarchical linear models. The first model evaluated the influence of only psychiatric comorbidities (ASD, anxiety, ADHD, lifetime mood disorder, and psychotic disorder), regardless of Group. A second model was computed with those same predictors, as well as the predictor of Group, and the results were compared via a third linear model. In the first regression model, ASD diagnosis was the only significant predictor of our main functional connectivity effect (β = 0.32, p = 0.010). In the second model, Group is the only predictor found to be significant (β = 0.55, p = 4.0 × 10−5), and this model predicted significantly more variance than the model that did not include the Group factor (Δ adjusted R2 = 0.166, F(79,79) = 19.09, p = 4.0 × 10−5). This is consistent with the interpretation that the 22q11DS diagnosis is predictive of the observed rs-fcMRI differences, regardless of additional comorbid diagnoses. Computing the hierarchical model in the opposite order (first, testing the effect of Group alone, then Group plus the additional comorbidities as predictors) shows that the psychiatric diagnostic categories do not explain variance beyond that already explained by Group, indicating that the 22q11DS diagnosis accounts for most between-group variance. There was no significant difference found between the two regression models in this scenario (Δ adjusted R2 = −0.014, F(79,79) = 0.70, p = 0.627). The only category of medical comorbidity present in a sufficient number of patients to examine statistically was congenital cardiac defects. The effect of Group (22q11DS vs HCs) on the combined thalamic and hippocampal functional connectivity score was evaluated in the context of this medical comorbidity using the same hierarchical linear model approach applied to psychiatric comorbidities. The model including both Group and Cardiac Defect explained significantly more of the variance in our observed functional connectivity effect, compared with the model with only Cardiac Defect as a predictor (Δ adjusted R2 = 0.202, F(79,79) = 25.19, p = 3.2 × 10−6). It is also worth noting that the model accounting for both Group and Cardiac Defect did not explain significantly more variance compared with modeling Group alone (Δ adjusted R2 = 0.017, F(79,79) = 3.06, p = 0.084). Finally, to evaluate the extent to which the Group main effects depend on the variance explained by IQ, the combined thalamic and hippocampal functional connectivity score was entered into a two-step hierarchical linear regression. First, only the influence of IQ was modeled; then IQ and Group were both modeled, and the results were compared via a third linear model. Accounting for IQ alone explained ∼14% of the variance in the functional connectivity score. Modeling Group in addition to IQ explained a significantly greater proportion of the variance, at ∼31% (Δ Adjusted R2 = 0.167, F(76,76) = 19.38, p = 3.49 × 10−5).
While our general focus was to characterize thalamic-hippocampal circuitry in humans with 22q11DS as a group, as an exploratory analysis, we examined whether the 22q11DS-driven effect may be related to psychotic symptoms that are present in some of the individuals in the final sample. Notably, in the current sample, 35.7% had symptoms in the prodromal range but only 7.1% had overt psychosis. We quantified the level of psychosis symptom severity via the SIPS-positive symptom score (Tang et al., 2014), which we related to the derived thalamic and hippocampal effects. No relationships were significant (all p values >0.2).
Discussion
22q11DS is associated with notable neural alterations and presents a compelling genetic high-risk model in which anomalous circuitry can be investigated before development of overt psychiatric illness. Yet, there is a knowledge gap in our understanding of translational neuromarkers in genetic risk models, such as 22q11DS. The thalamocortical system presents a unique leverage point for investigations of brainwide dysconnectivity given its central functional role (Behrens et al., 2003; Zhang et al., 2010). Similarly, the hippocampus exhibits distinct brainwide rs-fcMRI patterns relative to the thalamus in healthy humans (Stein et al., 2000), and structural and functional hippocampal alterations feature prominently across neuropsychiatric illness (Tamminga et al., 2010). Notably, disruptions of this circuitry have been identified in a mouse model of the 22q11.2 deletion (Sigurdsson et al., 2010; Chun et al., 2014). Here, we identified dissociable functional disruptions of thalamic versus hippocampal systems in human 22q11.2 deletion carriers. Specifically, findings revealed dissociable thalamic versus hippocampal connectivity patterns with bilateral SOM regions, including auditory cortex, in 22q11DS versus typically developing controls, with the opposing effects in select cerebellar regions. Notably, the hippocampal and thalamic dysconnectivity were functionally related in 22q11DS across individuals. The overall effect was verified via a priori functional networks (Buckner et al., 2011; Yeo et al., 2011; Choi et al., 2012). The 22q11DS group showed significantly increased thalamic and decreased hippocampal connectivity to the SOM network, and the opposite effect in the FPN network. Data-driven clustering indicated punctate disruptions within specific thalamic and hippocampal sectors in 22q11DS. This result is in concert with the view that these are heterogeneous structures, which can be divided into multiple nuclei with distinct connectivity profiles. Finally, machine learning revealed above-chance classification of 22q11DS patients versus controls, based on thalamohippocampal dysconnectivity. These effects implicate disrupted thalamohippocampal dysconnectivity in 22q11DS and its functional role in neuropsychiatric illness.
