Short abstract
Male infertility is a complex multifactorial disease affecting approximately 10% of couples who want to have children. Some cases of infertility can be explained by genetic factors. Septins are members of the GTPase superfamily, which are involved in diverse biological processes including morphogenesis, compartmentalization, cytokinesis, and apoptosis. The septin 12 gene, SEPT12, is expressed exclusively in post-meiotic male germ cells and is considered as a critical gene for spermatogenesis. In this study, we evaluated 200 patients with non-obstructive azoospermia and detected mutations of 25 spermatogenesis-associated genes by targeted exome sequencing. We report a missense SEPT12 variant, c.673C>A/p.Gln225Lys, in an infertile man with non-obstructive azoospermia. The variation was located inside the GTPase domain and had a SIFT score of 0.02 (<0.50) and was considered to be ‘probably damaging’ by PolyPhen. This case may provide clues to help establish the relationship between SEPT12 gene alterations and some cases of idiopathic male infertility. The role of this variant should thus be investigated further.
Keywords: SEPT12, single nucleotide polymorphism, spermatogenesis failure, male infertility, azoospermia, septin
Introduction
Male infertility is a complex multifactorial disease that affects approximately 10% of couples wanting to have children, with consequently substantial clinical and social impacts.1 However, partly due to the nature of the condition, the causes of male infertility remain largely unexplained.2 Approximately 10% to 15% of infertility cases are known to be due to genetic causes, including chromosomal abnormalities and single-gene mutations.3,4 A large proportion of infertile men do not receive a clear diagnosis. However, aberrations in many additional genes are likely to underlie a significant proportion of cases of male infertility/subfertility, given that spermatogenesis requires the coordinated action of thousands of genes,2 alterations in the structure or expression of any of which might result in male infertility.4–10 Genetic polymorphisms may also increase the susceptibility to some forms of male infertility.11,12
Few causal mutations have been associated with spermatogenesis defects in recent decades; however, the use of parallel sequencing technologies has allowed the identification of increasing numbers of genes essential for spermatogenesis and mutations involved in spermatogenesis failure.13
We aimed to monitor genetic variations in some genes associated with spermatogenic defects in infertile Chinese men with non-obstructive azoospermia. We also present a patient in whom next generation sequencing suggested the existence of a single nucleotide polymorphism (SNP) variation in the septin 12 gene (SEPT12).
Septins are a family of polymerizing GTP-binding proteins.1 SEPT12 encodes a testis-specific septin and is considered as a critical gene for spermatogenesis.14 Septins are members of the GTPase superfamily, which are involved in diverse biological processes including morphogenesis, compartmentalization, cytokinesis, and apoptosis.15 In association with other septins, septin 12 is an essential annulus component in mature mammalian sperm.16 High expression levels of SEPT12 mRNA have been observed exclusively in the testis of sexually mature male humans and mice, but not in men who were sterile as a result of the inability to produce mature spermatozoa.7 SEPT12 knock out in mouse embryonic stem cells resulted in chimeric mice with severe spermatogenic defects. SEPT12 has thus been suggested to play a crucial role in the process of spermatogenesis.14
This evidence suggests that SEPT12 may be a potential sterility gene in humans, and genetic mutations or polymorphisms of SEPT12 may participate in male infertility. In this study and case report, we aimed to provide clues to establish the existence of any relationship between SEPT12 gene alterations and idiopathic male infertility.
Methods and case report
This study was reviewed and approved by the Medical Ethics Committee of First Hospital of Jilin University (No. 2014-321) and performed according to the tenets of the Declaration of Helsinki. All the patients were Chinese and provided written informed consent. Written informed consent for specific patient information and images to be published was provided by the Medical Ethics Committee of First Hospital of Jilin University. We evaluated a cohort of 200 patients who consulted for sterility and displayed non-obstructive azoospermia. They underwent comprehensive examination, including a detailed medical history, physical examination, hormone profiles, chromosome analysis, and molecular testing for Y-chromosome microdeletions. Patients without Y-chromosome microdeletions were included in the study and were subjected to analysis of spermatogenesis-associated genes.
