Abstract
Recent studies have shown that long non-coding RNAs (lncRNAs) play a pivotal role in the pathogenesis and progression of hepatocellular carcinoma (HCC). However, the biological action and potential mechanism of liver cancer cell drug resistance have not been clearly clarified. In this study, lncRNAs were screened and differentially expressed in parental and cisplatin-resistant cell lines (HepG2 and HepG2/CDDP). A novel lncRNA, termed NRAL (Nrf2 regulation-associated lncRNA), was identified, and the initial results indicated that it was highly expressed in HepG2 cisplatin resistant cell lines compared to their parental counterparts. Functionally, NRAL depletion significantly enhanced CDDP-mediated cytotoxicity and apoptosis in two cisplatin-resistant HCC cell lines. Mechanistically, the results indicated that NRAL regulates Nrf2 expression through miR-340-5p serving as a competing endogenous RNA (ceRNA), thus influencing the CDDP-induced phenotype in HCC. Collectively, the present investigation suggest that the NRAL/miR-340-5p/Nrf2 axis mediates cisplatin resistance in HCC, which may provide novel targets for overcoming cisplatin resistance in hepatocellular carcinoma cells.
Key words: lncRNA· miR-340-5p· Nrf2· Cisplatin·chemoresistance· Hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers due to its persistently increasing incidence and mortality rates (Cucchetti et al. 2017; Wang et al. 2017). Conventional therapies, including tumour resection and liver transplantation, are not available for HCC patients diagnosed at advanced stages; instead, these patients can turn only to chemotherapy to slow down the neoplastic transformation and progression. These patients also have an extremely poor prognosis, which may be attributed to the enhanced occurrence of acquired drug resistance(Terashima et al. 2016). Several mechanisms are associated with cancer-specific drug resistance, such as increased drug detoxification, gene function alterations, and DNA tolerance. Recent evidence has indicated that epigenetic alterations also play a key role in acquired clinical resistance following anticancer therapy(Takahashi et al. 2014). The detailed molecular mechanism underlying acquired drug resistance in HCC remains to be clarified.
Keap1(Kelch-like ECH-associated protein 1) -NFE2 L2 or Nrf2 (nuclear factor erythroid 2-related factor 2) signalling axis acts as “cellular defence machinery” to resist oxidative/electrophilic stimuli and chemical insults. Keap1 serves as a substrate adaptor protein for Nrf2 and the ubiquitin-linked ligand cullin-3 (Cul3); it also accelerates proteasomal Nrf2 degradation. However, the modification of specific thiols hampers Keap1-mediated proteasomal degradation; consequently, Nrf2 is dissociated from Keap1 and transferred to the nucleus, resulting in the subsequent transactivation of a wide array of downstream genes associated with the metabolism and detoxification of free radicals. Several groups have demonstrated that the constitutive activation of Nrf2 promotes tumour cell growth and survival, and this “dark” side of Nrf2 confers chemo- and/or radio-resistance during anti-cancer therapies.
More than 70 percent of the human genome is ncRNAs which can generally be divided into siRNAs, miRNAs, and lncRNAs according to their size. Research on siRNAs and microRNAs has been carried out extensively in the past 10 years(Vinogradova et al. 2013). lncRNAs are currently defined as a transcript longer than 200 nucleotides and do not contain obvious open reading frames. With the rapid development of bioinformatics and high-throughput sequencing technology, more and more literatures have demonstrated that lncRNAs can dysregulate gene expression and multiple biological processes in various types of human cancers. In addition, several lncRNAs, such as HEIH (high expression in liver cancer), lncRNA-ATB (invasion-metastasis and prognosis), lncRNA-UFC1 (proliferation and invasion), MVIH (microvascular invasion), and DANCR (differentiation antagonizing non-protein coding RNA), have been discovered to be aberrantly expressed and play functional roles in HCC (Zhao et al. 2014). There have been many reports of differential expression of lncRNAs in some tumors, but the effect of differential expression of lncRNAs on cisplatin resistance, however, is unclear in HCC cells.
Recently, a novel regulatory mechanism has been hypothesised in which crosstalk occurs between lncRNAs and mRNAs by competing specifically for shared miRNA response elements (MREs); these molecules act as competing endogenous RNAs (ceRNAs) to sponge miRNAs, thereby regulating each other’s expression levels and imposing an additional level of post-transcriptional regulation. Increasing experimental evidence has shown that ceRNAs play a vital role in the pathogenesis of common human cancers (Klahan et al. 2014). For example, H19 has been reported to be a sponge for let-7 that suppresses its target gene in modulating muscle differentiation; the H19/miR-148a-3p/DNMT1 axis is involved in the migration, proliferation and invasion of laryngeal squamous cell carcinoma (LSCC); a muscle-specific lncRNA, linc-MD1, acts as a ceRNA that suppresses MyoD messenger RNA (mRNA) degradation; HULC may contain miR-372 binding sites and down-regulate the activity of a series of miRNAs; and lncRNA-ATB competitively binds to miR-200 family members that up-regulate ZEB1 and ZEB2 to mediate epithelial-mesenchymal transition (EMT) and invasion in HCC(Yuan et al. 2014; Cui et al. 2015; Wu et al. 2016).
In this investigation, the phenotype of HepG2 resistant to cisplatin were established, microarray and q-PCR assays of lncRNAs were performed to analyse differential lncRNA expression between HepG2/CDDP and HepG2 cells. A novel lncRNA, NRAL, was obviously up-regulated in cisplatin-resistant HepG2 cells. Furthermore, NRAL was verified to serve as a ceRNA that sequesters miR-340-5p to manipulate its target gene, Nrf2; this action significantly affected cisplatin-mediated resistance in HCC. These findings may shed new light on targeting cisplatin resistance in HCC.
Materials and methods
Cell lines and cell culture
SMMC-7721 and HepG2 cell lines were obtained from the CCTCC (China Center for Type Culture Collection, Wuhan, China). HepG2 cells were cultured in DMEM(Dulbecco’s modified Eagle’s medium, Gibco Company, USA), and SMMC-7721 cells were maintained in RPMI 1640 (Gibco Company, USA). All culture media contained 10% foetal bovine serum (FBS, Sijiqing, China), and the cells were grown at 37°C and contain 5% carbon dioxide in moisture. Cisplatin was obtained from Sigma-aldrich Company (USA) and dissolved in double-distilled water for this research. The drug-resistant phenotypes HepG2/CDDP, SMMC-7721/CDDP, and Huh7/CDDP are developed by their parental cell exposure to progressively increasing doses of CDDP in cell culture medium. The drug-resistant phenotypes were maintained in no drug medium for two weeks before the experiments. Logarithmically growing cells were used in all experiments.
Microarray assay
Microarray analysis was performed by an array analysis platform (Agilent, USA) . Briefly, purified mRNAs were amplified and transcribed into cDNA, and the cDNA was labelled and hybridized onto a Human LncRNA Array v3.0 following the previous manufacturer’s guidence.
Human Specimens
Thirty HCC tissues were derived from the First Affiliated Hospital of Wenzhou Medical University from 2016 to 2017. All patients had not received preoperative radiotherapy or chemotherapy before tissue resection. HCC was diagnosed by three pathologists based on the WHO classification criteria. The tumor specimen was frozen in liquid nitrogen after excision and was immediately stored in the -80 °C refrigerator. This investigation was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University, and all patients signed the informed consent prior to tissue resection.
