Abstract
Brown adipose tissue (BAT) dissipates energy to produce heat. Thus, it has the potential to regulate body temperature by thermogenesis. For the last decade, BAT has been in the spotlight due to its rediscovery in adult humans. This is evidenced by over a hundred clinical trials that are currently registered to target BAT as a therapeutic tool in the treatment of metabolic diseases, such as obesity or diabetes. The goal of most of these trials is to activate the BAT thermogenic program via several approaches such as adrenergic stimulation, natriuretic peptides, retinoids, capsinoids, thyroid hormones, or glucocorticoids. However, the impact of BAT activation on total body energy consumption and the potential effect on weight loss is still limited. Other studies have focused on increasing the mass of thermogenic BAT. This can be relevant in obesity, where the activity and abundance of BAT have been shown to be drastically reduced. The aim of this review is to describe pathological processes associated with obesity that may influence the correct differentiation of BAT, such as catecholamine resistance, inflammation, oxidative stress, and endoplasmic reticulum stress. This will shed light on the thermogenic potential of BAT as a therapeutic approach to target obesity-induced metabolic diseases.
Keywords: differentiation, BAT recruitment, preadipocyte, obesity, catecholamine, inflammation, oxidative stress, endoplasmic reticulum stress
Brown Adipose Tissue and Obesity
In recent years, adipose tissue has become a central focus of studies on the mechanisms involved in obesity-related diseases. In humans, this tissue is composed mainly of white adipose tissue (WAT), which stores energy in the form of triglycerides, and brown adipose tissue (BAT), which is responsible for thermogenesis. Much has been learned in the past decades about the pathophysiology of obesity in relation to WAT molecular deregulation (Gesta et al., 2007; Shoelson et al., 2007; de Heredia et al., 2012). However, little was known about these processes in BAT during obesity progression (Villarroya et al., 2018). With the rediscovery of BAT in 2009, and the fact that its mass and activity is reduced in obese and diabetic patients, a door opened for the treatment of obesity and its associated disorders (Cypess et al., 2009; Saito et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009; Zingaretti et al., 2009).
BAT mass and activity also change with age. In newborns, an increased BAT mass at birth has been related to decreased body fat accumulation during the first 6 months of life (Entringer et al., 2017). In adulthood, a decline of BAT mass and activity has been observed in males and females with age, and may have an impact on the accumulation of body fat (Pfannenberg et al., 2010; Yoneshiro et al., 2011).
Although a few studies have described some characteristics of BAT in a mouse model of diet-induced obesity, the molecular mechanisms involved remain unclear. Recently, it was demonstrated that BAT from obese and hyperglycemic mice shows higher levels of inflammation (macrophages and T cell infiltration), endoplasmic reticulum (ER) stress, oxidative damage, and enhanced mitochondrial respiration activity (Calderon-Dominguez et al., 2016; Alcala et al., 2017). The results of transcriptomic studies have reported several BAT molecular networks modulated in a time-dependent mode in response to a high-fat diet (HFD). The molecular networks are associated with skeletal muscle development, regulation of ion transport, neurotransmitter secretion, the immune system, and lipid metabolism (McGregor et al., 2013; Cao et al., 2018). In addition, several microRNAs (miR) have been identified (miR-491, miR-455, miR-423-5p, miR-132-3p, miR-365-3p, and miR-30b) in obese BAT and could be novel potential pharmacological targets (Gottmann et al., 2018).
BAT Origin and Differentiation
Most brown adipocytes originate from precursor mesenchymal stem cells (MSC) in the somites during embryonic development. These somatic multipotent precursor cells are characterized by the expression of certain transcription factors such as myogenic factor 5 (Myf5), paired box protein 7 (Pax7), and engrailed-1 (En1) (Atit et al., 2006; Lepper and Fan, 2010; Sanchez-Gurmaches and Guertin, 2014; Wang and Scherer, 2014; Ishibashi and Seale, 2015). Skeletal myocytes, dorsal dermis, and a subset of white adipocytes in certain fat depots also arise from this lineage. Genetic lineage tracing denotes that a multistage process involves the serial activation and repression of transcription factors, co-activators, co-repressors, and cell-cycle regulatory molecules during brown fat adipogenesis (Tapia et al., 2018).
Although the upstream factors that determine brown fat lineage remain unclear, the developmental origin of classical brown adipocytes has been studied in depth (Carobbio et al., 2013; Rosenwald and Wolfrum, 2014). Many transcription factors have been described as core regulators of brown fat development and function, such as peroxisome proliferator-activated receptor γ (PPARγ), CCAAT/enhancer binding proteins (C/EBPα, C/EBPβ, C/EBPδ), PPARγ coactivator 1 alpha (PGC-1α), PRD1-BF1-RIZ1 homologous domain-containing 16 (PRDM16), and even microRNAs. These factors are individually described below.
PPARγ has been described as a master transcription factor in the general differentiation program of brown adipocytes and induces uncoupling protein 1 (Ucp1) expression during adipogenesis (Tontonoz and Spiegelman, 2008). Adipocyte-specific Pparγ-/- animals and the identification of mutations in the Pparγ gene in lipodystrophic patients have verified the key role of PPARγ in adipogenesis in vivo (Barak et al., 1999; Rosen et al., 1999; Agarwal and Garg, 2002; He et al., 2003). The second group of essential adipogenic transcription factors is the CCAAT/enhancer binding proteins (C/EBPs) (Lane et al., 1999). These transcription factors control the differentiation of a range of cell types and are expressed in early adipogenesis. C/EBPβ and C/EBPδ regulate the expression of C/EBPα and PPARγ, which are involved in the last stages of adipogenic differentiation. However, the mechanism that regulates brown cell lineage determination is not completely clear. Neither PPARγ nor C/EBPs are sufficient to induce and complete the brown adipogenic transcriptional program, although they are considered crucial transcription factors in this process (Tapia et al., 2018).
PRDM16 configures a transcriptional complex with C/EBP-β, which controls the cell fate switch from Myf5+ cells to brown preadipocytes (Kajimura et al., 2009; Sanchez-Gurmaches and Guertin, 2014). This transcriptional regulator can activate Pparγ expression and induce the thermogenic program. PRDM16 was considered critical for embryonic brown fat development (Sanchez-Gurmaches and Guertin, 2014). Nevertheless, recent studies have shown that brown fat appears in the absence of Prdm16 expression, because of independent activation of Pparγ and C/ebpα/β genes during brown fat development (Ishibashi and Seale, 2015). Hence, the participation of PRDM16 in early brown adipogenesis remains unclear.
PGC-1α was initially characterized as a cold-inducible co-activator of PPARγ (Puigserver et al., 1998). This transcription factor was indicated as a key regulator of mitochondrial biogenesis and adaptive thermogenesis. However, the expression of several brown fat-selective genes and the mass of BAT are not affected by genetic ablation of PGC-1α. Hence, PGC-1α does not determine the cellular specification of BAT (Sharma et al., 2014). There has been increasing interest in the differentiation of brown adipocyte by non-coding RNAs (Zhou and Li, 2014; Kajimura et al., 2015). Certain miRs, including miR-378, miR-30, and miR-26, induce brown or beige adipocyte differentiation (Karbiener et al., 2014; Pan et al., 2014; Hu et al., 2015).
Recently, the helix-loop-helix transcription factor, early B-cell factor 2 (EBF2), has been described as an essential mediator of brown adipocyte commitment and terminal differentiation (Rajakumari et al., 2013; Wang W. et al., 2014; Stine et al., 2016; Shapira et al., 2017). EBF2 has been proposed as one of the initial markers in the embryological development of brown fat cells. Moreover, this transcription factor is essential for adequate binding of PPARγ to Ucp1 and other thermogenic genes (Rajakumari et al., 2013). However, the mechanism by which EBF2 activates the brown fat transcriptional program remains poorly defined.
