Fungi are ubiquitous in the biosphere with an estimate of around two to five million species (Blackwell 2011; Hawksworth and Lucking 2017). This diverse group of organisms occupies most, if not all, environmental niches and plays many roles vital to our daily life (see the 2018 State of the World’s Fungi report for interesting fungal facts – https://stateoftheworldsfungi.org). One indispensable role of fungi is their contribution to the ecosystem; they are crucial in decomposing complex carbon compounds, recycling nutrients, and facilitating nutrient exchanges between organisms (Mohan et al. 2014; Treseder and Lennon 2015). There is a long history of fungi used in food production (e.g., fermentation), and some of them can be taken directly as food for nutritional or medicinal purposes (Dupont et al. 2017). Fungi are also renowned for their drug potentials with the most famous example being the antibiotic penicillin that has saved countless lives since its discovery. Analysis of hundreds of fungal genome sequences indicates a large repertoire of novel secondary metabolites waiting to be discovered for therapeutic uses (Khaldi et al. 2010; de Vries et al. 2017). Moreover, certain fungi are also employed by the Biotech industry as “factories” for drugs, enzymes, and biofuel production (Baker 2013; Buijs et al. 2013; Money 2016; Cairns et al. 2018).
Fungi also have their dark side. Fungal pathogens pose increasing threats to plants and animals including humans (Fisher et al. 2012). Mild superficial fungal infections of humans are very common, but more importantly, life-threatening systemic fungal infections have risen over the past decade as a result of increasing application of immuno-suppressive therapy for various diseases. In fact, preventing and treating fungal diseases have become a major clinical challenge. Similarly, plant fungal pathogens have always been a big problem in agriculture; e.g., fungal infections of food crops not only reduce yields but can also cause mycotoxin contamination that may have adverse effects on humans such as the cancer-causing aflatoxin (Gruber-Dorninger et al. 2017).
Growth and metabolic flexibility and ability to quickly sense, respond, and adapt to constantly changing environments are essential for the success of fungi in colonizing almost any environments. For example, human fungal pathogens need to adapt to the host body temperature as well as to respond and overcome attacks from the immune system during infection (Braunsdorf et al. 2016; Köhler et al. 2017). Similarly, saprophytic fungi have to constantly compete with other microbes for limiting nutrients in harsh environments. Therefore, most fungal physiological processes and responses must be highly dynamic.
Transcription is the first essential step in retrieving genome information for all biological processes. It is expected that genes required (or detrimental) for growth and survival of fungi (in fact all organisms) under a specific condition must be tightly regulated in response to constantly changing environments in order to maintain cellular homeostasis. As such, knowing what and when genes are expressed under particular conditions of interest can reveal the physiological pathways necessary for fungi to survive and grow under that condition. Microarray and RNA-seq are two commonly used methods for determining genome-wide gene expression. These methods are based on calculating the steady-state mRNA level, which is a composite measure of both mRNA synthesis and mRNA degradation rates. While useful for estimating gene expression, they cannot reliably reflect the actual level of transcription activities and are not suitable for mapping dynamic transcription responses, especially over very short time scales. This is due to the fact that mRNA degradation rate can differ between genes and even among transcript isoforms of the same gene (Geisberg et al. 2014; Gupta et al. 2014) and that mRNA stability may also vary between different physiological conditions (Maekawa et al. 2015; Miller et al. 2018). These would then affect steady-state mRNAs levels. Therefore, the classical profiling methods like microarray and RNA-seq have apparent shortcomings in studying fungal transcriptional responses.
ChIP-seq can map genome-wide active and dynamic transcription responses
Chromatin immuno-precipitation (ChIP) measures in vivo association of proteins with chromatin. It involves crosslinking (usually by formaldehyde) of proteins to chromatin, followed by pulling down a protein-of-interest (e.g., transcription factor) along with any associated DNA using either a protein-specific antibody or an epitope-specific antibody if the protein-of-interest is tagged with an epitope. The level of protein occupancy on DNA can be measured by a number of downstream methods. For a small number of known or candidate targets, the pulled-down DNAs can be quantified by PCR or real-time PCR. Alternatively, microarray (ChIP) or next generation sequencing (seq) may be applied for mapping protein occupancy at the genome-wide level as well as for identifying unknown targets bound by the protein-of-interest. ChIP, ChIP-chip, and ChIP-seq have been extensively applied to studying various biological processes that happen on chromatin (e.g., transcription, chromatin remodeling, and DNA repair) and are arguably the most powerful techniques in the transcription and chromatin field.