Plasmid construction
NRAL, which contained the target sequences of hsa-miR-340-5p, was amplified and cloned into the pcDNA3.1 plasmid (Invitrogen) between the BamHI and EcoRI restriction sites. The recombinant vector was named wild-type pcDNA3.1/NRAL. The NRAL segment with mutations in hsa-miR-340-5p, which contained the putative target site, was also inserted into the pcDNA3.1 vector at the same restriction sites and named pcDNA3.1/NRALmut. The annealed complementary shRNA oligonucleotides of NRAL ligated into the KpnI and XhoI sites of the pYr-LVX-shRNA vector (Invitrogen) to construct the silencing plasmids. The DNA segments that contained pri-miR-340-5p were inserted into pcDNA3.1 at the BamHI and EcoRI sites to construct the miR-340-5p-expressing plasmid. Oligonucleotides included the target sites of the NRAL and Nrf2 3’UTRs were cloned into pmirGLO plasmids (Promega, E5320) to generate the wild-type (WT) plasmid. To get mutant plasmids, The target area of the mutation of the NRAL and Nrf2 3’UTRs were synthesized by Worship Company (Shanghai, China) according to the methods described above. The empty vector of pmirGLO was used as a negative control. The above vectors were all validated by gene sequencing. Oligonucleotide of ASO-miR-340-5p and negative control (ASO-NC) were obtained from Sagene Technology Company (China).
qRT-PCR
The procedure of qRT-PCR has been stated previously. Total RNA from each of the cell lines was extracted with Trizol reagent (Thermo Fisher Scientific, 15596026) according to the protocol of manufacturer. To quantify the expression of miRNA, RNA was isolated using a mirVana™ miRNA Isolation Kit (Ambion, AM1560). cDNA was reverse transcribed from 1 mg of RNA using a TUREscript 1st Strand cDNA Synthesis Kit (KALANG, KL101-252). qPCR was performed using SYBR FASTqPCR Kit Master Mix (2X) (KAPA, KR0389). The primers for qRT-PCR are listed in Table 1. Either β-actin or U6 snRNA was used for mRNA and miRNA normalization. The relative expression of the transcripts were assessed by 2-△△Ct method.
Table 1.
Primers and oligonucleotides used in this work
| LncRNA | Forward and Reverse Primer | Product length |
|---|---|---|
| ENST00000412153 | CTGCAGACTTGCTCTTTGTACC ATGCTCCCATACTCCACTCC |
380 |
| XLOC_004474 | GCCTTGGTTAGCAATGCCTTAG GGACCTCGGTTTGGATCAGTT |
234 |
| AL590303.1 | CCGCCGTGGGTTAGGTTTA GGGGATGATAAGGCTTGGATG |
106 |
| LOC101928766 | CTGACAGCGTTGTGGGATT GGCAAATATGGGTAGTGGTG |
173 |
| LINC00442 | AGTGCTACAGACCCACAAACA CCTCCAAAGACATTGCTCCTAAA |
224 |
| ENST00000497718 | CCAGTGGACGGACATGCTTT CACAGAGTTTGTGAGGGAGT |
295 |
| bA305P22.4 | CGGAAGGGAGCAAGTCAA CCAGCCATGTTTCGCAGT |
384 |
| RP5-837M10.4 | CACGATGTTGTCACTGGGTG TGCTTGTAGGAGCTTGCTGT |
330 |
| AP001469.9 | ATACAATCACTTCCCACCAG TGACAACAAAGCAAGACCCT |
376 |
| AC144835.1 | TGGCTTAGTTAGACCAACCG CCAGCTTTCCCTCCAATCAC |
177 |
Dual-luciferase reporter assay
HepG/CDDP cells were co-transfected with miR-340-5p plasmids or empty pcDNA3, HepG2 cells were co-transfected with Anti-miR-340-5p or Anti-NC together with the pmirGLO-NRAL-WT plasmid or mutant plasmid using Lipofectamine 3000 (Invitrogen, L3000-015) in 48-well plates. The empty pmirGLO plasmid was used as a negative control. After 48 hrs of transfection, the relative luciferase activity was detected using the dual luciferase reporting and analysis system in a luminometer (Promega, E3710). Three independent experiments were carried out.
RIP assay
RNA immunoprecipitation(RIP) assays were performed using an EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore,17–701) following the manufacturer’s instructions. Briefly, cells were lysed completely in the buffer of RIP lysis and then incubated with RIP buffer containing magnetic beads labelled with an anti-AGO2 antibody (Abcam, ab 31251). Isotype-matching IgG served as a negative control. The immunoprecipitated RNA in the samples was isolated after incubation with proteinase K, and the levels of NRAL and miR-340-5p in the precipitates were measured by qRT-PCR.
Cell viability assay
A total of 5x103 cells were seeded into 96-well plates. After 24 hrs, cells were incubated with fresh medium containing or without different doses of CDDP for 48 hrs. Growth viability was measured by CCK-8 (Toyobo, Japan) assays following the manufacturer’s instructions as described previously. The IC50 value of CDDP was measured using GraphPad Prism 7.0 software. Three experiments were performed independently.
Apoptosis assay
Cells were harvested by trypsinization after 48 hrs of adherent growth and incubated with 0.5 mL binding buffer; next, the cells were subjected to double staining with annexin-V-FITC/propidium iodide for 20 min in the darkness and then measured with a FCM (flow cytometer, Beckman Coulter).
Western blot analysis
Cells were lysed in RIPA buffer. The same amount of protein were isolated by SDS-PAGE, transferred onto PVDF membranes and incubated with primary antibodies. The membranes were incubated with anti-Keap1 (1:2000 dilution, Abcam, UK), anti-Nrf2 (1:4000 dilution, Abcam, UK), and anti-actin (1:6000, Santa Cruz, CA) primary antibodies and then with a secondary horseradish peroxidase-labeled goat anti-mouse IgG or goat anti-rabbit IgG antibody for 1 hr. Antigen-antibody complexes were measured using a chemiluminescent kit (Pierce). The the normalized expression of the target protein were calculated with Quantity One software (Bio-Rad Life Science, Shanghai, China).
Statistical analysis
All statistical analyses were carried out using GraphPad Prism 7.0 software. The experiments were repeated independently in triplicate, the data are presented as the mean ± SD. Student’s t-test was performed for comparing between two groups. Comparisons between multiple groups with single controls were carried out using ANOVA.