Finally, bone morphogenetic protein 7 (BMP7) is a new critical candidate for progenitor cells to commit to brown fat lineage (Tseng et al., 2008; Schulz et al., 2013; Chen and Yu, 2018). BMP7 is expressed during the early phase of adipogenesis, and several studies revealed that it induces expression of the early regulator of brown fat fate Pgc-1α, Prdm16 (Puigserver et al., 1998), as well as the brown adipocyte-specific genes Ucp1, Dio2, Cidea, Zic1, Tfam, and Nrf-1 in vivo and in vitro (Chen and Yu, 2018). Although other BMP family members can enhance adipogenesis in vitro, only BMP7 initiates the brown adipogenic program. In fact, MSC fate depends on levels of BMP7 (Tseng et al., 2008). A multifunctional protein EWS (Ewing sarcoma), coupled with its binding partner Y-box binding protein 1 (YBX1), induces Bmp7 transcription. These results indicate that EWS is also essential for early brown adipocyte lineage determination (Park et al., 2013).
BAT Quantification in Humans
An increase in BAT mass could emerge as a promising strategy against obesity and related metabolic diseases. Approaches could entail increasing the mass of active cells by promoting differentiation and proliferation or reducing apoptosis of precursor cells.
Prior to the analysis of factors that regulate brown adipocyte recruitment in obese patients, it is important to note that we lack imaging techniques to unequivocally detect the presence of BAT in humans. The traditional combination of 18F-fluorodeoxyglucose and computed tomography (18F-FDG PET-CT) allows visualization of the tissue from a functional perspective, since the technique is based on detecting radioactive-labeled glucose uptake by the active tissue. Other imaging techniques, recently reviewed in Ong et al. (2018), are also based on functional studies. This implies that patients classified as BAT-negative could be better defined as BAT-inactive. However, the question of whether increased BAT detection is related to preexisting BAT activation or enhanced BAT recruitment still needs to be addressed. Other techniques, such as magnetic resonance imaging (MRI) (Deng et al., 2018) or Xenon-CT (Branca et al., 2018) should be further explored (Sampath et al., 2016). MRI may have benefits over the classic PET-CT approach. For instance, PET-CT is ethically limited in the pediatric population due to ionizing radiation. In addition, the uptake of FDG might be accidentally modified by the room temperature or anesthesia. MRI measurements depend on the hydration state of BAT, which in an obese state is similar to that of WAT and presents high intra- and inter-individual variability (Hu et al., 2013). Inert lipophilic xenon gas in Xenon-CT specifically detects BAT with a high resolution regardless of its activation state. However, it requires the use of ionizing radiation and the implementation of xenon inhalation protocols for its use in humans. Finally, validation of these new imaging methods requires the use of larger cohorts of patients to assess specificity and sensitivity.
Mechanisms Involved in BAT Expansion
Cold/Adrenergic Stimulation
Cold-induced adrenergic stimulation is the best-studied intervention for activating the thermogenic program of BAT. Noradrenaline release stimulates UCP-1 expression and WAT lipolysis, which, together with glucose, supplies BAT with energy-rich substrates that are easily oxidable. KO mice models lacking key genes of BAT lipolysis have been used recently to demonstrate that cold-induced thermogenesis requires WAT lipolysis rather than BAT lipolysis (Cannon and Nedergaard, 2017; Schreiber et al., 2017; Shin et al., 2017).
Coupled with this observation, an increase in brown adipocyte recruitment has been reported in rodents since the 1960s (Cameron and Smith, 1964) and more recently suggested in humans. Mild exposure to cold in humans (10°C, 2 h daily for 4 weeks or 15°C, 6 h daily for 10 days) increases the volume of active tissue as reported by 18F-FDG PET-CT (van der Lans et al., 2013; Blondin et al., 2014). Even in patients with non-detectable BAT prior to cold intervention, mild exposure to 17°C, 2 h daily for 6 weeks was enough to increase 2-deoxyglucose uptake and BAT activity (Yoneshiro et al., 2013). A similar result was found in a pilot study on young obese patients (Hanssen et al., 2016), although the report has two major drawbacks: a low number of subjects (n = 5) and BAT activity was measured rather than BAT volume, as discussed above.
The molecular mechanism of cold-induced BAT recruitment has been thoroughly reviewed (Nedergaard et al., 2018). Briefly, the authors summarize how BAT recruitment in rodents exposed to cold is due to enhanced proliferation of a group of MSC within the tissue, in addition to a reduction in apoptosis (Lee et al., 2015). Unlike white subcutaneous adipocytes, which can activate a thermogenic program by sensing cold without the strict action of adrenergic stimuli (Ye et al., 2013), brown adipocyte proliferation is exclusively linked to the presence of β1 adrenoreceptors, which is the only subtype that is expressed in brown preadipocytes (Bronnikov et al., 1999). In addition, mature adipocyte proliferation can be stimulated by β3 agonists (Fukano et al., 2016). Intracellular signaling involves activation of the cAMP pathway, mediated by well-known mitogen regulators such as phosphatidylinositol-3-kinase (PI3K) (Hinoi et al., 2014), mammalian target of rapamycin (mTOR) (Labbe et al., 2016), and extracellular signal-regulated kinase (ERK1/2) (Fredriksson and Nedergaard, 2002). However, in vitro, the addition of inhibitors of these pathways was unable to completely inhibit cAMP-mediated cell proliferation (Wang Y. et al., 2014).
Another interesting field of research is how cold affects brown preadipocyte differentiation. In a recent report, the authors describe an in vitro model to enhance brown adipocyte differentiation from an immortalized line of mouse MSC by reducing the incubation temperature from 37 to 32°C for 9 days (Velickovic et al., 2018). This could indicate that differentiation can be independent of adrenergic stimulation. However, in this model, differentiated cold-induced cells resemble a beige phenotype according to the expression levels of several beige/brown feature transcription factors.
The key role of adrenergic innervation to enhance BAT recruitment is potentially one of the reasons for the reduced amount of BAT in obese patients. Central obesity has been inversely related to plasma catecholamine levels (Wang et al., 2011). In addition, obesity is characterized by a catecholamine-resistant state, at least in WAT, with reduced expression of adrenergic receptors and a reduced response to noradrenaline-induced lipolysis (Arner, 1999; Guo et al., 2014). We suggest that this situation could also affect BAT, by hampering brown adipocyte proliferation and differentiation.
Adipokines/Batokines
Immune cells and inflammatory cytokines play a key role in regulation of the thermogenic program of BAT, although knowledge of this process is not as well-known as in WAT. Some of the latest evidence has been thoughtfully reviewed in van den Berg et al. (2017). For instance, alternatively activated M2 macrophages have been reported to be necessary to sustain a cold-adaptive thermogenic program in BAT, probably due to the ability to synthesize catecholamines (Nguyen et al., 2011). Regulatory T cells are also required to maintain a proper adaptive response to cold. Genetic ablation of this type of immune cells impaired the expression of thermogenic markers and promoted the invasion of proinflammatory macrophages (Medrikova et al., 2015).
In obesity, BAT shows a low-degree of inflammation characterized by the M1 macrophage, T cell infiltration, regulatory T cell decline and cytokine release. However, it takes longer to appear and has a more limited extension than in white adipose depots (Fitzgibbons et al., 2011; Alcala et al., 2017). Time-course microarrays on HFD-fed mice revealed that the upregulation of immune cell trafficking genes begins after week 8 and spikes by week 20, together with an inflammatory response (McGregor et al., 2013). The infiltration of M1 macrophages and the proinflammatory cytokines that are released promotes a decline in UCP-1 expression, which alters thermogenic activity (Sakamoto et al., 2016).