Implications for the neurobiology of psychosis
Thalamic overconnectivity with SOM regions and cerebellar underconnectivity in 22q11DS are in line with prior observations in patients with established schizophrenia (Anticevic et al., 2014) and those at clinical high-risk for the disorder (Anticevic et al., 2015b). Notably, the previously reported thalamic effect was driven by those clinical high-risk youth who subsequently converted to psychosis, which would suggest that these network-level disturbances are present before onset of overt illness. In 22q11DS, hypoconnectivity with broader FPN executive regions was supported by a priori network-level analysis. Our findings showing distinct thalamic versus hippocampal effects are notable, given that the nucleus reuniens of the thalamus directly innervates the hippocampus (Herkenham, 1978; Lisman, 2012), and was recently determined to play a key role in regulating bidirectional communication between the dorsal hippocampus and mPFC (Hallock et al., 2016). This hypothesis is further supported by the k-means solutions, which implicate the key functional roles of “anterior” subdivisions for both thalamic and hippocampal seeds.
Nevertheless, BOLD rs-fcMRI is an indirect observational neuroimaging measure, and thus cannot address underlying cellular mechanisms. However, these processes can be investigated in translational studies in animal (Hiroi et al., 2013) and in vitro models (Brennand et al., 2012) as well as computational modeling studies, which can generate testable predictions at the circuit level (Anticevic et al., 2015a). Theoretical models of psychosis implicate alterations in glutamatergic, dopaminergic, and inhibitory GABAergic neurotransmission, which may be relevant to the observed disruptions of thalamo-striatal-cortical circuitry (Gonzalez-Burgos and Lewis, 2012; Lewis et al., 2012; Woodward et al., 2012). At present, the origin of the widespread thalamic-hippocampal disruption is not fully understood. However, investigation of this circuitry in the context of a well-characterized genetic etiology, as demonstrated in the current study, is a key advantage and a path forward. One possibility may involve dysfunction of NMDARs (Javitt, 2007; Loh et al., 2007), which may impact excitatory-inhibitory balance in cortical circuits and lead to large-scale disturbances in thalamocortical information flow. Notably, this hypothesis is supported by data from the 22q11.2 mouse model, as discussed below. Alternatively, it is possible that a local “hotspot” of dysfunction (e.g., the nucleus reuniens of the thalamus) emerges, via confluence of polygenic risk (Anticevic and Lisman, 2017).
Convergence with 22q11.2 mouse model
In a mouse model of the 22q11.2 deletion, Chun et al. (2014) reported disrupted glutamatergic synaptic transmission at thalamic inputs to the auditory cortex, suggesting that thalamocortical disruption could be a pathogenic mechanism that mediates susceptibility to positive psychotic symptoms in 22q11DS. Furthermore, it was determined that thalamocortical disruption in 22q11DS mice was caused by abnormal elevation of dopamine D2 (DRD2) receptors in the thalamus. Increased DRD2 in the thalamus and other brain regions has been reported in antipsychotic naive schizophrenia patients (Oke et al., 1988; Cronenwett and Csernansky, 2010). The dgcr8 gene, which encodes part of the microprocessor complex that mediates microRNA (miRNA) biogenesis, was pinpointed as being responsible for this neuronal phenotype in the 22q11DS mouse model. Consequently, reduced dosage of dgcr8 in 22q11DS may lead to miRNA dysregulation, and downstream disruption of synaptic function and proper neural circuit development (Earls and Zakharenko, 2014).