Results
An up-regulated lncRNA, NRAL is differentially expressed in CDDP-resistant HCC cell lines
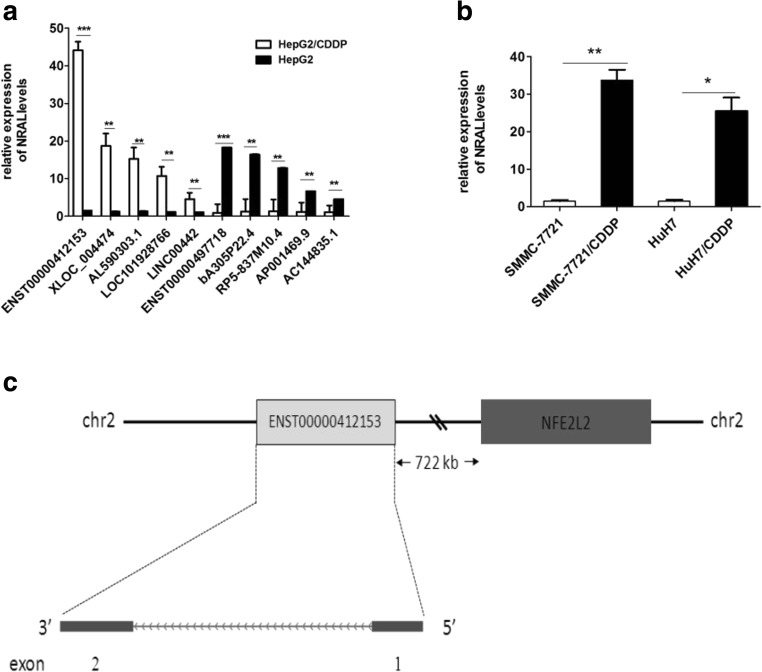
To validate the differential expression profiles of lncRNAs between HepG2 and HepG2/CDDP cells, lncRNA microarrays were performed to analyse the correlation of lncRNAs with CDDP resistance. A total of 1138 lncRNAs, including 853 up-regulated and 285 down-regulated lncRNAs (fold change ≥ 2.0, P < 0.05), were differentially expressed in HepG2/CDDP cells compared with those in HepG2 cells. To confirm the microarray data results, 5 significantly up- and down-regulated lncRNAs (Table 2) were selected and verified by qRT-PCR in HepG2 and HepG2/CDDP cells (Fig. 1a). The results of qRT-PCR were highly agree well with the microarray data. In a variety of differentially expressed lncRNAs, an obviously up-regulated lncRNA, lncRNA-ENST00000412153, was selected for further study; the expression of this lncRNA was 29.38-fold higher in HepG2/CDDP cells than in HepG2 cells. lncRNA-ENST00000412153 is an annotated 495 nt transcript, which is located on chromosome 2, 722 Kb from the NFE2L2 (Nrf2) gene (Fig. 1c); because it may regulate Nrf2 gene expression, the lncRNA was termed NRAL (Nrf2 regulation-associated lncRNA), and qRT-PCR was used to detect the levels of NRAL in two CDDP-resistant HCC cell line variants, including SMMC-7721/CDDP and HuH7/CDDP cells developed from their parental cells. NRAL was validated to be 21.6-fold and 16.4-fold up-regulated in SMMC-7721/CDDP and HuH7/CDDP cells compared with their parental counterparts, which suggested that the link between NRAL and CDDP resistance is not a single cell-specific effect (Fig. 1b).
Table 2.
Five significant up- and down-regulated lncRNAs in HepG2/CDDP cell-lines as compared to its parental counterpart
| Name | Chromosome | regulation | Fold change P value | |
|---|---|---|---|---|
| ENST00000412153 | 2 | Up | 29.38 | 0.006 |
| XLOC_004474 | 5 | Up | 14.37 | 0.013 |
| AL590303.1 | 6 | Up | 10.87 | 0.016 |
| LOC101928766 | 17 | Up | 8.91 | 0.020 |
| LINC00442 | 13 | Up | 4.11 | 0.040 |
| ENST00000497718 | 19 | Down | 20.32 | 0.009 |
| bA305P22.4 | 20 | Down | 12.67 | 0.019 |
| RP5-837M10.4 | 1 | Down | 9.17 | 0.026 |
| AP001469.9 | 21 | Down | 5.51 | 0.037 |
| AC144835.1 | 15 | Down | 4.11 | 0.012 |
Fig. 1.
Comprehensive differentially expressed lncRNAs in cisplatin-resistant HepG2/CDDP cells. (a) The relative levels of 10 significantly deregulated lncRNAs were validated in HepG2/CDDP and HepG2 cells using real-time PCR, **p < 0.01, ***p < 0.01. (b) qRT-PCR assays were used to detect NRAL expression levels in SMMC-7721/CDDP and HuH7/CDDP cells and their parental counterparts. β-Actin was used for normalization, *p < 0.05, **p < 0.01. (c) Genomic location of NRAL and the relationship with its target gene, NFE2L2(Nrf2)
NRAL enhances HCC cell phenotypes resistant to cisplatin
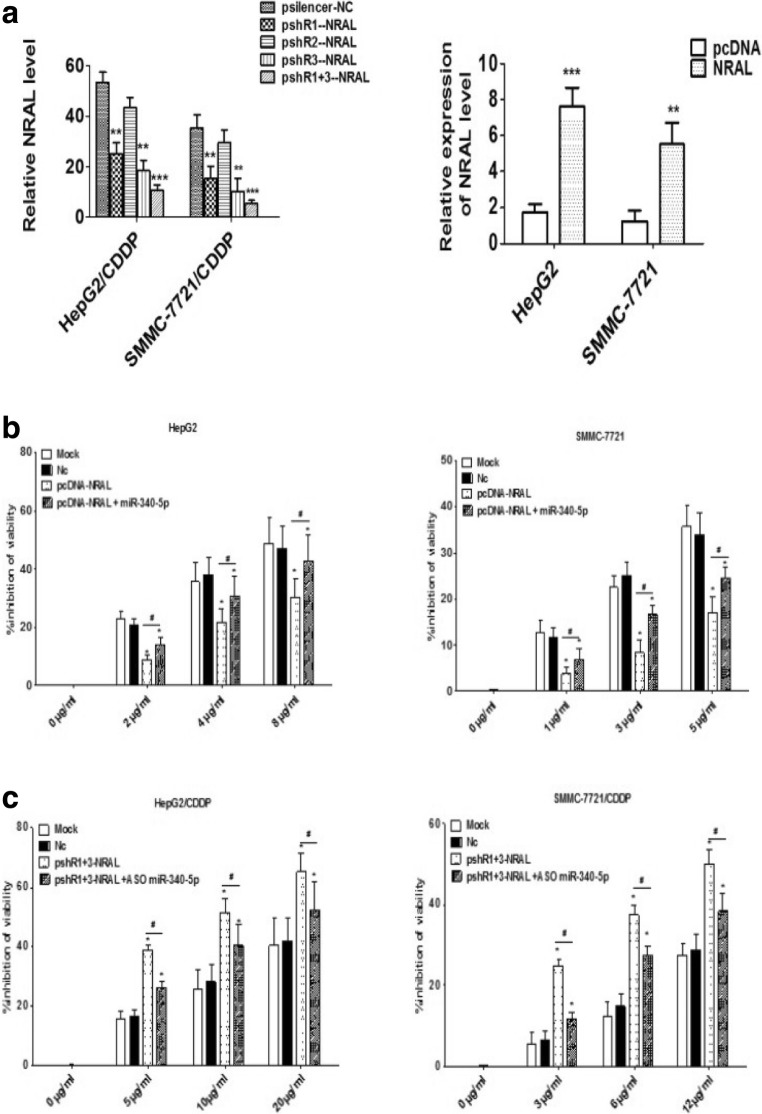
Considering that NRAL was significantly upregulated in cisplatin - resistant cell lines of HCC, the hypothesis is that NRAL confers HCC cell resistance to CDDP; first, three pSilencer 2.1-U6 neo silencing plasmids (pshR1-NRAL, pshR2-NRAL and pshR3-NRAL) and a pcDNA3.1/NRAL overexpressing plasmid (pcDNA-NRAL) were generated, and the efficiency of the plasmids was validated in two parental and resistant HCC cell lines by qRT-PCR. As shown in Fig. 2a, double knockdown (pshR1+3-NRAL) decreased NRAL expression in both HepG2/CDDP and SMMC-7721/CDDP cell lines, while NRAL was overexpressed in HepG2 and SMMC-7721 cell lines transfected with the pcDNA-NRAL plasmid. Next, the effects of NRAL expression on HCC cell phenotypes resistant to cisplatin were investigated. Interestingly, CCK-8 assays showed that the overexpression of NRAL obviously impaired the CDDP-induced cytotoxicity and strengthened CDDP resistance in both HepG2 and SMMC-7721 cells (Fig. 2b), whereas knocking down NRAL expression in the HepG2/CDDP and SMMC-7721/CDDP cell lines led to the opposite effects (Fig. 2c). Figure 2 data related to miR-340-5p is explained elsewhere.