In addition, infiltrated immune cells with pro-inflammatory potential and both circulating and self-synthesized chemokines can inhibit BAT recruitment during obesity. During normal brown preadipocyte differentiation, there is a time-dependent downregulation of the expression of pattern recognition receptors such as NOD2 and TLR2, both upstream of the NF-kB proinflammatory pathway (Bae et al., 2014). When these receptors are activated by their corresponding agonists, brown preadipocyte differentiation and adipogenesis are inhibited in a NF-κB-dependent mechanism (Bae et al., 2015). Similarly, exposure to pro-inflammatory molecules such as TNF-α, IL-1, LPS, or Oncostatin M, secreted by T cells and macrophages, inhibits brown differentiation in vitro (Mracek et al., 2004; Zoller et al., 2016; Sanchez-Infantes et al., 2017). This is achieved by downregulating key adipogenic factors such as PPARγ, which reproduces effects that were previously observed in white preadipocytes (Ron et al., 1992).
Inflammatory signals can also promote cellular apoptosis, which impedes the expansion of BAT. For instance, the induction of apoptosis by TNF-α has been traditionally described in white (Furuoka et al., 2016; Zoller et al., 2016) and brown adipocytes (Valladares et al., 2000; Miranda et al., 2010).
Finally, inflammation can inhibit brown adipocyte proliferation indirectly by inhibiting catecholamine signaling (Villarroya et al., 2018). As mentioned above, noradrenaline promotes brown adipocyte proliferation and preadipocyte differentiation. However, obesity-induced inflammation may reduce the noradrenergic tone by several potential mechanisms:
-
simple (1)
Interrupting cAMP intracellular signaling. IKK𝜀 overexpression drives the activation of NF-κB and phosphodiesterases, which reduces the availability of cAMP (Mowers et al., 2013).
-
simple (2)
Reducing the synthesis of catecholamines. Traditionally, it was claimed that the obesity-induced phenotype shift from M2 anti-inflammatory macrophage to M1 pro-inflammatory macrophage was accompanied by loss of the capacity to express tyrosine hydroxylase (Nguyen et al., 2011), the rate-limiting step in the synthesis of noradrenaline. However, recent studies question the initial capacity of M2 macrophages to express TH (Fischer et al., 2017).
-
simple (3)
Enhancing the clearance of catecholamines. Growth differentiation factor-3 (GDF3), a member of the TGF-β family, enhances the activity of monoamine oxidase in macrophages through activation of the inflammasome system. This promotes noradrenaline uptake and degradation (Guo et al., 2014).
Oxidative Stress
During obesity development, mitochondrial dysfunction due to increased substrate oxidation, together with the action of other oxidases, increases the production of reactive oxygen species (ROS). As a compensatory mechanism, the expression of antioxidant enzymes is upregulated via NRF2 and FOXO, which maintains the intracellular redox state. Oxidative stress occurs when excessive production of ROS overrides the antioxidant defense, causing macromolecule oxidation. In WAT, oxidative stress is one of the mechanisms that accounts for malfunction of the adipocyte, since it has an impact on insulin signaling or inflammation, among other factors (Furukawa et al., 2004; Jankovic et al., 2014; Alcala et al., 2015). Obese mouse BAT presented the same signs of oxidative stress: increased ROS production and a decline in antioxidant capacity (Alcala et al., 2017).
The relevant role of oxidative stress in BAT recruitment lies in the dual effect of ROS as pro-oxidative molecules at high pathological concentrations when they surpass the antioxidant defense, and their action as second messengers in cell signaling processes at physiological levels (Jones and Sies, 2015; Castro et al., 2016).
Unfortunately, little is known about the effect of ROS on BAT differentiation. A recent report describes an increase in mRNA and protein levels of antioxidant enzymes during murine brown preadipocyte differentiation (Rebiger et al., 2016). The same was observed in white adipocytes, in which the expression of antioxidant enzymes during differentiation is also increased to prevent oxidative stress (Adachi et al., 2009; Higuchi et al., 2013). In fact, Furukawa et al. (2004) described for the first time that in vitro differentiation of 3T3-L1 adipocytes was accompanied by an increase in ROS formation. These results have been reproduced in other in vitro models of white preadipocytes (Kanda et al., 2011). The addition of extracellular H2O2 shows a concentration-dependent effect on differentiation in the micromolar range. This enhances adipogenesis by acting on PPARγ and the C/EBP family of transcription factors (Tormos et al., 2011; Higuchi et al., 2013) to accommodate the excess of fat and accelerating cell proliferation (Lee et al., 2009).
However, to the best of our knowledge, there is little evidence of the role of ROS in the regulation of differentiation and adipogenesis in BAT, although some reports point to the role of Wnt signaling. BAT expresses Wnt10a, which is upregulated, at least in vitro, by the addition of H2O2 (Yasuniwa et al., 2010) and Wnt10b. Wnt activation leads to impaired brown preadipocyte differentiation and whitening of the mature brown adipocyte (Kang et al., 2005).
The incubation of preadipocytes with antioxidants led to reduced ROS formation and impaired differentiation (Calzadilla et al., 2011; Hou et al., 2012), which reflects the need to maintain a proper intracellular redox balance. Similarly, when BSO, a glutathione quencher, was added to mimic an oxidative stress situation, ROS production increased but differentiation was inhibited (Findeisen et al., 2011).
If these molecular pathways were common between white and brown preadipocytes, they would lead to changes in BAT phenotype during obesity. Changes would range from “whitening” as ROS secretion begins to a complete loss of the ability to differentiate new cells when oxidative stress is established. As a result, thermogenic capability would be completely lost.
Endoplasmic Reticulum Stress
During obesity, the overload of protein folding requirement can trigger ER stress to activate the unfolded protein response (UPR). Briefly, the UPR tries to restore the function of the ER through three pathways: decreasing protein translation, enhancing protein folding, and triggering cellular apoptosis if the repair process fails. Key markers of UPR such as Bip, Chop, Atf4, or Atf6 expression have been found to be overexpressed in BAT from HFD-fed obese mice (Alcala et al., 2017; Liu et al., 2017). However, caspase 3 was not overexpressed, which indicates that the UPR was activated for a reparative rather than a proapoptotic end. Actually, when HFD-fed mice were further induced ER stress with the administration of thapsigargin or tunicamycin, gene expression of caspase 3 and 12 and Bax was upregulated (Liu et al., 2017).
A recent report uses two approaches to inhibit UPR in brown preadipocytes: incubation with 4-phenyl butyric acid (4-PBA), an inhibitor of ER stress, and siRNA for Xbp1. Both approaches drastically reduced differentiation and adipogenesis, which indicates the key role of the activation of UPR during differentiation. Furthermore, the incubation of brown preadipocytes with capsaicin, a component of red chili peppers, stimulates brown adipogenesis as well as the expression of UPR genes such as Xbp1 or Chop (Kida et al., 2018). More recently, Ju et al. (2018) described the role of a member of the Bcl2 family, Bcl2l13, in the differentiation of brown preadipocytes. Bcl2l13 play a key role in the regulation of mitochondrial dynamics. Silencing Bcl2l13 expression resulted in hampered brown preadipocyte differentiation due to decreased expression of mitochondrial fusion genes, biogenesis and respiratory chain complexes.
These results may suggest that reparative branches of the UPR must be activated for brown preadipocyte differentiation, in a similar manner to that previously observed in white differentiation (Sha et al., 2009). However, when ER stress is strongly induced (by chemical inducers or by long-term obesity), the apoptotic pathway can be triggered, which participates partially in BAT atrophy. BAT from obese rats has lower Bcl-2/Bax mRNA and protein ratios than the BAT of their lean littermates (Briscini et al., 1998).
miRNA
miRNAs are a type of single-stranded mRNA with a variable size ranging from 21 to 25 nucleotides. They play a key role in the regulation of gene expression, mainly by binding to the 3′UTR of target mRNAs and blocking their translation. In the last few years, the role of several miRNAs in adipose tissue biology has been revealed (Brandao et al., 2017). More specifically, miRNA arrays were used to detect at least 25 BAT-enriched miRNA genes targeting up to 788 genes involved in brown adipocyte growth, proliferation and differentiation (Guller et al., 2015; Price and Fernandez-Hernando, 2016). However, the regulation of these miRNAs in obesity has not yet been determined. Findings regarding miRNAs and BAT expansion in obesity are summarized in Table 1.