More recently, Chun et al. (2017) further established a thalamus-enriched miRNA in the 22q11DS mouse model, which specifically targets DRD2 (miRNA 338–3p). This may be a key mediator of the disruption of synaptic transmission at thalamocortical projections and the late adolescent/early adult onset of auditory perceptual anomalies in individuals with 22q11DS (Chun et al., 2017).
Although, to our knowledge, interactive disruption of the hippocampal-thalamic circuit has not yet been directly probed in this mouse model, there is complementary evidence for impaired synchronization of neural activity between the hippocampus and prefrontal cortex. Specifically, Sigurdsson et al. (2010) found that, while hippocampal-prefrontal synchrony increased during working memory performance in wild-type mice, this phase-locking did not occur in the 22q11DS mice. Further, the magnitude of baseline hippocampal-prefrontal coherence was predictive of how long it took the mice to learn the task. These findings suggest that observations of disrupted large-scale network coherence in 22q11DS are recapitulated in the animal model. Recent studies from rodent models have also revealed a broader role of the thalamus in higher-order cognitive functions (e.g., working memory). Indeed, working memory maintenance required mediodorsal thalamic inputs, suggesting a causal role for mediodorsal dysfunction in cognitive deficits associated with schizophrenia (Bolkan et al., 2017).
Findings in 22q11DS patients, and in other highly penetrant “neuropsychiatric” CNVs provide an opportunity for mechanistic exploration in cross-species translational studies; for example, a recent imaging study of the 16p11.2 deletion, a mutation that confers high risk for autism, found thalamoprefrontal alterations and impaired prefrontal connectivity in both mouse and human carriers, which were associated with sociocognitive impairments (Bertero et al., 2018). Thus, rescue experiments in the 22q11DS mouse model are warranted to determine whether the observed thalamic-hippocampal dysfunction can be “normalized” via overexpression of specific genes within the locus.
Pitfalls and future solutions
Notably, only a minority of 22q11DS participants were taking medications at the time of the scan; thus, it is unlikely that medication effects played a role in the observed findings. Moreover, we found no significant differences in the main rs-fcMRI effects when comparing medicated and unmedicated subsets of the 22q11DS cohort (Table 3). Another concern, present across rs-fcMRI studies in clinical populations, relates to head movement. We movement-scrubbed all data and tested movement (% frames scrubbed) as a covariate in our formal GLM, which did not alter the observed findings. Furthermore, motion parameters did not significantly differ between 22q11DS and HCs (Table 1), and rs-fcMRI effects were not related to head movement or SNR (Table 2). Finally, we studied subjects who, by virtue of a highly penetrant CNV, were at elevated risk for psychosis (and other neuropsychiatric symptoms). Given the young age of many of the study participants, current findings cannot address the question of whether the magnitude of thalamic-hippocampal dysconnectivity is indeed associated with subsequent risk for the development of psychosis. We attempted to relate derived effects to SIPS-positive symptoms, which did not yield significant results. This may be due to limited power in this small and not acutely symptomatic sample. Importantly, the classification results indicate robust sensitivity-specificity, which may aid future prediction research. Prospective longitudinal studies are currently underway to address this key knowledge gap and to establish the link to more acute psychiatric clinical symptom presentation.
In conclusion, this study leverages the genetic etiology of 22q11DS, thus providing a robust high-penetrance model to guide and test mechanistic hypotheses regarding disrupted neurodevelopment and subsequent consequences for circuit dysfunction leading to neuropsychiatric symptoms. Our findings offer evidence for distinct thalamic and hippocampal functional dysconnectivity with cortical regions in this genetic risk model. Notably, the observed findings pinpoint an anterior axis of thalamic-hippocampal systems in line with animal models, which yield a robust classifier that could be refined for longitudinal risk prediction. These findings suggest that focus on thalamic-hippocampal circuit interactions in 22q11DS patients and in animal models can guide translation of neurobiologically informed markers for therapeutic targets.