Fig. 2.
Effects of NRAL on HCC phenotypes resistant to cisplatin. (a) qRT-PCR was performed to analyse the expression levels of NRAL in HCC cell lines after transfection with pshR-NRAL and pcDNA-NRAL plasmids. **p < 0.01, ***p < 0.001 vs the NC or pcDNA group. (b-c) Cell viability inhibition was assessed by CCK-8 assays in HCC cell lines transfected with pshR1+3-NRAL or pcDNA-NRAL plasmids; rescue experiments were also performed in pshR-NRAL plus ASO miR-340-5p or pcDNA-NRAL plus miR-340-5p plasmid groups. *p < 0.05 vs the NC group, #p < 0.05
NRAL contributes to reduced chemo-sensitivity to CDDP-induced apoptosis
It is known that CDDP could cause apoptosis in tumor cells by depletion of their antioxidant capacity; thus, we speculated that NRAL might inhibit CDDP-induced apoptosis to confer drug resistance. To verify this assumption, HepG2 and SMMC-7721 cells were transfected with pcDNA-NRAL or its negative controls and were treated with different doses of CDDP. The apoptosis rates significantly decreased in the pcDNA-NRAL-transfected cells than in the negative controls according to double annexin V-PI staining. Lower cleaved caspase 3 activity was also observed in NRAL-overexpressing cells than in the control cells after CDDP treatment for 48 h (Fig. 3a-b). Conversely, apoptotic cells and cleaved caspase 3 activity were significantly increased when NRAL was silenced in HepG2/CDDP and SMMC-7721/CDDP cells transfected with pshR1+3-NRAL compared to the negative controls. These data indicated that NRAL promotes CDDP resistance in HCC cells by blocking CDDP-induced apoptosis (Fig. 3a-b). Figure 3 data related to miR-340-5p is explained elsewhere.
Fig. 3.
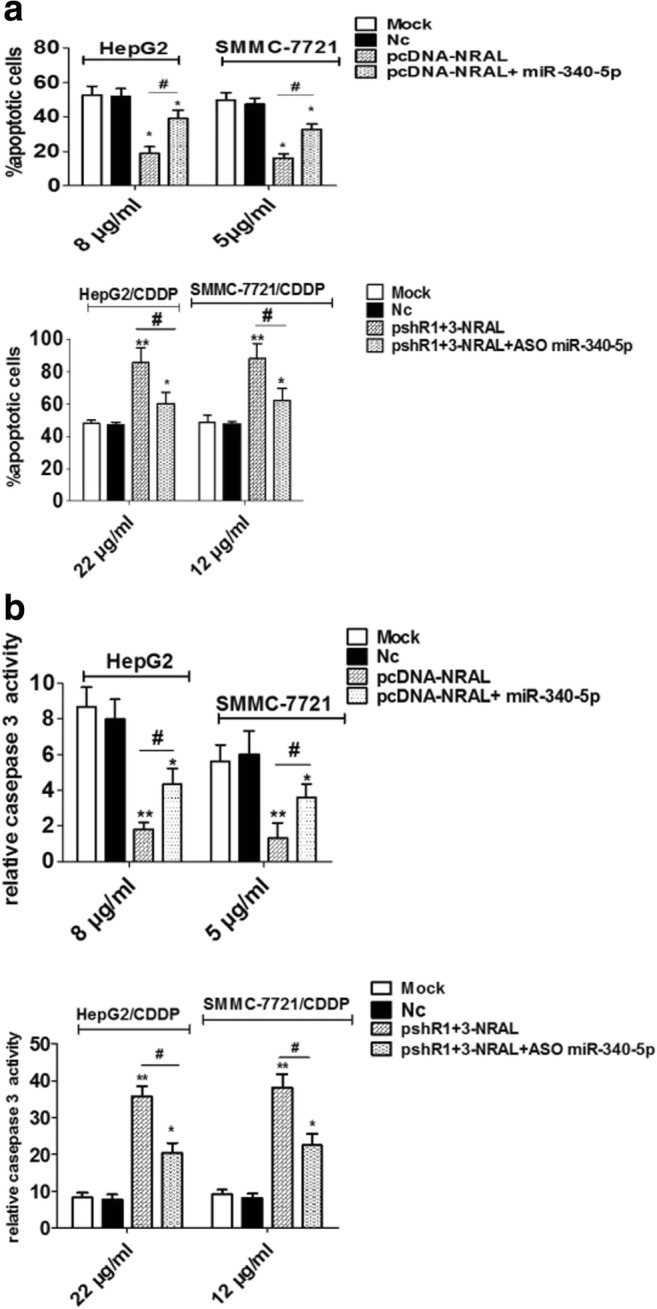
HepG2 and SMMC-7721 cells were transfected with the pcDNA-NRAL plasmid and its negative control, while HepG2/CDDP and SMMC-7721/CDDP cells were transfected with pshR1+3-NRAL and its negative control. pcDNA-NRAL plus miR-340-5p-expressing plasmids or pshR-NRAL plus ASO miR-340-5p were used for rescue experiments in both parental and resistant cells. (a) Transfected cells were seeded into a 96-well plate at a density of 5×103 cells per well in triplicate. After 8 hrs, the cells were treated with the indicated doses of CDDP for 48 hrs. Then, the cells were harvested for apoptosis analysis by double annexin-V/PI staining and flow cytometry. *p < 0.05, **p < 0.01 vs the NC group, #p < 0.05. (b) A caspase-Glo3/7 assay kit (Promega) was used for detecting cleaved caspase3 activity. *p < 0.05, **p < 0.01 vs the NC group, #p < 0.05
NRAL directly interacts with miR-340-5p
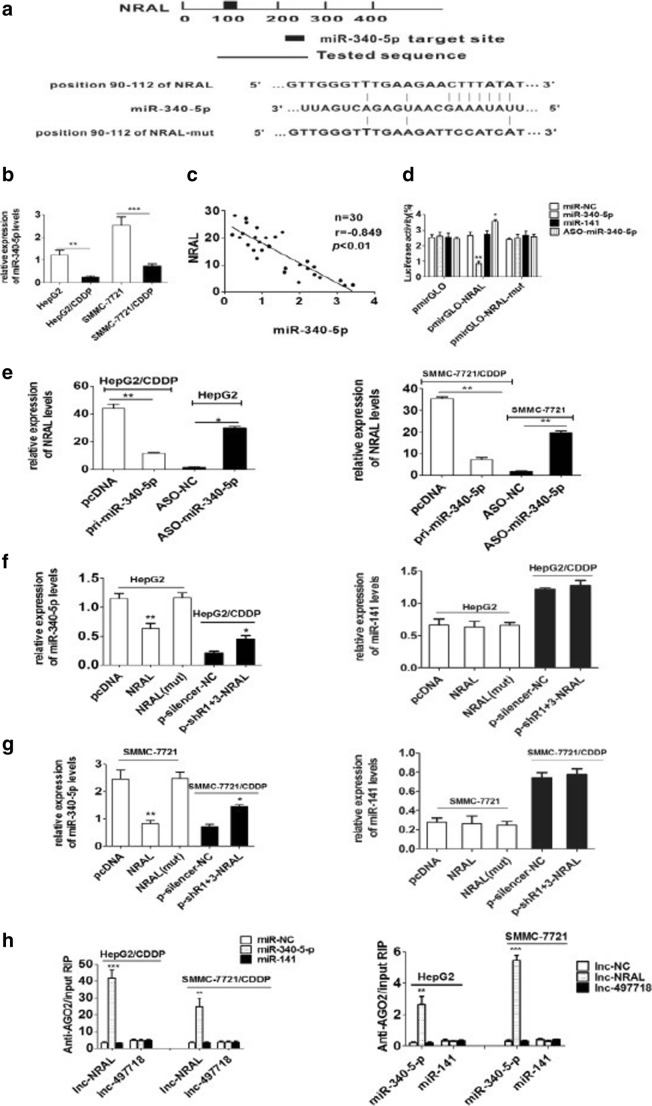
Recently, accumulating investigation have shown that lncRNAs might act as ceRNAs to indirectly modulate miRNAs, hence functionally regulating the expression of other targeted RNA transcripts. To explore whether NRAL can serve as a ceRNA in HCC cells resistant to cisplatin, a bio-informatics database (https://cm.jefferson.edu/rna22/Interactive/) was used to predict the putative miR-340-5p binding sites in NRAL (Fig. 4a). According to the known profiles and functions of miR-340-5p expression that have been reported previously(Shi et al. 2014), miR-340-5p was selected for further study.