Table 1.
miRNA involved in BAT expansion.
| miRNA | Effect in differentiation | Target gene | Obesity | Reference |
|---|---|---|---|---|
| miR-27 | ↓ | Prdm16 Creb Pparα Pgc-1β | miR-27 expression correlates with BMI | Yu et al., 2018 Sun and Trajkovski, 2014 |
| miR-34a | ↓ | Fgf21 Sirt1 | miR-34a expression is increased in obesity. miR-34a overexpression protected against HFD-induced obesity | Fu et al., 2014 |
| miR-133 | ↓ | Prdm16 | Antagonist miR-133 treatment protected against HFD-induced obesity | Yin et al., 2013 |
| miR-155 | ↓ | Cebpβ | miR-155 KO protected against HFD-induced obesity | Gaudet et al., 2016 |
| miR-199a-214 | ↓ | Prdm16 Pgc-1α | miR-199a-214 expression reduced in genetic models of obesity. Anti-miR-199a-214 injection protected against body weight gain | He et al., 2018 |
| miR-328 | ↑ | Bace1 | HFD-induced obesity increased miRNA-328 expression in BAT | Oliverio et al., 2016 |
| miR-378 | ↑ | Pde1b | miR-378 promoted BAT expansion, protecting against genetic and HFD-induced obesity | Pan et al., 2014 |
| miR-455 | ↑ | Runxlt1 Necdin | miRNA-455 transgenic mice (FAT455) protected against HFD-induced obesity | Zhang et al., 2015 |
| miR-93-106b | ↓ | Ppparα | HFD-induced obesity increased miRNA-93-106b expression in BAT | Wu et al., 2013 |
Role in obesity.
Conclusion
Although not completely understood yet, brown adipocyte differentiation involves a complex network of transcription factors, genes, and miRNAs that are apparently interrupted in obesity.
The relations between the pathological basis of obesity and strategies to recruit active BAT are summarized in Figure 1. Resistance to catecholamines and inflammatory processes directly reduce adipocyte differentiation and proliferation and can promote apoptosis. In addition, a moderate increase in ROS generation (before antioxidant defense is surpassed and oxidative stress is established) and ER stress favors brown preadipocyte differentiation and adipogenesis. The goal would most likely be to accumulate an excess of fat, which involves brown adipocytes changing to acquire a white phenotype, and thus losing their thermogenic potential. If the obesity challenge continues and oxidative stress and ER stress are completely established, both differentiation and proliferation are inhibited, and cellular apoptosis is triggered. The decrease of BAT mass and activity in obese individuals indicates that any strategy leading to an enhancement of preadipocyte differentiation and proliferation or a reduction of apoptosis could be potentially added to the therapeutic arsenal against obesity.
FIGURE 1.
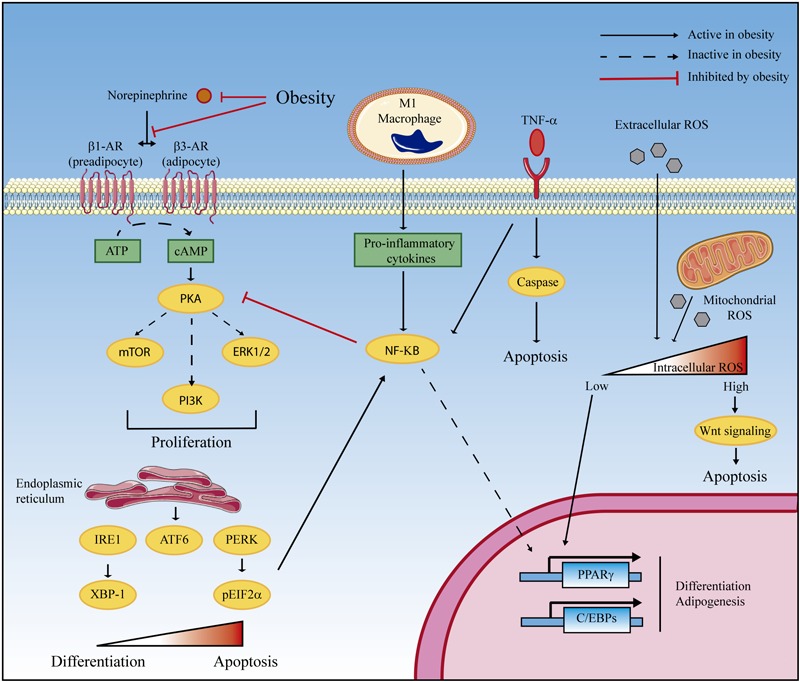
Proposed mechanisms of obesity-induced BAT depletion. BAT mass and activity are minimized in obese patients due to reduced cell proliferation and preadipocyte differentiation and increased apoptosis. At least four obesity-related mechanisms can be involved. (1) Catecholamine resistance in obesity is characterized by decreased synthesis of norepinephrine and beta-adrenergic (β-AR) receptors and by defective intracellular signaling, which impedes PKA-mediated cell proliferation. (2) Obesity promotes the infiltration of M1 macrophages that participate in norepinephrine clearance and contribute to the synthesis of proinflammatory cytokines. NF-κB-mediated signaling inhibits the PKA proliferation pathway and represses PPARγ and C/EBPs gene expression, which inhibits differentiation and adipogenesis. In addition, TNF-α overexpression triggers cellular apoptosis. (3) The unfolded protein response (UPR) in the endoplasmic reticulum plays a dual role in brown adipocyte differentiation according to the intensity of the signal. While activation of the three branches of UPR (IRE-1, ATF6, and PERK) is required for differentiation, excessive UPR activation (that can be found in severe obesity) triggers proapoptotic mechanisms. (4) In a similar manner, reactive oxygen species (ROS) are also hormetic regulators of cell differentiation and apoptosis. Physiological ROS concentrations promote C/EBP expression leading to differentiation, whereas supraphysiological concentration leads to oxidative stress and apoptosis via Wnt signaling. Artwork was obtained from Servier Medical Art, licensed under a Creative Common Attribution 3.0 Generic License (http://smart.servier.com/).
Author Contributions
MA, MC-D, and MV participated in the conception of the study and in the preparation of the manuscript. LH and DS participated in the preparation of the manuscript and critically reviewed the final draft.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Artwork was obtained from Servier Medical Art, licensed under a Creative Common Attribution 3.0 Generic License (http://smart.servier.com/).