Mutation screening of spermatogenesis-associated genes was performed by targeted exome sequencing. Genomic DNA was isolated from blood lymphocytes and subjected to exome capture using an in-house targeted gene panel (Peking Medriv Academy of Genetics and Reproduction, Peking, China), followed by next generation sequencing on the Illumina MiSeq sequencing platform (HiSeq2000, Illumina, San Diego, CA, USA), which includes 25 spermatogenesis-associated genes. Capture probes were also prepared based on the 25 spermatogenesis-associated genes. Sequence reads were mapped to the human reference genome assembly (NCBI build 37/hg19) using the Burrow-Wheeler Aligner (BWA version 0.7.12). Duplicated reads from the library and polymerase chain reaction (PCR) preparation were removed using Picard tools. After alignment to the human reference genome (GRCh37/hg19), the most likely non-deleterious variants were filtered as described previously.17 Variants with minor allele frequencies >1% in the dbSNP, 1000 Genomes Project, Exome Aggregation Consortium, and Exome Variant Server databases were excluded because they were unlikely to be deleterious. Intronic variants and synonymous variants located within intronic regions, regulatory regions, and non-regulatory intergenic regions were also excluded. Nonsynonymous variants and splice site variants were retained.
Possible pathogenic effects of the candidate variants were evaluated using PolyPhen2 (https://genetics.bwh.harvard.edu/pph2/, version 2.2), with probabilistic scores set at <0.15, 0.15 to 0.85, and >0.85 for benign, possibly damaging, and probably damaging, respectively,18 and SIFT (https://sift.jcvi.org, version 1.03) with a cut-off score of ≤0.05 for deleterious variants.19
A missense variant was confirmed by conventional PCR and Sanger sequencing (ABI 3730XL, Applied Biosystems, Carlsbad, CA, USA) carried out by BGI Genomics, Beijing Genomics Institute, Shenzhen, China. The primers for the detected missense variant c.673C>A/p.Gln225Lys in SEPT12 (GenBank accession number NM_144605.4) were 5′-CCTT GAGCTTGTGGATGAGTAGA-3′ (forward) and 5′-GGAAATGGTGTCAGGT GGTGC-3′ (reverse).
Case report
A 29-year-old Chinese man was shown to have the SEPT12 c.673C>A/p.Gln225Lys variant. He and his partner had been attempting to conceive for 1 year. Azoospermia was confirmed. The patient reported no genital traumas and there was no sign of varicocele. His testicular volumes were 8 mL left and 10 mL right. He had a high serum concentration of follicle-stimulating hormone (23.88 IU/L) and slightly elevated luteinizing hormone (10.9 IU/L), and a normal level of free testosterone (10.79 pmol/L). Chromosome analysis showed a normal 46,XY karyotype and no Y-chromosome microdeletions. The definitive histological results showed Sertoli cell-only syndrome (SCOS).
Results
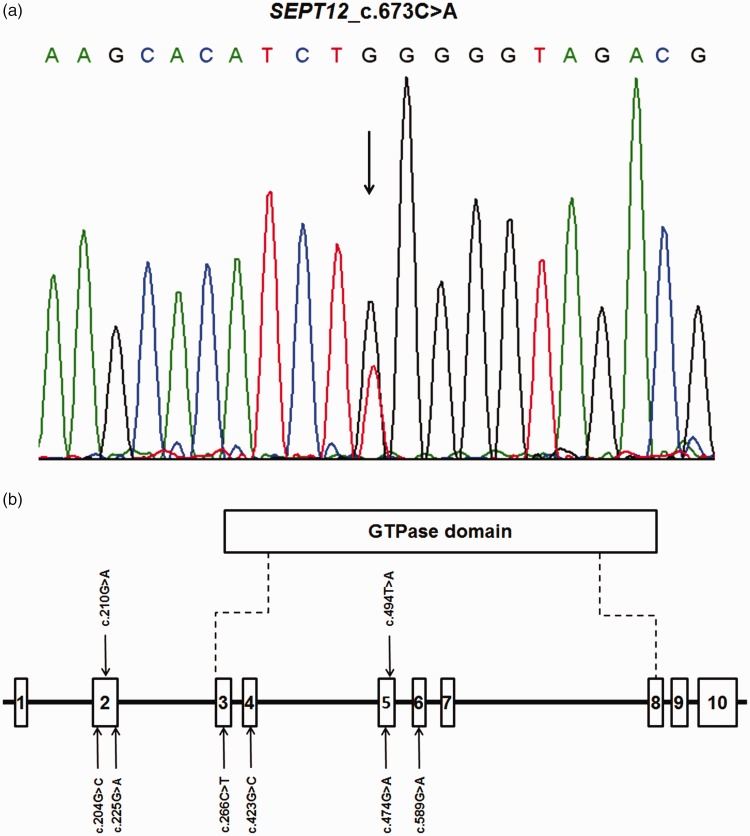
Targeted exome sequencing for mutations in 25 spermatogenesis-associated genes in 200 individuals (average age 30.25±5.24 years, range 19 to 47 years) with non-obstructive azoospermia detected a SEPT12 variant in an infertile man. The variant was heterozygous, and involved a single nucleotide change located within exon 7. The SNP was not novel (rs771771089), according to the Single Nucleotide Polymorphism database (dbSNP), but it had not previously been reported in association with spermatogenesis failure. This SNP was nonsynonymous (c.673C>A/p.Gln225Lys) and had a SIFT score of 0.02 (<0.50), and was considered to be ‘probably damaging’ by PolyPhen. PCR and Sanger sequencing confirmed the heterozygous missense variant in the patient (Figure 1a).