Footnotes
This work was supported by the National Institutes of Health Awards DP5-OD012109, R01-MH108590, and R01-MH112189 to A.A., Awards R01-MH085953 and U54-EB020403 to C.E.B., and Award T32MH073526 to A.L.; Brain and Behavior Foundation (National Alliance for Research on Schizophrenia and Depression) Independent Investigator Grant to A.A.; and the Joanne and George Miller Family Endowed Term Chair to C.E.B. Aspects of the data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 National Institutes of Health Institutes and Centers that support the National Institutes of Health Blueprint for Neuroscience Research; and the McDonnell Center for Systems Neuroscience at Washington University.
A.A. consults and is a Scientific Advisory Board member for BlackThorn Therapeutics Inc. The remaining authors declare no competing financial interests.
References
- Anticevic A, Lisman J (2017) How can global alteration of excitation/inhibition balance lead to the local dysfunctions that underlie schizophrenia? Biol Psychiatry 81:818–820. 10.1016/j.biopsych.2016.12.006 [DOI] [PubMed] [Google Scholar]
- Anticevic A, Cole MW, Repovs G, Murray JD, Brumbaugh MS, Winkler AM, Savic A, Krystal JH, Pearlson GD, Glahn DC (2014) Characterizing thalamo-cortical disturbances in schizophrenia and bipolar illness. Cereb Cortex 24:3116–3130. 10.1093/cercor/bht165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anticevic A, Murray JD, Barch DM (2015a) Bridging levels of understanding in schizophrenia through computational modeling. Clin Psychol Sci 3:433–459. 10.1177/2167702614562041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anticevic A, Haut K, Murray JD, Repovs G, Yang GJ, Diehl C, McEwen SC, Bearden CE, Addington J, Goodyear B, Cadenhead KS, Mirzakhanian H, Cornblatt BA, Olvet D, Mathalon DH, McGlashan TH, Perkins DO, Belger A, Seidman LJ, Tsuang MT, et al. (2015b) Association of thalamic dysconnectivity and conversion to psychosis in youth and young adults at elevated clinical risk. JAMA Psychiatry 72:882–891. 10.1001/jamapsychiatry.2015.0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW (2008) Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep 10:148–157. 10.1007/s11920-008-0026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Johansen-Berg H, Woolrich MW, Smith SM, Wheeler-Kingshott CA, Boulby PA, Barker GJ, Sillery EL, Sheehan K, Ciccarelli O, Thompson AJ, Brady JM, Matthews PM (2003) Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci 6:750–757. 10.1038/nn1075 [DOI] [PubMed] [Google Scholar]
- Bertero A, Liska A, Pagani M, Parolisi R, Masferrer ME, Gritti M, Pedrazzoli M, Galbusera A, Sarica A, Cerasa A, Buffelli M, Tonini R, Buffo A, Gross C, Pasqualetti M, Gozzi A (2018) Autism-associated 16p11. 2 microdeletion impairs prefrontal functional connectivity in mouse and human. Brain 141:2055–2065. 10.1093/brain/awy111 [DOI] [PubMed] [Google Scholar]
- Bolkan SS, Stujenske JM, Parnaudeau S, Spellman TJ, Rauffenbart C, Abbas AI, Harris AZ, Gordon JA, Kellendonk C (2017) Thalamic projections sustain prefrontal activity during working memory maintenance. Nat Neurosci 20:987–996. 10.1038/nn.4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Tran N, Gage FH (2012) Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry 17:1239–1253. 10.1038/mp.2012.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EC, Clark DL, Hassel S, MacQueen G, Ramasubbu R (2017) Thalamocortical connectivity in major depressive disorder. J Affect Disord 217:125–131. 10.1016/j.jad.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT (2011) The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol 106:2322–2345. 10.1152/jn.00339.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EY, Yeo BT, Buckner RL (2012) The organization of the human striatum estimated by intrinsic functional connectivity. J Neurophysiol 108:2242–2263. 10.1152/jn.00270.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Westmoreland JJ, Bayazitov IT, Eddins D, Pani AK, Smeyne RJ, Yu J, Blundon JA, Zakharenko SS (2014) Specific disruption of thalamic inputs to the auditory cortex in schizophrenia models. Science 344:1178–1182. 