Fig. 4.
NRAL is targeted by miR-340-5p (a) Schematic diagram of the miR-340-5p putative binding site in NRAL according to RNA22 software; the mutant seed sequence of miR-340-5p is shown in Fig. 4. (b) qRT-PCR was performed to validate the expression levels of miR-340-5p in HepG2/CDDP and SMMC-7721/CDDP cells and their parental counterparts. U6 was used as an internal control for normalizing the data, **p < 0.01, ***p < 0.001. (c) Correlations between the levels of miR-340-5p and NRAL in 30 HCC tissue samples according to Pearson’s correlation analysis (r =-0.849, P< 0.01). (d) Relative luciferase intensities of the luciferase constructs harbouring wild-type or mutant NRAL were measured in 293T cells 48 hrs after transfection with miR-340-5p or ASO-miR-340-5p, n=3, *p < 0.05, **p < 0.01 vs the miR-NC group. (e) Relative NRAL gene expression was assessed by qRT-PCR in HCC cell lines transfected with miR-340-5p or ASO-miR-340-5p, n=3, *p < 0.05, **p < 0.01. (f-g) Relative miR-340-5p gene expression was analysed by qRT-PCR assays in HCC cell lines with NRAL overexpression or suppression. (*P < 0.05; **P < 0.01 vs the NC group) (h) RIP experiments were performed to detect the interaction between NRAL and miR-340-5p. The relative RNA levels of NRAL and miR-340-5p in the immunoprecipitants were determined by qRT-PCR. NRAL and miR-340-5p data are presented as the fold enrichment in AGO2 cells relative to IgG immunoprecipitants. The columns are the mean of three independent experiments; the data are expressed as the mean±SD. **p < 0.01, ***p < 0.001 vs the other group
The level of miR-340-5p was first examined in HepG2 and SMMC-7721 cells and their respective cisplatin-resistant phenotypes. The present investigation revealed that the level of miR-340-5p was much lower in HepG2/CDDP and SMMC-7721/CDDP cells than in their respective parental phenotypes (Fig. 4b). Furthermore, the expression of mir-340-5p and NRAL was negatively correlated in 30 HCC tissue samples (Fig. 4c).
To substantiate the lncRNA-miRNA interaction experimentally, PmirGLO-NRAL and pmirGLO-NRAL-mut were co-transfected into 293T cells with miR-340-5p plasmids or ASO-miR-340-5p; it was shown that miR-340-5p reduced and ASO-miR-340-5p increased the relative luciferase activity in the pmirGLO-NRAL group. In contrast, no notable change was observed in the luciferase intensity after the co-transfection of the PmirGLO-NRAL-mut vector with the miR-340-5p plasmid or ASO-miR-340-5p into 293T cells (Fig. 4d).
To further validate these results, qRT-PCR was employed, It was found that overexpression of mir-340-5p resulted in a significantly decrease in NRAL expression in HepG2/CDDP and SMMC-7721/CDDP cells transfected with the miR-340-5p plasmid (Fig. 4e), while However, the effect is opposite in ASO-miR-340-5p transfected cells of HepG2 and SMMC-7721.
Since miR-340-5p can attenuate NRAL expression, it is speculated that NRAL can also reduce miR-340-5p expression in a similar way. miR-141, which contains no binding sites in NRAL, was selected as a negative control. Overexpression of NRAL significantly inhibited the miR-340-5p levels in HepG2 and SMMC-7721 cells, while the silenced NRAL notably elevated the miR-340-5p levels in HepG2/CDDP and SMMC-7721/CDDP cells. The expression level of mir-141 is not regulated by NRAL (Fig. 4f-g), suggesting that NRAL is specifically related to the expression level of miR-340-5p. However, no significant difference was observed in miR-340-5p levels following transfection with NRAL mut (Fig. 4f-g).
Consequently, the direct interaction between NRAL and miR-340-5p was determined by Argonaute-2 (AGO2) assays. Argonaute-2 (AGO2) is considered to be the core component of inducible RNA silencing complexes. Therefore, RNA binding protein immunoprecipitation (RIP) experiments were conducted with an anti-AGO2 antibody. Endogenous lncRNA-NRAL pull-down by AGO2 was enriched specifically in miR-340-5p, but not in miR-141- or NC-transfected HepG2/CDDP and SMMC-7721/CDDP cells (Fig. 4h). Similarly, miR-340-5p was preferentially enriched in lncRNA-NRAL WT but not mutant- or lncRNA-497718-transfected HepG2 and SMMC-7721 cells (Fig. 4h). Thus, NRAL is present in AGO2-containing RNA-induced silencing complexes, likely through interacting with miR-340-5p; this result is consistent with our analysis of bioinformatics and luciferase assays.
In summary, these findings indicate that mir-340-5p interacts directly with NRAL, and there is mutual inhibition between NRAL and mir-340-5p.