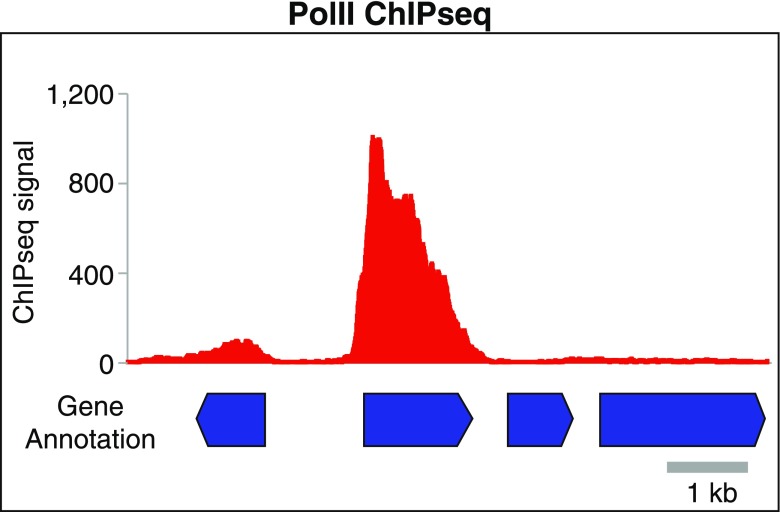
Using ChIP-chip or ChIP-seq against RNA polymerase II (PolII), actively transcribing genes, can be mapped and the transcription level of each gene can be measured simply by quantifying PolII occupancy (e.g., the amount of DNA pulled down by PolII) on the gene (Fig. 1). As with RNA-seq, PolII ChIP-seq/ChIP-chip readouts are also quantitative measurements of gene expression, although they are somewhat less accurate for genes that are subjected to transcript stability controls. Importantly, as formaldehyde crosslinking is instantaneous, transcription levels determined by PolII ChIP-seq present a snapshot of active transcription events occurring at the time of crosslinking. This feature has made the ChIP-based technique extremely powerful in capturing dynamic transcription responses in time-course experiments with timescales as short as a few minutes (Mason and Struhl 2005; Proft et al. 2006; Wong and Struhl 2011; Wong et al. 2014). Such high temporal resolution cannot be achieved by methods relying on steady-state mRNA levels.
Fig. 1.
Genome-browser view of PolII ChIP-seq data. PolII ChIP-seq can be used to profile genome-wide transcription activities by measuring PolII occupancies (e.g., ChIP-seq signals) over gene bodies. This powerful genome-wide transcription profiling method can capture active transcription events and global dynamic transcription changes in time-course experiments
Commercial antibodies specific to the highly conserved C-terminal domain of PolII are available and can be used for fungi
Availability, specificity, and affinity of antibodies to proteins-of-interest not only dictate whether ChIP/ChIP-seq assays can be applied but also govern whether a ChIP experiment would be successful and determine data quality. The C-terminal domain (CTD) of the largest subunit of the PolII complex is highly conserved among eukaryotes and is present in all eukaryotic PolII, differing mainly in the numbers of heptad repeats (e.g., YSPTSPS) between different species (Eick and Geyer 2013; Yang and Stiller 2014). For example, Saccharomyces cerevisiae and human PolII have 26 and 52 heptad repeats, respectively, with many of the repeats being identical although minor sequence variation is observed for some heptads. Because of this, antibodies specific for heptad repeats (e.g., 8WG16) or their post-translationally modified forms (e.g., 3E8 and 3E10) can be used to immuno-precipitate PolII from different eukaryotic species. Some of these antibodies are commercially available and have been extensively used in PolII ChIP and ChIP-seq experiments of various organisms from humans to worms, flies, as well as fungi (e.g., S. cerevisiae, Schizosaccharomyces pombe, Candida albicans, and Aspergillus nidulans).