Figure 1.
(a) SEPT12 c.673C>A/p.Gln225Lys heterozygous variant in an infertile patient was confirmed by Sanger sequencing. Arrow indicates location of the variation. (b) Previously reported genetic variants of SEPT12 significantly associated with male infertility.15,20–22 Open bars indicate exons. Exon 3 to exon 8 encodes the GTPase domain of SEPT12.
Discussion
Previous studies have indicated that spermatogenesis failure in patients with azoospermia may result from chromosomal abnormalities, Y-chromosome microdeletions, or single gene variants.20,21 However, the underlying genes and molecular mechanisms remain poorly known.
The septins are a family of polymerizing GTP-binding proteins, originally discovered in a group of cell cycle mutants of budding yeast with defects in cytokinesis.22 The functions of septins include cytoskeletal remodeling, mitosis, vesicle trafficking, and membrane compartmentalization. Septins have also been implicated in the pathogenesis of many diseases, including hereditary neuralgic amyotrophy, leukemia, breast cancer, ovarian tumors, Alzheimer’s disease, and male infertility.23
The roles of septins in spermatogenesis have begun to be revealed. Among the 14 septin family members, septins 4, 7, and 12 are major constituents of the annulus,24 a submembrane ring structure demarcating the principle and mid-piece of the sperm tail.15 Septin 4 plays important roles in maintaining the proper mitochondrial architecture and establishing the annulus during spermatogenesis.23 Male Sept4 knockout mice were sterile due to immotile sperm with a defective annulus, and the immotile sperm showed dis-localization of septins 1, 6, and 7 from the annulus.25 Annulus (septins 1/4/6/7) complexes tend to be absent in mature spermatozoa from men with asthenozoospermia.22 Investigation of expression patterns suggested that septin 7 may be involved in the regulation of subcellular-compartment formation during spermiogenesis, and that the absence of a septin 7 signal correlated with multiple sperm defects.26
In this study, a SEPT12 genetic variant (c.673C>A/p.Gln225Lys) was detected in an infertile man with non-obstructive azoospermia. This point mutation was detected in exon 7 and located in the GTPase domain, resulting in the replacement of glutamine with lysine. The mutant protein (septin 12Q225K) may lose its GTP-hydrolysis ability, thus adversely affecting septin filament formation compared with wild-type septin 12.15 The GTPase domain is critical for the polymerization of septin subunits into filaments, which are required for formation of the sperm annulus/septin ring, which is in turn involved in the normal structure and function of sperm and in male fertility.27
Some SEPT12 gene variants have been associated with male infertility (Figure 1b). Several recent studies showed that some SEPT12 SNPs may predispose men to azoospermia.28,29 Miyamoto et al. carried out mutational analysis of SEPT12 in 30 men with azoospermia due to meiotic arrest and 140 fertile male controls, and found that the frequencies of the c.204G>C allele and CC genotype were significantly higher in the cases compared with the controls. This SEPT12 variant previously demonstrated a relationship with increased susceptibility to azoospermia caused by meiotic arrest.28 Miyakawa et al. investigated 100 Japanese patients with azoospermia caused by SCOS by direct sequencing of the coding regions of the SEPT12 gene. They detected eight coding SNPs, some of which had notably higher genotype and allele frequencies in SCOS patients compared with the control group.29 These findings suggested that SEPT12 and its mutations might be associated with human spermatogenesis. In the current study, we reported a SEPT12 variant, c.673C>A/p.Gln225Lys, in a man with non-obstructive azoospermia caused by SCOS. Overall, these results suggest that the c.673C>A/p.Gln225Lys variant was likely to be related to spermatogenesis failure in this patient.