Abbreviations
- 4-PBA
4-phenyl butyric acid
- ATF
Activating transcription factor
- BAT
Brown adipose tissue
- BIP
Binding immunoglobulin protein
- BMP7
Bone morphogenic protein 7
- C/EBP
CCAAT-enhancer-binding proteins
- CHOP
C/EBP homologous protein
- EBF2
Early B cell factor 2
- EN1
Homeobox protein engrailed 1
- ER
Endoplasmic reticulum
- ERK
Extracellular signal-regulated kinase
- EWS
Ewing sarcoma protein
- FOXO
(Forkhead box) proteins
- GDF-3
Growth differentiation factor 3
- HFD
High-fat diet
- IL-1
Interleukin 1
- LPS
Lipopolysaccharide
- MSC
Mesenchymal cells
- mTOR
Mammalian target of rapamycin
- MYF5
Myogenic factor 5
- NF-κB
Nuclear factor kappa B
- NOD
Nucleotide-binding oligomerization domain-containing protein
- NRF2
Nuclear factor (erythroid-derived 2)-like 2
- PAX7
Paired box-protein 7
- PI3K
Phosphatidylinositol-3-kinase
- PPAR
Peroxisome proliferator-activated receptor
- PRDM16
PR domain containing 16
- TLR
Toll-like receptor
- TNF-α
Tumor necrosis factor Alpha
- UCP1
Uncoupling protein 1
- UPR
Unfolded protein response
- WAT
White adipose tissue
- XBP1
X-box binding protein 1
- YBX1
Y-box binding protein 1
Footnotes
Funding. This study was supported by the Universidad CEU San Pablo-Banco Santander (FUSP-BS-PPC-USP02/2017) to MA, the Ministry of Spain (MINECO) (Grant SAF2017-83813-C3-1-R to DS and LH cofunded by the European Regional Development Fund [ERDF]), the Centro de Investigación Biomédica en Red de Fisiopatología de la Obesidad y la Nutrición (CIBEROBN) (Grant CB06/03/0001 to DS), and the Fundació La Marató de TV3 (Grant 87/C/2016 to DS).
References
- Adachi T., Toishi T., Wu H., Kamiya T., Hara H. (2009). Expression of extracellular superoxide dismutase during adipose differentiation in 3T3-L1 cells. Redox Rep. 14 34–40. 10.1179/135100009X392467 [DOI] [PubMed] [Google Scholar]
- Agarwal A. K., Garg A. (2002). A novel heterozygous mutation in peroxisome proliferator-activated receptor-gamma gene in a patient with familial partial lipodystrophy. J. Clin. Endocrinol. Metab. 87 408–411. 10.1210/jcem.87.1.8290 [DOI] [PubMed] [Google Scholar]
- Alcala M., Calderon-Dominguez M., Bustos E., Ramos P., Casals N., Serra D., et al. (2017). Increased inflammation, oxidative stress and mitochondrial respiration in brown adipose tissue from obese mice. Sci. Rep. 7:16082. 10.1038/s41598-017-16463-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala M., Sanchez-Vera I., Sevillano J., Herrero L., Serra D., Ramos M. P., et al. (2015). Vitamin E reduces adipose tissue fibrosis, inflammation, and oxidative stress and improves metabolic profile in obesity. Obesity 23 1598–1606. 10.1002/oby.21135 [DOI] [PubMed] [Google Scholar]
- Arner P. (1999). Catecholamine-induced lipolysis in obesity. Int. J. Obes. Relat. Metab. Disord. 23(Suppl. 1) 10–13. 10.1038/sj.ijo.0800789 [DOI] [PubMed] [Google Scholar]
- Atit R., Sgaier S. K., Mohamed O. A., Taketo M. M., Dufort D., Joyner A. L., et al. (2006). Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev. Biol. 296 164–176. 10.1016/j.ydbio.2006.04.449 [DOI] [PubMed] [Google Scholar]
- Bae J., Chen J., Zhao L. (2015). Chronic activation of pattern recognition receptors suppresses brown adipogenesis of multipotent mesodermal stem cells and brown pre-adipocytes. Biochem. Cell Biol. 93 251–261. 10.1139/bcb-2014-0139 [DOI] [PubMed] [Google Scholar]
- Bae J., Ricciardi C. J., Esposito D., Komarnytsky S., Hu P., Curry B. J., et al. (2014). Activation of pattern recognition receptors in brown adipocytes induces inflammation and suppresses uncoupling protein 1 expression and mitochondrial respiration. Am. J. Physiol. Cell Physiol. 306 C918–C930. 10.1152/ajpcell.00249.2013 [DOI] [PubMed] [Google Scholar]
- Barak Y., Nelson M. C., Ong E. S., Jones Y. Z., Ruiz-Lozano P., Chien K. R., et al. (1999). PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 4 585–595. 10.1016/S1097-2765(00)80209-9 [DOI] [PubMed] [Google Scholar]
- Blondin D. P., Labbe S. M., Tingelstad H. C., Noll C., Kunach M., Phoenix S., et al. (2014). Increased brown adipose tissue oxidative capacity in cold-acclimated humans. J. Clin. Endocrinol. Metab. 99 E438–E446. 10.1210/jc.2013-3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branca R. T., McCallister A., Yuan H., Aghajanian A., Faber J. E., Weimer N., et al. (2018). Accurate quantification of brown adipose tissue mass by xenon-enhanced computed tomography. Proc. Natl. Acad. Sci. U.S.A. 115 174–179. 10.1073/pnas.1714431115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao B. B., Guerra B. A., Mori M. A. (2017). Shortcuts to a functional adipose tissue: the role of small non-coding RNAs. Redox Biol. 12 82–102. 10.1016/j.redox.2017.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscini L., Tonello C., Dioni L., Carruba M. O., Nisoli E. (1998). Bcl-2 and Bax are involved in the sympathetic protection of brown adipocytes from obesity-linked apoptosis. FEBS Lett. 431 80–84. 10.1016/S0014-5793(98)00730-3 [DOI] [PubMed] [Google Scholar]
- Bronnikov G., Bengtsson T., Kramarova L., Golozoubova V., Cannon B., Nedergaard J. (1999). beta1 to beta3 switch in control of cyclic adenosine monophosphate during brown adipocyte development explains distinct beta-adrenoceptor subtype mediation of proliferation and differentiation. Endocrinology 140 4185–4197. 10.1210/endo.140.9.6972 [DOI] [PubMed] [Google Scholar]
- Calderon-Dominguez M., Mir J. F., Fucho R., Weber M., Serra D., Herrero L. (2016). Fatty acid metabolism and the basis of brown adipose tissue function. Adipocyte 5 98–118. 10.1080/21623945.2015.1122857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzadilla P., Sapochnik D., Cosentino S., Diz V., Dicelio L., Calvo J. C., et al. (2011). N-acetylcysteine reduces markers of differentiation in 3T3-L1 adipocytes. Int. J. Mol. Sci. 12 6936–6951. 10.3390/ijms12106936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron I. L., Smith R. E. (1964). Cytological Responses of Brown Fat Tissue in Cold-Exposed Rats. J. Cell Biol. 23 89–100. 10.1083/jcb.23.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B., Nedergaard J. (2017). What Ignites UCP1? Cell Metab. 26 697–698. 10.1016/j.cmet.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Cao J., Zhu Q., Liu L., Glazier B. J., Hinkel B. C., Liang C., et al. (2018). Global transcriptome analysis of brown adipose tissue of diet-induced obese mice. Int. J. Mol. Sci. 19:E1095. 10.3390/ijms19041095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carobbio S., Rosen B., Vidal-Puig A. (2013). Adipogenesis: new insights into brown adipose tissue differentiation. J. Mol. Endocrinol. 51 T75–T85. 10.1530/JME-13-0158 [DOI] [PubMed] [Google Scholar]
- Castro J. P., Grune T., Speckmann B. (2016). The two faces of reactive oxygen species (ROS) in adipocyte function and dysfunction. Biol. Chem. 397 709–724. 10.1515/hsz-2015-0305 [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Yu Y. H. (2018). The potential of brown adipogenesis and browning in porcine bone marrow-derived mesenchymal stem cells. J. Anim. Sci. 10.1093/jas/sky230 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess A. M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A. B., et al. (2009). Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360 1509–1517. 10.1056/NEJMoa0810780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Heredia F. P., Gomez-Martinez S., Marcos A. (2012). Obesity, inflammation and the immune system. Proc. Nutr. Soc. 71 332–338. 10.1017/S0029665112000092 [DOI] [PubMed] [Google Scholar]