PolII ChIP-seq is much more affordable than RNA-seq
The advent of next generation sequencing has opened up genome-wide profiling studies to any organisms, but experimental cost is still a major consideration. It is noteworthy that a number of unique features of fungal genomes have rendered genome-wide profiling experiments relatively less expensive for fungi as compared to higher eukaryotes. First, fungal genomes are very compact with around 6000–15,000 genes arranged within genomes of ~ 10–30 Mb in size on average. Second, the majority of genes are relatively short (e.g., around 1–2 kb), as compared to genes in higher eukaryotes. Third, unlike in humans, fungal introns are usually small with respect to exons, and certain species such as S. cerevisiae do not even have introns for most genes (Goffeau et al. 1996; Stajich et al. 2007; Hooks et al. 2014).
Although these genomic features equally apply to both RNA-seq and PolII ChIP-seq, PolII ChIP-seq has an additional and rather significant cost advantage in that PolII ChIP-seq only captures active transcription events (i.e., genes/DNA regions with elongating PolII), while RNA-seq measures total steady-state mRNAs that include stable mRNAs from previous transcription events. For example, most genes have some level of mRNA detected in RNA-seq experiments, while analysis of a significant number of fungal PolII ChIP-seq data reveals that in general, only about 10% of genes are actually engaged in the transcription process at a given time and condition. Therefore, far fewer sequencing reads are actually needed for a genome-wide transcription profile by PolII ChIP-seq as compared to RNA-seq. This provides a significant cost saving advantage for PolII Chipseq. Based on our experience, when using similar amounts of fungal cells or biomass as starting materials, around two to three millions raw reads from the Illumina platform are sufficient for high-quality PolII ChIP-seq profiles for fungi (e.g., S. cerevisiae, C. albicans, and A. nidulans), while standard RNA-seq datasets typically require ~ 20 to 30 million raw reads (assuming a mapping rate of about 80%). In other words, the sequencing cost for transcription profiles using PolII ChIP-seq is at least ten times cheaper than by RNA-seq.
In addition, experimental cost for a PolII ChIP-seq experiment (antibody, DNA detection and purification reagents, ChIP-seq library preparation kit, etc.) is only approximately half of that for a standard RNA-seq pipeline (RNA extraction, RNA quality assessment, mRNA purification or enrichment, RNA-seq library preparation, etc.). Currently, the overall cost for one PolII ChIP-seq profile generated using a multiplexing system (Wong et al. 2013) is around USD 50–60. At this cost, ChIP DNA detection through sequencing is actually much more cost-effective than by real-time PCR, because the amount is merely sufficient for quantifying a handful of genes using real-time PCR as opposed to genome-wide information by sequencing. This already extremely affordable cost is expected to further reduce as NGS technology improves and sequencing costs continues to drop. Therefore, global transcription profiling by PolII ChIP-seq is likely to become a routine method in fungal studies.
Concluding remarks
With the advent of NGS, genome-wide transcription profiling can now be readily performed for any organism. RNA-seq is a powerful method and often is the method-of-choice for global gene expression studies, but it has shortcomings in studying fungal physiologies and transcriptional responses that are rapid and highly dynamic. To this end, the ability to map active transcription changes over short temporal time scales and highly affordable experimental cost render PolII ChIP-seq a more attractive profiling method for fungi (and in fact, also for other organisms, especially those with a small genome size). It has been proven useful in understanding various key fungal functions (Leach et al. 2016; Xie et al. 2017; Veri et al. 2018), and we believe that PolII ChIP-seq is a powerful approach for studying this group of fascinating organisms.
Acknowledgements
We thank members of the Wong lab for sharing their experience with PolII ChIP-seq and Prof. Michael J. Hynes and Prof. Richard B. Todd for comments and suggestions on the manuscript.
Funding information
We acknowledge the Science and Technology Development Fund of Macau S.A.R (grant number: FDCT085/2014/A2), Research Services and Knowledge Transfer Office of the University of Macau (grant numbers: MYRG2015-00186-FHS and MYRG2016-00211-FHS), and the Faculty of Health Sciences for their funding supports.