NRAL regulates the target gene of an endogenous miR-340-5p, Nrf2
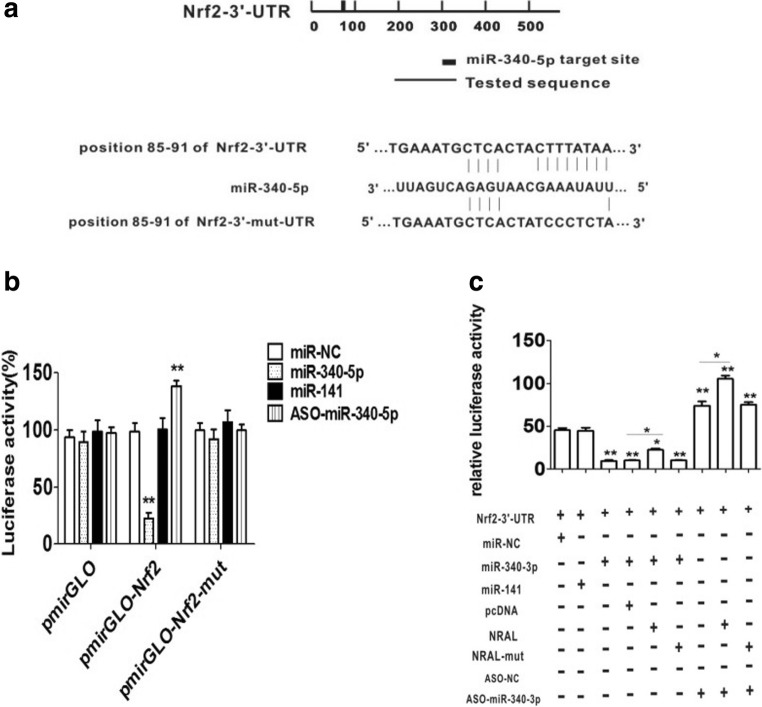
Among the the numerous target genes of miR-340-5p, Nrf2 is of interest because it plays a key role in cellular defense against oxidative and electrophilic insults. Previously, Our study suggests that mir-340-5p regulates cisplatin hepatocellular carcinoma resistance at least in part by inhibiting Nrf2(Shi et al. 2014). Next, miRNA databases (TargetScan, Pictar, and MicroRNA) were employed to predict a conserved putative sequence for miR-340-5p in the human Nrf2 3’UTR at position 85-91 (Fig. 5a). To validate whether miR-340-5p interacts with Nrf2 mRNA, H293T cells were cotransfected with the pmirGLO-Nrf2-3’UTR vector containing the putative target site for miR-340-5p and plasmids encoding miR-340-5p or ASO-miR-340-5p; miRNA-141 was used as a negative control. The results showed that compared with miR-NC, miR-340-5p decreased and ASO-miR-340-5p elevated luciferase activity. These effects disappear when the presumptive Nrf2 3'UTR binding site mutates (Fig. 5b). Collectively, the present findings revealed that Nrf2 is a direct target gene of miR-340-5p.
Fig. 5.
NRAL controls the target of miR-340-5p. (a) Schematic diagram of the miR-340-5p binding site in the Nrf2 3’UTR or its mutant sequences based on TargetScan software. (b) Relative luciferase intensities of the luciferase constructors harbouring the wild-type or mutant Nrf2 3’UTRs were measured in 293T cells 48 h after transfection with the pri-miR-340-5p plasmid or ASO-miR-340-5p, n=3, **p < 0.01 vs the other group. (c) pmirGLO-Nrf2-3’-UTR and pri-miR-340-5p plasmids were co-transfected into HEK293T cells with plasmids expressing NRAL (pcDNA/NRAL) or NRAL- mut (pcDNA/NRAL-mut) with a control vector to verify the ceRNA activity of NRAL. miRNA-141 was used as a negative control. The histogram indicates the luciferase values 48 h after transfection
In addition, the pmirGLO-Nrf2-3’UTR vector was then co-transfected with plasmids encoding miR-340-5p and NRAL (pcDNA/NRAL). Luciferase tests showed that in the presence of NRAL, luciferase activity repression was partly restored compared with the control group. Enhanced luciferase activity was also promoted in the presence of NRAL in the ASO-miR-340-5p-transfected group (Fig. 5c).
All the results indicate that NRAL binds to miR-340-5p via an endogenous ‘sponge’, thereby weakening the miRNA-induced repressive activity of the Nrf2 3’UTR.
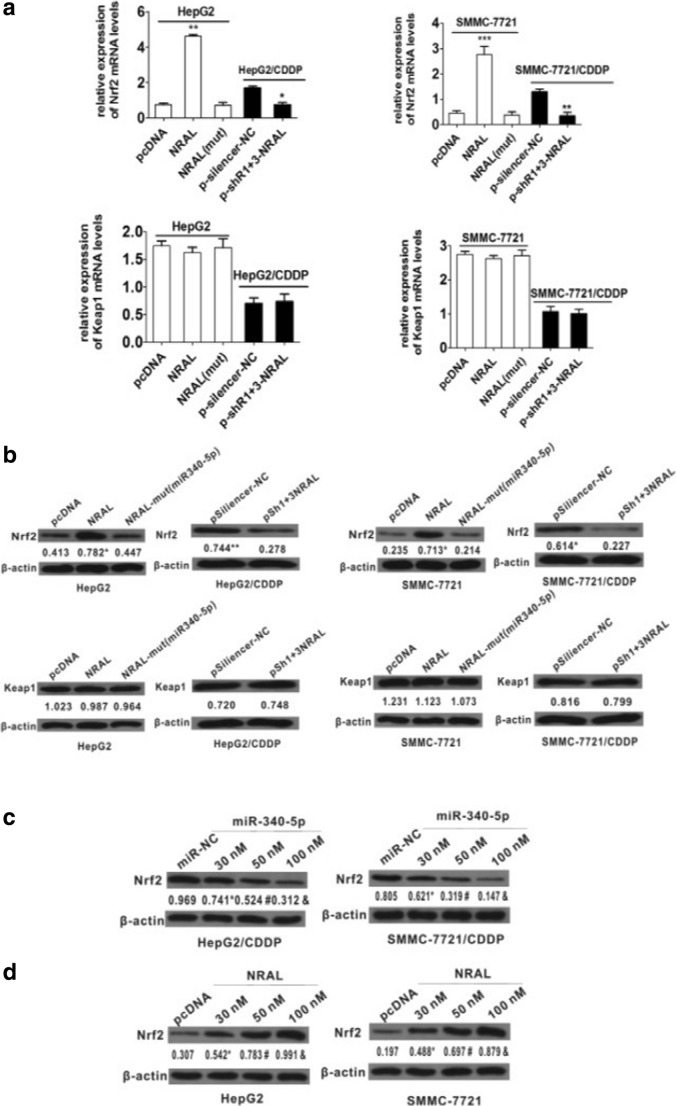
The qRT-PCR and western blot results also revealed that increased Nrf2(but not Keap1) mRNA and protein levels were observed in pcDNA3.1/NRAL (but not in mutant)-transfected HepG2 and SMMC-7721 cells; decreased Nrf2(but not Keap1) mRNA and protein levels were detected in pSilencer 2.1-U6 neo-NRAL (but not in mutant)-transfected HepG2/CDDP and SMMC-7721/CDDP cells (Fig. 6a-b).
Fig. 6.
(a-b) mRNA (A, *p < 0.05 vs the NC group, **p < 0.01, ***p < 0.001 vs the pcDNA group) and protein (b) levels of Nrf2 and Keap1 when the NRAL expression level was altered. β-actin was used as a reference gene, and the normalized values represent the means from at least 3 separate experiments, *p < 0.05 vs the pcDNA group, **p < 0.01 vs the NC group. (c-d) miR-340-5p-expressing plasmids cause a dose-dependent decrease in the protein expression of Nrf2. pcDNA/NRAL caused a dose-dependent increase in the protein expression of Nrf2. β-actin was used as a reference gene, and the normalized values represent the means from at least 3 separate experiments, *p < 0.05 vs the miR NC group,#p < 0.05 vs the 30 nm group, &p < 0.05 vs the 50 nm group
To further ascertain whether NRAL regulates Nrf2 expression by binding to miR-340-5p, qRT-PCR and western blot analyses were employed; in the presence of the same concentration of pcDNA3.1/NRAL, the protein level of Nrf2 was decreased dose-dependently in the miR-340-5p-expressing plasmid-transfected HepG2/CDDP and SMMC-7721/CDDP cells (Fig. 6c). At the same concentration of miR-340-5p-expressing plasmid, the protein level of Nrf2 was increased dose-dependently in pcDNA3.1/NRAL-transfected HepG2 and SMMC-7721 cells (Fig. 6d).
Collectively, these data uphold the conclusion that NRAL serves as a ceRNA for Nrf2 by directly interacting with miR-340-5p and abolishes the miRNA-induced repressing activity of the Nrf2-3’UTR.