- Deng J., Neff L. M., Rubert N. C., Zhang B., Shore R. M., Samet J. D., et al. (2018). MRI characterization of brown adipose tissue under thermal challenges in normal weight, overweight, and obese young men. J. Magn. Reson. Imaging 47 936–947. 10.1002/jmri.25836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entringer S., Rasmussen J., Cooper D. M., Ikenoue S., Waffarn F., Wadhwa P. D., et al. (2017). Association between supraclavicular brown adipose tissue composition at birth and adiposity gain from birth to 6 months of age. Pediatr. Res. 82 1017–1021. 10.1038/pr.2017.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen H. M., Pearson K. J., Gizard F., Zhao Y., Qing H., Jones K. L., et al. (2011). Oxidative stress accumulates in adipose tissue during aging and inhibits adipogenesis. PLoS One 6:e18532. 10.1371/journal.pone.0018532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Ruiz H. H., Jhun K., Finan B., Oberlin D. J., van der Heide V., et al. (2017). Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat. Med. 23 623–630. 10.1038/nm.4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbons T. P., Kogan S., Aouadi M., Hendricks G. M., Straubhaar J., Czech M. P. (2011). Similarity of mouse perivascular and brown adipose tissues and their resistance to diet-induced inflammation. Am. J. Physiol. Heart Circ. Physiol. 301 H1425–H1437. 10.1152/ajpheart.00376.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson J. M., Nedergaard J. (2002). Norepinephrine specifically stimulates ribonucleotide reductase subunit R2 gene expression in proliferating brown adipocytes: mediation via a cAMP/PKA pathway involving Src and Erk1/2 kinases. Exp. Cell Res. 274 207–215. 10.1006/excr.2002.5470 [DOI] [PubMed] [Google Scholar]
- Fu T., Seok S., Choi S., Huang Z., Suino-Powell K., Xu H. E., et al. (2014). MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell Biol. 34 4130–4142. 10.1128/MCB.00596-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukano K., Okamatsu-Ogura Y., Tsubota A., Nio-Kobayashi J., Kimura K. (2016). Cold exposure induces proliferation of mature brown adipocyte in a ss3-adrenergic receptor-mediated pathway. PLoS One 11:e0166579. 10.1371/journal.pone.0166579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y., et al. (2004). Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 114 1752–1761. 10.1172/JCI21625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuoka M., Ozaki K., Sadatomi D., Mamiya S., Yonezawa T., Tanimura S., et al. (2016). TNF-alpha induces caspase-1 activation independently of simultaneously induced NLRP3 in 3T3-L1 cells. J. Cell Physiol. 231 2761–2767. 10.1002/jcp.25385 [DOI] [PubMed] [Google Scholar]
- Gaudet A. D., Fonken L. K., Gushchina L. V., Aubrecht T. G., Maurya S. K., Periasamy M., et al. (2016). miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 6:22862. 10.1038/srep22862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S., Tseng Y. H., Kahn C. R. (2007). Developmental origin of fat: tracking obesity to its source. Cell 131 242–256. 10.1016/j.cell.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Gottmann P., Ouni M., Saussenthaler S., Roos J., Stirm L., Jahnert M., et al. (2018). A computational biology approach of a genome-wide screen connected miRNAs to obesity and type 2 diabetes. Mol. Metab. 11 145–159. 10.1016/j.molmet.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guller I., McNaughton S., Crowley T., Gilsanz V., Kajimura S., Watt M., et al. (2015). Comparative analysis of microRNA expression in mouse and human brown adipose tissue. BMC Genomics 16:820. 10.1186/s12864-015-2045-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Marmol P., Moliner A., Bjornholm M., Zhang C., Shokat K. M., et al. (2014). Adipocyte ALK7 links nutrient overload to catecholamine resistance in obesity. eLife 3:e03245. 10.7554/eLife.03245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen M. J., van der Lans A. A., Brans B., Hoeks J., Jardon K. M., Schaart G., et al. (2016). Short-term cold acclimation recruits brown adipose tissue in obese humans. Diabetes 65 1179–1189. 10.2337/db15-1372 [DOI] [PubMed] [Google Scholar]
- He L., Tang M., Xiao T., Liu H., Liu W., Li G., et al. (2018). Obesity-associated miR-199a/214 cluster inhibits adipose browning via PRDM16-PGC-1alpha transcriptional network. Diabetes 67 2585–2600. 10.2337/db18-0626 [DOI] [PubMed] [Google Scholar]
- He W., Barak Y., Hevener A., Olson P., Liao D., Le J., et al. (2003). Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. U.S.A. 100 15712–15717. 10.1073/pnas.2536828100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Dusting G. J., Peshavariya H., Jiang F., Hsiao S. T., Chan E. C., et al. (2013). Differentiation of human adipose-derived stem cells into fat involves reactive oxygen species and Forkhead box O1 mediated upregulation of antioxidant enzymes. Stem Cells Dev. 22 878–888. 10.1089/scd.2012.0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E., Iezaki T., Fujita H., Watanabe T., Odaka Y., Ozaki K., et al. (2014). PI3K/Akt is involved in brown adipogenesis mediated by growth differentiation factor-5 in association with activation of the Smad pathway. Biochem. Biophys. Res. Commun. 450 255–260. 10.1016/j.bbrc.2014.05.108 [DOI] [PubMed] [Google Scholar]
- Hou Y., Xue P., Bai Y., Liu D., Woods C. G., Yarborough K., et al. (2012). Nuclear factor erythroid-derived factor 2-related factor 2 regulates transcription of CCAAT/enhancer-binding protein beta during adipogenesis. Free Radic Biol. Med. 52 462–472. 10.1016/j.freeradbiomed.2011.10.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Wang M., Xiao T., Yin B., He L., Meng W., et al. (2015). miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 64 2056–2068. 10.2337/db14-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H. H., Perkins T. G., Chia J. M., Gilsanz V. (2013). Characterization of human brown adipose tissue by chemical-shift water-fat MRI. AJR Am. J. Roentgenol. 200 177–183. 10.2214/AJR.12.8996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J., Seale P. (2015). Functions of Prdm16 in thermogenic fat cells. Temperature 2 65–72. 10.4161/23328940.2014.974444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic A., Korac A., Srdic-Galic B., Buzadzic B., Otasevic V., Stancic A., et al. (2014). Differences in the redox status of human visceral and subcutaneous adipose tissues-relationships to obesity and metabolic risk. Metabolism 63 661–671. 10.1016/j.metabol.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Jones D. P., Sies H. (2015). The redox code. Antioxid. Redox Signal. 23 734–746. 10.1089/ars.2015.6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L., Chen S., Alimujiang M., Bai N., Yan H., Fang Q., et al. (2018). A novel role for Bcl2l13 in promoting beige adipocyte biogenesis. Biochem. Biophys. Res. Commun. 506 485–491. 10.1016/j.bbrc.2018.10.034 [DOI] [PubMed] [Google Scholar]
- Kajimura S., Seale P., Kubota K., Lunsford E., Frangioni J. V., Gygi S. P., et al. (2009). Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 460 1154–1158. 10.1038/nature08262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S., Spiegelman B. M., Seale P. (2015). Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22 546–559. 10.1016/j.cmet.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y., Hinata T., Kang S. W., Watanabe Y. (2011). Reactive oxygen species mediate adipocyte differentiation in mesenchymal stem cells. Life Sci. 89 250–258. 10.1016/j.lfs.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Kang S., Bajnok L., Longo K. A., Petersen R. K., Hansen J. B., Kristiansen K., et al. (2005). Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha. Mol. Cell Biol. 25 1272–1282. 10.1128/MCB.25.4.1272-1282.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbiener M., Pisani D. F., Frontini A., Oberreiter L. M., Lang E., Vegiopoulos A., et al. (2014). MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 32 1578–1590. 10.1002/stem.1603 [DOI] [PubMed] [Google Scholar]