10.1126/science.1253895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Du F, Westmoreland JJ, Han SB, Wang YD, Eddins D, Bayazitov IT, Devaraju P, Yu J, Mellado Lagarde MM, Anderson K, Zakharenko SS (2017) Thalamic miR-338–3p mediates auditory thalamocortical disruption and its late onset in models of 22q11.2 microdeletion. Nat Med 23:39–48. 10.1038/nm.4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronenwett WJ, Csernansky J (2010) Thalamic pathology in schizophrenia. Curr Top Behav Neurosci 4:509–528. 10.1007/7854_2010_55 [DOI] [PubMed] [Google Scholar]
- Debbané M, Lazouret M, Lagioia A, Schneider M, Van De Ville D, Eliez S (2012) Resting-state networks in adolescents with 22q11.2 deletion syndrome: associations with prodromal symptoms and executive functions. Schizophr Res 139:33–39. 10.1016/j.schres.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Earls LR, Zakharenko SS (2014) A synaptic function approach to investigating complex psychiatric diseases. Neuroscientist 20:257–271. 10.1177/1073858413498307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund A, Nichols TE, Knutsson H (2016) Cluster failure: why fMRI inferences for spatial extent have inflated false-positive rates. Proc Natl Acad Sci U S A 113:7900–7905. 10.1073/pnas.1602413113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JB (1996) Structured Clinical Interview for DSM-IV Axis I Disorders, Ed 2.0. New York: Biomedical Research Department, New York State Psychiatric Institute. [Google Scholar]
- Geschwind DH, Flint J (2015) Genetics and genomics of psychiatric disease. Science 349:1489–1494. 10.1126/science.aaa8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M (2013) The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80:105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, Smith SM, Van Essen DC (2016) A multi-modal parcellation of human cerebral cortex. Nature 536:171–178. 10.1038/nature18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Bijsterbosch JD, Harrison SJ, Harms MP, Anticevic A, Van Essen DC, Smith SM (2018) Using temporal ICA to selectively remove global noise while preserving global signal in functional MRI data. Neuroimage 181:692–717. 10.1016/j.neuroimage.2018.04.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Lewis DA (2012) NMDA receptor hypofunction, parvalbumin-positive neurons and cortical gamma oscillations in schizophrenia. Schizophr Bull 38:950–957. 10.1093/schbul/sbs010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T, Gothelf D, Glaser B, Debbané M, Frisch A, Kotler M, Weizman A, Eliez S (2009) Psychiatric disorders and intellectual functioning throughout development in velocardiofacial (22q11.2 deletion) syndrome. J Am Acad Child Adolesc Psychiatry 48:1060–1068. 10.1097/CHI.0b013e3181b76683 [DOI] [PubMed] [Google Scholar]
- Guo ZV, Inagaki HK, Daie K, Druckmann S, Gerfen CR, Svoboda K (2017) Maintenance of persistent activity in a frontal thalamocortical loop. Nature 545:181–186. 10.1038/nature22324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber S, McFarland NR (2001) The place of the thalamus in frontal cortical-basal ganglia circuits. Neuroscientist 7:315–324. 10.1177/107385840100700408 [DOI] [PubMed] [Google Scholar]
- Hallock HL, Wang A, Griffin AL (2016) Ventral midline thalamus is critical for hippocampal-prefrontal synchrony and spatial working memory. J Neurosci 36:8372–8389. 10.1523/JNEUROSCI.0991-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M. (1978) The connections of the nucleus reuniens thalami: evidence for a direct thalamo-hippocampal pathway in the rat. J Comp Neurol 177:589–610. 10.1002/cne.901770405 [DOI] [PubMed] [Google Scholar]