Mir-340-5pmiR-340-5p is involved in NRAL regulation of HCC resistance to cisplatin
In the present study, rescue experiments were conducted to investigate whether miR-340-5p is required for HCC phenotypes resistant to cisplatin via NRAL.
The results shown in Fig. 2-3 indicate that compared with control cells, HepG2 and SMMC-7721 cells with NRAL overexpression showed much higher cisplatin resistance, with a prominent decrease in cytotoxicity and cellular apoptosis, while ectopically expressing miR-340-5p partially rescued this effect. In addition, compared with their respective control cells, NRAL-silenced HepG2/CDDP and SMMC-7721/CDDP cells exhibited a reversal in cisplatin resistance, with a significant increase in cytotoxicity and cellular apoptosis, while ectopically expressing ASO miR-340-5p partially abolished this NRAL knockdown-mediated promotion.
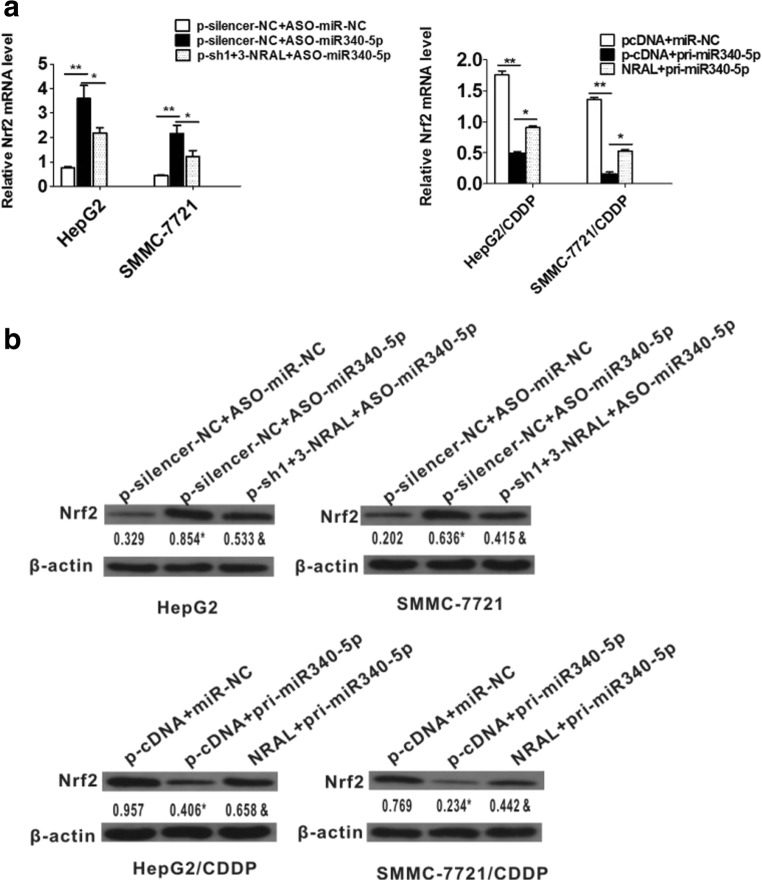
Subsequently, NRAL was knocked down in ASO-miR-340-5p-transfected HepG2 and SMMC-7721 cells. The depletion of miR-340-5p resulted in a significant increase in the mRNA and protein levels of Nrf2; this effect was partially attenuated by knocking down NRAL. In contrast, NRAL overexpression led to the opposite effect in miR-340-5p-expressing plasmids transfected HepG2/CDDP and SMMC-7721/CDDP cells (Fig. 7a and b).
Fig. 7.
(a, b) NRAL reverted the effects of miR-340-5p on HCC phenotypes resistant to cisplatin. mRNA (a, *p < 0.05, **p < 0.01) and protein (b) levels of Nrf2 in HepG and SMMC-7721 cells after co-transfection with ASO-miR-340-5p and pshR1+3-NRAL or in HepG2/CDDP and SMMC-7721/CDDP cells after co-transfection with pri-miR-340-5p and NRAL. β-actin was used as a reference gene, and the normalized values represent the means from at least 3 separate experiments, *p < 0.05 vs the NC group, &p < 0.05 vs the pcDNA group
In summary, the current findings strongly indicate that NRAL directly interacts with miR-340-5p to up-regulate the expression of its target, Nrf2, to mediate cisplatin-resistant HCC phenotypes.
Discussion
Emerging evidence has revealed important protagonists of lncRNAs in cellular functions, although recent studies have identified only a few functional lncRNAs in HCC(Zhang et al. 2016). However, the exact role of lncRNAs underlying the transformation of cancer cells from chemotherapy-sensitive to chemotherapy-resistant in HCC has not been well established.
Nuclear factor E2-related factor 2 (NFE2L2 or Nrf2) plays a pivotal role in the process of anti-oxidation and detoxification. Previous investigations have indicated that the continued activation of the Nrf2-related signalling pathway is associated with chemotherapy resistance in most types of cancer cells (Furfaro et al. 2016). However, possible modulation mechanism of Nrf2 expression during drug resistance development needs to be further elucidated
In the present report, lncRNA expression profiles were screened via lncRNA microarrays in HepG2 and HepG2/CDDP cells; a significantly up-regulated lncRNA, lncRNA-ENST00000412153, was selected for further study (Fig. 1a). The qRT-PCR analysis data validated that lncRNA-ENST00000412153 was significantly up-regulated in several HCC phenotypes resistant to cisplatin (Fig. 1b). lncRNA-ENST00000412153 is an annotated 495 nt transcript, which is located on chromosome 2, 722 Kb from the NFE2L2 (Nrf2) gene (Fig. 1c); because it may regulate Nrf2 gene expression, it has been called NRAL (Nrf2 regulation-associated lncRNA). Further studies applying methods of gain and loss of function will be necessary to identify the functions of NRAL in HCC cells.
Recently, it has been reported that deregulated lncRNA and miRNA expression is usually associated with the acquisition of resistance to various chemotherapeutic agents, including CDDP(Ayers and Vandesompele 2017). In the present study, CCK-8 and apoptotic assays indicated that NRAL overexpression significantly inhibited CDDP-induced cytotoxicity (decreased apoptotic cells) and enhanced CDDP resistance, while NRAL knockdown resulted in an opposite effect (Fig. 2-3). These results suggest that NRAL might act as a regulator in HCC chemoresistance.
lncRNAs have a variety of molecular regulatory mechanisms, including chromatin remodeling and RNA transcriptional or post-transcriptional splicing. As a method kind of post-transcriptional regulation, lncRNAs can act as ceRNAs to decoy/spongy adsorption of miRNAs, thus resulting in the de-repression of miRNA targets(Lu et al. 2016). Yu, et al, for example, demonstrated that HOTAIR down-regulates miR-29b and attenuates its control of DNMT3b, leading to DNMT3b restoration and PTEN methylation enhancement, which contribute to liver fibrosis. As a ceRNA, HOTAIR can effectively adsorb mir-331-3 in the form of post-transcriptional regulation, so as to eliminate its inhibiting effect on HER2 in gastric cancers(Lai et al. 2017). As a ceRNA, long non-coding RNA SNHG6-003 can promote the progression of hepatocellular carcinoma (Cao et al. 2017). To clarify whether NRAL acts as a miRNA sponge, bioinformatics analysis and luciferase assays were employed to analyse the predicted miRNA binding sites in the full-length NRAL transcript. As expected, miR-340-5p was discovered to have complementary base pairing with NRAL; for this reason, considerable attention has been focused on the NRAL regulation of miR-340-5p (Fig. 4a). PmirGLO-NRAL luciferase reporter and qRT-PCR assays demonstrate that miR-340-5p interacts directly with NRAL and that there is reciprocal repression between NRAL and miR-340-5p (Fig. 4d-g). Additionally, NRAL and miR-340-5p co-immunoprecipitation with anti-AGO2 demonstrated that NRAL is present in AGO2-containing RNA-induced silencing complexes, likely through interaction with miR-340-5p; this finding further validates NRAL’s miRNA sequestering activity (Fig. 4h). All of these results suggest that NRAL works as a ceRNA to sponge and suppress miR-340-5p.