- Kida R., Noguchi T., Murakami M., Hashimoto O., Kawada T., Matsui T., et al. (2018). Supra-pharmacological concentration of capsaicin stimulates brown adipogenesis through induction of endoplasmic reticulum stress. Sci. Rep. 8:845. 10.1038/s41598-018-19223-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe S. M., Mouchiroud M., Caron A., Secco B., Freinkman E., Lamoureux G., et al. (2016). mTORC1 is required for brown adipose tissue recruitment and metabolic adaptation to cold. Sci. Rep. 6:37223. 10.1038/srep37223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. D., Tang Q. Q., Jiang M. S. (1999). Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 266 677–683. 10.1006/bbrc.1999.1885 [DOI] [PubMed] [Google Scholar]
- Lee H., Lee Y. J., Choi H., Ko E. H., Kim J. W. (2009). Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J. Biol. Chem. 284 10601–10609. 10.1074/jbc.M808742200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Petkova A. P., Konkar A. A., Granneman J. G. (2015). Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J. 29 286–299. 10.1096/fj.14-263038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Fan C. M. (2010). Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48 424–436. 10.1002/dvg.20630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gu H., Gan L., Xu Y., Feng F., Saeed M., et al. (2017). Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget 8 9267–9279. 10.18632/oncotarget.14035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor R. A., Kwon E. Y., Shin S. K., Jung U. J., Kim E., Park J. H., et al. (2013). Time-course microarrays reveal modulation of developmental, lipid metabolism and immune gene networks in intrascapular brown adipose tissue during the development of diet-induced obesity. Int. J. Obes. 37 1524–1531. 10.1038/ijo.2013.52 [DOI] [PubMed] [Google Scholar]
- Medrikova D., Sijmonsma T. P., Sowodniok K., Richards D. M., Delacher M., Sticht C., et al. (2015). Brown adipose tissue harbors a distinct sub-population of regulatory T cells. PLoS One 10:e0118534. 10.1371/journal.pone.0118534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda S., Gonzalez-Rodriguez A., Revuelta-Cervantes J., Rondinone C. M., Valverde A. M. (2010). Beneficial effects of PTP1B deficiency on brown adipocyte differentiation and protection against apoptosis induced by pro- and anti-inflammatory stimuli. Cell Signal. 22 645–659. 10.1016/j.cellsig.2009.11.019 [DOI] [PubMed] [Google Scholar]
- Mowers J., Uhm M., Reilly S. M., Simon J., Leto D., Chiang S. H., et al. (2013). Inflammation produces catecholamine resistance in obesity via activation of PDE3B by the protein kinases IKKepsilon and TBK1. eLife 2:e01119. 10.7554/eLife.01119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mracek T., Cannon B., Houstek J. (2004). IL-1 and LPS but not IL-6 inhibit differentiation and downregulate PPAR gamma in brown adipocytes. Cytokine 26 9–15. 10.1016/j.cyto.2003.12.001 [DOI] [PubMed] [Google Scholar]
- Nedergaard J., Wang Y., Cannon B. (2018). Cell proliferation and apoptosis inhibition: essential processes for recruitment of the full thermogenic capacity of brown adipose tissue. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864 51–58. 10.1016/j.bbalip.2018.06.013 [DOI] [PubMed] [Google Scholar]
- Nguyen K. D., Qiu Y., Cui X., Goh Y. P., Mwangi J., David T., et al. (2011). Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature 480 104–108. 10.1038/nature10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio M., Schmidt E., Mauer J., Baitzel C., Hansmeier N., Khani S., et al. (2016). Dicer1-miR-328-Bace1 signalling controls brown adipose tissue differentiation and function. Nat. Cell Biol. 18 328–336. 10.1038/ncb3316 [DOI] [PubMed] [Google Scholar]
- Ong F. J., Ahmed B. A., Oreskovich S. M., Blondin D. P., Haq T., Konyer N. B., et al. (2018). Recent advances in the detection of brown adipose tissue in adult humans: a review. Clin. Sci. 132 1039–1054. 10.1042/CS20170276 [DOI] [PubMed] [Google Scholar]
- Pan D., Mao C., Quattrochi B., Friedline R. H., Zhu L. J., Jung D. Y., et al. (2014). MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 5:4725. 10.1038/ncomms5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Kang H. J., Kang S. I., Lee J. E., Hur J., Ge K., et al. (2013). A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev. Cell 26 393–404. 10.1016/j.devcel.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannenberg C., Werner M. K., Ripkens S., Stef I., Deckert A., Schmadl M., et al. (2010). Impact of age on the relationships of brown adipose tissue with sex and adiposity in humans. Diabetes 59 1789–1793. 10.2337/db10-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. L., Fernandez-Hernando C. (2016). miRNA regulation of white and brown adipose tissue differentiation and function. Biochim. Biophys. Acta 1861(12 Pt B) 2104–2110. 10.1016/j.bbalip.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998). A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92 829–839. 10.1016/S0092-8674(00)81410-5 [DOI] [PubMed] [Google Scholar]
- Rajakumari S., Wu J., Ishibashi J., Lim H. W., Giang A. H., Won K. J., et al. (2013). EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17 562–574. 10.1016/j.cmet.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebiger L., Lenzen S., Mehmeti I. (2016). Susceptibility of brown adipocytes to pro-inflammatory cytokine toxicity and reactive oxygen species. Biosci. Rep. 36:e00306. 10.1042/BSR20150193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Brasier A. R., McGehee R. E., Jr., Habener J. F. (1992). Tumor necrosis factor-induced reversal of adipocytic phenotype of 3T3-L1 cells is preceded by a loss of nuclear CCAAT/enhancer binding protein (C/EBP). J. Clin. Invest. 89 223–233. 10.1172/JCI115566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E. D., Sarraf P., Troy A. E., Bradwin G., Moore K., Milstone D. S., et al. (1999). PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 4 611–617. 10.1016/S1097-2765(00)80211-7 [DOI] [PubMed] [Google Scholar]
- Rosenwald M., Wolfrum C. (2014). The origin and definition of brite versus white and classical brown adipocytes. Adipocyte 3 4–9. 10.4161/adip.26232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J., et al. (2009). High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58 1526–1531. 10.2337/db09-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Nitta T., Maruno K., Yeh Y. S., Kuwata H., Tomita K., et al. (2016). Macrophage infiltration into obese adipose tissues suppresses the induction of UCP1 level in mice. Am. J. Physiol. Endocrinol. Metab. 310 E676–E687. 10.1152/ajpendo.00028.2015 [DOI] [PubMed] [Google Scholar]
- Sampath S. C., Sampath S. C., Bredella M. A., Cypess A. M., Torriani M. (2016). Imaging of brown adipose tissue: state of the art. Radiology 280 4–19. 10.1148/radiol.2016150390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Guertin D. A. (2014). Adipocyte lineages: tracing back the origins of fat. Biochim. Biophys. Acta 1842 340–351. 10.1016/j.bbadis.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Infantes D., Cereijo R., Peyrou M., Piquer-Garcia I., Stephens J. M., Villarroya F. (2017). Oncostatin m impairs brown adipose tissue thermogenic function and the browning of subcutaneous white adipose tissue. Obesity 25 85–93. 10.1002/oby.21679 [DOI] [PubMed] [Google Scholar]