Conflict of interest
Kaeling Tan declares that she has no conflict of interest. Koon Ho Wong declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Big Data’ edited by Joshua WK Ho and Eleni Giannoulatou.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Baker SE. Fungi and industrial biotechnology – a special issue for an amazing kingdom. Ind Biotechnol. 2013;9:105–107. doi: 10.1089/ind.2013.1576. [DOI] [Google Scholar]
- Blackwell M. The fungi: 1, 2, 3 ... 5.1 million species? Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- Braunsdorf C, et al. Fungal sensing of host environment. Cell Microbiol. 2016;18:1188–1200. doi: 10.1111/cmi.12610. [DOI] [PubMed] [Google Scholar]
- Buijs NA, et al. Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr Opin Chem Biol. 2013;17:480–488. doi: 10.1016/j.cbpa.2013.03.036. [DOI] [PubMed] [Google Scholar]
- Cairns TC, et al. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol and Biotechnol. 2018;5:13. doi: 10.1186/s40694-018-0054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18:28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J et al (2017) Fungi as a source of food. In: Heitman J, Howlett B, Crous P, Stukenbrock E, James T, Gow N (eds) The fungal kingdom. ASM Press, Washington, DC, p 1063–1085
- Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- Fisher MC, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, et al. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274(5287):546–567 [DOI] [PubMed]
- Gruber-Dorninger C, et al. Emerging mycotoxins: beyond traditionally determined food contaminants. J Agric Food Chem. 2017;65:7052–7070. doi: 10.1021/acs.jafc.6b03413. [DOI] [PubMed] [Google Scholar]
- Gupta I, et al. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol Syst Biol. 2014;10:719. doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Lucking R (2017) Fungal diversity revisited: 2.2 to 3.8 million species. In: Heitman J, Howlett B, Crous P, Stukenbrock E, James T, Gow N (eds) The fungal kingdom. ASM Press, Washington, DC, p 79–95
- Hooks KB, et al. Intron evolution in Saccharomycetaceae. Genome Biol Evol. 2014;6:2543–2556. doi: 10.1093/gbe/evu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler JR et al (2017) Fungi that infect humans. In: Heitman J, Howlett B, Crous P, Stukenbrock E, James T, Gow N (eds) The fungal kingdom. ASM Press, Washington, DC, p 813–843
- Leach MD, et al. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Commun. 2016;7:11704. doi: 10.1038/ncomms11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, et al. Analysis of RNA decay factor mediated RNA stability contributions on RNA abundance. BMC Genomics. 2015;16:154. doi: 10.1186/s12864-015-1358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Miller D, et al. Systematic identification of factors mediating accelerated mRNA degradation in response to changes in environmental nitrogen. PLoS Genet. 2018;14:e1007406. doi: 10.1371/journal.pgen.1007406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan JE, et al. Mycorrhizal fungi mediation of terrestrial ecosystem responses to global change: mini-review. Fungal Ecol. 2014;10:3–19. doi: 10.1016/j.funeco.2014.01.005. [DOI] [Google Scholar]
- Money NP, et al. Chapter 12 - fungi and biotechnology. In: Watkinson SC, et al., editors. The Fungi. 3. Boston: Academic Press; 2016. pp. 401–424. [Google Scholar]
- Proft M, et al. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Stajich JE, et al. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev. 2015;79:243. doi: 10.1128/MMBR.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veri AO, et al. Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet. 2018;14:e1007270. doi: 10.1371/journal.pgen.1007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Struhl K. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 2011;25:2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, et al. Multiplex Illumina sequencing using DNA barcoding. Curr Protoc Mol Biol. 2013;Chapter 7:Unit 7 11. doi: 10.1002/0471142727.mb0711s101. [DOI] [PubMed] [Google Scholar]
- Wong KH, et al. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol Cell. 2014;54:601–612. doi: 10.1016/j.molcel.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JL, et al. The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat Commun. 2017;8:499. doi: 10.1038/s41467-017-00547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Stiller JW. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc Natl Acad Sci U S A. 2014;111:5920–5925. doi: 10.1073/pnas.1323616111. [DOI] [PMC free article] [PubMed] [Google Scholar]