- Hiroi N, Takahashi T, Hishimoto A, Izumi T, Boku S, Hiramoto T (2013) Copy number variation at 22q11.2: from rare variants to common mechanisms of developmental neuropsychiatric disorders. Mol Psychiatry 18:1153–1165. 10.1038/mp.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K, Bertolero MA, Liu WB, D'Esposito M (2017) The human thalamus is an integrative hub for functional brain networks. J Neurosci 37:5594–5607. 10.1523/JNEUROSCI.0067-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR. (2010) Rethinking schizophrenia. Nature 468:187–193. 10.1038/nature09552 [DOI] [PubMed] [Google Scholar]
- Jalbrzikowski M, Carter C, Senturk D, Chow C, Hopkins JM, Green MF, Galván A, Cannon TD, Bearden CE (2012) Social cognition in 22q11.2 microdeletion syndrome: relevance to psychosis. Schizophr Res 142:99–107. 10.1016/j.schres.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalbrzikowski M, Jonas R, Senturk D, Patel A, Chow C, Green MF, Bearden CE (2013) Structural abnormalities in cortical volume, thickness, and surface area in 22q11.2 microdeletion syndrome: relationship with psychotic symptoms. Neuroimage Clin 3:405–415. 10.1016/j.nicl.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalbrzikowski M, Villalon-Reina JE, Karlsgodt KH, Senturk D, Chow C, Thompson PM, Bearden CE (2014) Altered white matter microstructure is associated with social cognition and psychotic symptoms in 22q11.2 microdeletion syndrome. Front Behav Neurosci 8:393. 10.3389/fnbeh.2014.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MM, Ronnqvist KC, Tsanov M, Vann SD, Wright NF, Erichsen JT, Aggleton JP, O'Mara SM (2013) The anterior thalamus provides a subcortical circuit supporting memory and spatial navigation. Front Syst Neurosci 7:45. 10.3389/fnsys.2013.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javitt DC. (2007) Glutamate and schizophrenia: phencyclidine, N-methyl-D-aspartate receptors, and dopamine-glutamate interactions. Int Rev Neurobiol 78:69–108. 10.1016/S0074-7742(06)78003-5 [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S (2002) Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841. 10.1006/nimg.2002.1132 [DOI] [PubMed] [Google Scholar]
- Kates WR, Burnette CP, Jabs EW, Rutberg J, Murphy AM, Grados M, Geraghty M, Kaufmann WE, Pearlson GD (2001) Regional cortical white matter reductions in velocardiofacial syndrome: a volumetric MRI analysis. Biol Psychiatry 49:677–684. 10.1016/S0006-3223(00)01002-7 [DOI] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW (2012) Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci 35:57–67. 10.1016/j.tins.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J. (2012) Excitation, inhibition, local oscillations, or large-scale loops: what causes the symptoms of schizophrenia? Curr Opin Neurobiol 22:537–544. 10.1016/j.conb.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh M, Rolls ET, Deco G (2007) A dynamical systems hypothesis of schizophrenia. PLoS Comput Biol 3:e228. 10.1371/journal.pcbi.0030228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, Zackai EH, Emanuel BS, Vermeesch JR, Morrow BE, Scambler PJ, Bassett AS (2015) 22q11.2 deletion syndrome. Nat Rev Dis Primers 1:15071. 10.1038/nrdp.2015.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meechan DW, Tucker ES, Maynard TM, LaMantia AS (2012) Cxcr4 regulation of interneuron migration is disrupted in 22q11.2 deletion syndrome. Proc Natl Acad Sci U S A 109:18601–18606. 10.1073/pnas.1211507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meechan DW, Maynard TM, Tucker ES, Fernandez A, Karpinski BA, Rothblat LA, LaMantia AS (2015) Modeling a model: mouse genetics, 22q11.2 deletion syndrome, and disorders of cortical circuit development. Prog Neurobiol 130:1–28. 10.1016/j.pneurobio.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. (2010) From maps to mechanisms through neuroimaging of schizophrenia. Nature 468:194–202. 10.1038/nature09569 [DOI] [PubMed] [Google Scholar]
- Miller TJ, McGlashan TH, Rosen JL, Somjee L, Markovich PJ, Stein K, Woods SW (2002) Prospective diagnosis of the initial prodrome for schizophrenia based on the structured interview for prodromal syndromes: preliminary evidence of interrater reliability and predictive validity. Am J Psychiatry 159:863–865. 10.1176/appi.ajp.159.5.863 [DOI] [PubMed] [Google Scholar]