- Schreiber R., Diwoky C., Schoiswohl G., Feiler U., Wongsiriroj N., Abdellatif M., et al. (2017). Cold-induced thermogenesis depends on ATGL-mediated lipolysis in cardiac muscle, but not brown adipose tissue. Cell Metab. 26 753–763.e7. 10.1016/j.cmet.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T. J., Huang P., Huang T. L., Xue R., McDougall L. E., Townsend K. L., et al. (2013). Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature 495 379–383. 10.1038/nature11943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H., He Y., Chen H., Wang C., Zenno A., Shi H., et al. (2009). The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 9 556–564. 10.1016/j.cmet.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. N., Lim H. W., Rajakumari S., Sakers A. P., Ishibashi J., Harms M. J., et al. (2017). EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. 31 660–673. 10.1101/gad.294405.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B. K., Patil M., Satyanarayana A. (2014). Negative regulators of brown adipose tissue (BAT)-mediated thermogenesis. J. Cell Physiol. 229 1901–1907. 10.1002/jcp.24664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Ma Y., Chanturiya T., Cao Q., Wang Y., Kadegowda A. K. G., et al. (2017). Lipolysis in brown adipocytes is not essential for cold-induced thermogenesis in mice. Cell Metab. 26 764–777.e5. 10.1016/j.cmet.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson S. E., Herrero L., Naaz A. (2007). Obesity, inflammation, and insulin resistance. Gastroenterology 132 2169–2180. 10.1053/j.gastro.2007.03.059 [DOI] [PubMed] [Google Scholar]
- Stine R. R., Shapira S. N., Lim H. W., Ishibashi J., Harms M., Won K. J., et al. (2016). EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol. Metab. 5 57–65. 10.1016/j.molmet.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Trajkovski M. (2014). MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism 63 272–282. 10.1016/j.metabol.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Tapia P., Fernandez-Galilea M., Robledo F., Mardones P., Galgani J. E., Cortes V. A. (2018). Biology and pathological implications of brown adipose tissue: promises and caveats for the control of obesity and its associated complications. Biol. Rev. Camb. Philos. Soc. 93 1145–1164. 10.1111/brv.12389 [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Spiegelman B. M. (2008). Fat and beyond: the diverse biology of PPARgamma. Annu. Rev. Biochem. 77 289–312. 10.1146/annurev.biochem.77.061307.091829 [DOI] [PubMed] [Google Scholar]
- Tormos K. V., Anso E., Hamanaka R. B., Eisenbart J., Joseph J., Kalyanaraman B., et al. (2011). Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 14 537–544. 10.1016/j.cmet.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y. H., Kokkotou E., Schulz T. J., Huang T. L., Winnay J. N., Taniguchi C. M., et al. (2008). New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454 1000–1004. 10.1038/nature07221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares A., Alvarez A. M., Ventura J. J., Roncero C., Benito M., Porras A. (2000). p38 mitogen-activated protein kinase mediates tumor necrosis factor-alpha-induced apoptosis in rat fetal brown adipocytes. Endocrinology 141 4383–4395. 10.1210/endo.141.12.7843 [DOI] [PubMed] [Google Scholar]
- van den Berg S. M., van Dam A. D., Rensen P. C., de Winther M. P., Lutgens E. (2017). Immune modulation of brown(ing) adipose tissue in obesity. Endocr. Rev. 38 46–68. 10.1210/er.2016-1066 [DOI] [PubMed] [Google Scholar]
- van der Lans A. A., Hoeks J., Brans B., Vijgen G. H., Visser M. G., Vosselman M. J., et al. (2013). Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J. Clin. Invest. 123 3395–3403. 10.1172/JCI68993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt W. D., Vanhommerig J. W., Smulders N. M., Drossaerts J. M., Kemerink G. J., Bouvy N. D., et al. (2009). Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360 1500–1508. 10.1056/NEJMoa0808718 [DOI] [PubMed] [Google Scholar]
- Velickovic K., Lugo Leija H. A., Bloor I., Law J., Sacks H., Symonds M., et al. (2018). Low temperature exposure induces browning of bone marrow stem cell derived adipocytes in vitro. Sci. Rep. 8:4974. 10.1038/s41598-018-23267-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya F., Cereijo R., Gavalda-Navarro A., Villarroya J., Giralt M. (2018). Inflammation of brown/beige adipose tissues in obesity and metabolic disease. J. Intern. Med. 284 492–504. 10.1111/joim.12803 [DOI] [PubMed] [Google Scholar]
- Virtanen K. A., Lidell M. E., Orava J., Heglind M., Westergren R., Niemi T., et al. (2009). Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360 1518–1525. 10.1056/NEJMoa0808949 [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhang M., Ning G., Gu W., Su T., Xu M., et al. (2011). Brown adipose tissue in humans is activated by elevated plasma catecholamines levels and is inversely related to central obesity. PLoS One 6:e21006. 10.1371/journal.pone.0021006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. A., Scherer P. E. (2014). The adipochaser mouse: a model tracking adipogenesis in vivo. Adipocyte 3 146–150. 10.4161/adip.27656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Kissig M., Rajakumari S., Huang L., Lim H. W., Won K. J., et al. (2014). Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl. Acad. Sci. U.S.A. 111 14466–14471. 10.1073/pnas.1412685111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sato M., Guo Y., Bengtsson T., Nedergaard J. (2014). Protein kinase a-mediated cell proliferation in brown preadipocytes is independent of Erk1/2, PI3K and mTOR. Exp. Cell Res. 328 143–155. 10.1016/j.yexcr.2014.07.029 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zuo J., Zhang Y., Xie Y., Hu F., Chen L., et al. (2013). Identification of miR-106b-93 as a negative regulator of brown adipocyte differentiation. Biochem. Biophys. Res. Commun. 438 575–580. 10.1016/j.bbrc.2013.08.016 [DOI] [PubMed] [Google Scholar]
- Yasuniwa Y., Izumi H., Wang K. Y., Shimajiri S., Sasaguri Y., Kawai K., et al. (2010). Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS One 5:e15330. 10.1371/journal.pone.0015330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Wu J., Cohen P., Kazak L., Khandekar M. J., Jedrychowski M. P., et al. (2013). Fat cells directly sense temperature to activate thermogenesis. Proc. Natl. Acad. Sci. U.S.A. 110 12480–12485. 10.1073/pnas.1310261110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pasut A., Soleimani V. D., Bentzinger C. F., Antoun G., Thorn S., et al. (2013). MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 17 210–224. 10.1016/j.cmet.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y., et al. (2013). Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 123 3404–3408. 10.1172/JCI67803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T., Aita S., Matsushita M., Okamatsu-Ogura Y., Kameya T., Kawai Y., et al. (2011). Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity 19 1755–1760. 10.1038/oby.2011.125 [DOI] [PubMed] [Google Scholar]
- Yu J., Lv Y., Di W., Liu J., Kong X., Sheng Y., et al. (2018). MiR-27b-3p Regulation in Browning of Human Visceral Adipose Related to Central Obesity. Obesity 26 387–396. 10.1002/oby.22104 [DOI] [PubMed] [Google Scholar]
- Zhang H., Guan M., Townsend K. L., Huang T. L., An D., Yan X., et al. (2015). MicroRNA-455 regulates brown adipogenesis via a novel HIF1an-AMPK-PGC1alpha signaling network. EMBO Rep. 16 1378–1393. 10.15252/embr.201540837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. Y., Li L. (2014). MicroRNAs are key regulators of brown adipogenesis. Biochim. Biophys. Acta 1841 1590–1595. 10.1016/j.bbalip.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Zingaretti M. C., Crosta F., Vitali A., Guerrieri M., Frontini A., Cannon B., et al. (2009). The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 23 3113–3120. 10.1096/fj.09-133546 [DOI] [PubMed] [Google Scholar]
- Zoller V., Funcke J. B., Keuper M., Abd El Hay M., Debatin K. M., Wabitsch M., et al. (2016). TRAIL (TNF-related apoptosis-inducing ligand) inhibits human adipocyte differentiation via caspase-mediated downregulation of adipogenic transcription factors. Cell Death Dis. 7:e2412. 10.1038/cddis.2016.286 [DOI] [PMC free article] [PubMed] [Google Scholar]