To validate the miRNA-related functions of NRAL in HCC phenotypes resistant to cisplatin, miR-340-5p, which targets nuclear factor E2-related factor 2 (NFE2 L2 or Nrf2), was chosen as a model miRNA for further investigations. Our previous reports have suggested that low expression of mir-340-5p is closely related to the formation of cisplatin resistance in hepatocellular carcinoma, at least in part through regulation of Nrf2-dependent antioxidant pathways (Shi et al. 2014). In the present study, Nrf2 was confirmed to be a direct target gene of miR-340-5p (Fig. 5a-b). Due to the interaction of NRAL/miR-340-5p, we speculated that NRAL may also modulate Nrf2 expression in HCC phenotypes resistant to cisplatin, which would support the role of NRAL in the chemoresistance regulatory network. Consistent with NRAL’s sequestration of miR-340-5p, the present results indicated that NRAL depletion reduced the mRNA and protein levels of Nrf2 in HepG2/CDDP and SMMC-7721/CDDP cells, while its overexpression increased the expression levels of Nrf2 in HepG2 and SMMC-7721 cells (Fig. 6a-b). Additionally, with the same concentration of pcDNA3.1/NRAL, the up-regulation of miR-340-5p decreased the protein levels of Nrf2 dose-dependently in HepG2 and SMMC-7721 cells, while the protein levels of Nrf2 were increased dose-dependently in HepG2/CDDP and SMMC-7721/CDDP cells transfected with ASO-miR-340-5p (Fig. 6c-d). Moreover, CCK-8 and apoptotic assays revealed that miR-340-5p could largely rescue the effect of NRAL on HCC phenotypes resistant to cisplatin (Fig. 3). These data indicate that there is a specific regulatory mechanism between NRAL and Nrf2 mRNA that compete with miR-340-5p binding, which supports the hypothesis that miRNAs can be sequestered by ceRNA to de-repress their target RNAs.
In conclusion, a novel lncRNA, NRAL, has been identified, and it plays a critical role in HCC phenotypes resistant to cisplatin; NRAL competitively binds to miR-340-5p and then promotes the expression of its target gene Nrf2 to activate the Nrf2-dependent antioxidant pathway. This NRAL/miR-340-5p/Nrf2 axis may also be used as a therapeutic target to overcome cisplatin resistance in hepatocellular carcinoma.
Acknowledgments
This project was supported by the Natural Science Foundation of China (81501823 ), Zhejiang Provincial Natural Science Foundation of China (LY18H160049, LY17H200005 and LY16H160047), Zhejiang Provincial Medical and Health Research Project(2017KY459), Wenzhou municipal Science and Technology Bureau (Y20160077and Y20160071 ).
Abbreviations
- ANOVA
One-way analysis of variance
- HCC
Hepatocellular carcinoma
- lncRNA
Long noncoding RNA
- ceRNA
Competitive endogenous RNA
- Keap1
Kelch-like ECH-associated protein 1
- NFE2L2 or Nrf2
Nuclear factor erythroid 2-related factor2
- NRAL
Nrf2 regulation associated lncRNAs
- CCK-8
Cell counting kit
- SDS-PAGE
Sodium dodecyl sulphate polyacrylamide gel electrophoresis
- qRT-PCR
Real time quantitative reverse transcription PCR
- RIP
RNA immunoprecipitation
Contributor Information
Xiang-yang Lin, Email: linxy1968@126.com.
Liang Shi, Email: shiliang6666@126.com.
References
- Ayers D, Vandesompele J (2017). Influence of microRNAs and Long Non-Coding RNAs in Cancer Chemoresistance. Genes. 8. [DOI] [PMC free article] [PubMed]
- Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112–1122. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- Cucchetti A, Sposito C, Pinna AD, Citterio D, Cescon M, Bongini M, Ercolani G, Cotsoglou C, Maroni L, Mazzaferro V. Competing risk analysis on outcome after hepatic resection of hepatocellular carcinoma in cirrhotic patients. World J Gastroenterol. 2017;23:1469–1476. doi: 10.3748/wjg.v23.i8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- Furfaro AL, Traverso N, Domenicotti C, Piras S, Moretta L, Marinari UM, Pronzato MA, Nitti M. The Nrf2/HO-1 Axis in Cancer Cell Growth and Chemoresistance. Oxidative Med Cell Longev. 2016;2016:1958174. doi: 10.1155/2016/1958174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahan S, Wu MS, Hsi E, Huang CC, Hou MF, Chang WC. Computational analysis of mRNA expression profiles identifies the ITG family and PIK3R3 as crucial genes for regulating triple negative breast cancer cell migration. Biomed Res Int. 2014;2014:536591. doi: 10.1155/2014/536591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YJ, He S, Ma LM, Lin H, Ren BY, Ma J, Zhu XY, Zhuang SF. HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy. Mol Cell Biochem. 2017;432:179–187. doi: 10.1007/s11010-017-3008-y. [DOI] [PubMed] [Google Scholar]
- Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- Shi L, Chen ZG, Wu LL, Zheng JJ, Yang JR, Chen XF, Chen ZQ, Liu CL, Chi SY, Zheng JY, Huang HX, Lin XY, Zheng F. miR-340 Reverses Cisplatin Resistance of Hepatocellular Carcinoma Cell Lines by Targeting Nrf2-dependent Antioxidant Pathway. Asian Pac J Cancer P. 2014;15:10439–10444. doi: 10.7314/APJCP.2014.15.23.10439. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima T, Yamashita T, Arai K, Kawaguchi K, Kitamura K, Sakai Y, Mizukoshi E, Honda M, Kaneko S. Response to chemotherapy improves hepatic reserve for patients with hepatocellular carcinoma and Child-Pugh B cirrhosis. Cancer Sci. 2016;107:1263–1269. doi: 10.1111/cas.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova SV, Soldatov RA, Mironov AA. Genome-wide identification of functional noncoding RNAs. Mol Biol. 2013;47:689–696. doi: 10.1134/S002689331304016X. [DOI] [PubMed] [Google Scholar]
- Wang Y, Attar BM, Hinami K, Fuentes HE, Jaiswal P, Zhang H, Simons-Linares CS, Tafur AJ (2017). Characteristics and impacts of venous thromboembolism in patients with hepatocellular carcinoma. J Gastrointest Cancer. [DOI] [PubMed]
- Wu T, Qu L, He G, Tian L, Li L, Zhou H, Jin Q, Ren J, Wang Y, Wang J, Kan X, Liu M, Shen J, Guo M, Sun Y. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Matsuura K, Kleiner DE, Zamboni F, Alter HJ, Farci P. Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J Transl Med. 2016;14:328. doi: 10.1186/s12967-016-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Greene CM, Gray SG, Lawless MW. Long noncoding RNAs in liver cancer: what we know in 2014. Expert Opin Ther Targets. 2014;18:1207–1218. doi: 10.1517/14728222.2014.941285. [DOI] [PubMed] [Google Scholar]