- Mukai J, Tamura M, Fénelon K, Rosen AM, Spellman TJ, Kang R, MacDermott AB, Karayiorgou M, Gordon JA, Gogos JA (2015) Molecular substrates of altered axonal growth and brain connectivity in a mouse model of schizophrenia. Neuron 86:680–695. 10.1016/j.neuron.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio Y, Hashimoto M, Ishii K, Ito D, Mugikura S, Takahashi S, Mori E (2014) Multiple thalamo-cortical disconnections in anterior thalamic infarction: implications for thalamic mechanisms of memory and language. Neuropsychologia 53:264–273. 10.1016/j.neuropsychologia.2013.11.025 [DOI] [PubMed] [Google Scholar]
- Oke AF, Adams RN, Winblad B, von Knorring L (1988) Elevated dopamine/norepinephrine ratios in thalami of schizophrenic brains. Biol Psychiatry 24:79–82. 10.1016/0006-3223(88)90123-0 [DOI] [PubMed] [Google Scholar]
- Padula MC, Schaer M, Scariati E, Schneider M, Van De Ville D, Debbané M, Eliez S (2015) Structural and functional connectivity in the default mode network in 22q11.2 deletion syndrome. J Neurodev Disord 7:23. 10.1186/s11689-015-9120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronett EM, Meechan DW, Karpinski BA, LaMantia AS, Maynard TM (2015) Ranbp1, deleted in DiGeorge/22q11.2 deletion syndrome, is a microcephaly gene that selectively disrupts layer 2/3 cortical projection neuron generation. Cereb Cortex 25:3977–3993. 10.1093/cercor/bhu285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE (2012) Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59:2142–2154. 10.1016/j.neuroimage.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Plitt M, Laumann TO, Martin A (2017) Sources and implications of whole-brain fMRI signals in humans. Neuroimage 146:609–625. 10.1016/j.neuroimage.2016.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Schmansky NJ, Rosas HD, Fischl B (2012) Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage 61:1402–1418. 10.1016/j.neuroimage.2012.02.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudra N, Ivleva EI, Hubbard NA, Rypma B, Sweeney JA, Clementz BA, Keshavan MS, Pearlson GD, Tamminga CA (2015) Alterations in hippocampal connectivity across the psychosis dimension. Psychiatry Res 233:148–157. 10.1016/j.pscychresns.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Vandekar S, Yi J, Calkins ME, Ruparel K, Roalf DR, Whinna D, Souders MC, Sattertwaite TD, Prabhakaran K, McDonald-McGinn DM, Zackai EH, Gur RC, Emanuel BS, Gur RE (2015) Aberrant cortical morphometry in the 22q11.2 deletion syndrome. Biol Psychiatry 78:135–143. 10.1016/j.biopsych.2014.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Debbané M, Bassett AS, Chow EW, Fung WL, van den Bree M, Owen M, Murphy KC, Niarchou M, Kates WR, Antshel KM, Fremont W, McDonald-McGinn DM, Gur RE, Zackai EH, Vorstman J, Duijff SN, Klaassen PW, Swillen A, Gothelf D, et al. (2014) Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 171:627–639. 10.1176/appi.ajp.2013.13070864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner M, Forsyth JK, Karlsgodt KH, Anderson AE, Hirsh N, Kushan L, Uddin LQ, Mattiacio L, Coman IL, Kates WR, Bearden CE (2017) Intrinsic Connectivity Network-based classification and detection of psychotic symptoms in youth with 22q11.2 deletions. Cereb Cortex 27:3294–3306. 10.1093/cercor/bhx076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA (2010) Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature 464:763–767. 10.1038/nature08855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Nichols TE (2009) Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44:83–98. 10.1016/j.neuroimage.2008.03.061 [DOI] [PubMed] [Google Scholar]
- Steiger JH. (1980) Tests for comparing elements of a correlation matrix. Psychol Bull 87:245–251. 10.1037/0033-2909.87.2.245 [DOI] [Google Scholar]
- Stein T, Moritz C, Quigley M, Cordes D, Haughton V, Meyerand E (2000) Functional connectivity in the thalamus and hippocampus studied with functional MR imaging. AJNR Am J Neuroradiol 21:1397–1401. [PMC free article] [PubMed] [Google Scholar]