Other studies have also reported an association between SEPT12 mutations and oligoasthenoteratozoospermia.15,30 Lin et al. found that most individuals homozygous for the SEPT12 c.474G>A allele had teratozoospermia (14%), and their sperm showed bent tails and de-condensed nuclei, with significant DNA damage.30 Kuo et al. reported two missense mutations in SEPT12 in infertile men, c.266C>T/p.Thr89Met and c.589G>A/p.Asp197Asn, both of which were located inside the GTPase domain with potential effects on protein structure.15 Patients carrying the two variants presented with oligoasthenozoospermia and asthenoteratozoospermia, respectively. These results suggested that SEPT12 loss-of-function mutations can disrupt the structural integrity of sperm by perturbing septin filament formation.15 The variant c.673C>A/p.Gln225Lys detected in the current study was a missense mutation, also located inside the GTPase domain and therefore also with the potential to alter the protein structure. We therefore hypothesized that it may be associated with spermatogenesis failure. However, further functional assays are needed to verify this hypothesis and to provide new insights into the cause of spermatogenesis failure in humans.
In conclusion, the c.673C>A/p.Gln225Lys genetic variant detected in the SEPT12 gene in a patient with non-obstructive azoospermia suggests that this SNP may play a causative role in spermatogenesis failure. This SNP is not novel, but it has not previously been reported in association with spermatogenesis failure. Although this study was not conclusive, it provides the basis for further studies aimed at clarifying the relationship between SEPT12 gene alterations and idiopathic male infertility, with the potential for this variant to be considered as a biomarker for idiopathic male infertility in the future.
Acknowledgements
We are sincerely grateful to all the staff at the Center for Reproductive Medicine, Center for Prenatal Diagnosis, the First Hospital of Jilin University, Changchun, China, for their hard work and support.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81471515).
References
- 1.Shahhoseini M, Azad M, Sabbaghian M, et al. New single nucleotide polymorphism G5508A in the SEPT12 gene may be associated with idiopathic male infertility in Iranian men. Iran J Reprod Med 2015; 13: 503–506. [PMC free article] [PubMed] [Google Scholar]
- 2.Hotaling JM. Genetics of male infertility. Urol Clin North Am 2014; 41: 1–17. [DOI] [PubMed] [Google Scholar]
- 3.Pizzol D, Ferlin A, Garolla A, et al. Genetic and molecular diagnostics of male infertility in the clinical practice. Front Biosci 2014; 19: 291–303. [DOI] [PubMed] [Google Scholar]
- 4.Miyamoto T, Minase G, Shin T, et al. Human male infertility and its genetic causes. Reprod Med Biol 2017; 16: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gava MM, Chagas Ede O, Bianco B, et al. Methylenetetrahydrofolate reductase polymorphisms are related to male infertility in Brazilian men. Genet Test Mol Biomarkers 2011; 15: 153–157. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Pan J, Liu Q, et al. Glutathione S-transferases gene polymorphisms and risk of male idiopathic infertility: a systematic review and meta-analysis. Mol Biol Rep 2013; 40: 2431–2438. [DOI] [PubMed] [Google Scholar]
- 7.Lin YH, Lin YM, Teng YN, et al. Identification of ten novel genes involved in human spermatogenesis by microarray analysis of testicular tissue. Fertil Steril 2006; 86: 1650–1658. [DOI] [PubMed] [Google Scholar]
- 8.Schlecht U, Demougin P, Koch R, et al. Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol Biol Cell 2004; 15: 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A 2003; 100: 12201–12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Zhou Z, Xu M, et al. A spermatogenesis-related gene expression profile in human spermatozoa and its potential clinical applications. J Mol Med (Berl) 2004; 82: 317–324. [DOI] [PubMed] [Google Scholar]