- Tamminga CA, Stan AD, Wagner AD (2010) The hippocampal formation in schizophrenia. Am J Psychiatry 167:1178–1193. 10.1176/appi.ajp.2010.09081187 [DOI] [PubMed] [Google Scholar]
- Tang SX, Yi JJ, Moore TM, Calkins ME, Kohler CG, Whinna DA, Souders MC, Zackai EH, McDonald-McGinn DM, Emanuel BS, Bilker WB, Gur RC, Gur RE (2014) Subthreshold psychotic symptoms in 22q11.2 deletion syndrome. J Am Acad Child Adolesc Psychiatry 53:991–1000.e2. 10.1016/j.jaac.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorndike RL. (1953) Who belongs in the family? Psychometrika 18:267–276. 10.1007/BF02289263 [DOI] [Google Scholar]
- Tibshirani R, Walther G, Hastie T (2001) Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc B Stat Methodol 63:411–423. 10.1111/1467-9868.00293 [DOI] [Google Scholar]
- Toritsuka M, Kimoto S, Muraki K, Landek-Salgado MA, Yoshida A, Yamamoto N, Horiuchi Y, Hiyama H, Tajinda K, Keni N, Illingworth E, Iwamoto T, Kishimoto T, Sawa A, Tanigaki K (2013) Deficits in microRNA-mediated Cxcr4/Cxcl12 signaling in neurodevelopmental deficits in a 22q11 deletion syndrome mouse model. Proc Natl Acad Sci U S A 110:17552–17557. 10.1073/pnas.1312661110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH. (1963) Hierarchical grouping to optimize an objective function. J Am Statist Assoc 58:236–244. 10.1080/01621459.1963.10500845 [DOI] [Google Scholar]
- Weinberger DR. (1987) Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry 44:660–669. 10.1001/archpsyc.1987.01800190080012 [DOI] [PubMed] [Google Scholar]
- Welsh RC, Chen AC, Taylor SF (2010) Low-frequency BOLD fluctuations demonstrate altered thalamocortical connectivity in schizophrenia. Schizophr Bull 36:713–722. 10.1093/schbul/sbn145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE (2014) Permutation inference for the general linear model. Neuroimage 92:381–397. 10.1016/j.neuroimage.2014.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward ND, Karbasforoushan H, Heckers S (2012) Thalamocortical dysconnectivity in schizophrenia. Am J Psychiatry 169:1092–1099. 10.1176/appi.ajp.2012.12010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL (2011) The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106:1125–1165. 10.1152/jn.00338.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakowski W, Zawistowski P, Braszka L, Jurkowlaniec E (2017) The effect of pharmacological inactivation of the mammillary body and anterior thalamic nuclei on hippocampal theta rhythm in urethane-anesthetized rats. Neuroscience 362:196–205. 10.1016/j.neuroscience.2017.08.043 [DOI] [PubMed] [Google Scholar]
- Zhang D, Snyder AZ, Shimony JS, Fox MD, Raichle ME (2010) Noninvasive functional and structural connectivity mapping of the human thalamocortical system. Cereb Cortex 20:1187–1194. 10.1093/cercor/bhp182 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All Regions Showing Significant Interaction-Derived Effects. List of all region coordinates (x,y,z), locations (left/right hemisphere or subcortex), anatomical names, sizes, and 22q11DS vs. HCS comparative statistics (Cohen's d, t-value, and related p-value) for every brain region found to exhibit a significant Group x Seed interaction. Download Figure 2-1, DOCX file (48.7KB, docx)
Regions Only Showing Significant Interaction-Derived Effects Before GSR. Region coordinates (x,y,z), locations (left/right hemisphere or subcortex), anatomical names, sizes, and 22q11DS vs. HCS comparative statistics (Cohen's d, t-value, and related p-value) for every brain region found to exhibit a significant Group x Seed interaction only when the main whole-brain permuted type I error-protected results were re-computed as a follow-up analysis excluding global signal regression (GSR) as a preprocessing step. In other words, the whole-brain type I error protected effect was not significant for these ROIs after GSR was applied (see Figure 2). Download Figure 8-1, DOCX file (60.9KB, docx)