- 11.Ma J, Lu HY, Xia YK, et al. BCL2 Ala43Thr is a functional variant associated with protection against azoospermia in a Han-Chinese population. Biol Reprod 2010; 83: 656–662. [DOI] [PubMed] [Google Scholar]
- 12.Safarinejad MR, Shafiei N, Safarinejad S. The role of endothelial nitric oxide synthase (eNOS) T-786C, G894T, and 4a/b gene polymorphisms in the risk of idiopathic male infertility. Mol Reprod Dev 2010; 77: 720–727. [DOI] [PubMed] [Google Scholar]
- 13.Stouffs K, Seneca S, Lissens W. Genetic causes of male infertility. Ann Endocrinol (Paris) 2014; 75: 109–111. [DOI] [PubMed] [Google Scholar]
- 14.Lin YH, Lin YM, Wang YY, et al. The expression level of septin12 is critical for spermiogenesis. Am J Pathol 2009; 174: 1857–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo YC, Lin YH, Chen HI, et al. SEPT12 mutations cause male infertility with defective sperm annulus. Hum Mutat 2012; 33: 710–719. [DOI] [PubMed] [Google Scholar]
- 16.Steels JD, Estey MP, Froese CD, et al. Sept12 is a component of the mammalian sperm tail annulus. Cell Motil Cytoskeleton 2007; 64: 794–807. [DOI] [PubMed] [Google Scholar]
- 17.Kim YM, Lee YJ, Park JH, et al. High diagnostic yield of clinically unidentifiable syndromic growth disorders by targeted exome sequencing. Clin Genet 2017; 92: 594–605. [DOI] [PubMed] [Google Scholar]
- 18.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013; Chapter 7: Unit7 20. [DOI] [PMC free article] [PubMed]
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 20.Tuttelmann F, Rajpert-De Meyts E, Nieschlag E, et al. Gene polymorphisms and male infertility–a meta-analysis and literature review. Reprod Biomed Online 2007; 15: 643–658. [DOI] [PubMed] [Google Scholar]
- 21.Lee JY, Dada R, Sabanegh E, et al. Role of genetics in azoospermia. Urology 2011; 77: 598–601. [DOI] [PubMed] [Google Scholar]
- 22.Ihara M, Kinoshita A, Yamada S, et al. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev Cell 2005; 8: 343–352. [DOI] [PubMed] [Google Scholar]
- 23.Lin YH, Chou CK, Hung YC, et al. SEPT12 deficiency causes sperm nucleus damage and developmental arrest of preimplantation embryos. Fertil Steril 2011; 95: 363–365. [DOI] [PubMed] [Google Scholar]
- 24.Hosseinifar H, Shafipour M, Modarresi T, et al. Relationship between absence of annulus and asthenozoospermia in Iranian men. J Assist Reprod Genet 2014; 31: 1681–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kissel H, Georgescu MM, Larisch S, et al. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell 2005; 8: 353–364. [DOI] [PubMed] [Google Scholar]
- 26.Chao HC, Lin YH, Kuo YC, et al. The expression pattern of SEPT7 correlates with sperm morphology. J Assist Reprod Genet 2010; 27: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen YR, Wang HY, Kuo YC, et al. SEPT12 phosphorylation results in loss of the septin ring/sperm annulus, defective sperm motility and poor male fertility. PLoS Genet 2017; 13: e1006631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto T, Tsujimura A, Miyagawa Y, et al. Single nucleotide polymorphisms in the SEPTIN12 gene may be associated with azoospermia by meiotic arrest in Japanese men. J Assist Reprod Genet 2012; 29: 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyakawa H, Miyamoto T, Koh E, et al. Single-nucleotide polymorphisms in the SEPTIN12 gene may be a genetic risk factor for Japanese patients with Sertoli cell-only syndrome. J Androl 2012; 33: 483–487. [DOI] [PubMed] [Google Scholar]
- 30.Lin YH, Wang YY, Chen HI, et al. SEPTIN12 genetic variants confer susceptibility to teratozoospermia. PloS One 2012; 7: e34011. [DOI] [PMC free article] [PubMed] [Google Scholar]